Introduction
Genomic instability refers to genetic alterations
that occur at a higher-than-normal frequency and are caused by
dysfunctional genome maintenance programs. The term includes
changes at numerous levels, from single nucleotides to chromosomes
(1), mainly in the form of
microsatellite instability and chromosomal instability (CIN)
(2). Genomic instability, one of
the most prevalent features of human cancers, can cause cells to
exhibit the cancer phenotype through mutations in oncogenes and
cancer suppressor genes (3).
Persistent genomic instability allows cancer cells to survive under
selective pressure and adapt to their microenvironment by evolving
to resist different therapies (4),
and affects patient prognoses (5).
Although genomic instability can promote cancer development and
drug resistance, it can cause cancer cell death when genomic
instability continues to increase to a limiting level; thus,
genomic instability has therapeutic potential for treating cancer
(5–7), and elucidating the specific
regulatory mechanisms involved is of great significance. Several
molecular mechanisms work together to maintain genomic stability
under normal physiological conditions. For example, the precise
segregation of chromosomes during mitosis and the protection of
chromosome ends by telomeres ensures chromosomal stability
(8,9), whereas DNA repair, the most important
process in the DNA damage response, prevents genomic instability by
efficiently repairing DNA damage (3,10).
Dysregulation of these processes may lead to genomic instability
and the development of cancer. In addition, epigenetic aberrations
have been suggested as mechanisms underlying genomic instability
(11).
Recent studies have shown that long non-coding RNAs
(lncRNAs) are aberrantly expressed in various cancers and are
involved in regulating genomic instability in cancer cells
(12–14). LncRNAs are transcripts greater than
200 nt in length that do not encode proteins (15) and were considered to have no
biological function (16).
However, later studies have shown that some lncRNAs can encode
polypeptides (17) and interact
with proteins, DNA, and RNA to form functional complexes and
perform a variety of functions (18). Proteins are the main partners of
lncRNAs (16), and the proteins
that bind to RNAs are called RNA-binding proteins (RBPs), which
bind various RNAs, including lncRNAs, through their RNA-binding
domains (19). This interaction
between lncRNAs and RBPs plays a critical role in the genomic
instability of cancer cells, and by targeting the lncRNA-RBP axis,
cancer progression can be inhibited, showing some potential in
cancer therapy (20–25).
The present review summarizes the involvement of
lncRNAs in regulating genomic instability in cancer by binding
proteins that affect mitosis, telomere function, DNA repair, and
epigenetics. The study aimed to elucidate the regulatory networks
involved in genomic instability in cancer, which may contribute to
the development of novel cancer therapies. A systematic literature
search using PubMed was performed. The following key words were
used for the literature search: ‘lncRNA’, ‘RBP’, ‘genomic
instability’, ‘telomeres’, ‘mitosis’, ‘DNA repair’, and
‘epigenetic’. The articles in which lncRNAs regulate genomic
instability through RBPs in cancer cells were selected.
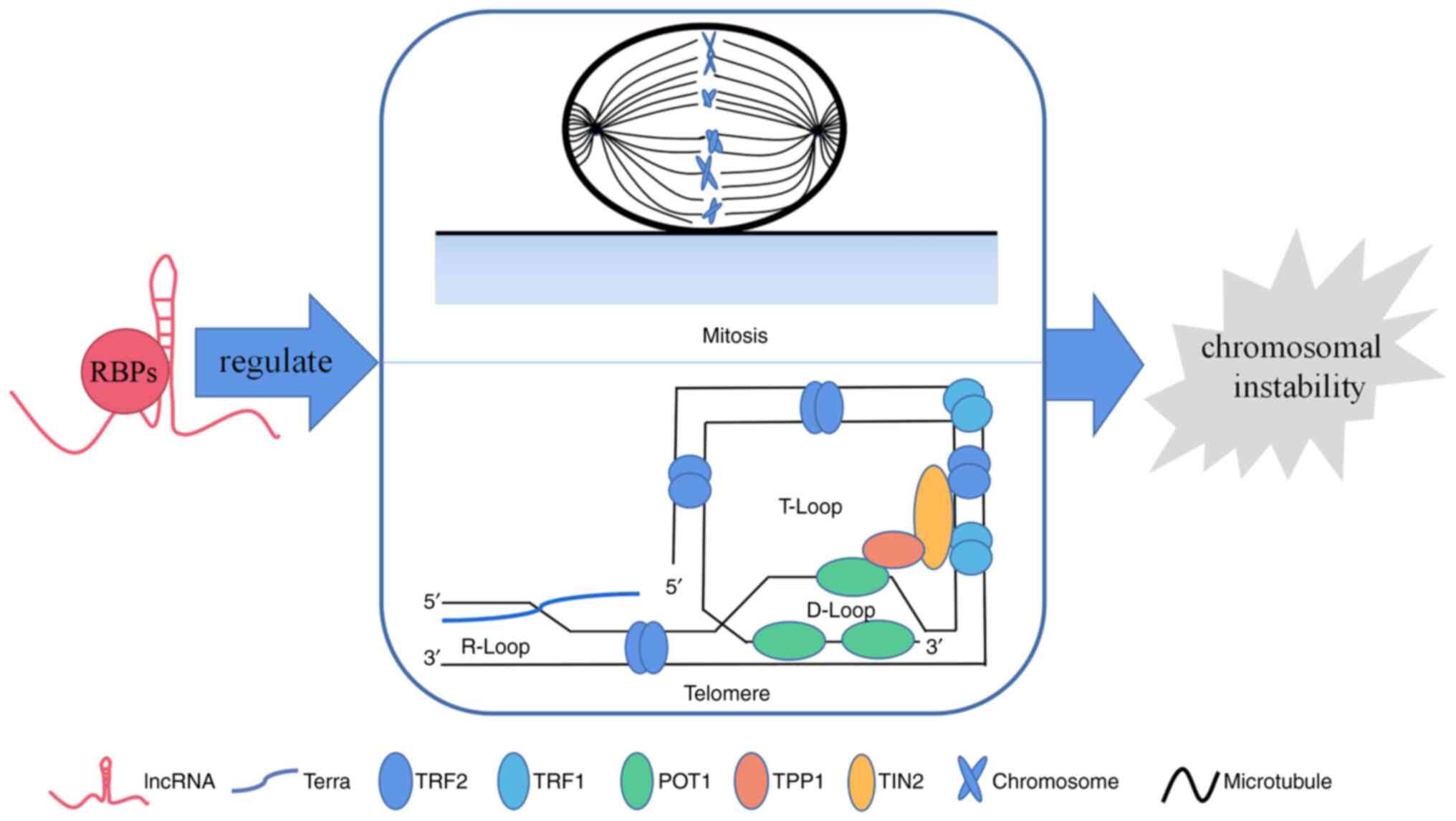
LncRNAs affect chromosome instability via
RBPs
As the most common form of genomic instability in
cancer, CIN is present in 60–80% of human tumors (26,27).
It is closely associated with the occurrence and development of
human cancers. On the one hand, CIN can promote tumor metastasis
and recurrence, accelerate the development of multi-drug resistance
in tumors, and be associated with poorer prognoses (28). On the other hand, exceedingly high
levels of CIN lead to sensitivity or even death of cancer cells
after exposure to cytotoxic drugs and radiotherapy (29). Both abnormal chromosome segregation
during mitosis and defects in telomere function contribute to CIN;
therefore, the role of lncRNA-protein binding in these processes is
reviewed (Fig. 1).
Mitosis
During mitosis, precise chromosome segregation
depends heavily on the precise binding of microtubules to each
sister chromatids (30). Ndc80 is
directly attached to microtubules and plays a central role in
stable kinetochore-microtubule junctions (31). In the case of incorrect
kinetochore-microtubule binding, Aurora B, phosphorylates Ndc80,
causing the kinetochore-microtubule binding to become unstable or
completely lose the ability to bind to microtubules (32). Concurrently, the spindle assembly
checkpoint detects the binding of the kinetochore and microtubules,
and transmits an unstable binding signal to the cell cycle. By
generating the mitotic checkpoint complex (MCC), it inhibits the
anaphase-promoting complex/cyclosome (APC/P). Through this
mechanism, mitotic cells are prevented from entering anaphase and
cell division until all kinetochore microtubules are stably bound
(33), thus preventing chromosome
missegregation and CIN.
LncRNAs can directly or indirectly affect proteins
involved in this sophisticated process through RBPs (Table I). The level of lncRNA CDKN2B-AS1
is markedly upregulated in renal clear cell carcinoma and is
significantly correlated with prognosis. Xie et al found
that CDKN2B-AS1 can bind directly to IGF2BP3 protein to stabilize
it while serving as a scaffold to bind to CBP and SMYD3 epigenetic
modification complexes to recruit them to the NUF2 promoter. This
mechanism stimulates NUF2 transcription and enhances its
cancer-promoting function (34).
NUF2 is a component of human Ndc80 that is required for stable
microtubule-positive end-binding sites in kinetochores; the precise
stoichiometry of the Ndc80 complex may play an important role in
microtubule binding (31,35). Stojic et al screened for
lncRNA linc00899 in HeLa cells by quantifying the effect of lncRNA
deletion on cell division. In this study it was determined that
linc00899 maintained genomic stability by inhibiting the expression
of microtubule-binding protein TPPP (a protein that stabilizes
microtubule networks and its overexpression inhibits microtubule
dynamics) through binding to chromatin-modifying complexes
(36). Moreover, an elevated level
of lncRNA CCAT2 in microsatellite stable colon cancer was revealed
to prolong the half-life of BOP1 by directly binding to BOP1;
overexpressed BOP1 increased the active form of Aurora B, and the
direct binding of CCAT2 to Aurora B also increased active Aurora B.
This was demonstrated to lead to incorrect segregation of
chromosomes and the occurrence of CIN, thus promoting the
progression of colon cancer. Colony formation ability and migration
ability of colon cancer cells were effectively inhibited by
knockout of BOP1 (20). Cdc20 and
Bub3 are components of MCC, and lncRNA CRYBG3 acts as a protein
decoy directly binding to Bub3, preventing Bub3 interaction with
CDC20, and thus activating APC/P and promoting abnormal mitosis.
This then leads to aneuploidy and the development of non-small cell
lung cancer. Inhibition of CRYBG3 was revealed to reduce the
ability of cancer to migrate in vitro and in vivo (21). Similarly, lncRNA NORAD induced
after DNA damage in HCT116 and other human cell lines can maintain
normal mitosis and chromosomal stability by binding to PUMILIO,
thereby interfering with PUMILIO binding and inhibiting its target
mRNAs (mainly including mRNAs such as chromosome cohesion complexes
and centromere complexes) (37).
In conclusion, the abovementioned evidence suggests that lncRNAs
are involved in the mitotic process by binding proteins that
regulate mitosis; however, the levels of induced CIN and the
specific biological functions of lncRNAs in cancer require further
validation.
 | Table I.LncRNAs interact with RBPs to affect
mitosis. |
Table I.
LncRNAs interact with RBPs to affect
mitosis.
| LncRNAs | RBPs | Mechanism | Effect | (Refs.) |
|---|
| CDKN2B-AS1 | SMYD3 and CBP | Promotes the
expression of NUF2 | Interferes with
normal mitosis | (34) |
| linc00899 | Chromatin-modifying
complexes | Inhibits TPPP
expression | Interferes with
normal mitosis | (36) |
| CCAT2 | BOP1 and Aurora
B | Increases the
active form of Aurora B | Interferes with
normal mitosis | (20) |
| CRYBG3 | Bub3 | Inhibits the
binding of Bub3 and CDC20 | Interferes with
normal mitosis | (21) |
| NORAD | PUMILIO | Inhibits the
binding of PUMILIO to its target mRNA | Protects normal
mitosis | (37) |
Telomeres
Human telomeres are DNA-protein complexes present at
the ends of chromosomes that consist of the non-coding DNA repeat
sequence TTAGGG and shelterin complexes (TRF1, TRF2, POT1, TIN2,
TPP1, and RAP1) to which they are bound. Its integrity is critical
to the stability of chromosomes (38). Telomeres are subsequently shortened
as cells divide due to end replication problems, and short
telomeres and defects in the shelterin complex fail to protect
chromosome ends leading to CIN. Cancer cells maintain telomere
length via the function of telomerase and use of the alternative
lengthening of telomeres (ALT) (9,39).
In addition, lncRNAs also play an important role in the maintenance
of telomeres via RBPs (Table
II).
 | Table II.LncRNAs regulate telomere function
through RBPs. |
Table II.
LncRNAs regulate telomere function
through RBPs.
| LncRNAs | RBPs | Mechanism | Effect | (Refs.) |
|---|
| TERRA | TRF2 | Promotes the
localization of TERRA in telomeres, and prevents TRF2 from binding
to telomeres | Regulates telomere
stability | (41,42) |
|
| TRF2, ORC | Promotes the
formation and maintenance of telomeric heterochromatin | Promotes telomere
elongation | (41) |
|
| TLS,
Histone-modifying enzyme, HP1α and β | Promotes telomeric
heterochromatin formation | Promotes telomere
elongation | (41,44) |
|
| HnRNP A1 | Regulates
telomerase activity in a dose-dependent manner | Regulates telomere
length | (48) |
|
| BRCA1 | Reduces telomeric
R-loop formation | Enhances telomere
stability | (49) |
|
| RAD51 | Promotes telomeric
R-loop formation | Reduces telomere
stability | (50) |
|
| TERT | Inhibits telomerase
activity | Prevents telomere
lengthening | (53) |
| HTR | TERT | Constitutes the
main active part of telomerase | Promotes telomere
elongation | (52) |
|
| Dyskerin, NOP10,
NHP2, TCAB1 and GAR1 | Components of
telomerase holoenzymes, maturation and localization of helper group
telomerase | Promotes telomere
elongation | (52) |
|
| PinX1 | Inhibits telomerase
activity | Prevents telomere
lengthening | (54) |
|
| Ku70/80 | Promotes telomere
capping | Enhances telomere
stability | (55,56) |
| CUDR | Cyclin D1 | Promotes telomerase
activity | Promotes telomere
elongation | (57) |
|
| P53
(N340Q/L344R) | Promotes telomerase
activity | Promotes telomere
elongation | (58) |
| HULC/MALAT1 | TRF2 | Promotes telomere
capping | Enhances telomere
stability | (59) |
| HULC | P53 | Inhibits telomere
capping | Reduces telomere
stability | (60) |
Telomeric repeat-containing RNA
(TERRA)
TERRA is a long non-coding RNA transcribed from
subtelomeric- and telomeric-derived sequences containing UUAGGG
repeats (40). Like telomeric DNA,
TERRA can also form a G-quadruplex structure and bind to the GAR
structural domain of TRF2, which is essential for TERRA
localization to telomeres. In the absence of the TERRA G-quadruplex
structure, telomeres bind more tightly to TRF2, which can promote
the formation of the telomeric T-loop. The quinoline derivative
CK-14 binds to the TERRA G-quadruplex to form a complex. This
complex binds to TRF2 and acts as an allosteric regulator of TRF2,
thereby preventing TRF2 from binding to telomeric DNA and
ultimately initiating the DNA damage response (DDR). In addition,
TERRA can bind to both the origin recognition complex (ORC) and
TRF2, enabling formation of a stable ternary complex, which is
involved in telomeric DNA replication and facilitates telomeric
heterochromatin formation and maintenance (41–43).
Similarly, translocated in liposarcoma (TLS) protein can bind to
both telomeric DNA and TERRA G-quadruplex structures, while TERRA
binds to histone-modifying enzymes, HP1α and β, and H3K9me3, which
play important roles in telomeric heterochromatin formation
(41,44). By contrast, telomeric
heterochromatin is a negative regulator of telomerase and ALT
elongation of telomeres (45,46).
Telomeric G-quadruplexes can also regulate telomere length by
preventing the binding of telomerase to telomeric DNA substrates.
Previous experiments have demonstrated that hnRNP A1 can facilitate
telomerase function by disrupting this high-level structure via
binding to telomeric DNA (47).
Interestingly, Redon et al determined that telomerase can
only function when TERRA and hnRNP A1 levels are balanced and bound
to form an inert complex. Moreover, excessive hnRNP A1 interferes
with telomerase activity by binding to telomeric DNA substrates
(48). In cancer cells lacking
telomerase, cells maintain telomere length primarily through ALT,
and the R-loop formed by telomeric DNA and TERRA facilitates this
process. BRCA1 binds directly to TERRA in an R-loop-dependent
manner and reduces R-loop formation; interference with its binding
leads to increased R-loops and telomere abnormalities. By contrast,
RAD51 can promote R-loop formation by binding TERRA (49,50).
Thus, TERRA plays an important role in various aspects of telomere
protein capping, heterochromatin formation, secondary structure
formation, and telomere lengthening. Owing to the complex role of
TERRA in telomeres, targeting its secondary structure or binding
proteins may be useful for cancer therapy.
hTR
Telomerase is a ribonucleoprotein complex consisting
of an RNA component (TERC) and the catalytic subunit of telomerase
reverse transcriptase (TERT) as the major active component. The
mature human TERC (hTR) is 451 nucleotides long and folds into a
highly conserved structural domain. It binds to TERT via CR4/CR5
and template/pseudoknot domains. The H/ACA structural domain of hTR
binds to a protein complex composed of dyskerin, NOP10, NHP2, and
GAR1, and facilitates hTR processing and maturation. TCAB1 binds to
the CR7 structural domain of hTR to localize telomerase (51,52).
TERRA can act as a natural ligand that binds directly to TERT and
hTR, thereby inhibiting telomerase activity. The binding of TERRA
to TERC does not depend on the presence of hTR. By contrast, PinX1,
a telomerase inhibitor, acts by binding directly to hTR and TERT,
but the binding of PinX1 to hTR in the intracellular environment is
dependent on the presence of TERT (53,54).
In addition to providing a template for telomeric DNA replication,
hTR plays an important role in telomere shelterin protein capping.
The Ku70/80 heterodimer and DNA-dependent protein kinase catalytic
subunit (DNA-PKcs) constitute the DNA-dependent protein kinase
holoenzyme. The CR7 motif of hTR interacts with KU70/80 to enhance
the phosphorylation activity of hnRNPA1. The phosphorylation of
hnRNPA1 increases its affinity for single-stranded telomeric DNA,
thereby replacing the replication protein A (RPA) at the telomere
ends. Subsequently, hnRNPA1 interacts with protein phosphorylase 2A
to undergo dephosphorylation, thereby stripping it from telomeres
and allowing POT1 to bind to telomeres. The binding of POT1 ensures
telomere capping and inhibits the DDR (55,56).
Other lncRNAs
In addition to TERRA and hTR, which play important
roles in telomere regulation, other lncRNAs also play a role in
telomere physiology. PTEN is one of the most lost tumor suppressors
in human cancers. In hepatocellular carcinoma stem cells, decreased
PTEN levels lead to increased binding of the lncRNA CUDR to the
cell cycle protein cyclin D1; the CUDR-cyclin D1 complex then loads
into the lncRNA H19 promoter region and reduces DNA methylation in
the H19 promoter region, thereby enhancing H19 expression. H19
overexpression increases TERT binding to TERC while reducing TERT
binding to TERRA. This process results in increased cellular
telomerase activity and extended telomere length and promoting the
malignant proliferation of hepatocellular carcinoma stem cells
(57). P53, another tumor
suppressor, is frequently mutated in cancer cells to promote cancer
progression. In hepatocellular carcinoma cells, the double mutant
p53 (N340Q/L344R) binds to CUDR and promotes telomerase activity
and lengthening of telomeres through a cascade reaction that
enhances TERT expression and reduces TERRA expression (58). In addition to regulating telomerase
activity, lncRNAs aberrantly expressed in cancer are involved in
telomeric protein capping. Overexpression of lncRNAs HULC and
MALAT1 results in increased RNApolII and P300 loading onto the TRF2
promoter region, enhancing TRF2 transcription at the
transcriptional level. The increased TRF2 binds to HULC and MALAT1
to form a complex that is loaded onto telomeres, replacing CST/AAF
and recruiting telomere-associated proteins, such as POT1, pPOT1,
ExoI, and SNM1B, to maintain telomere length and stability. By
contrast, lncRNA MEG3 promotes the binding of HULC to p53, thereby
inhibiting the binding of telomere-associated proteins to HULC and
decreasing telomere stability (59,60).
LncRNAs are involved in DNA repair through
RBPs
The genome of an organism is subjected to endogenous
and exogenous damage, causing each cell to produce up to
105 times the amount of DNA damage per cell per day.
Under normal physiological conditions, cells have six main DNA
repair pathways by which DNA damage can be precisely repaired to
maintain genomic stability (61–64).
DNA double-strand breaks (DSBs) are the most cytotoxic type of DNA
damage and require complex repair mechanisms. They are repaired by
homologous recombination (HR) and non-homologous end joining
(NHEJ). Both not repairing DSBs and selecting the wrong way to
repair DSBs lead to genomic instability (65–67).
Therefore, lncRNAs that regulate DSB repair by binding key proteins
during NHEJ and HR were mainly examined (Fig. 2; Table III).
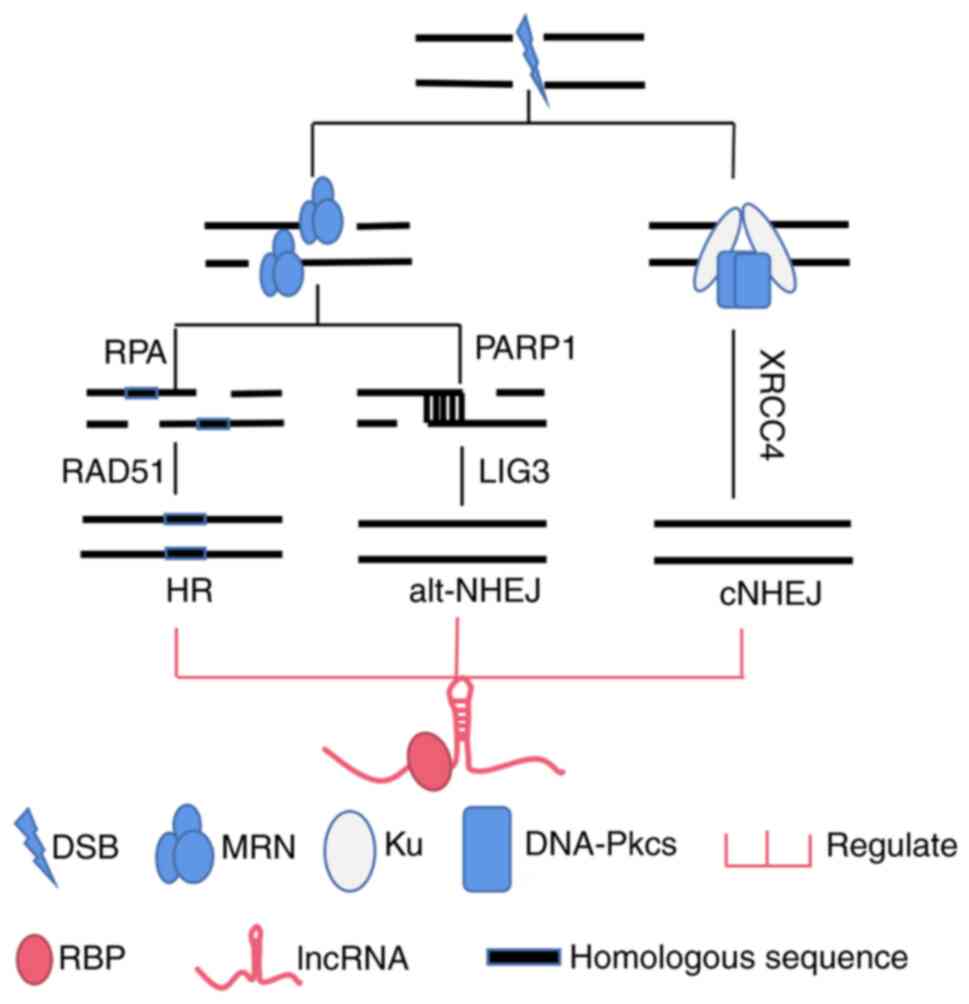 | Figure 2.LncRNAs are involved in the
regulation and selection of different DSB repair pathways by
binding RBPs. LncRNAs, long non-coding RNAs; DSB, DNA double-strand
break; RBPs, RNA-binding proteins; RPA, replication protein A;
PARP1, poly ADP ribose polymerase1; HR, homologous recombination;
alt-NHEJ, alternative nonhomologous end joining; cNHEJ, classical
nonhomologous end joining; MRN, MRE11-Rad50-Nbs1; DNA-Pkcs,
DNA-dependent protein kinase catalytic subunit. |
 | Table III.LncRNAs involved in DSB repair via
RBPs. |
Table III.
LncRNAs involved in DSB repair via
RBPs.
| LncRNAs | RBPs | Mechanism | Effect | (Refs.) |
|---|
| LRIK | Ku70 | Enhances the
binding of Ku heterodimer to DSB | Promotes cNHEJ | (72) |
| LINP1 | Ku80 and
DNA-PKcs | Enhances the
interaction between Ku heterodimers and DNA-PKcs | Promotes cNHEJ | (22,73) |
| Linc00312 | DNA-PKcs | Inhibits the
recruitment of Ku to DNA-PKcs | Inhibits cNHEJ | (74) |
| MALAT1 | PARP1 | Promotes
co-localization between LIG3 and γH2A.X | Activates
alt-NHEJ | (77) |
| PRLH1 | RNF169 | Promotes RNF169 to
replace 53BP1 | Promotes HR and
inhibits cNHEJ | (78) |
| SNHG17 | NONO | Promotes the
formation of the NHEJ repair complex | Promotes cNHEJ and
inhibits HR | (79) |
| HITTERS | MRE11 and
Rad50 | Promotes the
interaction between MRE11 and Rad50 | Promotes HR | (82) |
| HITT | ATM | Prevents ATM
recruitment by the MRN complex | Inhibits HR | (23) |
| GUARDIN | BRCA1 and
BARD1 | Enhances the
interaction between BRCA1 and BARD1 | Promotes HR | (84) |
| BGL3 | PARP1 and
BARD1 | Promotes
BRCA1-BARD1 retention at the DSB | Promotes HR | (85) |
| DDSR1 | BRCA1 and
hnRNPUL1 | Prevents the
formation of the BRCA1-RPA80 complex | Promotes HR | (86,87) |
NHEJ
Classical NHEJ (cNHEJ) is the primary repair
mechanism for DSBs. It does not require a homologous template,
requires minor or no processing of DSB ends, and is then directly
ligated by enzymatic action, making it an efficient but error-prone
repair modality (68,69). In this repair process, Ku70-Ku80
first binds to the DSB and acts as a recruitment platform for other
cNHEJ proteins, such as the XRCC4. Ku70/80 binds to DNA-PKcs
activating their kinase function, which leads to the
phosphorylation of Ku and other cNHEJ factors, such as Artemis. The
activated Artemis allows the processing of DNA ends. Finally, end
linkage is catalyzed by a complex consisting of LIG4 and XRCC4
(70,71).
Wang et al reported a novel lncRNA, LRIK,
induced by DSB in HeLa cells, which enhances the binding of the Ku
heterodimer to DSB through direct binding to the Ku70 subunit. This
process promotes assembly of downstream NHEJ factors and the
formation of γ-H2AX, ultimately promoting the efficiency of cNHEJ
(72). Similarly, lncRNA LINP1,
activated by the epidermal growth factor in triple-negative breast
cancer, can be recruited to the DSB by binding directly to Ku80.
LINP1 also binds to DNA-PKcs through a different region and acts as
a molecular scaffold to enhance the interaction between Ku
heterodimers and DNA-PKcs. This in turn enhances cNHEJ-mediated DNA
repair activity and reduces cancer sensitivity to radiotherapy.
Downregulation of LINP1 expression sensitizes cancer cells to
radiotherapy due to defective repair activity (22). Thapar et al performed
further studies and found that Ku binds to the LINP1 stem-loop and
G-quadruplex structures (73); the
Ku-LINP1 interaction replaces the NHEJ cofactor PAXX protein more
efficiently, increasing the stability and net concentration of NHEJ
factors at the DSB. Moreover, it bridges the Ku heterodimer at both
ends of the DSB to better promote DSB end-joining (71,73).
Conversely, lncRNA linc00312, which is expressed at low levels in
nasopharyngeal carcinoma, can act as a protein decoy to bind to
DNA-PKcs, thereby blocking the recruitment of Ku to DNA-PKcs and
inhibiting cNHEJ and resistance to radiotherapy (74).
In the case of cNHEJ damage, end linkage without the
involvement of cNHEJ core factors is referred to as alt-NHEJ, as an
alternate DNA repair pathway to cNHEJ that is more prone to
chromosomal alterations (75,76).
The alt-NHEJ pathway requires rapid recruitment of the MRN complex
by poly ADP ribose polymerase1 (PARP1), which triggers end
resection and is dependent on polymerase theta and LIG3 for
microhomologous sequence annealing and ligation. In multiple
myeloma, the lncRNA MALAT1 can bind directly to PARP1 and
indirectly to LIG3. MALAT1 knockdown did not affect LIG3/PARP1
co-localization but disrupted co-localization between LIG3 and
γH2A.X, suggesting that MALAT1 is important for PARP1/LIG3 complex
recognition of the γH2A.X on DSB and activating alt-NHEJ repair,
which promotes MM mutagenesis and drug resistance (77).
HR and NHEJ are two competing pathways in the early
stages of DSB repair. The selection and balance between the two
repair modalities are crucial for genomic stability (78), and lncRNAs are involved in the
selection between them. Infection of normal gastric epithelial
cells with Helicobacter pylori was demonstrated to induce high
expression of lncRNA SNHG17, and lncRNA SNHG17 in the nucleus
interacted directly with NONO, thus enhancing the interaction
between NONO and Ku, which promoted the formation of the NHEJ
repair complex. SNHG17 in the cytoplasm was shown to bind to
miR-3909 as a competing endogenous RNA (ceRNA), thereby inhibiting
HR, shifting the balance of DSB repair to NHEJ, and ultimately
promoting gastric cancer development (79). The E3 ubiquitin ligase RNF169 can
replace 53BP1, which inhibits end resection at DSB to promote NHEJ
to enhance HR. In hepatocellular carcinoma, lncRNA PRLH1 can bind
to RNF169 to form a stable complex that enhances the stability of
RNF19 and the affinity of this protein for DSB to replace 53BP1
more efficiently, shifting the balance of repair to HR (78).
HR
Because HR is performed using sister chromatids as
templates, the process occurs in the late S and G2 phases and
facilitates precise repairs. The starting step of HR is the sensing
of the damaged site by the MRE11-Rad50-Nbs1 (MRN) complex and
producing a free 3′ end single-strand overhang (80). Rad51 recombinase is the final
effector of the HR cascade reaction, and its binding to
single-stranded DNA depends on BRCA2 as well as the interaction of
the BRCA1-BARD1 complex and PALB2. Once Rad51 binds to
single-stranded DNA at the DSB, it begins the subsequent homology
search and strand invasion to initiate DNA repair (81).
The lncRNA HITTERS, which is highly expressed in
oral squamous cell carcinoma cells, induced by endoplasmic
reticulum stress, can directly bind MRE11 and Rad50, thus promoting
their interaction, while also increasing MRE11 and Nbs1 protein
levels and promoting the formation of the MRN complex. Ultimately,
the function of HITTERS facilitates DNA repair via multiple
pathways including HR (82). Like
DNA-PKcs in NHEJ, the capillary dilation ataxia mutated gene (ATM)
protein kinase is the apical kinase in HR; MRN serves as a protein
platform to promote autophosphorylation of ATM to stimulate its
activity (83). The lncRNA HITT,
which is expressed at low levels in several cancers due to hypoxic
contingency, binds directly to the binding site of ATM-binding Nbs1
and thereby prevents ATM recruitment by the MRN and antagonizes
HR-mediated DNA repair. Through this mechanism, overexpression of
HITT can enhance the sensitivity of cancer cells to genotoxic
therapies (23).
BRCA1 and BARD1 form a heterodimer, which plays a
role in HR. The lncRNA GUARDIN acts as a molecular scaffold that
directly binds to BRCA1 and BARD1, enhancing their interaction to
promote HR (84). Similarly,
lncRNA BGL3 is recruited to the DSB early by binding to PARP1,
whereas direct binding to BARD1 promotes BRCA1-BARD1 retention at
the DSB and facilitates the interaction between BARD1 and Rad51
(85). The lncRNA DDSR1 is known
to interact with BRCA1 and hnRNPUL1; Sharma et al suggested
that this interaction prevents the formation of the BRCA1-RPA80
complex and the binding of this complex at the DSB (86). In turn, binding of the BRCA1-RPA80
complex to the DSB restricts DNA end excision and thus limits HR
(87).
Thus, lncRNAs play an important role in the
different repair pathways of DSBs by binding proteins, whereas the
role of lncRNAs in other repair modalities is poorly understood and
warrants further investigation.
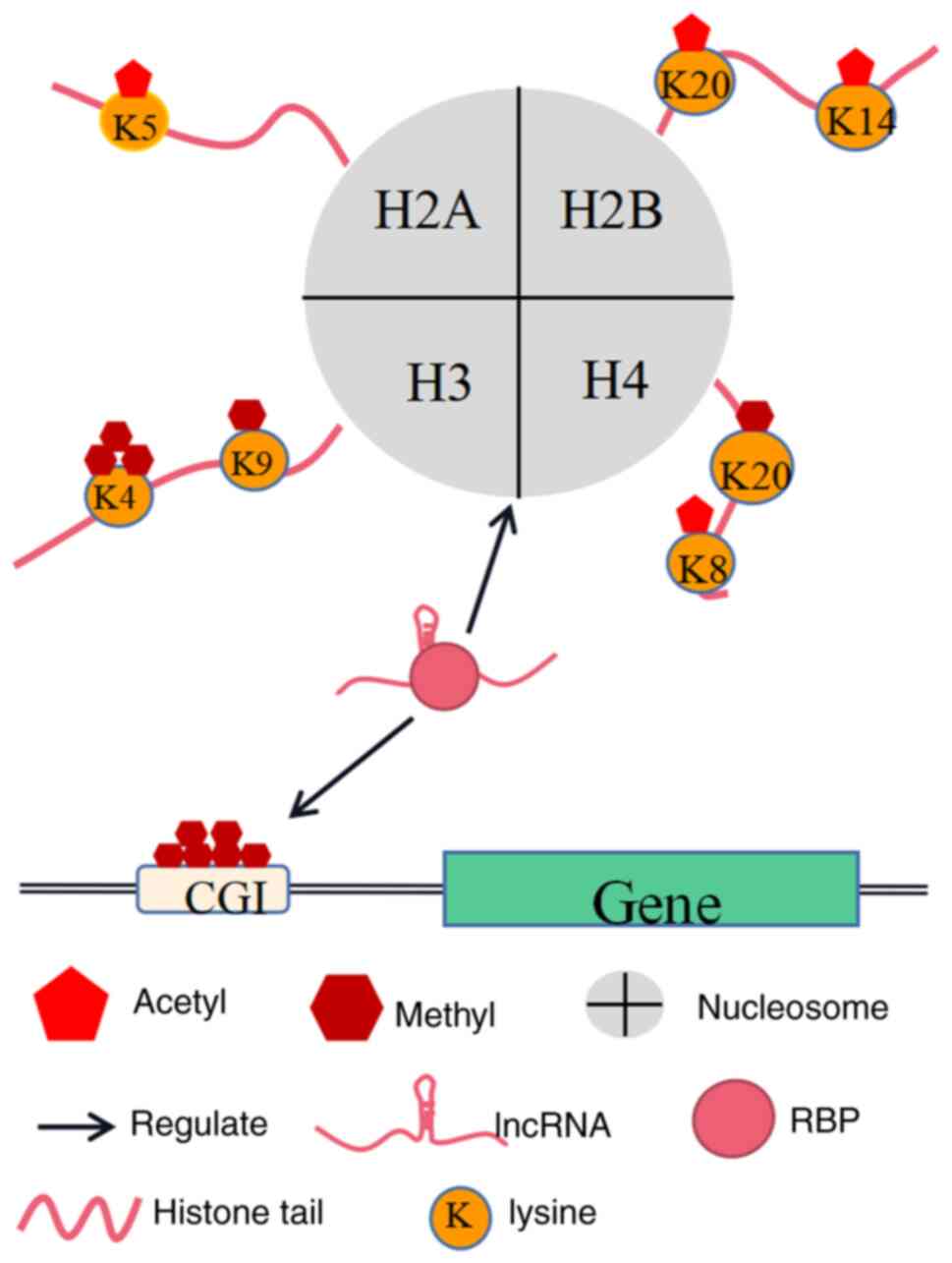
LncRNAs regulate other epigenetic modalities
through RBPs
Epigenetic inheritance refers to the production of
heritable changes in gene expression without changes to the DNA
nucleotide sequence, and such epigenetic aberrations are considered
a form of genomic instability. Like mutations, epigenetic
inheritance plays a key role in cancer development by altering the
expression of oncogenes and cancer suppressor genes (11). In addition, epigenetics may serve
as an advantageous biological marker for cancer diagnosis,
prognosis, and treatment. Epigenetic regulatory mechanisms mainly
include DNA methylation, chromatin remodeling, and non-coding RNA
(88,89). These regulatory mechanisms
crosstalk: for example, lncRNAs can act as miRNA sponges to inhibit
miRNAs (90). Therefore, the
regulation of chromatin remodeling and DNA methylation by lncRNAs
via binding to multiple enzymes (Fig.
3) was investigated.
Chromatin remodeling
The nucleosome is the basic unit of chromatin, and
consists mainly of the core histones H2A, H2B, H3, and H4 that form
an octamer wrapped around 147 base pairs of DNAs in humans
(91); nucleosomes further
assemble into higher-order chromatin (92). This highly folded state of
chromatin prevents the binding of DNA-binding proteins to
promoters, thereby inhibiting transcription (93). Histone-modifying enzymes and
ATP-dependent chromatin remodeling complexes mediate chromatin
remodeling, which controls gene expression by altering the
accessibility of local chromatin DNA (93–95).
Histone modification
The amino-terminal ends of core histones can extend
outside the nucleosome and be covalently modified by various
histone-modifying enzymes via methylation, acetylation,
ubiquitination, and phosphorylation. These modifications can affect
the affinity of histones for DNA duplexes, alter the loose or
condensed state of chromatin, and mediate DNA accessibility and
protein-chromatin interactions, ultimately affecting gene
expression (91,96,97).
Methylation and acetylation are among the most intensively studied
processes.
LncRNAs can play a role in cancer development by
directing histone-modifying enzymes to regulate the methylation and
acetylation status of histones and cis-regulating the expression of
nearby genes (24,98,99).
For example, lncRNA EZR-AS1 recruits H3K4 methyltransferase SMYD3
to catalyze Tri-methylation of lysine 4 on histone H3 (H3K4me3) at
the EZR promoter. This process promotes EZR transcription, thereby
enhancing the metastasis and invasion of the esophageal squamous
cell carcinoma. Conversely, interfering with the expression of
EZR-AS1 has an inhibitory effect on cancer cells (24). Similarly, lncRNAs, such as HOTAIR
and AS1DHRS4, can have a trans-regulatory role in gene expression
through histone modifications (100,101). LncRNAs can also serve as protein
scaffolds for histone-modifying enzymes. For example, lncRNA
AGAP2-AS1 acts as a protein scaffold and binds to the polycomb
repressive complex 2 (PRC2) core catalytic subunit EZH2 and
lysine-specific demethylase LSD1 to promote histone modifications
in pancreatic adenocarcinoma (102), glioma (103), non-small cell lung cancer
(104), and gastric cancer
(105). Through this mechanism
the expression of target genes is suppressed, and cancer
progression is promoted. The lncRNA CDKN2B-AS1 can bind to both
histone acetyltransferases CBP and SMYD3 to promote acetylation of
lysine 27 on histone 3 (H3K27ac) and H3K4me3 at the NUF2 promoter,
which further enhances NUF2 expression (34). Conversely, lncRNAs can act as a
protein decoy to regulate histone modification. For example,
LINC00261 binds to the acetylase P300/CBP complex, inhibiting its
binding to the c-Myc gene promoter to reduce H3K27ac and inhibit
pancreatic cancer progression by suppressing c-Myc expression
(106). Similarly, lncRNA DLEU2
can also act as a protein decoy for EZH2 (107).
ATP-dependent chromatin
remodeling
ATP-dependent chromatin remodeling complexes are
divided into four major classes: SWI/SNF, ISWI, CHD, and INO80
(108). The complexes can alter
the accessibility of transcription factors to DNA by disrupting the
interaction between DNA and histones using the energy generated by
ATP hydrolysis, altering the position of nucleosomes along the DNA
or replacing histones (94,95).
LncRNAs are also involved in this process. Subunits of the INO80
complex, INO80 and RUVBL2, bind to the lncRNA HAND2-AS1. This
process activates BMPR1A expression and ultimately stimulates the
self-renewal of hepatocellular carcinoma stem cells by recruiting
INO80 to the BMPR1A promoter and forming the formation complex
(109). Similarly, Tang et
al reported that many lncRNAs in cancer interact with SWI/SNF
and thus participate in the regulation of chromatin remodeling
(110).
DNA methylation
In humans, DNA methylation occurs mainly at the
cytosine 5′ carbon atom of CpG dinucleotides. DNA methylation is
catalyzed by the active DNA methyltransferases, DNMT1, DNMT3a, and
DNMT3b that add methyl groups to cytosine to form 5-methylcytosine
(5mC) (111,112). CpG dinucleotides usually exist in
CpG islands in the promoter region of the human genome (113). Methylation of CpG islands can
directly or indirectly inhibit the binding of transcription factors
to promoters (114). LncRNAs can
directly bind to these active DNA methylases and regulate gene
expression, thereby interfering with cancer development (115–118). LncRNA LINC01270 is highly
expressed in esophageal cancer and can act as a protein scaffold to
simultaneously bind to DNMT1, DNMT3a, and DNMT3b. This mechanism
mediates the hypermethylation of GSTP1 promoter, inhibiting its
expression and promoting esophageal cancer progression and drug
resistance (119). Interestingly,
in colon cancer cells, lncRNA lnc-LALC can also recruit DNMT1,
DNMT3a, and DNMT3b to the LZTS1 promoter simultaneously, but this
requires direct binding of lnc-LALC to EZH2 (120). The lncRNA PARTICLE, which is
highly expressed in response to low irradiation, promotes both DNA
and histone methylation by binding to DNMT1 and the PRC2 core
subunit SUZ12 and suppresses the expression of tumor suppressor
MAT2A in cis as well as the tumor suppressor WWOX in trans
(121,122).
DNA methylation is a stable modification process;
however, ten-eleven translocation (TET) family proteins catalyze
the conversion of 5mC to 5-hydroxymethylcytosine (5hmC),
5-formylcytosine (5fC), and 5-carboxycytosine (5caC). 5HmC is
diluted during DNA replication, while 5fC and 5caC are removed by
thymine DNA glycosylase (TDG), resulting in DNA demethylation
(112,123). LncRNA TARID can bind to growth
arrest and DNA damage-inducible 45A (GADD45A), a protein that
mediates DNA demethylation. Through this interaction, TARID directs
GADD45A cis to the tumor suppressor TCF21 promoter and indirectly
recruits TETs as well as TDG, which can bind to GADD45A and
co-mediate DNA demethylation (123). Similarly, lncRNA ZNF667-AS1 can
bind to TET1 and histone H3K27 demethylase to promote both DNA and
histone demethylation at the ZNF667 and E-calmodulin promoters,
thereby inhibiting the development of esophageal squamous
epithelial carcinoma (25).
Summary and prospects
Genomic instability is a feature of most cancers
that undoubtedly contributes to cancer progression and
heterogeneity through the accumulation of oncogenic and cancer
suppressor genic mutations, regardless of whether it acts as a
‘passenger’ or a ‘driver’ in cancer. Although the exact mechanism
is unknown, cancer cells have a tolerance limit to genomic
instability, which indicates the potential of genomic
destabilization as a therapeutic approach to cancer. Investigating
the mechanisms of tolerance to genomic instability in cancer cells
may provide new insights into cancer treatment. The aberrantly
expressed lncRNAs in cancer cells and their binding proteins form
networks that regulate genomic instability. Exploitation of
lncRNA-RBP networks may provide new biological markers for cancer
diagnosis and prognosis as well as new molecular targets and entry
points for driving genomic instability to the limit of cellular
tolerance or suppressing it in cancer therapies. However, the
causal relationship between dysregulated lncRNA expression and
genomic instability in many cancers has not yet been verified, and
the exact molecular mechanism between the two remains unclear,
which warrants further investigations.
Acknowledgements
Not applicable.
Funding
The present review was supported by the National Natural Science
Foundation of China (grant no. 82160575) and the Outstanding Young
Technological and Innovative Talent Cultivation Project of Zunyi
Municipal Science and Technology Bureau, 2021 (no. 10).
Availability of data and materials
Not applicable.
Authors' contributions
KY and KW conceived the study. KY drafted the
manuscript. KY, KW and XL made substantial contributions to the
interpretation, drafting the manuscript and revising it critically
for important intellectual content. Data authentication is not
applicable. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lee JK, Choi YL, Kwon M and Park PJ:
Mechanisms and consequences of cancer genome instability: Lessons
from genome sequencing studies. Annu Rev Pathol. 11:283–312. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aguilera A and Gómez-González B: Genome
instability: A mechanistic view of its causes and consequences. Nat
Rev Genet. 9:204–217. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Salmaninejad A, Ilkhani K, Marzban H,
Navashenaq JG, Rahimirad S, Radnia F, Yousefi M, Bahmanpour Z,
Azhdari S and Sahebkar A: Genomic instability in cancer: Molecular
mechanisms and therapeutic potentials. Curr Pharm Des.
27:3161–3169. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mehrotra S and Mittra I: Origin of genome
instability and determinants of mutational landscape in cancer
cells. Genes (Basel). 11:11012020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Andor N, Maley CC and Ji HP: Genomic
instability in cancer: Teetering on the limit of tolerance. Cancer
Res. 77:2179–2185. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abbas T, Keaton MA and Dutta A: Genomic
instability in cancer. Cold Spring Harb Perspect Biol.
5:a0129142013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
O'Connor MJ: Targeting the DNA damage
response in cancer. Mol Cell. 60:547–560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sansregret L, Vanhaesebroeck B and Swanton
C: Determinants and clinical implications of chromosomal
instability in cancer. Nat Rev Clin Oncol. 15:139–150. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maciejowski J and de Lange T: Telomeres in
cancer: Tumour suppression and genome instability. Nat Rev Mol Cell
Biol. 18:175–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lord CJ and Ashworth A: The DNA damage
response and cancer therapy. Nature. 481:287–294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Choi JD and Lee JS: Interplay between
epigenetics and genetics in cancer. Genomics Inform. 11:164–173.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang W, Guan X and Tang J: The long
non-coding RNA landscape in triple-negative breast cancer. Cell
Prolif. 54:e129662021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang L, Xie Y, Jiang S, Han W, Zeng F and
Li D: The lncRNA signatures of genome instability to predict
survival in patients with renal cancer. J Healthc Eng.
2021:10906982021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yin T, Zhao D and Yao S: Identification of
a genome instability-associated LncRNA signature for prognosis
prediction in colon cancer. Front Genet. 12:6791502021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Matsumoto A, Pasut A, Matsumoto M,
Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI,
Clohessy JG and Pandolfi PP: mTORC1 and muscle regeneration are
regulated by the LINC00961-encoded SPAR polypeptide. Nature.
541:228–232. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hentze MW, Castello A, Schwarzl T and
Preiss T: A brave new world of RNA-binding proteins. Nat Rev Mol
Cell Biol. 19:327–341. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen B, Dragomir MP, Fabris L, Bayraktar
R, Knutsen E, Liu X, Tang C, Li Y, Shimura T, Ivkovic TC, et al:
The long noncoding RNA CCAT2 induces chromosomal instability
through BOP1-AURKB signaling. Gastroenterology. 159:2146–2162.e33.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo Z, Dai Y, Hu W, Zhang Y, Cao Z, Pei W,
Liu N, Nie J, Wu A, Mao W, et al: The long noncoding RNA CRYBG3
induces aneuploidy by interfering with spindle assembly checkpoint
via direct binding with Bub3. Oncogene. 40:1821–1835. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang
Z, Yuan J, Shan W, Li C, Hu X, et al: Long noncoding RNA LINP1
regulates repair of DNA double-strand breaks in triple-negative
breast cancer. Nat Struct Mol Biol. 23:522–530. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao K, Wang X, Xue X, Li L and Hu Y: A
long noncoding RNA sensitizes genotoxic treatment by attenuating
ATM activation and homologous recombination repair in cancers. PLoS
Biol. 18:e30006662020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang XD, Huang GW, Xie YH, He JZ, Guo JC,
Xu XE, Liao LD, Xie YM, Song YM, Li EM and Xu LY: The interaction
of lncRNA EZR-AS1 with SMYD3 maintains overexpression of EZR in
ESCC cells. Nucleic Acids Res. 46:1793–1809. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dong Z, Li S, Wu X, Niu Y, Liang X, Yang
L, Guo Y, Shen S, Liang J and Guo W: Aberrant
hypermethylation-mediated downregulation of antisense lncRNA
ZNF667-AS1 and its sense gene ZNF667 correlate with progression and
prognosis of esophageal squamous cell carcinoma. Cell Death Dis.
10:9302019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Carter SL, Cibulskis K, Helman E, McKenna
A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et
al: Absolute quantification of somatic DNA alterations in human
cancer. Nat Biotechnol. 30:413–421. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Negrini S, Gorgoulis VG and Halazonetis
TD: Genomic instability-an evolving hallmark of cancer. Nat Rev Mol
Cell Biol. 11:220–228. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jo M, Kusano Y and Hirota T: Unraveling
pathologies underlying chromosomal instability in cancers. Cancer
Sci. 112:2975–2983. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Piemonte KM, Anstine LJ and Keri RA:
Centrosome aberrations as drivers of chromosomal instability in
breast cancer. Endocrinology. 162:bqab2082021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hara M and Fukagawa T: Dynamics of
kinetochore structure and its regulations during mitotic
progression. Cell Mol Life Sci. 77:2981–2995. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Monda JK and Cheeseman IM: The
kinetochore-microtubule interface at a glance. J Cell Sci.
131:jcs2145772018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Welburn JP, Vleugel M, Liu D, Yates JR
III, Lampson MA, Fukagawa T and Cheeseman IM: Aurora B
phosphorylates spatially distinct targets to differentially
regulate the kinetochore-microtubule interface. Mol Cell.
38:383–392. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sacristan C and Kops GJ: Joined at the
hip: Kinetochores, microtubules, and spindle assembly checkpoint
signaling. Trends Cell Biol. 25:21–28. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xie X, Lin J, Fan X, Zhong Y, Chen Y, Liu
K, Ren Y, Chen X, Lai D, Li X, et al: LncRNA CDKN2B-AS1 stabilized
by IGF2BP3 drives the malignancy of renal clear cell carcinoma
through epigenetically activating NUF2 transcription. Cell Death
Dis. 12:2012021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
DeLuca JG, Dong Y, Hergert P, Strauss J,
Hickey JM, Salmon ED and McEwen BF: Hec1 and nuf2 are core
components of the kinetochore outer plate essential for organizing
microtubule attachment sites. Mol Biol Cell. 16:519–531. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Stojic L, Lun ATL, Mascalchi P, Ernst C,
Redmond AM, Mangei J, Barr AR, Bousgouni V, Bakal C, Marioni JC, et
al: A high-content RNAi screen reveals multiple roles for long
noncoding RNAs in cell division. Nat Commun. 11:18512020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee S, Kopp F, Chang TC, Sataluri A, Chen
B, Sivakumar S, Yu H, Xie Y and Mendell JT: Noncoding RNA NORAD
regulates genomic stability by sequestering PUMILIO proteins. Cell.
164:69–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schmidt JC and Cech TR: Human telomerase:
Biogenesis, trafficking, recruitment, and activation. Genes Dev.
29:1095–1105. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Frias C, Pampalona J, Genesca A and Tusell
L: Telomere dysfunction and genome instability. Front Biosci
(Landmark Ed). 17:2181–2196. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Schoeftner S and Blasco MA:
Developmentally regulated transcription of mammalian telomeres by
DNA-dependent RNA polymerase II. Nat Cell Biol. 10:228–236. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Deng Z, Norseen J, Wiedmer A, Riethman H
and Lieberman PM: TERRA RNA binding to TRF2 facilitates
heterochromatin formation and ORC recruitment at telomeres. Mol
Cell. 35:403–413. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mei Y, Deng Z, Vladimirova O, Gulve N,
Johnson FB, Drosopoulos WC, Schildkraut CL and Lieberman PM: TERRA
G-quadruplex RNA interaction with TRF2 GAR domain is required for
telomere integrity. Sci Rep. 11:35092021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Y, Zeng D, Cao J, Wang M, Shu B,
Kuang G, Ou TM, Tan JH, Gu LQ, Huang ZS and Li D: Interaction of
Quindoline derivative with telomeric repeat-containing RNA induces
telomeric DNA-damage response in cancer cells through inhibition of
telomeric repeat factor 2. Biochim Biophys Acta Gen Subj.
1861:3246–3256. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Takahama K, Takada A, Tada S, Shimizu M,
Sayama K, Kurokawa R and Oyoshi T: Regulation of telomere length by
G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem
Biol. 20:341–350. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Blasco MA: Telomeres and human disease:
Ageing, cancer and beyond. Nat Rev Genet. 6:611–622. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Benetti R, García-Cao M and Blasco MA:
Telomere length regulates the epigenetic status of mammalian
telomeres and subtelomeres. Nat Genet. 39:243–250. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang QS, Manche L, Xu RM and Krainer AR:
hnRNP A1 associates with telomere ends and stimulates telomerase
activity. RNA. 12:1116–1128. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Redon S, Zemp I and Lingner J: A
three-state model for the regulation of telomerase by TERRA and
hnRNPA1. Nucleic Acids Res. 41:9117–9128. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Vohhodina J, Goehring LJ, Liu B, Kong Q,
Botchkarev VV Jr, Huynh M, Liu Z, Abderazzaq FO, Clark AP, Ficarro
SB, et al: BRCA1 binds TERRA RNA and suppresses R-Loop-based
telomeric DNA damage. Nat Commun. 12:35422021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Feretzaki M, Pospisilova M, Valador
Fernandes R, Lunardi T, Krejci L and Lingner J: RAD51-dependent
recruitment of TERRA lncRNA to telomeres through R-loops. Nature.
587:303–308. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gala K and Khattar E: Long non-coding RNAs
at work on telomeres: Functions and implications in cancer therapy.
Cancer Lett. 502:120–132. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Podlevsky JD and Chen JJ: Evolutionary
perspectives of telomerase RNA structure and function. RNA Biol.
13:720–732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Redon S, Reichenbach P and Lingner J: The
non-coding RNA TERRA is a natural ligand and direct inhibitor of
human telomerase. Nucleic Acids Res. 38:5797–5806. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Banik SS and Counter CM: Characterization
of interactions between PinX1 and human telomerase subunits hTERT
and hTR. J Biol Chem. 279:51745–51748. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Raghunandan M and Decottignies A: The
multifaceted hTR telomerase RNA from a structural perspective:
Distinct domains of hTR differentially interact with protein
partners to orchestrate its telomerase-independent functions.
Bioessays. 43:e21000992021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sui JD, Tang Z, Chen BPC, Huang P, Yang
MQ, Wang NH, Yang HN, Tu HL, Jiang QM, Zhang J, et al: Protein
phosphatase 2A-dependent mitotic hnRNPA1 dephosphorylation and
TERRA formation facilitate telomere capping. Mol Cancer Res.
20:583–595. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Pu H, Zheng Q, Li H, Wu M, An J, Gui X, Li
T and Lu D: CUDR promotes liver cancer stem cell growth through
upregulating TERT and C-Myc. Oncotarget. 6:40775–40798. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wu M, An J, Zheng Q, Xin X, Lin Z, Li X,
Li H and Lu D: Double mutant P53 (N340Q/L344R) promotes
hepatocarcinogenesis through upregulation of Pim1 mediated by PKM2
and LncRNA CUDR. Oncotarget. 7:66525–66539. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wu M, Lin Z, Li X, Xin X, An J, Zheng Q,
Yang Y and Lu D: HULC cooperates with MALAT1 to aggravate liver
cancer stem cells growth through telomere repeat-binding factor 2.
Sci Rep. 6:360452016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jiang X, Wang L, Xie S, Chen Y, Song S, Lu
Y and Lu D: Long noncoding RNA MEG3 blocks telomerase activity in
human liver cancer stem cells epigenetically. Stem Cell Res Ther.
11:5182020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hoeijmakers JH: DNA damage, aging, and
cancer. N Engl J Med. 361:1475–1485. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhao H, Fuemmeler BF and Shen J: DNA
repair in cancer development and aging. Aging (Albany NY).
13:23435–23436. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Tian H, Gao Z, Li H, Zhang B, Wang G,
Zhang Q, Pei D and Zheng J: DNA damage response-a double-edged
sword in cancer prevention and cancer therapy. Cancer Lett.
358:8–16. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bever KM and Le DT: DNA repair defects and
implications for immunotherapy. J Clin Invest. 128:4236–4242. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhao Y and Chen S: Targeting DNA
double-strand break (DSB) repair to counteract tumor
radio-resistance. Curr Drug Targets. 20:891–902. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ceccaldi R, Rondinelli B and D'Andrea AD:
Repair pathway choices and consequences at the double-strand break.
Trends Cell Biol. 26:52–64. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Scully R, Panday A, Elango R and Willis
NA: DNA double-strand break repair-pathway choice in somatic
mammalian cells. Nat Rev Mol Cell Biol. 20:698–714. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Burma S, Chen BP and Chen DJ: Role of
non-homologous end joining (NHEJ) in maintaining genomic integrity.
DNA Repair (Amst). 5:1042–1048. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Shibata A, Conrad S, Birraux J, Geuting V,
Barton O, Ismail A, Kakarougkas A, Meek K, Taucher-Scholz G,
Löbrich M and Jeggo PA: Factors determining DNA double-strand break
repair pathway choice in G2 phase. EMBO J. 30:1079–1092. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Stinson BM and Loparo JJ: Repair of DNA
double-strand breaks by the nonhomologous end joining pathway. Annu
Rev Biochem. 90:137–164. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ghosh D and Raghavan SC: Nonhomologous end
joining: New accessory factors fine tune the machinery. Trends
Genet. 37:582–599. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang D, Zhou Z, Wu E, Ouyang C, Wei G,
Wang Y, He D, Cui Y, Zhang D, Chen X, et al: LRIK interacts with
the Ku70-Ku80 heterodimer enhancing the efficiency of NHEJ repair.
Cell Death Differ. 27:3337–3353. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Thapar R, Wang JL, Hammel M, Ye R, Liang
K, Sun C, Hnizda A, Liang S, Maw SS, Lee L, et al: Mechanism of
efficient double-strand break repair by a long non-coding RNA.
Nucleic Acids Res. 48:10953–10972. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Guo Z, Wang YH, Xu H, Yuan CS, Zhou HH,
Huang WH, Wang H and Zhang W: LncRNA linc00312 suppresses
radiotherapy resistance by targeting DNA-PKcs and impairing DNA
damage repair in nasopharyngeal carcinoma. Cell Death Dis.
12:692021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Decottignies A: Alternative end-joining
mechanisms: A historical perspective. Front Genet. 4:482013.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Chiruvella KK, Liang Z and Wilson TE:
Repair of double-strand breaks by end joining. Cold Spring Harb
Perspect Biol. 5:a0127572013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Hu Y, Lin J, Fang H, Fang J, Li C, Chen W,
Liu S, Ondrejka S, Gong Z, Reu F, et al: Targeting the
MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in
multiple myeloma. Leukemia. 32:2250–2262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Deng B, Xu W, Wang Z, Liu C, Lin P, Li B,
Huang Q, Yang J, Zhou H and Qu L: An LTR retrotransposon-derived
lncRNA interacts with RNF169 to promote homologous recombination.
EMBO Rep. 20:e476502019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Han T, Jing X, Bao J, Zhao L, Zhang A,
Miao R, Guo H, Zhou B, Zhang S, Sun J and Shi J: H. pylori
infection alters repair of DNA double-strand breaks via SNHG17. J
Clin Invest. 130:3901–3918. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ranjha L, Howard SM and Cejka P: Main
steps in DNA double-strand break repair: An introduction to
homologous recombination and related processes. Chromosoma.
127:187–214. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yamamoto H and Hirasawa A: Homologous
recombination deficiencies and hereditary tumors. Int J Mol Sci.
23:3482021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wu C, Chen W, Yu F, Yuan Y, Chen Y, Hurst
DR, Li Y, Li L and Liu Z: Long noncoding RNA HITTERS protects oral
squamous cell carcinoma cells from endoplasmic reticulum
stress-induced apoptosis via promoting MRE11-RAD50-NBS1 complex
formation. Adv Sci (Weinh). 7:20027472020. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Paull TT: Mechanisms of ATM activation.
Annu Rev Biochem. 84:711–738. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Hu WL, Jin L, Xu A, Wang YF, Thorne RF,
Zhang XD and Wu M: GUARDIN is a p53-responsive long non-coding RNA
that is essential for genomic stability. Nat Cell Biol. 20:492–502.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Hu Z, Mi S, Zhao T, Peng Y, Chen L, Zhu W,
Yao Y, Song Q, Li X, Li X, et al: BGL3 lncRNA mediates retention of
the BRCA1/BARD1 complex at DNA damage sites. EMBO J.
39:e1041332020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sharma V, Khurana S, Kubben N, Abdelmohsen
K, Oberdoerffer P, Gorospe M and Misteli T: A BRCA1-interacting
lncRNA regulates homologous recombination. EMBO Rep. 16:1520–1534.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Coleman KA and Greenberg RA: The
BRCA1-RAP80 complex regulates DNA repair mechanism utilization by
restricting end resection. J Biol Chem. 286:13669–13680. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Costa-Pinheiro P, Montezuma D, Henrique R
and Jerónimo C: Diagnostic and prognostic epigenetic biomarkers in
cancer. Epigenomics. 7:1003–1015. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Kim JJ, Lee SY and Miller KM: Preserving
genome integrity and function: The DNA damage response and histone
modifications. Crit Rev Biochem Mol Biol. 54:208–241. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Iacobuzio-Donahue CA: Epigenetic changes
in cancer. Annu Rev Pathol. 4:229–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Luo RX and Dean DC: Chromatin remodeling
and transcriptional regulation. J Natl Cancer Inst. 91:1288–1294.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wang Z, Liu S and Tao Y: Regulation of
chromatin remodeling through RNA polymerase II stalling in the
immune system. Mol Immunol. 108:75–80. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Koreman E, Sun X and Lu QR: Chromatin
remodeling and epigenetic regulation of oligodendrocyte myelination
and myelin repair. Mol Cell Neurosci. 87:18–26. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Strahl BD and Allis CD: The language of
covalent histone modifications. Nature. 403:41–45. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Fang K, Huang W, Sun YM, Chen TQ, Zeng ZC,
Yang QQ, Pan Q, Han C, Sun LY, Luo XQ, et al: Cis-acting lnc-eRNA
SEELA directly binds histone H4 to promote histone recognition and
leukemia progression. Genome Biol. 21:2692020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Wang YQ, Jiang DM, Hu SS, Zhao L, Wang L,
Yang MH, Ai ML, Jiang HJ, Han Y, Ding YQ and Wang S: SATB2-AS1
suppresses colorectal carcinoma aggressiveness by inhibiting
SATB2-dependent snail transcription and epithelial-mesenchymal
transition. Cancer Res. 79:3542–3556. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Chu C, Qu K, Zhong FL, Artandi SE and
Chang HY: Genomic maps of long noncoding RNA occupancy reveal
principles of RNA-chromatin interactions. Mol Cell. 44:667–678.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Li Q, Su Z, Xu X, Liu G, Song X, Wang R,
Sui X, Liu T, Chang X and Huang D: AS1DHRS4, a head-to-head natural
antisense transcript, silences the DHRS4 gene cluster in cis and
trans. Proc Natl Acad Sci USA. 109:14110–14115. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Hui B, Ji H, Xu Y, Wang J, Ma Z, Zhang C,
Wang K and Zhou Y: RREB1-induced upregulation of the lncRNA
AGAP2-AS1 regulates the proliferation and migration of pancreatic
cancer partly through suppressing ANKRD1 and ANGPTL4. Cell Death
Dis. 10:2072019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Luo W, Li X, Song Z, Zhu X and Zhao S:
Long non-coding RNA AGAP2-AS1 exerts oncogenic properties in
glioblastoma by epigenetically silencing TFPI2 through EZH2 and
LSD1. Aging (Albany NY). 11:3811–3823. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Li W, Sun M, Zang C, Ma P, He J, Zhang M,
Huang Z, Ding Y and Shu Y: Upregulated long non-coding RNA
AGAP2-AS1 represses LATS2 and KLF2 expression through interacting
with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death
Dis. 7:e22252016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Qi F, Liu X, Wu H, Yu X, Wei C, Huang X,
Ji G, Nie F and Wang K: Long noncoding AGAP2-AS1 is activated by
SP1 and promotes cell proliferation and invasion in gastric cancer.
J Hematol Oncol. 10:482017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Liu S, Zheng Y, Zhang Y, Zhang J, Xie F,
Guo S, Gu J, Yang J, Zheng P, Lai J, et al: Methylation-mediated
LINC00261 suppresses pancreatic cancer progression by
epigenetically inhibiting c-Myc transcription. Theranostics.
10:10634–10651. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Salerno D, Chiodo L, Alfano V, Floriot O,
Cottone G, Paturel A, Pallocca M, Plissonnier ML, Jeddari S,
Belloni L, et al: Hepatitis B protein HBx binds the DLEU2 lncRNA to
sustain cccDNA and host cancer-related gene transcription. Gut.
69:2016–2024. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Hota SK and Bruneau BG: ATP-dependent
chromatin remodeling during mammalian development. Development.
143:2882–2897. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wang Y, Zhu P, Luo J, Wang J, Liu Z, Wu W,
Du Y, Ye B, Wang D, He L, et al: LncRNA HAND2-AS1 promotes liver
cancer stem cell self-renewal via BMP signaling. EMBO J.
38:e1011102019. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Tang Y, Wang J, Lian Y, Fan C, Zhang P, Wu
Y, Li X, Xiong F, Li X, Li G, et al: Linking long non-coding RNAs
and SWI/SNF complexes to chromatin remodeling in cancer. Mol
Cancer. 16:422017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Ma X and Kang S: Functional implications
of DNA methylation in adipose biology. Diabetes. 68:871–878. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Nishiyama A and Nakanishi M: Navigating
the DNA methylation landscape of cancer. Trends Genet.
37:1012–1027. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Schübeler D: ESCI award lecture:
Regulation, function and biomarker potential of DNA methylation.
Eur J Clin Invest. 45:288–293. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Nan X, Ng HH, Johnson CA, Laherty CD,
Turner BM, Eisenman RN and Bird A: Transcriptional repression by
the methyl-CpG-binding protein MeCP2 involves a histone deacetylase
complex. Nature. 393:386–389. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zhang Y, Yan H, Jiang Y, Chen T, Ma Z, Li
F, Lin M, Xu Y, Zhang X, Zhang J and He H: Long non-coding RNA
IGF2-AS represses breast cancer tumorigenesis by epigenetically
regulating IGF2. Exp Biol Med (Maywood). 246:371–379. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Ma F, Lei YY, Ding MG, Luo LH, Xie YC and
Liu XL: LncRNA NEAT1 interacted with DNMT1 to regulate malignant
phenotype of cancer cell and cytotoxic T cell infiltration via
epigenetic inhibition of p53, cGAS, and STING in lung cancer. Front
Genet. 11:2502020. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Tang J, Xie Y, Xu X, Yin Y, Jiang R, Deng
L, Tan Z, Gangarapu V, Tang J and Sun B: Bidirectional
transcription of Linc00441 and RB1 via H3K27 modification-dependent
way promotes hepatocellular carcinoma. Cell Death Dis. 8:e26752017.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Feng H and Liu X: Interaction between
ACOT7 and LncRNA NMRAL2P via methylation regulates gastric cancer
progression. Yonsei Med J. 61:471–481. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Li N, Zhao Z, Miao F, Cai S, Liu P, Yu Y
and Wang B: Silencing of long non-coding RNA LINC01270 inhibits
esophageal cancer progression and enhances chemosensitivity to
5-fluorouracil by mediating GSTP1methylation. Cancer Gene Ther.
28:471–485. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zhang C, Wang L, Jin C, Zhou J, Peng C,
Wang Y, Xu Z, Zhang D, Huang Y, Zhang Y, et al: Long non-coding RNA
Lnc-LALC facilitates colorectal cancer liver metastasis via
epigenetically silencing LZTS1. Cell Death Dis. 12:2242021.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
O'Leary VB, Ovsepian SV, Smida J and
Atkinson MJ: PARTICLE-the RNA podium for genomic silencers. J Cell
Physiol. 234:19464–19470. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
O'Leary VB, Hain S, Maugg D, Smida J,
Azimzadeh O, Tapio S, Ovsepian SV and Atkinson MJ: Long non-coding
RNA PARTICLE bridges histone and DNA methylation. Sci Rep.
7:17902017. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Arab K, Park YJ, Lindroth AM, Schäfer A,
Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, et
al: Long noncoding RNA TARID directs demethylation and activation
of the tumor suppressor TCF21 via GADD45A. Mol Cell. 55:604–614.
2014. View Article : Google Scholar : PubMed/NCBI
|