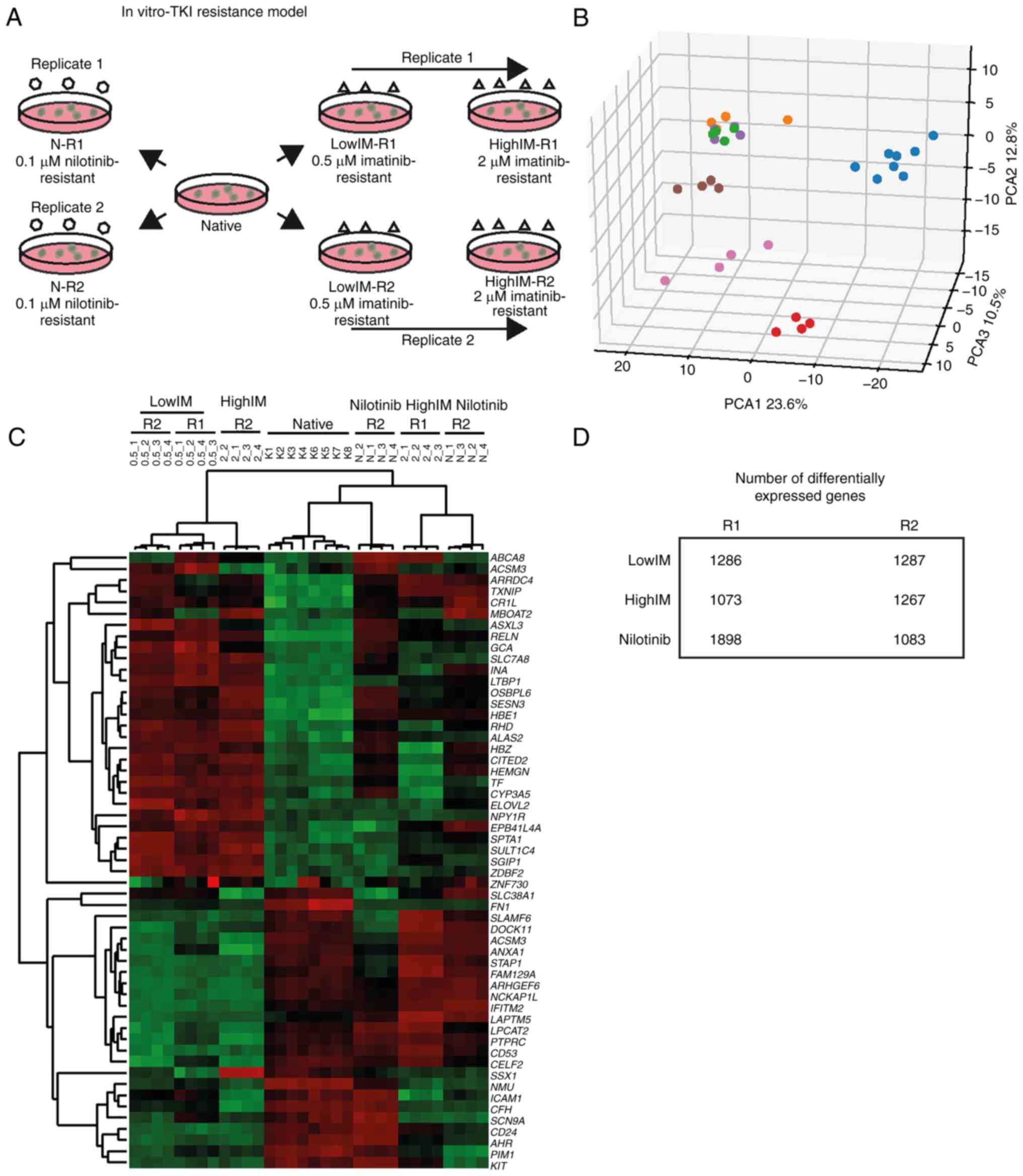
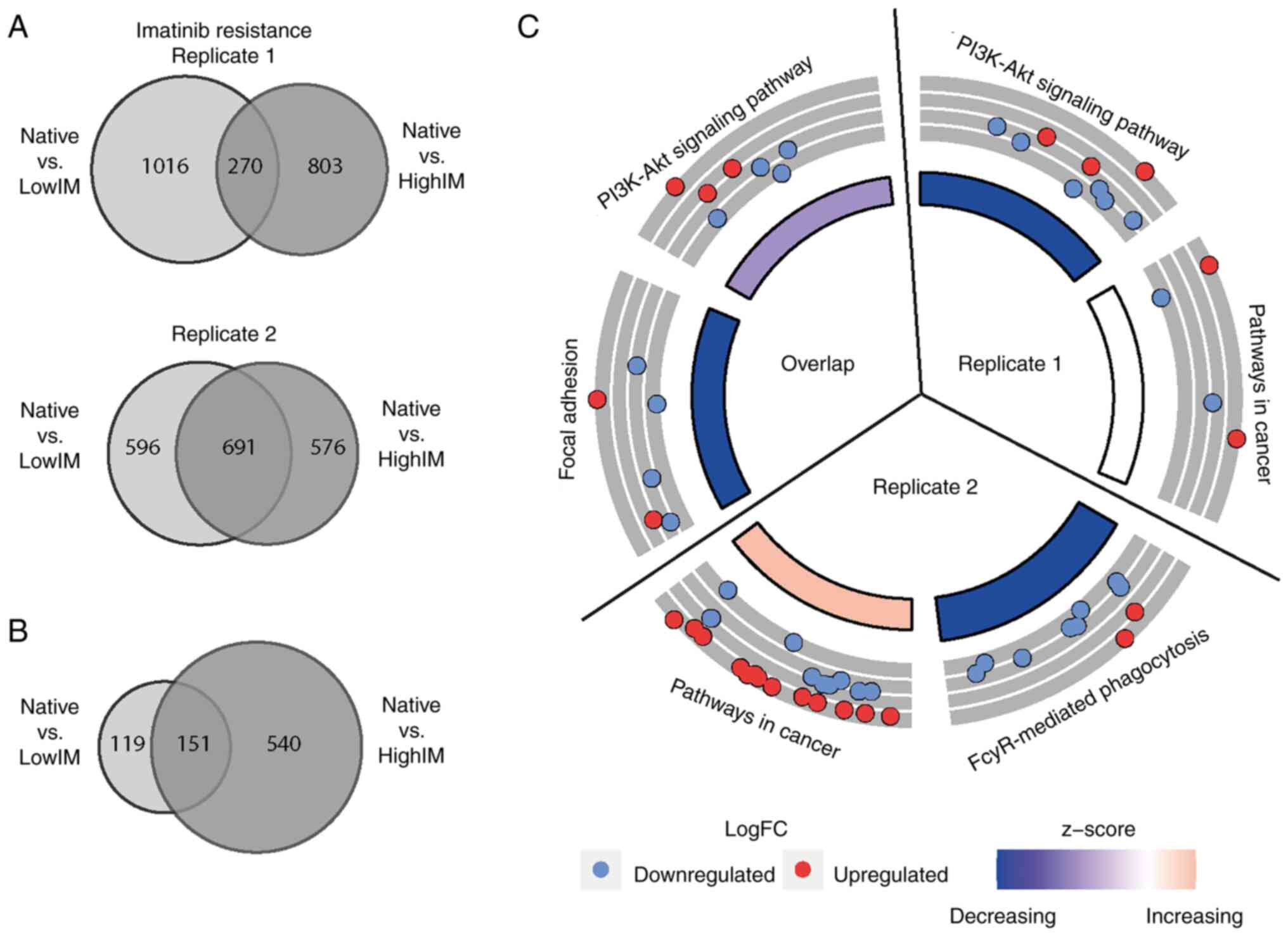
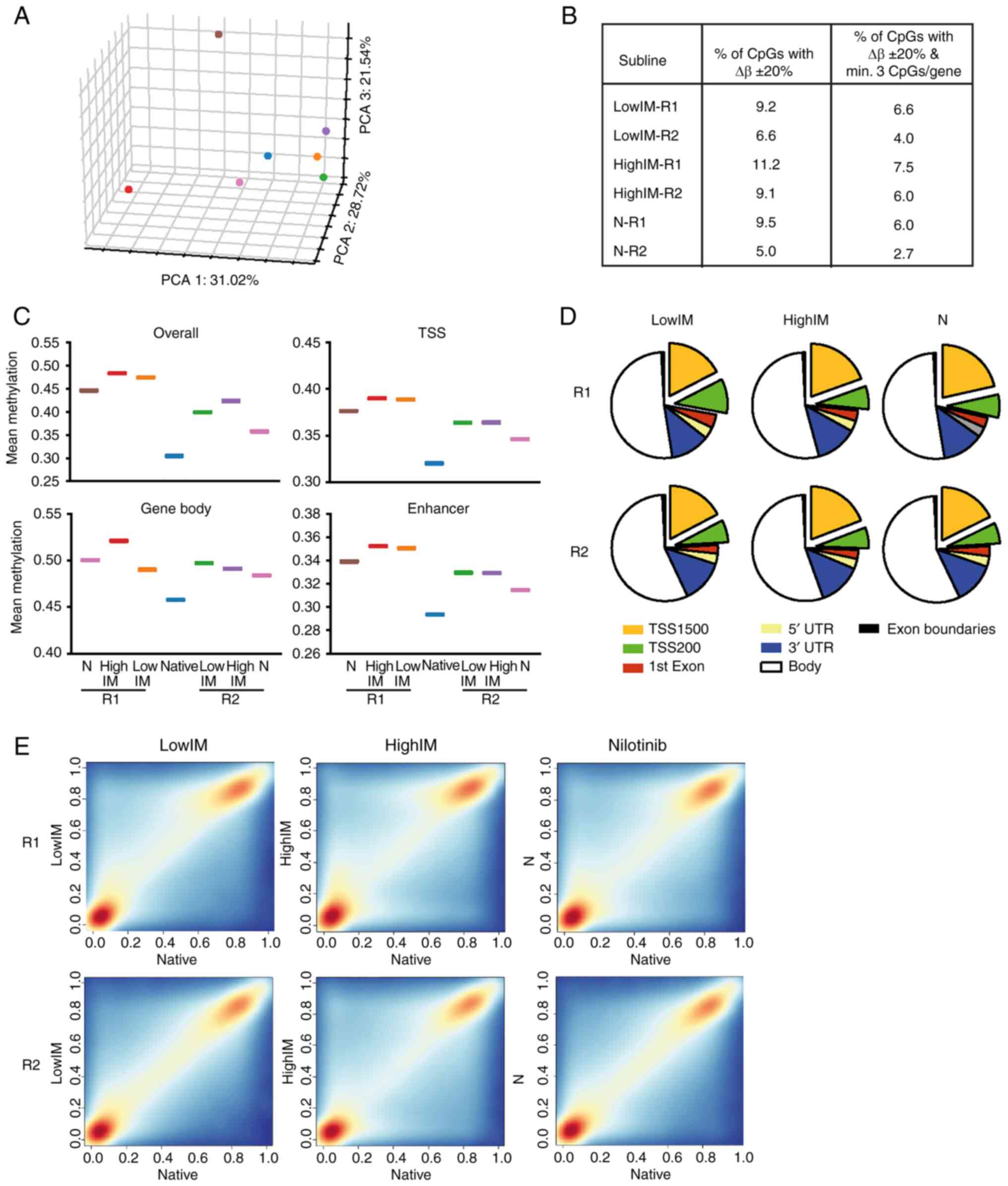
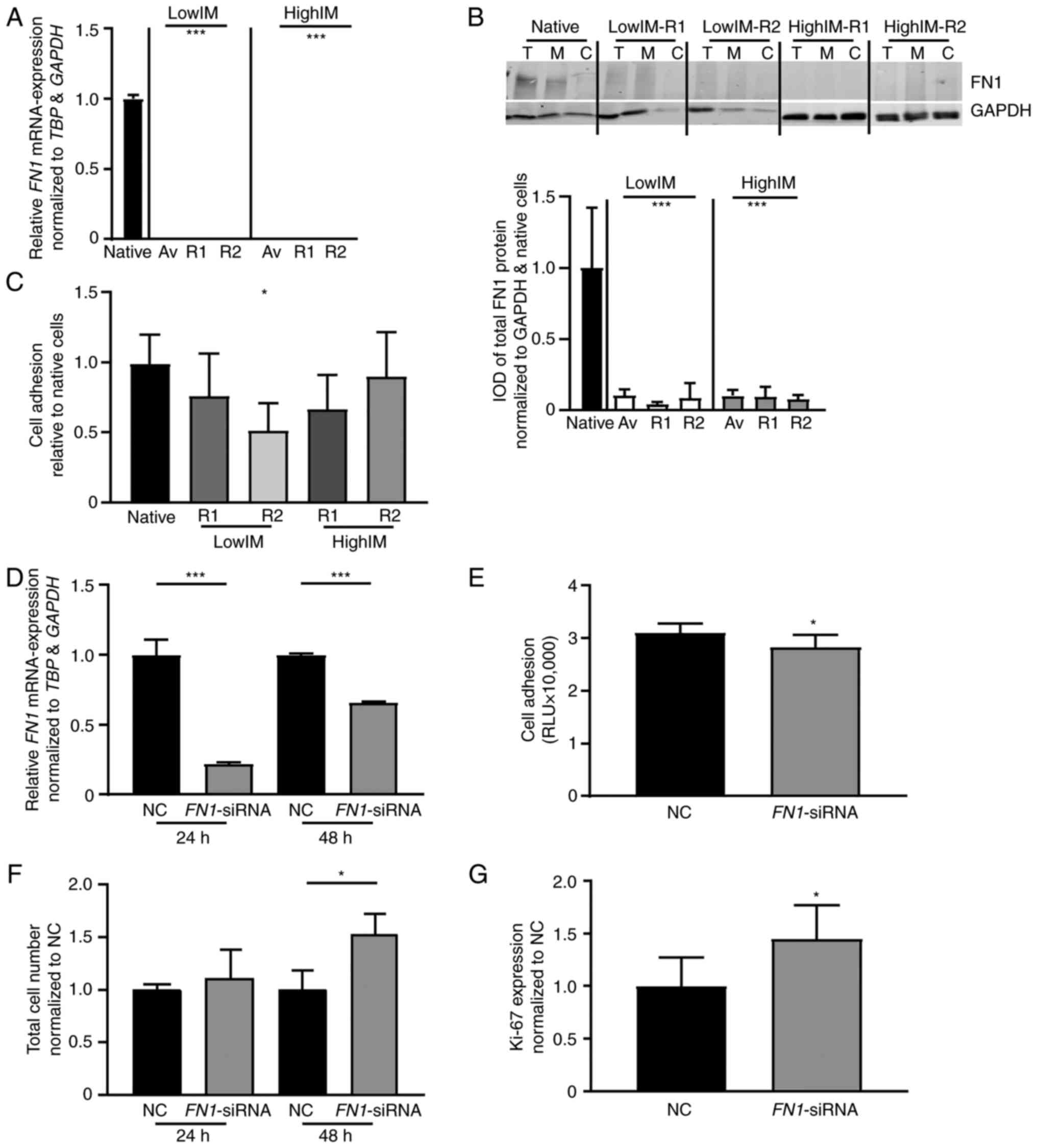
|
1
|
Deininger MW, Goldman JM and Melo JV: The
molecular biology of chronic myeloid leukemia. Blood. 96:3343–3356.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Quintas-Cardama A and Cortes J: Molecular
biology of bcr-abl1-positive chronic myeloid leukemia. Blood.
113:1619–1630. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Druker BJ, Tamura S, Buchdunger E, Ohno S,
Segal GM, Fanning S, Zimmermann J and Lydon NB: Effects of a
selective inhibitor of the Abl tyrosine kinase on the growth of
Bcr-Abl positive cells. Nat Med. 2:561–566. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baccarani M, Deininger MW, Rosti G,
Hochhaus A, Soverini S, Apperley JF, Cervantes F, Clark RE, Cortes
JE, Guilhot F, et al: European LeukemiaNet recommendations for the
management of chronic myeloid leukemia: 2013. Blood. 122:872–884.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hochhaus A, Larson RA, Guilhot F, Radich
JP, Branford S, Hughes TP, Baccarani M, Deininger MW, Cervantes F,
Fujihara S, et al: Long-term outcomes of imatinib treatment for
chronic myeloid leukemia. N Engl J Med. 376:917–927. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Milojkovic D and Apperley J: Mechanisms of
resistance to imatinib and second-generation tyrosine inhibitors in
chronic myeloid leukemia. Clin Cancer Res. 15:7519–7527. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Soverini S, De Benedittis C, Papayannidis
C, Paolini S, Venturi C, Iacobucci I, Luppi M, Bresciani P,
Salvucci M, Russo D, et al: Drug resistance and BCR-ABL kinase
domain mutations in Philadelphia chromosome-positive acute
lymphoblastic leukemia from the imatinib to the second-generation
tyrosine kinase inhibitor era: The main changes are in the type of
mutations, but not in the frequency of mutation involvement.
Cancer. 120:1002–1009. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zabriskie MS, Eide CA, Tantravahi SK,
Vellore NA, Estrada J, Nicolini FE, Khoury HJ, Larson RA, Konopleva
M, Cortes JE, et al: BCR-ABL1 compound mutations combining key
kinase domain positions confer clinical resistance to ponatinib in
Ph chromosome-positive leukemia. Cancer Cell. 26:428–442. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Lavallade H and Kizilors A: The
importance of mutational analyses in chronic myeloid leukaemia for
treatment choice. Eur Med J Oncol. 4:86–95. 2016.
|
|
10
|
Jelinek J, Gharibyan V, Estecio MR, Kondo
K, He R, Chung W, Lu Y, Zhang N, Liang S, Kantarjian HM, et al:
Aberrant DNA methylation is associated with disease progression,
resistance to imatinib and shortened survival in chronic
myelogenous leukemia. PLoS One. 6:e221102011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Turrini E, Haenisch S, Laechelt S, Diewock
T, Bruhn O and Cascorbi I: MicroRNA profiling in K-562 cells under
imatinib treatment: Influence of miR-212 and miR-328 on ABCG2
expression. Pharmacogenet Genomics. 22:198–205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu Y, Song Y, Ma W, Zheng W and Yin H:
Decreased microRNA-30a levels are associated with enhanced ABL1 and
BCR-ABL1 expression in chronic myeloid leukemia. Leuk Res.
37:349–356. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shibuta T, Honda E, Shiotsu H, Tanaka Y,
Vellasamy S, Shiratsuchi M and Umemura T: Imatinib induces
demethylation of miR-203 gene: An epigenetic mechanism of
anti-tumor effect of imatinib. Leuk Res. 37:1278–1286. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cortes J and O'Dwyer ME: Clonal evolution
in chronic myelogenous leukemia. Hematol Oncol Clin North Am.
18:671–684. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Corbin AS, Agarwal A, Loriaux M, Cortes J,
Deininger MW and Druker BJ: Human chronic myeloid leukemia stem
cells are insensitive to imatinib despite inhibition of BCR-ABL
activity. J Clin Invest. 121:396–409. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hamad A, Sahli Z, El Sabban M, Mouteirik M
and Nasr R: Emerging therapeutic strategies for targeting chronic
myeloid leukemia stem cells. Stem Cells Int. 2013:7243602013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kaehler M, Ruemenapp J, Gonnermann D,
Nagel I, Bruhn O, Haenisch S, Ammerpohl O, Wesch D, Cascorbi I and
Bruckmueller H: MicroRNA-212/ABCG2-axis contributes to development
of imatinib-resistance in leukemic cells. Oncotarget.
8:92018–92031. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Oliveros JC: An interactive tool for
comparing lists with Venn's diagrams. 2007-2015.https://bioinfogp.cnb.csic.es/tools/venny/index.html
|
|
19
|
Walter W, Sanchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
RStudio Team (2020), . RStudio: Integrated
Development for R. RStudio. PBC; Boston, MA: http://www.rstudio.com/
|
|
21
|
Pedregosa F, Varoquaux G, Gramfort A,
Michel V and Thirion B: Scikit-learn: Machine learning in python. J
Machine Learning Research. 12:2825–2830. 2011.
|
|
22
|
Kaehler M, Dworschak M, Rodin JP,
Ruemenapp J, Vater I, Penas EMM, Liu C, Cascorbi I and Nagel I:
Zfp36l1 plays an ambiguous role in the regulation of cell expansion
and negatively regulates Cdkn1a in chronic myeloid leukemia cells.
Exp Hematol. 99:54–64. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Waetzig V, Haeusgen W, Andres C, Frehse S,
Reinecke K, Bruckmueller H, Ruwen Boehm H, Herdegen T and Cascorbi
I: Retinoic acid-induced survival effects in SH-SY5Y neuroblastoma
cells. J Cell Biochem. 120:5974–5986. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bruhn O, Lindsay M, Wiebel F, Kaehler M,
Nagel I, Böhm R, Röder C and Cascorbi I: Alternative
polyadenylation of ABC transporters of the C-family (ABCC1, ABCC2,
ABCC3) and implications on posttranscriptional Micro-RNA
regulation. Mol Pharmacol. 97:112–122. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Edeki C: Comparative study of microarray
and next generation sequencing technologies. IJCSMC. 1:15–20.
2012.
|
|
26
|
Pop L, Zanoaga O, Chiroi P, Nutu A, Korban
SS, Stefan C, Irimie A and Berindan-Neagoe I: Microarrays and NGS
for drug discovery. Drug Design. Parikesit AA: IntechOpen; 2021
|
|
27
|
Bakay M, Chen YW, Borup R, Zhao P,
Nagaraju K and Hoffman EP: Sources of variability and effect of
experimental approach on expression profiling data interpretation.
BMC Bioinformatics. 3:42002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bammler T, Beyer RP, Bhattacharya S,
Boorman GA, Boyles A, Bradford BU, Bumgarner RE, Bushel PR,
Chaturvedi K, Choi D, et al: Standardizing global gene expression
analysis between laboratories and across platforms. Nat Methods.
2:351–356. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Draghici S, Khatri P, Eklund AC and
Szallasi Z: Reliability and reproducibility issues in DNA
microarray measurements. Trends Genet. 22:101–109. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chung YJ, Kim TM, Kim DW, Namkoong H, Kim
HK, Ha SA, Kim S, Shin SM, Kim JH, Lee YJ, et al: Gene expression
signatures associated with the resistance to imatinib. Leukemia.
20:1542–1550. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim TM, Ha SA, Kim HK, Yoo J, Kim S, Yim
SH, Jung SH, Kim DW, Chung YJ and Kim JW: Gene expression
signatures associated with the in vitro resistance to two tyrosine
kinase inhibitors nilotinib and imatinib. Blood Cancer J.
1:e322011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Krause DS and Scadden DT: A hostel for the
hostile: The bone marrow niche in hematologic neoplasms.
Haematologica. 100:1376–1387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bazzoni G, Carlesso N, Griffin JD and
Hemler ME: Bcr/Abl expression stimulates integrin function in
hematopoietic cell lines. J Clin Invest. 98:521–528. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nishihara T, Miura Y, Tohyama Y, Mizutani
C, Hishita T, Ichiyama S, Uchiyama T and Tohyama K: Effects of the
tyrosine kinase inhibitor imatinib mesylate on a Bcr-Abl-positive
cell line: Suppression of autonomous cell growth but no effect on
decreased adhesive property and morphological changes. Int J
Hematol. 78:233–240. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Y, Clough N, Sun X, Yu W, Abbott BL,
Hogan CJ and Dai Z: Bcr-Abl induces abnormal cytoskeleton
remodeling, beta1 integrin clustering and increased cell adhesion
to fibronectin through the Abl interactor 1 pathway. J Cell Sci.
120:1436–1446. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Windisch R, Pirschtat N, Kellner C,
Chen-Wichmann L, Lausen J, Humpe A, Krause DS and Wichmann C:
Oncogenic deregulation of cell adhesion molecules in leukemia.
Cancers (Basel). 11:3112019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Damiano JS, Hazlehurst LA and Dalton WS:
Cell adhesion-mediated drug resistance (CAM-DR) protects the K562
chronic myelogenous leukemia cell line from apoptosis induced by
BCR/ABL inhibition cytotoxic drugs and gamma-irradiation. Leukemia.
15:1232–1239. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Obr A, Roselova P, Grebenova D and
Kuželová K: Real-time analysis of imatinib- and dasatinib-induced
effects on chronic myelogenous leukemia cell interaction with
fibronectin. PLoS One. 9:e1073672014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kumar R, Pereira RS, Zanetti C, Minciacchi
VR, Merten M, Meister M, Niemann J, Dietz MS, Rüssel N, Schnütgen
F, et al: Specific targetable interactions with the
microenvironment influence imatinib-resistant chronic myeloid
leukemia. Leukemia. 34:2087–2101. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wlodek J and Pituch-Noworolska A: The
influence of fibronectin on proliferation and apoptosis of acute
lymphoblastic leukaemia cells in vitro. Pol J Pathol. 69:62–66.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Baharudin R, Ab Mutalib NS, Othman SN,
Sagap I, Rose IM, Mokhtar NM and Jamal R: Identification of
predictive DNA methylation biomarkers for chemotherapy response in
colorectal cancer. Front Pharmacol. 8:472017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo R, Wu G, Li H, Qian P, Han J, Pan F,
Li W, Li J and Ji F: Promoter methylation profiles between human
lung adenocarcinoma multidrug resistant A549/cisplatin (A549/DDP)
cells and its progenitor A549 cells. Biol Pharm Bull. 36:1310–1316.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Romero-Garcia S, Prado-Garcia H and
Carlos-Reyes A: Role of DNA methylation in the resistance to
therapy in solid tumors. Front Oncol. 10:11522020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lebecque B, Bourgne C, Vidal V and Berger
MG: DNA methylation and intra-clonal heterogeneity: The chronic
myeloid leukemia model. Cancers (Basel). 13:35872021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Amabile G, Di Ruscio A, Müller F, Welner
RS, Yang H, Ebralidze AK, Zhang H, Levantini E, Qi L, Martinelli G,
et al: Dissecting the role of aberrant DNA methylation in human
leukaemia. Nat Commun. 6:70912015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jose-Eneriz ES, Agirre X, Jimenez-Velasco
A, Cordeu L, Martín V, Arqueros V, Gárate L, Fresquet V, Cervantes
F, Martínez-Climent JA, et al: Epigenetic down-regulation of BIM
expression is associated with reduced optimal responses to imatinib
treatment in chronic myeloid leukaemia. Eur J Cancer. 45:1877–1889.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Elias MH, Baba AA, Husin A, Sulong S,
Hassan R, Sim GA, Wahid SFA and Ankathil R: HOXA4 gene promoter
hypermethylation as an epigenetic mechanism mediating resistance to
imatinib mesylate in chronic myeloid leukemia patients. Biomed Res
Int. 2013:1297152013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Nishioka C, Ikezoe T, Yang J, Udaka K and
Yokoyama A: Imatinib causes epigenetic alterations of PTEN gene via
upregulation of DNA methyltransferases and polycomb group proteins.
Blood Cancer J. 1:e482011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Borisov N, Tkachev V, Suntsova M,
Kovalchuk O, Zhavoronkov A, Muchnik I and Buzdin A: A method of
gene expression data transfer from cell lines to cancer patients
for machine-learning prediction of drug efficiency. Cell Cycle.
17:486–491. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mirabelli P, Coppola L and Salvatore M:
Cancer cell lines are useful model systems for medical research.
Cancers. 11:10982019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rumjanek VM, Vidal RS and Maia RC:
Multidrug resistance in chronic myeloid leukaemia: How much can we
learn from MDR-CML cell lines? Biosci Rep. 33:e000812013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
McDermott M, Eustace AJ, Busschots S,
Breen L, Crown J, Clynes M, O'Donovan N and Stordal B: In vitro
development of chemotherapy and targeted therapy drug-resistant
cancer cell lines: A practical guide with case studies. Front
Oncol. 4:402014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Peng B, Lloyd P and Schran H: Clinical
pharmacokinetics of imatinib. Clin Pharmacokinet. 44:879–894. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
de Kogel CE and Schellens JH: Imatinib.
Oncologist. 12:1390–1394. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Picard S, Titier K, Etienne G, Teilhet E,
Ducint D, Bernard MA, Lassalle R, Marit G, Reiffers J, Begaud B, et
al: Trough imatinib plasma levels are associated with both
cytogenetic and molecular responses to standard-dose imatinib in
chronic myeloid leukemia. Blood. 109:3496–3499. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Gromicho M, Dinis J, Magalhães M,
Fernandes AR, Tavares P, Laires A, Rueff J and Rodrigues AS:
Development of imatinib and dasatinib resistance: dynamics of
expression of drug transporters ABCB1, ABCC1, ABCG2, MVP, and
SLC22A1. Leuk Lymphoma. 52:1980–1990. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Eadie LN, Hughes TP and White DL: ABCB1
overexpression is a key initiator of resistance to tyrosine kinase
inhibitors in CML cell lines. PLoS One. 11:e01614702016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
de Lima LT, Vivona D, Bueno CT, Hirata
RDC, Hirata MH, Luchessi AD, de Castro FA, de Lourdes F,
Chauffaille M, Zanichelli MA, Chiattone CS, et al: Reduced ABCG2
and increased SLC22A1 mRNA expression are associated with imatinib
response in chronic myeloid leukemia. Med Oncol. 31:8512014.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Nies AT, Schaeffeler E, van der Kuip H,
Cascorbi I, Bruhn O, Kneba M, Pott C, Hofmann U, Volk C, Hu S, et
al: Cellular uptake of imatinib into leukemic cells is independent
of human organic cation transporter 1 (OCT1). Clin Cancer Res.
20:985–994. 2014. View Article : Google Scholar : PubMed/NCBI
|