Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
sixth most frequent cancer globally, with an incidence of 890,000
new cases in 2018 (1). For
oropharyngeal squamous cell carcinoma (OPSCC), a rising incidence
rate has been reported over recent decades (2). This rising incidence has been
observed in particular in the sub-group of patients with disease
related to human papilloma virus (HPV) infection (2). Classically, treatment of OPSCC mainly
involves chemo- and/or radiotherapy (3). However, a primary surgical treatment
strategy has been recently established for early-stage primary
disease and recurrent cases (3).
Molecular imaging as a form of applied health technology is a field
that is developing rapidly. Optimization of existing modalities or
translation of novel modalities is currently in progress, with the
aim of improving imaging for the staging and treatment of cancer
(4). In particular, targeted
optical imaging has been explored intensively over the past decade,
since this modality allows for the real-time intraoperative
visualization of tumor extension and margins (5). This in turn may aid in addressing the
major challenge of inadequate tumor margins across a range of solid
cancers that are treated surgically (5). For surgically treated OPSCC, the
reported pooled rate of tumor-positive margins is 8% (6). Identification of molecular targets
that are highly expressed in cancer and absent or minimally
expressed in adjacent healthy tissues is key to the efficacy of
molecular imaging (7). To evaluate
potential targets for imaging purposes, studies investigating the
exact expression pattern within the tumor compartment and the
demarcation boundary with the non-cancerous tissue at the margins
in appropriately-sized cohorts of patients are required (8).
Key parameter for the applicability of a target are
tumor specificity and the expression rate in most if not all types
of cancer. The present study investigated the expression pattern of
urokinase-type plasminogen activator receptor (uPAR; also known as
CD87), EGFR and tissue factor (TF; also known as CD142) in OPSCC.
These are membrane-bound receptors that are considered to be
candidate targets for imaging purposes in several solid cancers,
including HNSCC (9–11) For uPAR and EGFR, numerous clinical
studies with targeted probes for positron emission tomography and
optical imaging are currently in progress (ClinicalTrials.gov Identifiers: NCT02945826,
NCT02965001, NCT02960724, NCT02755675 and NCT03134846).
The plasminogen activator (PA) system serves a
central role in cancer invasion and metastasis, which is mediated
through pericellular proteolytic activity to degrade the
extracellular matrix at the invasive front of a tumor. uPAR is
expressed by both cancer cells and tumor-associated stromal cells.
High expression levels in tumors and limited expression of uPAR in
non-cancerous tissues have been previously reported in various
cancers, including oral cavity squamous cell carcinoma (OSCC)
(12–15) In addition, higher expression levels
of uPAR have been associated with poorer survival and more
aggressive disease in numerous types of cancer including breast,
bladder and colorectal cancer (16–18).
The role of EGFR in HNSCC has been extensively
explored, primarily for antibody-based targeted therapy. However,
it also emerged as a candidate for targeted imaging over the past
decade (19,20) EGFR is closely associated with tumor
growth, since it is expressed by tumor cells in addition to various
normal tissues in the human body (21).
An established relationship between cancer and
thrombosis and venous thromboembolic events has been extensively
reported. TF form part of the extrinsic coagulation cascade and it
has been reported to be highly expressed in several solid tumors
(22). In the tumor
microenvironment, TF participates in multiple oncogenic signaling
pathways that stimulate tissue remodeling and tumor angiogenesis in
the invasive process; TF also modulates the immune response within
the tumor compartment and assists tumor cells to escape the host
immune system (23). Preclinical
data has been reported to support the rationale for targeting TF
using imaging and therapy (11,24).
Therefore, the present study aimed to investigate
the expression patterns of uPAR, EGFR and TF by
immunohistochemistry (IHC) to evaluate their potential for targeted
imaging and possible prognostic value in OPSCC.
Materials and methods
Patients and tumor
characteristics
From an established retrospective database of
patients with primary OPSCC diagnosed between January 2000 and
January 2012 in the department of Otolaryngology, Head & Neck
surgery & Audiology at Rigshospitalet (Copenhagen, Denmark), a
balanced cohort of 48 (52%) HPV-positive and 46 (48%) HPV-negative
patients were randomly assembled. The inclusion criteria were
primary OPSCC with preserved large biopsies or diagnostic resection
specimens being available for IHC expression analysis. Exclusion
criteria were insufficient tumor tissue for IHC analysis or
incomplete dataset in the database. All specimens were collected
prior to non-surgical treatment if this was indicated. Because HPV
is a well-known powerful prognostic marker of OPSCC outcome, a
balanced selection based on HPV status was performed. Both p16 and
HPV-DNA status was available. The dataset in this current study has
been part of previous publications and assays for the analysis of
p16 and HPV-DNA have previously been described (25). Briefly, for p16 detection by IHC,
the anti-P16 clone E6HA OptiView DAB IHC Detection Kit (Roche
Diagnostics); for HPV-DNA analysis, DNA was isolated using the
QIAamp DNA FFPE Tissue Kit (Qiagen, Inc.) and PCR was performed
using General Primers GP5+/GP6+. The 8th
version of the TNM Union For International Cancer Control (UICC8)
staging manual was applied (26).
The recorded clinicopathological characteristics and treatment
modalities are provided in Table
I. The present study was approved by the Ethical Committee of
the Capital Region of Denmark (protocol H-15016322; Copenhagen,
Denmark) and was conducted in accordance with The Declaration of
Helsinki.
 | Table I.Clinicopathological characteristics
of the patients with oropharyngeal squamous cell carcinoma. |
Table I.
Clinicopathological characteristics
of the patients with oropharyngeal squamous cell carcinoma.
| Clinicopathological
characteristic | N (%) or median
(range) |
|---|
| Sex |
|
|
Female | 24 (26) |
|
Male | 69 (74) |
| Age, years | 60 (46–84) |
| Tumor location |
|
| Pharynx
wall | 8 (9) |
|
Palatine tonsils | 70 (75) |
| Lingual
tonsils | 12 (13) |
| Soft
palate | 3 (3%) |
| HPV-DNA |
|
|
Positive | 45 (48) |
|
Negative | 48 (52) |
| Smoking status |
|
| Never
smoker | 16 (17) |
|
Previous | 38 (41) |
| Current
smoker | 39 (42) |
| Tumor
differentiation |
|
|
Non-keratinizing | 23 (25) |
|
High | 3 (3) |
|
Modrate | 37 (40) |
|
Low | 30 (32) |
| T stage |
|
| T1 | 16 (17) |
| T2 | 45 (48) |
| T3 | 14 (15) |
| T4 | 18 (19) |
| N stage |
|
| N0 | 24 (26) |
| N1 | 43 (46) |
|
N2a/2b/2c | 22 (24) |
| N3 | 4 (4) |
| UICC8 TMN
stage |
|
| S1 | 36 (39) |
| S2 | 20 (22) |
| S3 | 16 (17) |
| S4 | 21 (23) |
| Treatment |
|
| RT | 33 (35) |
|
RCT | 46 (49) |
|
Surgery | 8 (9) |
| Surgery
+ RT | 1 (2) |
| Surgery
+ RCT | 1 (2) |
|
Palliation | 4 (4) |
| Recurrence |
|
| No | 74 (80) |
|
Yes | 19 (20) |
|
Local | 6 (6) |
|
Regional | 6 (6) |
|
Locoregional | 1 (2) |
|
Distant | 6 (6) |
| Second primary |
|
| No | 79 (85) |
|
Yes | 14 (15) |
IHC
At the time of collection, fresh tissue was fixed in
10% buffered formalin at room temperature for 24 h and then
embedded in paraffin. The paraffin-embedded tissue blocks were
sliced into 4-µm thick serial sections. Deparaffinization was
performed with xylene for 15 min followed by dehydration using an
ethanol gradient (from 99 to 70%, then demineralized water). IHC
staining was performed on a Ventana Benchmark Ultra semi-automated
autostainer (Roche Diagnostics). All antibody protocols have been
previously optimized on positive and negative control tissues. The
following antibodies were used: anti-cytokeratin (CK) AE1/AE3+8/18
(Pan Cytokeratin Plus; cat. no. 162; 1:200; Biocare Medical, LLC),
mouse anti-human uPAR R2 (1:20,000; in-house antibody; Finsen
Laboratory), anti-EGFR (Confirm anti-EGFR 5B7; cat. no. 790-4347;
ready-to-use; Ventana Medical Systems, Inc.; Roche Diagnostics) and
anti-TF (anti-TF; cat. no. 4509; 1:150; American Diagnostica,
Inc.). Antigen retrieval for uPAR was performed with proteinase K
(1:50 in Tris-HCl; cat. no. S3004; DAKO, Agilent Technologies,
Inc.) for 8 min at room temperature, followed by 100°C heating with
Cell Conditioning 1 buffer (CC1; Ventana Medical Systems, Inc.;
Roche Diagnostics) for 16 min. For TF, EGFR and CK, heat-induced
epitope retrieval was performed at 100°C in CC1 buffer for 32 min.
The blocking step was performed using Peroxidase Blocking Solution
(cat. no. S2023; DAKO, Agilent Technologies, Inc.) for 8 min at
room temperature, followed by 2% BSA (cat. no. A7906;
Sigma-Aldrich; Merck KGaA) blocking for 10 min at room temperature.
For uPAR and TF, EnVision mouse HRP-conjugated EnVision FLEX+
secondary antibodies were used at room temperature for 40 min (cat.
no. K8002; ready-to-use; DAKO; Agilent Technologies, Inc.),
followed by detection with DAB and DAB+ Chromogen Solution (cat.
no. K4065; diluted according to manufacturer's instructions; DAKO,
Agilent Technologies, Inc.). For EGFR and CK, ultraView Universal
DAB Detection Kit combined with ultraView Kit+ amplification (cat.
nos. 760-500 and 760-080, respectively; Roche Diagnostics) and
OptiView DAB IHC Detection Kit (cat. no. 760-700; Roche
Diagnostics), respectively, were applied for detection.
In addition, separate sections were also stained
with H&E for a 60 min protocol including deparaffinization,
dehydration as described above, then stained in hematoxylin (cat.
no. 854183; Capital Region, Hospital Farmacy) for five minutes,
rinsed and stained with eosin (cat. no. 854653; Capital Region,
Hospital Farmacy) for three minutes at room temperature. Digital
images of all IHC slides were acquired using a Axio Scan.Z1 (Carl
Zeiss AG) brightfield microscope slide scanner.
Expression analysis and scoring
system
Five slides from each individual tumor were reviewed
and scored by two experienced head and neck pathologists (KK and
GL) blinded to the clinical dataset. In case of any discrepant
assessment of IHC target expression, individual slides would be
reviewed together to produce a consensus score. For each tumor, the
extent of tumor compartmentalization and differentiation was
evaluated using H&E and CK staining images. For uPAR, TF and
EGFR respectively, IHC staining was semi-quantitatively assessed
for intensity (I-score) and proportional expression (P-score; that
is, the proportion of cells with positive biomarker expression)
within the tumor compartment by assessment of the whole tumor
section at lowest magnification. The I-score was stratified as 0–3
(none, weak, moderate and strong intensity, respectively) whereas
the P-score was stratified as 0–3 (0–10, 11–50, 51–75 and 76–100%,
respectively). The P-score was estimated based on the observed
target expression on tumor cells, stromal cells and inflammatory
cells within the tumor compartment. By adding the I-score and
P-score, a composite semi-quantitative score (PI-score) was
calculated, as proposed by Allred et al (27). The PI-score is a 7-point system
stratified from 0 to 6. If present, IHC expression in adjacent
non-cancerous tissues would also be qualitatively characterized. To
estimate the expression rate of uPAR, TF and EGFR within the
cohort, expression would be considered positive if the PI-score was
>1. To assess the possible prognostic value of the three
targets, the PI-score was dichotomized into high and low expression
based on the mean value for each target for association analyses of
associations. PI-scores <3 was used for the dichotomization of
uPAR and TF, whereas PI-scores <4 was used for EGFR.
Statistical analysis
For associations between biomarker expression and
each of the clinicopathological variables, Pearson's χ2
test or Fisher's exact test (for low number of events) was applied;
for age comparison, an unpaired t-test was performed. For survival
statistics, overall survival (OS) was defined as time from the date
of diagnosis until death of any cause. Recurrence-free survival
(RFS) was defined as time from the date of diagnosis until the date
of recurrence confirmed by biopsy, death by any cause. Kaplan-Meier
curves were applied for survival plots and 5-year survival
estimates, where log-rank test was used for comparison between the
groups. Using the Cox proportional hazards model, univariate and
multivariate estimates of hazard ratios (HRs) were calculated for
OS and RFS following adjustment for sex, age, smoking status, HPV
status, tumor location, T-site, T-stage, N-stage, UICC8 TNM-stage,
tumor differentiation and biomarker expression. P<0.05 was
considered to indicate a statistically significant difference. Data
analysis was performed in the SAS software package, version 6.1
(SAS Institute, Inc.) and R statistics (version 3.6.1).
Results
Basal patient characteristics
In this cohort of 93 patients, the median age at
diagnosis was 60 years (range, 40–84 years), where 69 of the
patients (74%) were men. The median follow-up calculated from the
day of diagnosis was 4.8 years (range, 0.03-14.2 years). The tumor
subsites were palatine tonsils (75%), base of the tongue (13%),
pharyngeal wall (9%) and soft palate (3%). At follow up, January
2020, 51 patients were alive, and 42 patients were deceased, 25
(27%) of whom succumbed to OPSCC. During follow-up, 19 (20%)
patients had recurrence confirmed by biopsy after primary
treatment. The median time to recurrence was 9 months.
Target expression analysis
The positive expression rates in tumors based on the
PI-score (PI>1) for uPAR, TF and EGFR were calculated to be
98.9, 76.3 and 98.9%, respectively. For the I-score, positive
staining (I>0) for uPAR, TF and EFGR was present in 100, 89.3
and 100% of the specimens. The exact extension of the tumor
compartment, and the margin demarcation from adjacent non-cancerous
tissues were evaluated on the H&E and CK stainings and then
compared to the topographic distribution of the IHC stainings for
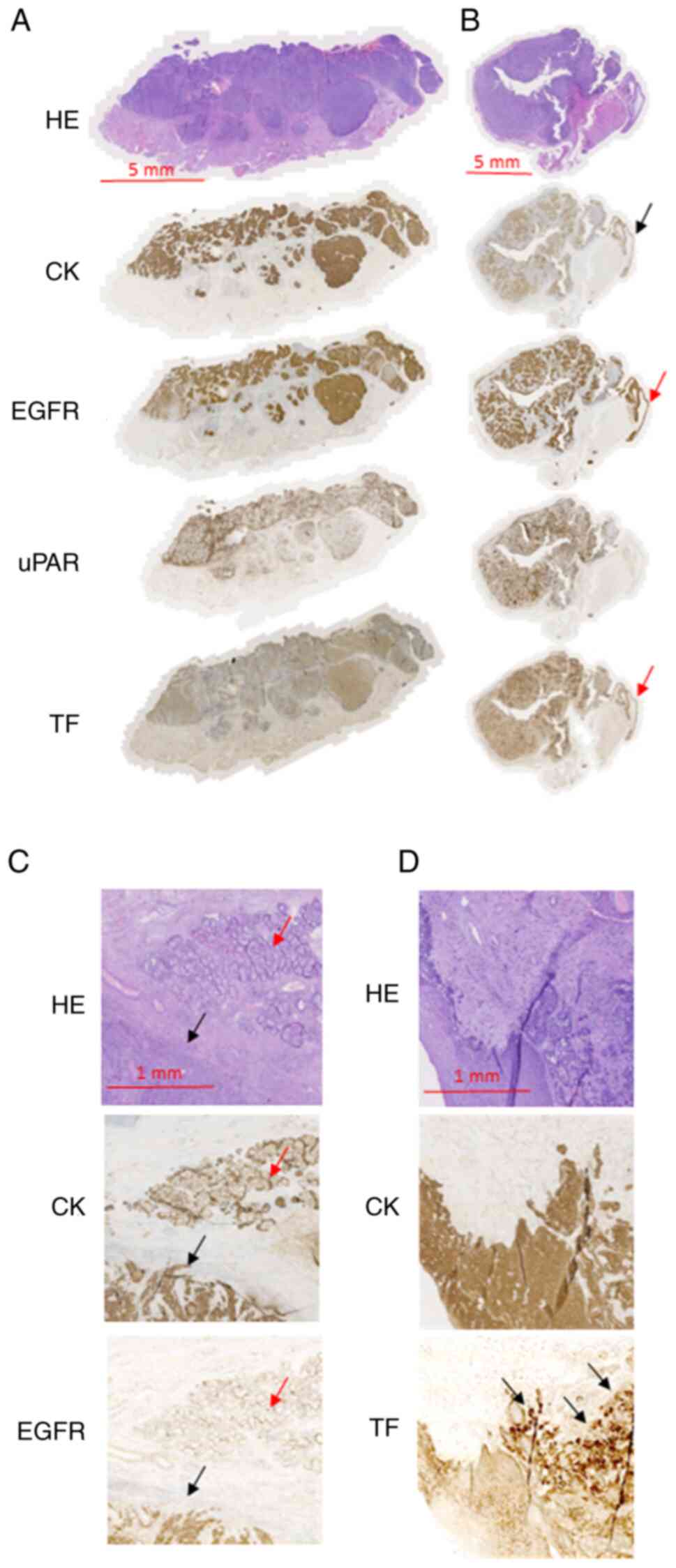
each of the three biomarkers. For uPAR a tumor-specific staining
pattern was observed, with enhanced expression within the tumor
compartment but limited or absent expression in the adjacent
non-cancerous tissues (Fig. 1). In
the tumor compartment, membranous uPAR staining was seen on tumor
cells as well as on stromal cells. Tumor-associated stromal uPAR
expression was also observed on macrophages, neutrophils,
fibroblasts. At the mucosal margin, there was a sharp demarcation
between carcinoma and adjacent epithelium, with the absence of uPAR
expression in the unaffected epithelial lining. At the deep
margins, uPAR staining delineated the tumor compartment. In
addition, in the majority of cases, a narrow rim of expression was
present in the stroma at the invasive border. Intense uPAR staining
was frequently observed at the invasive front of the tumor and in
the microscopic tumor buds separated from the primary tumor. In 10
(11%) of the specimens, uPAR positivity was also found on
neutrophilic granulocytes, where pronounced inflammatory
infiltration or abscess formation was discovered outside the tumor
compartment.
TF demonstrated a tumor-specific expression pattern
when positive, though in some cases adjacent normal epithelium also
demonstrated weak positivity. As a general pattern, more intense TF
staining was typically seen at the invasive front at the deep
margin (Fig. 1).
For EGFR, strong and homogeneous expression was
observed in the tumor compartment, but high expression could also
be observed in the normal mucosal epithelium (Fig. 1). EGFR expression in the salivary
tissue was consistently positive in the ductal epithelium (Fig. 1).
Survival and recurrence analysis
A Cox proportional hazards univariate and
multivariate analysis was performed for RFS and OS (Table II). Age, HPV status and UICC8 TNM
stage were identified to be independent prognostic factors for OS
according to the univariate analysis, but only age and HPV status
maintained significance in the multivariate analysis (Table II). Only increase in TNM staging
for more advanced stage 3 and 4 disease was associated with RFS
according to univariate analysis. For the three biomarkers, high
uPAR PI-scores had a non-significant trend towards reduced OS
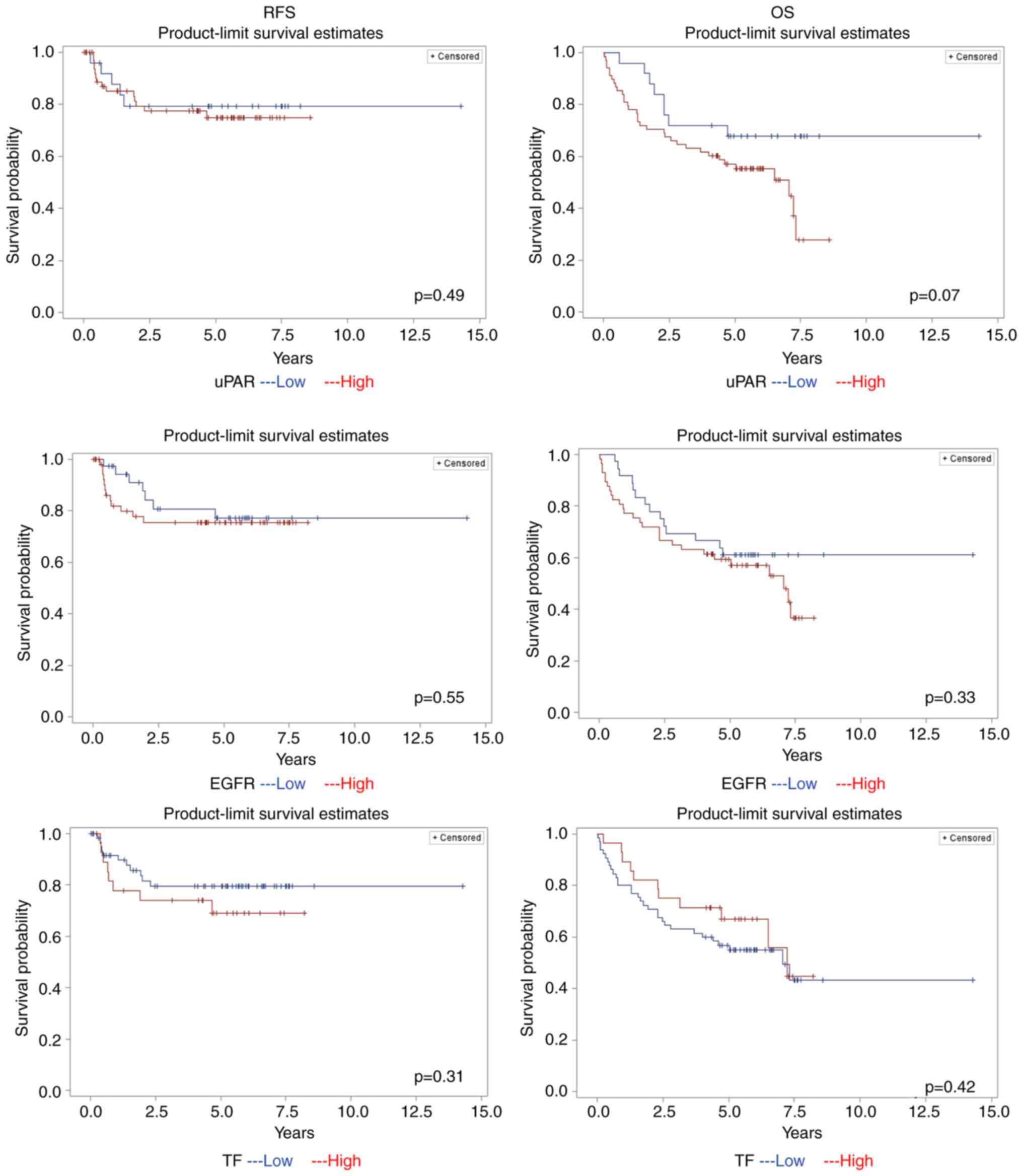
according to univariate analysis only (Table II). In addition, Kaplan-Meier
survival curves were generated with univariate log-rank tests
performed to assess the association of the PI-scores of the three
biomarkers with the OS and RFS (Fig.
2). For the entire cohort, the 5-year OS and RFS was 59 and
76%, respectively. For HPV-positive and HPV-negative patients, the
respective 5-year OS was 77 and 41%, whereas the respective 5-year
RFS was 81 and 67%. For patients with a high and low PI-scores for
uPAR, the 5-year OS was 57 and 68%, respectively, whereas the RFS
was 74 and 79% for patients with high and low PI-scores,
respectively (Fig. 2). In
addition, separate tests were performed for the P-score and I-score
but no statistically significant association with OS or RFS could
be detected (data not shown).
 | Table II.Cox proportional hazards model on
survival outcome from univariate and multivariate statistics. |
Table II.
Cox proportional hazards model on
survival outcome from univariate and multivariate statistics.
|
| Overall
survival | Recurrence-free
survival |
|---|
|
|
|
|
|---|
|
| Univariate | Multivariate | Univariate | Multivariate |
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
characteristic | HR (CI) | P-value | HR (CI) | P-value | HR (CI) | P-value | HR (CI) | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
|
|
Female | Ref. |
| Ref. |
| Ref. |
| Ref. |
|
|
Male | 1.03
(0.51-2.05) | 0.94 | 1.16
(0.46-2.91) | 0.75 | 0.91
(0.33-2.54) | 0.86 | 0.75
(0.18-3.05) | 0.69 |
| Age |
|
|
|
|
|
|
|
|
| Risk
per 1-year increase in age | 1.06
(1.03-1.10) | <0.01 | 1.08
(1.03-1.13) | <0.01 | 1.05
(1.00-1.10) | 0.05 | 1.05
(0.98-1.13) | 0.14 |
| Tumor location |
|
|
|
|
|
|
|
|
|
Palatine and lingual
tonsils | Ref. |
| Ref. |
| Ref. |
| Ref. |
|
|
Other | 1.57
(0.70-3.54) | 0.28 | 1.51
(0.48-4.79) | 0.48 | 0.41
(0.06-3.09) | 0.39 | 0.51
(0.05-5.20) | 0.57 |
| Human
papillomavirus status |
|
|
|
|
|
|
|
|
|
Positive | Ref. |
| Ref. |
| Ref. |
| Ref. |
|
|
Negative | 4.23
(2.08-8.63) | <0.01 | 5.48
(1.11-27.05) | 0.04 | 1.73
(0.70-4.32) | 0.24 | 1.31
(0.17-10.21) | 0.8 |
| Smoking status |
|
|
|
|
|
|
|
|
| Never
smoker | Ref. |
| Ref. |
| Ref. |
| Ref. |
|
|
Previous or current
smoker | 2.29
(0.82-6.43) | 0.11 | 0.95
(0.24-3.81) | 0.94 | 1.96
(0.45-8.48) | 0.37 | 5.21
(0.44-62.31) | 0.19 |
| T-stage |
|
|
|
|
|
|
|
|
| T1 | Ref. |
| Ref. |
| Ref. |
| Ref. |
|
| T2 | 0.80
(0.33-1.91) | 0.61 | 2.11
(0.60-7.38) | 0.24 | 0.87
(0.23-3.29) | 0.84 | 0.99
(0.21-4.76) | 0.99 |
| T3 | 0.26
(0.05-1.23) | 0.09 | 0.73
(0.12-4.23) | 0.72 | 0.67
(0.11-4.01) | 0.66 | 0.68
(0.06-7.09) | 0.75 |
| T4 | 3.17
(1.28-7.85) | 0.01 | 2.64
(0.66-10.56) | 0.17 | 3.54
(0.87-14.38) | 0.08 | 10.27
(0.50-210.62) | 0.13 |
| N-stage |
|
|
|
|
|
|
|
|
| N0 | Ref. |
| Ref. |
| Ref. |
| Ref. |
|
| N1 | 0.72
(0.33-1.57) | 0.41 | 0.93
(0.31-2.78) | 0.9 | 1.19
(0.37-3.86) | 0.78 | 1.23
(0.22-6.81) | 0.81 |
| N2 | 1.42
(0.63-3.19) | 0.39 | 0.73
(0.20-2.66) | 0.64 | 1.58
(0.42-5.87) | 0.5 | 1.56
(0.24-10.07) | 0.64 |
| N3 | 3.72
(1.02-13.56) | 0.05 | 2.57
(0.42-15.74) | 0.31 | 3.22
(0.36-28.83) | 0.3 | 1.79
(0.09-33.93) | 0.7 |
| UICC8 –TNM
stage |
|
|
|
|
|
|
|
|
| 1 | Ref. |
| Ref. |
| Ref. |
| Ref. |
|
| 2 | 0.60
(0.19-1.91) | 0.39 | 0.19
(0.03-1.15) | 0.07 | 1.00
(0.24-4.17) | 1 | 0.73
(0.10-5.25) | 0.75 |
| 3 | 3.21
(1.35-7.62) | <0.01 | 1.68
(0.36-7.84) | 0.51 | 3.71
(1.17-11.70) | 0.03 | 2.64
(0.28-24.56) | 0.4 |
| 4 | 4.45
(2.03-9.76) | <0.01 | 0.95
(0.12-7.44) | 0.96 | 2.17
(0.58-8.12) | 0.25 | 0.15
(0.00-7.73) | 0.35 |
| Urokinase-type
plasminogen activator receptor |
|
|
|
|
|
|
|
|
|
Low | Ref. |
| Ref. |
| Ref. |
| Ref. |
|
|
High | 2.01
(0.92-4.37) | 0.07 | 0.87
(0.32-2.31) | 0.77 | 1.20
(0.43-3.34) | 0.72 | 0.88
(0.27-2.91) | 0.83 |
| Tissue factor |
|
|
|
|
|
|
|
|
|
Low | Ref. |
| Ref. |
| Ref. |
| Ref. |
|
|
High | 0.75
(0.38-1.50) | 0.42 | 1.17
(0.50-2.76) | 0.72 | 1.60
(0.64-3.97) | 0.31 | 2.39
(0.67-8.51) | 0.18 |
| EGFR |
|
|
|
|
|
|
|
|
|
Low | Ref. |
| Ref. |
| Ref. |
| Ref. |
|
|
High | 1.37
(0.72-2.61) | 0.34 | 1.55
(0.73-3.28) | 0.25 | 1.33
(0.52-3.37) | 0.55 | 2.11
(0.68-6.53) | 0.19 |
Biomarker association analysis
Possible associations between clinicopathological
parameters and high or low PI-scores of uPAR, EGFR and TF were next
investigated by univariate analysis (Table III). No statistically significant
relationships were be detected.
 | Table III.Univariate analysis of the level of
biomarker expression and clinicopathological characteristics.
Biomarker expression based on PI-score. |
Table III.
Univariate analysis of the level of
biomarker expression and clinicopathological characteristics.
Biomarker expression based on PI-score.
|
| Urokinase-type
plasminogen activator receptor | Tissue factor | EGFR |
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristic | High, n (%) | Low, n (%) | P-value | High, n (%) | Low, n (%) | P-value | High, n (%) | Low, n (%) | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
|
|
|
Male | 51 (55) | 18 (19) |
| 22 (24) | 47 (51) |
| 42 (45) | 27 (29) |
|
|
Female | 17 (18) | 7 (8) | 0.77 | 6 (6) | 18 (19) | 0.53 | 15 (16) | 9 (10) | 0.89 |
| Age |
|
| 0.29 |
|
| 0.44 |
|
| 0.37 |
| Tumor location |
|
|
|
|
|
|
|
|
|
| Pharynx
wall | 1 (1) | 7 (7) |
| 8 (9) | 0 (0) |
| 3 (3) | 5 (5) |
|
|
Palatine tonsils | 18 (19) | 52 (56) |
| 48 (52) | 22 (24) |
| 26 (28) | 44 (47) |
|
| Lingual
tonsils | 4 (4) | 8 (9) |
| 7 (8) | 5 (5) |
| 7 (7) | 5 (5) |
|
| Soft
palate | 2 (2) | 1 (1) | 0.31 | 2 (2) | 1 (1) | 0.23 | 0 (0) | 3 (3) | 0.27 |
| Human papilloma
virus-DNA |
|
|
|
|
|
|
|
|
|
|
Positive | 35 (49) | 15 (16) |
| 16 (17) | 32 (34) |
| 28 (30) | 20 (22) |
|
|
Negative | 35 (38) | 10 (11) | 0.33 | 12 (13) | 33 (35) | 0.48 | 29 (31) | 16 (17) | 0.55 |
| Smoking status |
|
|
|
|
|
|
|
|
|
|
Never | 27 (29) | 8 (9) |
| 14 (15) | 21 (23) |
| 21 (23) | 14 (15) |
|
|
Ever | 41 (44) | 17 (18) | 0.5 | 14 (15) | 44 (47) | 0.11 | 39 (63) | 22 (24) | 0.84 |
| Tumor
differentiation |
|
|
|
|
|
|
|
|
|
|
G1-G2 | 17 (18) | 9 (10) |
| 6 (6) | 20 (22) |
| 12 (13) | 14 (15) |
|
|
G3-G4 | 51 (55) | 16 (17) | 0.29 | 22 (24) | 45 (48) | 0.36 | 45 (48) | 22 (24) | 0.06 |
| T-stage |
|
|
|
|
|
|
|
|
|
|
T1-T2 | 42 (45) | 19 (20) |
| 17 (18) | 44 (47) |
| 61 (66) | 26 (28) |
|
|
T3-T3 | 26 (28) | 6 (6) | 0.2 | 11 (12) | 21 (23) | 0.52 | 22 (24) | 11 (28) | 0.28 |
| N-stage |
|
|
|
|
|
|
|
|
|
| N0 | 16 (17) | 8 (9) |
| 6 (6) | 18 (19) |
| 15 (16) | 9 (10) |
|
| N+ | 52 (56) | 17 (18) | 0.41 | 22 (24) | 47 (51) | 0.53 | 42 (45) | 27 (29) | 0.89 |
| TNM stage |
|
|
|
|
|
|
|
|
|
|
S1-S2 | 39 (42) | 17 (18) |
| 21 (23) | 35 (38) |
| 34 (37) | 22 (24) |
|
|
S3-S4 | 29 (31) | 9 (32) | 0.35 | 7 (8) | 30 (32) | 0.06 | 23 (25) | 14 (15) | 0.89 |
| Recurrence |
|
|
|
|
|
|
|
|
|
| No | 54 (58) | 20 (22) |
| 20 (22) | 54 (58) |
| 45 (48) | 29 (31) |
|
|
Yes | 14 (15) | 5 (5) | 0.95 | 8 (9) | 11 (12) | 0.2 | 12 (13) | 7 (8) | 0.85 |
Discussion
The present study investigated the expression
patterns and applicability of three candidate receptor targets in
OPSCC for targeted molecular imaging based on IHC staining in large
biopsy or tumor resection specimens. To the best of our knowledge,
such topographic data on uPAR and TF expression for this large
sub-site type of HNSCC has not been previously reported. Expression
of EGFR in OPSCC has been previously described in a number of other
studies (28–30) An important finding from the present
study was that uPAR in particular appeared to be a promising target
in OPSCC due to its highly tumor-specific expression profile with
limited expression in the non-cancerous regions adjacent to the
tumor compartment. Although a tumor-specific expression pattern
could also be observed for TF, the positive expression rate in the
whole cohort was lower (76%). EGFR exhibited a high positive
expression rate and a homogeneous expression pattern within the
tumor compartment, equivalent to high receptor densities, rendering
it a potential marker for targeted strategies. However, EGFR did
not show a tumor-specific expression pattern, instead having
regular expression levels in the adjacent normal epithelium and
salivary gland tissues. Therefore, EGFR may be a less attractive
target for molecular imaging. Our group previously performed a
similar study of the expression of uPAR, TF and EFGR in OSCC
(31); in comparison, the
microscopic expression patterns and tumor-specificity of these
three biomarkers appears very similar to the findings in OPSCC.
This may indicate that individual targeted molecular strategies
against either uPAR, TF or EGFR may be applied in OSCC and OPSCC,
and possibly in HNSCC in general, which expands the utility of
individual-targeted imaging agents or therapeutics.
It has been reported previously that targeted
intraoperative optical imaging allows for a lower detection
threshold for tumor deposits in the µm-range (10). Therefore, it is highly likely that
this technology can be used to guide resection with adequate
margins using biomarkers with tumor-specific expression patterns.
In a clinical study using optical imaging in HNSCC by coupling with
optically labeled antibodies against EGFR, Gao et al
(32) previously demonstrated the
potential use of targeted optical imaging for margin evaluation in
the post-operative histopathological work-up. In addition, this
previous study also reported tracer uptake in non-cancerous tissues
outside the tumor compartment, such as the salivary gland tissues
(32). The challenge of EGFR
expression in normal tissues compared with targeted tumor imaging
has been discussed by several previous studies, which underlines
the need for exploring novel biomarkers in different types of
cancer that exhibit highly tumor-specific expression (33–35)
The identification of biomarkers for targeting
purposes with a high positive expression rate is of importance for
ensuring applicability and reproducibility in highly heterogenous
populations. Van Oosten et al (8) previously suggested a lower expression
rate of 80% for qualifying a given biomarker as clinically relevant
for a targeted imaging strategy. Collecting preoperative biopsies
for assessing biomarker expression in individual patients, which is
already performed during immunotherapeutic procedures, is another
alternative to imaging (36).
However, it is a less feasible strategy if the targeted imaging of
biomarkers with lower positive expression rates is anticipated.
Baart et al (33) proposed
the simultaneous use of several targeting agents against different
biomarkers to achieve maximal agent-to-target binding in the same
patient.
The role of the PA system in HNSCC has been previous
reported, especially for OSCC and to a lesser extent for laryngeal
SCC (37,38) However, it has not been previously
described for OPSCC (37,38). In addition, little is known about
TF in HNSCC, though a tumor-specific expression pattern in OSCC,
very similar to the expression pattern found in this study, has
been reported previously by our group (31). Since the mucosal lining in both the
oral cavity and the pharynx is non-keratinizing squamous
epithelium, it is likely that the same biomarker expression
profiles are present in both locations (39). Depending on the IHC scoring system
used, the positive expression rate of uPAR in OSCC has been
reported to be between 39 and 100% (38,40)
which is in line with a positivity rate of 98.9% in the present
study. The positive expression rate for TF in OSCC was reported to
be 58% (31), which is lower
compared with the positive expression rate of 76.9% in OPSCC seen
in the current study.
The present study did not detect an association
between the expression levels of uPAR, TF and EFGR with HPV status,
where the microscopic distribution of these three biomarkers did
not reveal any differences between sections with and without HPV.
It is well documented that HPV-positive and -negative tumors are
two distinctly different diseases in terms of pathogenesis and
prognosis, such that the carcinogenesis process in the presence of
HPV infection can interact with the immune (41,42)
Both uPAR and TF have been reported to serve a role in
immunological regulation and responses in the tumor
microenvironment (17,23) However, the expression of only three
receptors within a highly complex biological system was studied in
the present study, which may explain why no relationship with HPV
status could be observed.
In the present cohort, HPV-positive patients had
significantly more favorable survival outcome. In the survival
analysis, the level of uPAR, TF and EGFR expression did not
significantly associate with any of the survival outcomes tested.
Only high uPAR expression had a non-significant trend towards
reduced OS. Other previous studies of OSCC and other types of
cancer, like breast or bladder cancer, have found that high uPAR or
uPA expression significantly associated with reduced survival and
more aggressive histopathological tumor physiology (43,44)
A possible reason for the lack of prognostic significance in terms
of uPAR in OPSCC expression in the present study may be the small
sample size and the inherent lack of granularity and accuracy in
the IHC-based scoring. Nevertheless, these methodological
shortcomings should equally influence all three molecular targets.
Therefore, they indicate that of the three markers tested, uPAR is
the most prognostic. However, the present study was limited by a
relatively small sample size to perform robust association analyses
on survival statistics.
In conclusion, results from the present study
suggest that uPAR, TF and EGFR are suitable targets for molecular
imaging and therapy in OPSCC. uPAR in particular may be an
attractive target owing to its highly tumor-specific expression
pattern and association with prognosis. For TF, preselection of
patients may be necessary due to a lower positivity rate.
Acknowledgements
Not applicable.
Funding
Funding was obtained from University of Copenhagen, The Agnes
& Poul Friis Foundation and The Haboe Foundation.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AC and CG designed the study in collaboration with
KK, GL, KJ, BWC, JM, AK and CvB. CG and JSJ collected tissue
specimens and established the dataset. AC and CG have assessed the
authenticity of the raw data. KK, GL and KJ conducted the
immunohistochemistry. KK and GL performed the histopathological
scoring. AC, CG and JSJ performed the statistical data analysis.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Approval for this study was obtained from the
Ethical Committee of the Capital Region of Denmark (protocol
H-15016322; Copenhagen, Denmark). The Dataset was anonymized prior
to analysis. The Ethical Committee waived the need to obtain
consent to participate from the patients due to the circumstance
that a great proportion of the cohort had succumbed to disease or
were severely ill, where revealing that information about this
project would lead to more distress in the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EGFR
|
epidermal growth factor receptor
|
|
HNSCC
|
head and neck squamous cell
carcinoma
|
|
IHC
|
immunohisto-chemistry
|
|
OPSCC
|
oropharyngeal squamous cell
carcinoma
|
|
OS
|
overall survival
|
|
RFS
|
recurrence-free survival
|
|
TF
|
tissue factor
|
|
uPAR
|
urokinase-type plasminogen activator
receptor
|
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Mathers C, Parkin DM, Piñeros M, Znaor A and Bray F: Estimating the
global cancer incidence and mortality in 2018: GLOBOCAN sources and
methods. Int J cancer. 144:1941–1953. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mehanna H, Beech T, Nicholson T, El-Hariry
I, McConkey C, Paleri V and Roberts S: Prevalence of human
papillomavirus in oropharyngeal and nonoropharyngeal head and neck
cancer-systematic review and meta-analysis of trends by time and
region. Head Neck. 35:747–755. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Virgilio A, Costantino A, Mercante G,
Pellini R, Ferreli F, Malvezzi L, Colombo G, Cugini G, Petruzzi G
and Spriano G: Transoral robotic surgery and intensity-modulated
radiotherapy in the treatment of the oropharyngeal carcinoma: A
systematic review and meta-analysis. Eur Arch Otorhinolaryngol.
278:1321–1335. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Visgauss JD, Eward WC and Brigman BE:
Innovations in intraoperative tumor visualization. Orthop Clin
North Am. 47:253–264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nguyen QT and Tsien RY:
Fluorescence-guided surgery with live molecular navigation-a new
cutting edge. Nat Rev Cancer. 13:653–662. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gorphe P and Simon C: A systematic review
and meta-analysis of margins in transoral surgery for oropharyngeal
carcinoma. Oral Oncol. 98:69–77. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boonstra MC, De Geus SWL, Prevoo HAJM,
Hawinkels LJAC, van de Velde CJH, Kuppen PJK, Vahrmeijer AL and
Sier CFM: Selecting targets for tumor imaging: An overview of
cancer–associated membrane proteins. Biomark Cancer. 8:119–133.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van Oosten M, Crane LMA, Bart J, van
Leeuwen FW and van Dam GM: Selecting potential targetable
biomarkers for imaging purposes in colorectal cancer using target
selection criteria (TASC): A novel target identification tool.
Transl Oncol. 4:71–82. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boonstra MC, van Driel PBAA, van Willigen
DM, Stammes MA, Prevoo HAJM, Tummers QRJG, Mazar AP, Beekman FJ,
Kuppen PJK, van de Velde CJH, et al: uPAR-targeted multimodal
tracer for pre- and intraoperative imaging in cancer surgery.
Oncotarget. 6:14260–14273. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Christensen A, Juhl K, Persson M, Charabi
BW, Mortensen J, Kiss K, Lelkaitis G, Rubek N, von Buchwald C and
Kjær A: uPAR-targeted optical near-infrared (NIR) fluorescence
imaging and PET for image-guided surgery in head and neck cancer:
Proof-of-concept in orthotopic xenograft model. Oncotarget.
8:15407–15419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nielsen CH, Jeppesen TE, Kristensen LK,
Jensen MM, Ali HHE, Madsen J, Wiinberg B, Petersen LC and Kjaer A:
PET imaging of tissue factor in pancreatic cancer using
64Cu-labeled active site-inhibited factor VII. J Nucl Med.
57:1112–1119. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Geus SWL, Baart VM, Boonstra MC, Kuppen
PJK, Prevoo HA, Mazar AP, Bonsing BA, Morreau H, van de Velde CJH,
Vahrmeijer AL and Sier CF: Prognostic impact of urokinase
plasminogen activator receptor expression in pancreatic cancer:
Malignant versus stromal cells. Biomark Insights.
12:11772719177154432017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boonstra MC, Verspaget HW, Ganesh S,
Kubben FJGM, Vahrmeijer AL, van de Velde CJH, Kuppen PJK, Quax PHA
and Sier CFM: Clinical applications of the urokinase receptor
(uPAR) for cancer patients. Curr Pharm Des. 17:1890–1910. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Noh H, Hong S and Huang S: Role of
urokinase receptor in tumor progression and development.
Theranostics. 3:487–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hundsdorfer B, Zeilhofer HF, Bock KP,
Dettmar P, Schmitt M, Kolk A, Pautke C and Horch HH:
Tumour-associated urokinase-type plasminogen activator (uPA) and
its inhibitor PAI-1 in normal and neoplastic tissues of patients
with squamous cell cancer of the oral cavity-clinical relevance and
prognostic value. J Craniomaxillofac Surg. 33:191–196. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bharadwaj AG, Holloway RW, Miller VA and
Waisman DM: Plasmin and plasminogen system in the tumor
microenvironment: Implications for cancer diagnosis, prognosis, and
therapy. Cancers (Basel). 13:18382021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Madunić J: The urokinase plasminogen
activator system in human cancers: An overview of its prognostic
and predictive role. Thromb Haemost. 118:2020–2036. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Serpa MS, Mafra RP, Queiroz SIML, da Silva
LP, de Souza LB and Pinto LP: Expression of urokinase-type
plasminogen activator and its receptor in squamous cell carcinoma
of the oral tongue. Braz Oral Res. 32:e932018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van Keulen S, van den Berg NS, Nishio N,
Birkeland A, Zhou Q, Lu G, Wang HW, Middendorf L, Forouzanfar T,
Martin BA, et al: Rapid, non-invasive fluorescence margin
assessment: Optical specimen mapping in oral squamous cell
carcinoma. Oral Oncol. 88:58–65. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weele EJT, Van Scheltinga AGT, Linssen MD,
Nagengast WB, Lindner I, Jorritsma-Smit A, de Vries EGE, Kosterink
JGW and Hooge MN: Development, preclinical safety, formulation, and
stability of clinical grade bevacizumab-800CW, a new near infrared
fluorescent imaging agent for first in human use. Eur J Pharm
Biopharm. 104:226–234. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zaryouh H, De Pauw I, Baysal H, Peeters M,
Vermorken JB, Lardon F and Wouters A: Recent insights in the
PI3K/Akt pathway as a promising therapeutic target in combination
with EGFR-targeting agents to treat head and neck squamous cell
carcinoma. Med Res Rev. 42:112–155. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Beleva E and Grudeva-Popova J: From
Virchow's triad to metastasis: circulating hemostatic factors as
predictors of risk for metastasis in solid tumors. J BUON.
18:25–33. 2013.PubMed/NCBI
|
|
23
|
Han X, Guo B, Li Y and Zhu B: Tissue
factor in tumor microenvironment: A systematic review. J Hematol
Oncol. 7:542014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Theunissen JW, Cai AG, Bhatti MM, Cooper
AB, Avery AD, Dorfman R, Guelman S, Levashova Z and Migone TS:
Treating tissue factor-positive cancers with antibody-drug
conjugates that do not affect blood clotting. Mol Cancer Ther.
17:2412–2426. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zamani M, Grønhøj C, Jensen DH, Carlander
AF, Agander T, Kiss K, Olsen C, Baandrup L, Nielsen FC, Andersen E,
et al: The current epidemic of HPV-associated oropharyngeal cancer:
An 18-year Danish population-based study with 2,169 patients. Eur J
Cancer. 134:52–59. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brierley JD, Gospodarowicz MK and
Wittekind C: TNM classification of malignant tumours. 8th edition.
Wiley Blackwell; 2017
|
|
27
|
Allred DC, Harvey JM, Berardo M and Clark
GM: Prognostic and predictive factors in breast cancer by
immunohistochemical analysis. Mod Pathol. 11:155–168.
1998.PubMed/NCBI
|
|
28
|
Bossi P, Resteghini C, Paielli N, Licitra
L, Pilotti S and Perrone F: Prognostic and predictive value of EGFR
in head and neck squamous cell carcinoma. Oncotarget.
7:74362–74379. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Preuss SF, Weinell A, Molitor M, Semrau R,
Stenner M, Drebber U, Wedemeyer I, Hoffmann TK, Guntinas-Lichius O
and Klussmann JP: Survivin and epidermal growth factor receptor
expression in surgically treated oropharyngeal squamous cell
carcinoma. Head Neck. 30:1318–1324. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chandarana SP, Lee JS, Chanowski EJP,
Sacco AG, Bradford CR, Wolf GT, Prince ME, Moyer JS, Eisbruch A,
Worden FP, et al: Prevalence and predictive role of p16 and
epidermal growth factor receptor in surgically treated
oropharyngeal and oral cavity cancer. Head Neck. 35:1083–1090.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Christensen A, Kiss K, Lelkaitis G, Juhl
K, Persson M, Charabi BW, Mortensen J, Forman JL, Sørensen AL,
Jensen DH, et al: Urokinase-type plasminogen activator receptor
(uPAR), tissue factor (TF) and epidermal growth factor receptor
(EGFR): Tumor expression patterns and prognostic value in oral
cancer. BMC Cancer. 17:5722017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gao RW, Teraphongphom NT, van den Berg NS,
Martin BA, Oberhelman NJ, Divi V, Kaplan MJ, Hong SS, Lu G, Ertsey
R, et al: Determination of tumor margins with surgical specimen
mapping using near-infrared fluorescence. Cancer Res. 78:5144–5154.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Baart VM, van Duijn C, van Egmond SL,
Dijckmeester WA, Jansen JC, Vahrmeijer AL, Sier CFM and Cohen D:
EGFR and αvβ6 as promising targets for molecular imaging of
cutaneous and mucosal squamous cell carcinoma of the head and neck
region. Cancers (Basel). 12:14742020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
de Boer E, Warram JM, Tucker MD, Hartman
YE, Moore LS, de Jong JS, Chung TK, Korb ML, Zinn KR, van Dam GM,
et al: In vivo fluorescence immunohistochemistry: Localization of
fluorescently labeled cetuximab in squamous cell carcinomas. Sci
Rep. 5:101692015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Voskuil FJ, de Jongh SJ, Hooghiemstra WTR,
Linssen MD, Steinkamp PJ, de Visscher SAHJ, Schepman KP, Elias SG,
Meersma GJ, Jonker PKC, et al: Fluorescence-guided imaging for
resection margin evaluation in head and neck cancer patients using
cetuximab-800CW: A quantitative dose-escalation study.
Theranostics. 10:3994–4005. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cramer JD, Burtness B and Ferris RL:
Immunotherapy for head and neck cancer: Recent advances and future
directions. Oral Oncol. 99:1044602019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wojtukiewicz MZ, Sierko E, Zacharski LR,
Rózanska-Kudelska M and Zimnoch L: Occurrence of components of
fibrinolytic pathways in situ in laryngeal cancer. Semin Thromb
Hemost. 29:317–320. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bacchiocchi R, Rubini C, Pierpaoli E,
Borghetti G, Procacci P, Nocini PF, Santarelli A, Rocchetti R,
Ciavarella D, Lo Muzio L and Fazioli F: Prognostic value analysis
of urokinase-type plasminogen activator receptor in oral squamous
cell carcinoma: An immunohistochemical study. BMC Cancer.
8:2202008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
van der Waal I: Potentially malignant
disorders of the oral and oropharyngeal mucosa; terminology,
classification and present concepts of management. Oral Oncol.
45:317–323. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Curino A, Patel V, Nielsen BS, Iskander
AJ, Ensley JF, Yoo GH, Holsinger FC, Myers JN, El-Nagaar A, Kellman
RM, et al: Detection of plasminogen activators in oral cancer by
laser capture microdissection combined with zymography. Oral Oncol.
40:1026–1032. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lechien JR, Seminerio I, Descamps G, Mat
Q, Mouawad F, Hans S, Julieron M, Dequanter D, Vanderhaegen T,
Journe F and Saussez S: Impact of HPV infection on the immune
system in oropharyngeal and non-oropharyngeal squamous cell
carcinoma: A systematic review. Cells. 8:10612019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Subbarayan RS, Arnold L, Gomez JP and
Thomas SM: The role of the innate and adaptive immune response in
HPV-associated oropharyngeal squamous cell carcinoma. Laryngoscope
Investig Otolaryngol. 4:508–512. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gonias SL and Zampieri C: Plasminogen
receptors in human malignancies: Effects on prognosis and
feasibility as targets for drug development. Curr Drug Targets.
21:647–656. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Magnussen S, Rikardsen OG, Hadler-Olsen E,
Uhlin-Hansen L, Steigen SE and Svineng G: Urokinase plasminogen
activator receptor (uPAR) and plasminogen activator inhibitor-1
(PAI-1) are potential predictive biomarkers in early stage oral
squamous cell carcinomas (OSCC). PLoS One. 9:e1018952014.
View Article : Google Scholar : PubMed/NCBI
|