Introduction
A total of 3 homologous genes of opiorphin, OPRPN
(known as ProL1 previously), submaxillary gland androgen-regulated
protein 3A (SMR3A) and SMR3B (1)
are known in humans. OPRPN-derived opiorphin was reported to act as
a potent inhibitor of two cell membrane-bound
enkephalin-inactivating peptidases, neutral endopeptidase (NEP;
CD10) and aminopeptidase N (APN; CD13) (2). CD10 and CD13 are widely distributed
among a wide range of tissues and organs, which regulate signaling
pathways mediating cell proliferation, survival and migration
(3–5). Dysregulated expression of both
proteins has been identified in various human tumor entities, such
as pancreas, gastric, prostate, breast, lung and oral carcinomas
(6–9).
Head and neck squamous cell carcinoma (HNSCC) is one
of the most common human malignancies worldwide with a yearly
incidence of 600,000 cases (10).
The standard pillars of therapy are surgery, irradiation (IR),
chemotherapy or a combination of these (11). Although treatment strategies have
improved in the last decades, the global 5-year-survival rate for
all HNSCC sites remains low at only 40–50% (10). Tumor recurrence after radiotherapy
frequently develops and significantly hampers rehabilitation
(12). The identification of
valuable biomarkers and radioresistant molecules contributing to
poor clinical outcomes may enable patient stratification to
properly select a therapeutic regimen (13). Our previous data revealed variable
protein expression patterns for both CD10 and CD13 in a cohort of
patients with oropharyngeal squamous cell carcinoma (OPSCC). In
addition, strong SMR3A protein expression was also found in 36% of
all primary OPSCC in a tissue microarray which served as an
unfavorable risk factor for clinical prognosis (14). Notably, an enrichment of
SMR3A-positive cells was observed in the fraction of vital HNSCC
cells after fractionated IR which was dependent on estrogen
receptor 2 (ESR2) signaling (15).
Moreover, dysregulated OPRPN was reported to be associated with
invasion in breast cancer (16).
To date, the understanding of the clinical relevance of opiorphin
proteins in head and neck cancer is limited.
In the present study, OPRPN protein levels were
investigated by immunohistochemical (IHC) staining on primary tumor
samples of OPSCC patients. The association between expression
patterns of OPRPN and SMR3A with clinical and histopathological
features as well as progression-free (PFS) and disease-specific
survival (DSS) were addressed. A potential function of OPRPN and
SMR3A after fractionated IR, in part mimicking a clinically applied
treatment protocol, were also highlighted.
Materials and methods
HNSCC cell lines
Human HNSCC cell lines FaDu and Cal27 were purchased
from the American Type Culture Collection (https://www.lgcstandards-atcc.org). FaDu cells were
established from a hypopharyngeal SCC. Cal27, derived from the
tongue, was described earlier as adenosquamous carcinoma (17), a rather aggressive subtype of oral
SCC (18). Detroit562 cells
originating from a metastatic pharyngeal carcinoma were purchased
from CLS (CLS Cell Lines Service GmbH). By this selection of HNSCC
cell lines depicting different origins/localizations the intention
was to approximate the heterogeneous features of HNSCC. Cells were
maintained in Dulbecco's Modified Eagle's Medium (DMEM)
supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc.), 2 mM L-glutamine and 50 µg/ml
penicillin-streptomycin antibiotics and sterile conditions with 6%
CO2 at 37°C. Cell cultures were regularly screened to
exclude mycoplasma contamination (Venor®GeM Classic
Mycoplasma Detection Kit; Minerva Biolabs GmbH) according to
manufacturer's recommendation, and the authentication of all cell
lines was confirmed by the Multiplex Human Cell Line Authentication
Test.
IR of cell cultures and
immunofluorescence (IF)
A total of 10,000 cells were seeded on sterile
coverslips in a 12-well plate and cultured for 24 h. Cells were
irradiated on four consecutive days with a daily dose of 2 gray
(Gy) using X-RAD 320 (Precision X-Ray). Controls were mock-treated.
Cells were fixed with 4% paraformaldehyde for 15 min at room
temperature. After being washed with phosphate-buffered saline
(PBS) three times, the cells were permeabilized with 0.5% Triton
X-100 buffer in PBS for 30 min, and after being rewashed three
times with PBS, they were blocked with 1% BSA (Sigma-Aldrich; Merck
KGaA)/0.2% Tween-20 (GERBU Biotechnik Gmbh) in 1X PBS for 30 min at
room temperature. The primary antibodies anti-PROL1 (1:200; cat.
no. ab169504) and anti-SMR3A (1:100; cat. no. ab97942; both from
Abcam) were diluted in T-buffer with indicated concentrations and
were incubated with cells for 1 h at room temperature or overnight
at 4°C. Following washing with PBS three times, the secondary
antibody (1:200; cat. no. SAB4600234; Sigma-Aldrich; Merck KGaA),
conjugated with Hoechst 33342 (BIOMOL International), was diluted
in T-buffer and was added for 30 min at room temperature in the
dark. Finally, cells were again washed with PBS three times and
embedded on glass slides with Mowiol (Sigma-Aldrich; Merck KGaA).
The glass slides were kept in the dark at 4°C for at least 12 h
before images were captured. Images were captured using a
fluorescence microscope (model, BX50F), Olympus XC30 Camera and
cellSens Entry imaging software (Olympus Soft Imaging Solutions
GmbH). Images were acquired under identical imaging conditions.
Mean fluorescence intensity was quantified using the ImageJ
software version 1.8 (National Institutes of Health).
Patients
A total of 96 patients with primary OPSCC who were
diagnosed and treated between 1990 and 2009 were comprised in the
retrospective study cohort. The mean age of the cohort was 58.9
years, and 70 of the patients were male. Samples were obtained at
the Department of Otorhinolaryngology, Head and Neck Surgery of
Heidelberg University Hospital (Heidelberg, Germany) during
diagnostic or therapeutic procedures. Biopsies of non-surgically
treated patients, as well as samples of patients who underwent
tumor surgery, were included. All subjects provided written
informed consent for data collection as it is a standard procedure
in our department. Patients with suspicious clinical findings who
underwent diagnostic panendoscopy and/or patients before tumor
surgery with a histologically confirmed diagnosis of OPSCC were
asked to consent. The protocol was approved (approval no. 176/2002)
by the Ethics Committee of the Medical Faculty of the University of
Heidelberg (Heidelberg, Germany) in accordance with The Declaration
of Helsinki in existing version from 1996. Experimental treatment
procedures were not part of the present study. The patients were
treated according to the guidelines for head and neck cancer. The
final analysis was based on 96 patients with OPSCC who were treated
with either definitive or post-surgical radiotherapy with or
without adjuvant chemotherapy. Clinical and therapeutic follow-up
of the cohort was assessed retrospectively (Table SI).
Tissue microarrays (TMAs) and IHC
TMAs were produced as previously described (19–21).
In brief, H&E-stained sections were cut from each donor block
to define representative tumor regions. From selected areas of each
donor block, small tissue cylinders with a diameter of 0.6 mm were
received using a tissue chip microarrayer (Beecher Instruments
Inc.) and transferred to a recipient paraffin block. By using
standard techniques, 2 µm paraffin sections were cut from this
recipient paraffin block. TMAs were stained with an Anti-OPRPN
(1:300; cat. no. ab204562; Abcam) and immunostaining was visualized
with the TSA Amplification Kit (PerkinElmer, Inc.) and DAB
peroxidase substrate (Vector Laboratories, Inc.), according to the
manufacturer's instructions. Counterstaining was performed by
hematoxylin to visualize tissue integrity. Stained TMAs were
scanned using the Nanozoomer HT Scan System (Hamamatsu Photonics
K.K.) and were evaluated by three independent observers using the
NDP Viewer software (version 1.1.27; Hamamatsu Photonics K.K.).
Evaluation considered the relative amount of positive cancer cells
(score 1=no positive cell, score 2 ≤33%, 33%> score 3 ≤66%,
score 4 >66%) and the staining intensity (score 1=no, score
2=low, score 3=moderate, score 4=high). Both values were multiplied
to calculate the final immunoreactivity score (IRS; range 1–16),
and the cut-off value for further analysis was:
OPRPNhigh >9 and OPRPNlow ≤9. Data on the
IRS for SMR3A were available from previous studies (22,23).
Ex vivo culture
Fresh tumor samples from the oropharynx were
procured immediately after surgical resection at the Department of
Otorhinolaryngology, Head and Neck Surgery, Heidelberg University
Hospital (Heidelberg, Germany). Informed consent was obtained after
the review of the local ethics board (ethic vote S-396/2012).
Samples were processed as previously described (21). For ex vivo analysis of tumor
response to fractionated IR, tumor sections were maintained in
six-well plates with inserts (Thinsert; Greiner Bio-One) in DMEM,
supplemented with 10% fetal bovine serum and antibiotics
(penicillin 100 U/ml and streptomycin 100 µg/ml). After one day in
culture, samples were irradiated with an intensity of 2 Gy on four
consecutive days. Non-treated controls were processed in parallel.
The medium was changed every second day. The tissue slices were
harvested 72 h posttreatment to be evaluated for histopathological
and IHC features. The sample is exemplarily depicted in Fig. 1.
Statistical analysis
SPSS 22 for Windows (IBM Corp.) and GraphPad Prism
version 9.1 (GraphPad Software, Inc.) were used for statistical
analysis. Fluorescence intensities were quantified using the ImageJ
and compared by unpaired Student's t-test. Person and Spearman's
correlation analysis between score A and B were performed before
they were multiplied. Correlations between OPRPN expression and
clinical and histopathological parameters (sex, age, tumor size,
lymph node metastases, tumor grade) as well as risk factors
(smoking, alcohol consumption, HPV status) were calculated by the
cross table and chi-square test. P<0.05 was considered to
indicate a statistically significant difference. Disease-specific
survival (DSS) and progression-free survival (PFS) data were
plotted by Kaplan-Meier survival curves. Differences between groups
were assumed using log-rank testing. Univariate and multivariate
Cox proportional hazard models were applied to define the
interdependence between multiple parameters and prognosis (DSS,
PFS) by using the approach ‘enter’. The endpoint PFS was defined as
the first appearance of an event that was counted for PFS. These
are tumor progression, recurrence, metastasis and secondary
tumors.
Results
Expression of OPRPN and SMR3A in HNSCC
cell lines and ex vivo tumor tissues response to fractionated
IR
It was previously demonstrated that SMR3A is
expressed in HNSCC cell lines at low levels, but the relative
amount of SMR3A-positive cells was increased after fractionated IR
(15). To investigate whether
OPRPN is postradiogenically upregulated and if there is a
co-expression with SMR3A in HNSCC cell lines, double
co-immunofluorescence staining (co-IF) was performed in FaDu, Cal27
and Detroit562 cells. Analogously to SMR3A, basal OPRPN protein
expression was low and could be detected only in a sub-fraction of
three cell lines (Fig. 1A).
However, upon fractionated IR prominent OPRPN expression was
observed in the majority of cells and was co-expressed with SMR3A
(Fig. 1A). The quantitative
analysis revealed that OPRPN and SMR3A protein expression were
significantly induced upon fractionated IR in both FaDu and
Detroit562 cells. However, the SMR3A induction as compared with
control-treated cells was highly significant, while OPRPN was found
without significant changes in Cal27 cells (Fig. 1B and C). To adapt these findings to
the clinical setting, a rapid and cost-effective patient-derived
explant ex vivo culture technique was developed for
evaluating the therapeutic response of fresh tumor tissues from
surgical resection. Elevated levels of SMR3A and OPRPN protein
expression were observed by IHC staining post-fractionated IR
(Fig. 1D). Thus, our ex
vivo culture data confirmed the findings from the HNSCC cell
lines.
Expression of OPRPN in primary OPSCC
and correlation with clinicopathological features
Data on SMR3A staining were in part available from
previous retrospective studies (14,15).
To determine whether aberrant OPRPN expression is relevant for the
pathogenesis and/or the clinical outcome of OPSCC, the TMAs with
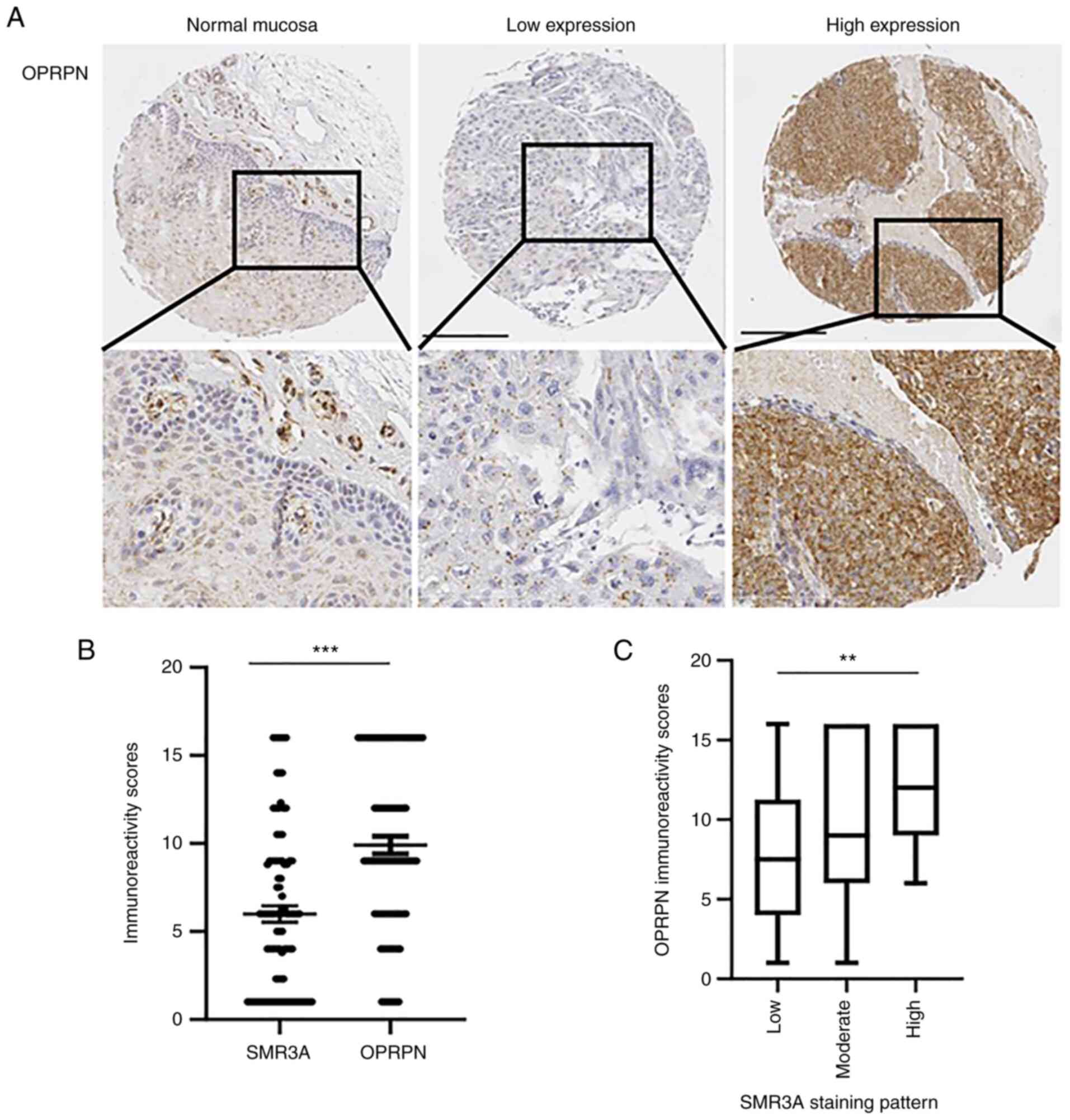
tissue samples of normal mucosa and OPSCC were stained by IHC.
Similarly, weak staining of OPRPN was observed in basal and
supra-basal keratinocytes of normal mucosa, which served as a
reference. In primary OPSCC, a heterogeneous staining pattern of
OPRPN protein was observed, which ranged from low to high
expression (Fig. 2A). The relative
number of positive tumor cells and the staining intensity were
estimated by three independent observers. Both scores revealed a
significant correlation (Spearman's correlation of 0.428 and
Pearson's correlation of 0.437) and were multiplied to obtain a
final OPRPN IRS for further analysis. The OPRPN IRS was
significantly higher than that of SMR3A protein levels (Fig. 2B). A strong expression of SMR3A was
significantly associated with a high OPRPN IRS, underlining the
strong correlation between the two proteins (Fig. 2C).
Subsequently, OPRPN and SMR3A expression patterns
and clinicopathological features were compared, including age, sex,
TNM status (AJCC Cancer Staging Manual 7th ed), pathological grade,
HPV status, smoking and alcohol consumption. However, these
parameters were not significantly correlated with OPRPN protein
levels (Table SI). These data are
in line with previous findings that SMR3A has no correlation with
clinicopathological features in OPSCC (14). Accordingly, the regulation of the
opiorphin gene family is independent of initial events during
neoplastic transformation and the malignant progression of
OPSCC.
Correlation of OPRPN and SMR3A
expression with disease-specific survival (DSS) and PFS
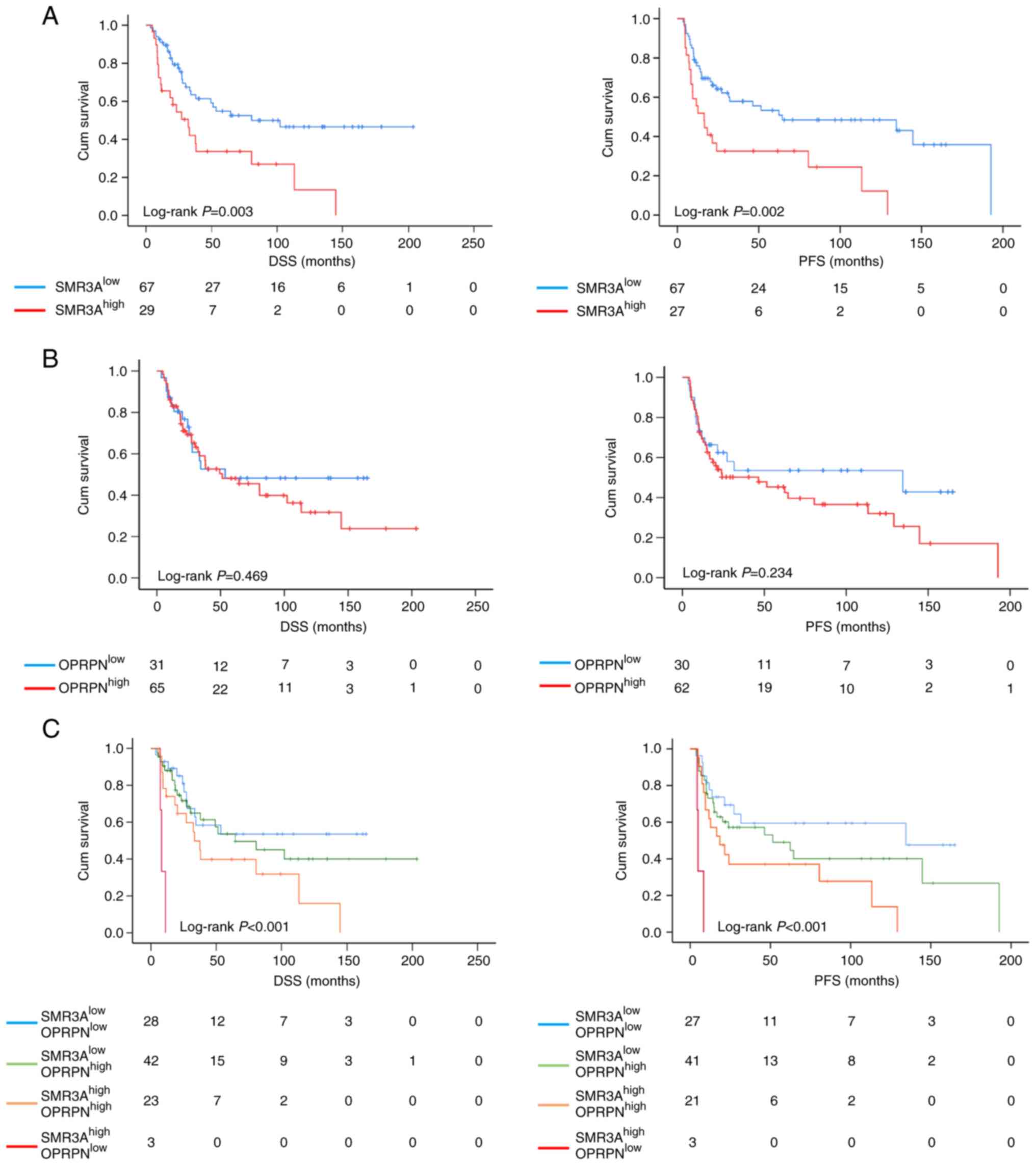
Next, to investigate the prognostic value of OPRPN
and SMR3A, patients were arranged into two categories according to
the IRS: patients with low protein expression of OPRPN
(OPRPNlow) and SMR3A (SMR3Alow) and those
with high protein expression of OPRPN (OPRPNhigh) and
SMR3A (SMR3Ahigh), respectively. Patients with high IRS
of SMR3A exhibited a poor DSS and PFS compared with patients with
low SMR3A levels, which was highly significant (DSS, P=0.003; PFS,
P=0.002) (Fig. 3A). This is in
line with a previous finding that apparently opiorphin family
members contribute to poor radiosensitivity in head and neck cancer
(15). SMR3A was considered as a
surrogate marker for the active signaling of ESR2. The prognostic
value of OPRPN expression for DSS and PFS of OPSCC patients was
analyzed by Kaplan-Meier plots and log-rank testing. However, no
statistically significant difference was observed for DSS and PFS
(Fig. 3B). Next, a combinatorial
analysis was accomplished in the subgroup of OPSCC patients, which
were treated with either definitive or post-surgical radiotherapy
with or without chemotherapy. Concerning DSS and PFS,
SMR3AhighOPRPNhigh staining pattern tumor
revealed an unfavorable clinical outcome as compared with other
subgroups (Fig. S1).
Notably, the subgroup of OPSCC patients with
SMR3Ahigh OPRPNlow staining pattern displayed
the worst clinical outcome in terms of the shortest DSS
(P<0.001) and PFS (P<0.001) (Fig. 3C).
SMR3A but not OPRPN serves as an
independent prognostic biomarker for the survival of OPSCC patients
with radiotherapy
Consequently, univariate analyses revealed that
SMR3Ahigh staining pattern significantly correlated with
shorter DSS [hazard ratio (HR), 2.286; 95% Confidence Interval (95%
CI), 1.293-4.041; P=0.004] and PFS (HR, 2.324; 95% CI, 1.327-4.071;
P=0.003), which suggested the patients with elevated SMR3A
expression are at increased risk for treatment failure, presumably
due to resistance against IR. It is worth noting that combinational
analysis with OPRPNhigh was mitigating the predictive
effect of SMR3A. OPRPNhighSMR3ahigh remained
significant on univariate analyses concerning DSS (HR, 1.826; 95%
CI, 1.019-3.272; P=0.043) and PFS (HR, 1.869; 95% CI, 1.047-3.338;
P=0.034) (Fig. 4). To adjust for
all available clinical parameters, a multivariate Cox regression
model was applied to confirm that patients with
SMR3Ahigh staining pattern had an unfavorable DSS (HR,
2.440; 95% CI, 1.187-5.018; P=0.015) and PFS (HR, 2.323; 95% CI,
1.181-4.570; P=0.015) (Fig. 5).
Notably, OPRPNlow SMR3Ahigh staining pattern
serves as the most unfavorable independent prognostic biomarker
(Table SII). These data indicated
that SMR3A and OPRPN serve as potential prognostic markers for
HNSCC after definitive surgery and adjuvant radiotherapy.
Discussion
The present study is the first report associating
elevated expression levels of opiorphin members with prognosis in
OPSCC cohorts with radiotherapy. The present data suggested that
the expression levels of OPRPN together with SMR3A correlate with
treatment failure after radiotherapy. At present, treatment of head
and neck cancer has been significantly improved by novel
radiotherapy techniques and protocols (24). Radiotherapy is applied as primary
or as adjuvant therapy after surgery and ~75% of HNSCC patients
will benefit (25). However,
intrinsic and acquired radioresistance remains a major barrier to
curative therapeutic approaches in HNSCC. It is crucial to unravel
the molecular mechanisms of radioresistance and to identify new
biomarkers for HNSCC patients at high risk for treatment
failure.
A previous study demonstrated that the mouse homolog
of human SMR3A gene, Smr1 is differentially expressed in primary
and recurrent tumors of an orthotopic mouse xenograft model for
oral cancer (26). Upon
fractionated IR SMR3A was shown to be prominently expressed in
vital tumor cells (15). It was
assumed as a surrogate for resistant tumor cells which are a
putative source for relapse after radiotherapy. High SMR3A
expression was furthermore described as a risk factor for
unfavorable PFS and overall survival in OPSCC patients (14). Nevertheless, no impact of ectopic
SMR3A expression was identified on tumor-relevant processes under
normal growth conditions, which suggested that SMR3A has no major
impact on tumor cell physiology under normal growth conditions. It
was hypothesized that SMR3A serve as a biomarker for a
subpopulation of resistant cells as a putative source for tumor
relapse after radiotherapy. The findings also explain that high
SMR3A expression revealed an unfavorable outcome in a cohort of
OPSCC patients.
It is worth noting that an increased transcript
level of Muc10, the mouse homolog of the human OPRPN gene, another
member of the opiorphin gene family, was detected in recurrent
tumors following surgical resection as compared with their matching
primaries. This finding indicated a general principle of regulation
and function of opiorphin family members in recurrence progression
and treatment failure (26). So
far, the opiorphin family members have been linked to various
physiological and pathological conditions, such as erectile
dysfunction (ED), colonic motility and nociception, pain and mood
disorders and hypoxic response (1,27–30).
For instance, it was reported that the pentapeptide opiorphin is a
potent analgesic as it inhibits pain perception. The
pain-suppressive efficacy is equal to morphine in the behavioral
rat model, suggesting opiorphin may act as a potential initiator to
develop a novel candidate drug for pain control (2). Opiorphin has been identified as a
potent inhibitor of enkephalin-degrading enzymes, namely CD10 and
CD13 (6,9,31–34).
Moreover, positive regulation of opiorphin family members by
hormone signaling has been reported in several studies (35,36).
This is in line with our previous findings that suggested ESR2
signaling regulates SMR3A expression and plays an important role in
radioresistance (15).
A cell culture model of fractionated IR was now
presented, providing evidence for the existence and expansion of a
subpopulation of tumor cells, which are characterized by IR-induced
OPRPN and SMR3A expression. The prognostic significance of another
member of the opiorphin gene family, OPRPN (formerly known as
ProL-1), and its potential role in mediating radioresistance were
also investigated. Therapy response is affected by pronounced
intratumorigenic heterogeneity in HNSCC. The selection of
radioresistant tumor cell subclones after fractionated radiotherapy
as clinically applied is thereby facilitated. However, numerous
previous studies made statements on molecular mechanisms of
radioresistance after single-dose IR considerably exceeding a
dosage of 2 Gy (37–40). The clonal selection of
radioresistant tumor cells under fractionated-IR is hereby not
taken into account. Therefore, a clinically applied fractionated-IR
protocol was mimicked. Norm fractionation with 5×2 Gy (37) applied over 6–7 weeks, considered
standard treatment in HNSCC OPRPN, was found to be distinctly
upregulated in both cell lines (FaDu and Detroit562) as well as in
the ex vivo tumor culture after fractionated IR. Notably,
the characteristics of the cell lines that affect the opiorphin
protein expression regulated by IR should be clarified in more head
and neck cancer cell lines.
Experimental data of our research group demonstrated
that SMR3A is expressed in HNSCC cell lines at low levels but was
postradiogenically upregulated (15). Similar to SMR3A, basal OPRPN
protein expression was low and detected only in a sub-fraction of
both cell lines. However, upon fractionated IR, prominent OPRPN
expression was identified in the majority of cells and was
co-expressed with SMR3A. In a recent study, OPRPN was regarded as a
favorable predictive factor in OPSCC as the expression of OPRPN was
associated with consistently increased survival rates (41), which is in line with the present
findings. The subgroup displaying high SMR3A but low OPRPN
expression levels is the one with the worst clinical outcome, while
patients whose tumors express both high are only the second-worst.
These data may indicate that SMR3A and OPRPN are inverse
prognosticators of the clinical outcome, but SMR3A is the more
powerful prognosticator. However, in our retrospective study
patient cohort is small. A larger number of patients would be
recruited in the future prospective study to confirm the predictive
value of human opiorphin proteins for HNSCC after radiotherapy.
Nevertheless, the predictive power of OPRPN appears
to be minor and mitigates the impact of SMR3A. Furthermore, an
upregulation of both the opiorphin family members was observed,
which means that both markers are affected by standard HNSCC
treatment such as fractionated IR and respond by upregulation of
expression levels. The correlation between OPRPN expression pattern
and clinical outcome of patients with OPSCC missed statistical
significance. However, combined expression of OPRPN and SMR3A was
significantly associated with unfavorable clinical prognosis post
definitive or adjuvant radiotherapy, indicating opiorphin-related
genes serve as a surrogate marker for HNSCC cells with intrinsic
radioresistance (Fig. 6).
In conclusion, to the best of our knowledge, this is
the first study to provide the experimental evidence for a
predictive but probably antagonistic role of opiorphin genes in
OPSCC after radiotherapy. Although the patient number was small,
the subgroup with low OPRPN and high SMR3A expression presented the
worst outcome in terms of DSS and PFS. SMR3A and OPRPN are likely
to serve as potential prognostic markers in HNSCC. As there is a
severe lack of stable and reliable predictive and prognostic
biomarkers in HNSCC due to the heterogeneity of this entity,
validating the impact of OPRPN proteins is also a worthy subject
for studies with larger patient collectives.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors gratefully acknowledge the excellent
technical support of Leoni Erdinger, Ines Kaden, Nataly Henfling,
Antje Lehmann and Ingeborg Vogt. The authors would also like to
thank the tissue bank of the National Center for Tumor Diseases
(Institute of Pathology, University Hospital Heidelberg) for
providing the tumor specimens of OPSCC patients.
Funding
The study was supported by the National Natural Science
Foundation of China (grant no. 82103121), the Natural Science
Foundation of Jiangsu Province (grant no. BK20200878), the Priority
Academic Program Development of Jiangsu Higher Education
Institutions (PAPD) and by a scholarship for physicians of
Heidelberg University's Medical Faculty.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CR, PKP and AA conceptualized and designed the
study. PKP and AA supervised the study. CR, JG, JT, CLP, GM, DH,
GD, JK, AL, CS, NR and AA curated raw data and wrote-reviewed and
edited the manuscript. CR and AA confirm the authenticity of all
the raw data. JG and JT conducted experimental investigation. CR
and AA performed data analyses and visualization. All authors have
read and agreed to the published version of the manuscript.
Ethics approval and consent to
participate
Informed consent was obtained from all patients
after approval (approval no. 176/2002) by the Ethics Committee of
the Medical Faculty of the University of Heidelberg (Heidelberg,
Germany).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tong Y, Tar M, Melman A and Davies K: The
opiorphin gene (ProL1) and its homologues function in erectile
physiology. BJU Int. 102:736–740. 2008. View Article : Google Scholar
|
|
2
|
Wisner A, Dufour E, Messaoudi M, Nejdi A,
Marcel A, Ungeheuer MN and Rougeot C: Human opiorphin, a natural
antinociceptive modulator of opioid-dependent pathways. Proc Natl
Acad Sci USA. 103:17979–17984. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mizerska-Dudka M and Kandefer-Szerszeń M:
Opioids, neutral endopeptidase, its inhibitors and cancer: Is there
a relationship among them? Arch Immunol Ther Exp (Warsz).
63:197–205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maguer-Satta V, Besançon R and
Bachelard-Cascales E: Concise review: Neutral endopeptidase (CD10):
A multifaceted environment actor in stem cells, physiological
mechanisms, and cancer. Stem Cells. 29:389–396. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen L, Lin YL, Peng G and Li F:
Structural basis for multifunctional roles of mammalian
aminopeptidase N. Proc Natl Acad Sci USA. 109:17966–17971. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sørensen KD, Abildgaard MO, Haldrup C,
Ulhøi BP, Kristensen H, Strand S, Parker C, Høyer S, Borre M and
Ørntoft TF: Prognostic significance of aberrantly silenced ANPEP
expression in prostate cancer. Br J Cancer. 108:420–428. 2013.
View Article : Google Scholar
|
|
7
|
Piattelli A, Fioroni M, Iezzi G, Perrotti
V, Stellini E, Piattelli M and Rubini C: CD10 expression in stromal
cells of oral cavity squamous cell carcinoma: A clinic and
pathologic correlation. Oral Dis. 12:301–304. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Erhuma M, Köbel M, Mustafa T, Wulfänger J,
Dralle H, Hoang-Vu C, Langner J, Seliger B and Kehlen A: Expression
of neutral endopeptidase (NEP/CD10) on pancreatic tumor cell lines,
pancreatitis and pancreatic tumor tissues. Int J Cancer.
120:2393–2400. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kawamura J, Shimada Y, Kitaichi H, Komoto
I, Hashimoto Y, Kaganoi J, Miyake M, Yamasaki S, Kondo K and
Imamura M: Clinicopathological significance of aminopeptidase
N/CD13 expression in human gastric carcinoma.
Hepatogastroenterology. 54:36–40. 2007.PubMed/NCBI
|
|
10
|
Leemans CR, Snijders PJF and Brakenhoff
RH: The molecular landscape of head and neck cancer. Nat Rev
Cancer. 18:269–282. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Higgins GS, O'Cathail SM, Muschel RJ and
McKenna WG: Drug radiotherapy combinations: Review of previous
failures and reasons for future optimism. Cancer Treat Rev.
41:105–113. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Castilho RM, Squarize CH and Almeida LO:
Epigenetic modifications and head and neck cancer: Implications for
tumor progression and resistance to therapy. Int J Mol Sci.
18:15062017. View Article : Google Scholar
|
|
13
|
Caudell JJ, Torres-Roca JF, Gillies RJ,
Enderling H, Kim S, Rishi A, Moros EG and Harrison LB: The future
of personalised radiotherapy for head and neck cancer. Lancet
Oncol. 18:e266–e273. 2017. View Article : Google Scholar
|
|
14
|
Koffler J, Holzinger D, Sanhueza GA,
Flechtenmacher C, Zaoui K, Lahrmann B, Grabe N, Plinkert PK and
Hess J: Submaxillary gland androgen-regulated protein 3A expression
is an unfavorable risk factor for the survival of oropharyngeal
squamous cell carcinoma patients after surgery. Eur Arch
Otorhinolaryngol. 270:1493–1500. 2013. View Article : Google Scholar
|
|
15
|
Grünow J, Rong C, Hischmann J, Zaoui K,
Flechtenmacher C, Weber KJ, Plinkert P and Hess J: Regulation of
submaxillary gland androgen-regulated protein 3A via estrogen
receptor 2 in radioresistant head and neck squamous cell carcinoma
cells. J Exp Clin Cancer Res. 36:252017. View Article : Google Scholar
|
|
16
|
Lang Z, Wu Y, Pan X, Qu G and Zhang T:
Study of differential gene expression between invasive
multifocal/multicentric and unifocal breast cancer. J BUON.
23:134–142. 2018.PubMed/NCBI
|
|
17
|
Jiang L, Ji N, Zhou Y, Li J, Liu X, Wang
Z, Chen Q and Zeng X: CAL 27 is an oral adenosquamous carcinoma
cell line. Oral Oncol. 45:e204–e207. 2009. View Article : Google Scholar
|
|
18
|
Sravya T, Rao GV, Kumar MP and
Sudheerkanth K: Oral adenosquamous carcinoma: Report of a rare
entity with a special insight on its histochemistry. J Oral
Maxillofac Pathol. 20:5482016. View Article : Google Scholar
|
|
19
|
Holzinger D, Schmitt M, Dyckhoff G, Benner
A, Pawlita M and Bosch FX: Viral RNA patterns and high viral load
reliably define oropharynx carcinomas with active HPV16
involvement. Cancer Res. 72:4993–5003. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karsai S, Abel U, Roesch-Ely M, Affolter
A, Hofele C, Joos S, Plinkert PK and Bosch FX: Comparison of
p16(INK4a) expression with p53 alterations in head and neck cancer
by tissue microarray analysis. J Pathol. 211:314–322. 2007.
View Article : Google Scholar
|
|
21
|
Nasser W, Flechtenmacher C, Holzinger D,
Hofele C and Bosch FX: Aberrant expression of p53, p16INK4a and
Ki-67 as basic biomarker for malignant progression of oral
leukoplakias. J Oral Pathol Med. 40:629–635. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Horn D, Freudlsperger C, Holzinger D,
Kunzmann K, Plinkert P, Dyckhoff G, Hoffmann J, Freier K and Hess
J: Upregulation of pAKT(Ser473) expression in progression of
HPV-positive oropharyngeal squamous cell carcinoma. Head Neck.
39:2397–2405. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Freudlsperger C, Horn D, Weißfuß S,
Weichert W, Weber KJ, Saure D, Sharma S, Dyckhoff G, Grabe N,
Plinkert P, et al: Phosphorylation of AKT(Ser473) serves as an
independent prognostic marker for radiosensitivity in advanced head
and neck squamous cell carcinoma. Int J Cancer. 136:2775–2785.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Grégoire V, Langendijk JA and Nuyts S:
Advances in radiotherapy for head and neck cancer. J Clin Oncol.
33:3277–3284. 2015. View Article : Google Scholar
|
|
25
|
Barton MB, Jacob S, Shafiq J, Wong K,
Thompson SR, Hanna TP and Delaney GP: Estimating the demand for
radiotherapy from the evidence: A review of changes from 2003 to
2012. Radiother Oncol. 112:140–144. 2014. View Article : Google Scholar
|
|
26
|
Acuña Sanhueza GA, Faller L, George B,
Koffler J, Misetic V, Flechtenmacher C, Dyckhoff G, Plinkert PP,
Angel P, Simon C and Hess J: Opposing function of MYBBP1A in
proliferation and migration of head and neck squamous cell
carcinoma cells. BMC Cancer. 12:722012. View Article : Google Scholar
|
|
27
|
Fu S, Tar MT, Melman A and Davies KP:
Opiorphin is a master regulator of the hypoxic response in corporal
smooth muscle cells. FASEB J. 28:3633–3644. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Javelot H, Messaoudi M, Garnier S and
Rougeot C: Human opiorphin is a naturally occurring antidepressant
acting selectively on enkephalin-dependent delta-opioid pathways. J
Physiol Pharmacol. 61:355–362. 2010.
|
|
29
|
Tian XZ, Chen J, Xiong W, He T and Chen Q:
Effects and underlying mechanisms of human opiorphin on colonic
motility and nociception in mice. Peptides. 30:1348–1354. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mukherjee A, Park A, Wang L and Davies KP:
Role of opiorphin genes in prostate cancer growth and progression.
Future Oncol. 17:2209–2223. 2021. View Article : Google Scholar
|
|
31
|
Carl-McGrath S, Lendeckel U, Ebert M and
Röcken C: Ectopeptidases in tumour biology: A review. Histol
Histopathol. 21:1339–1353. 2006.
|
|
32
|
Fujita S, Yamamoto S, Akasu T, Moriya Y,
Taniguchi H and Shimoda T: Quantification of CD10 mRNA in
colorectal cancer and relationship between mRNA expression and
liver metastasis. Anticancer Res. 27:3307–3311. 2007.PubMed/NCBI
|
|
33
|
Luo Y, Fujii K, Ohmori H, Sasahira T,
Moriwaka Y, Isobe M and Kuniyasu H: Antisense phosphorothioate
oligodeoxynucleic acid for CD10 suppresses liver metastasis of
colorectal cancer. Pathobiology. 76:267–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kuniyasu H, Luo Y, Fujii K, Sasahira T,
Moriwaka Y, Tatsumoto N, Sasaki T, Yamashita Y and Ohmori H: CD10
enhances metastasis of colorectal cancer by abrogating the
anti-tumoural effect of methionine-enkephalin in the liver. Gut.
59:348–356. 2010. View Article : Google Scholar
|
|
35
|
Chua RG, Calenda G, Zhang X, Siragusa J,
Tong Y, Tar M, Aydin M, DiSanto ME, Melman A and Davies KP:
Testosterone regulates erectile function and Vcsa1 expression in
the corpora of rats. Mol Cell Endocrinol. 303:67–73. 2009.
View Article : Google Scholar
|
|
36
|
Señorale-Pose M, Jacqueson A, Rougeon F
and Rosinski-Chupin I: Acinar cells are target cells for androgens
in mouse submandibular glands. J Histochem Cytochem. 46:669–678.
1998. View Article : Google Scholar
|
|
37
|
Derer A, Spiljar M, Bäumler M, Hecht M,
Fietkau R, Frey B and Gaipl US: Chemoradiation increases PD-L1
expression in certain melanoma and glioblastoma cells. Front
Immunol. 7:6102016. View Article : Google Scholar
|
|
38
|
Guglas K, Kolenda T, Teresiak A,
Kopczyńska M, Łasińska I, Mackiewicz J, Mackiewicz A and Lamperska
K: lncRNA expression after irradiation and chemoexposure of HNSCC
cell lines. Noncoding RNA. 4:332018.
|
|
39
|
Ricco N, Flor A, Wolfgeher D, Efimova EV,
Ramamurthy A, Appelbe OK, Brinkman J, Truman AW, Spiotto MT and
Kron SJ: Mevalonate pathway activity as a determinant of radiation
sensitivity in head and neck cancer. Mol Oncol. 13:1927–1943. 2019.
View Article : Google Scholar
|
|
40
|
Todorovic V, Prevc A, Zakelj MN, Savarin
M, Brozic A, Groselj B, Strojan P, Cemazar M and Sersa G:
Mechanisms of different response to ionizing irradiation in
isogenic head and neck cancer cell lines. Radiat Oncol. 14:2142019.
View Article : Google Scholar
|
|
41
|
Wu Q, Cao R, Chen J and Xie X: Screening
and identification of biomarkers associated with
clinicopathological parameters and prognosis in oral squamous cell
carcinoma. Exp Ther Med. 18:3579–3587. 2019.PubMed/NCBI
|