Introduction
Oral cancer is one of most common malignancies
worldwide, with an estimated 354,864 new cases and 177,384
cancer-related deaths occurring in 2018 (1). The major pathology of oral cancer is
squamous cell carcinoma (SCC) (2).
Despite advances in the diagnosis and treatment of oral squamous
cell carcinoma (OSCC), the prognosis of patients with advanced OSCC
has not been improved in the past four decades (3). Metastasis to the cervical lymph nodes
is an accurate prognostic factor for patients with OSCC (4–7);
however, its molecular mechanism remains unclear. Therefore, there
is a need for studies investigating crucial biomarkers and
therapeutic targets of metastasis in OSCC.
Src-associated in mitosis 68 kDa (Sam68) was
originally identified as the first mitotic substrate of the
tyrosine kinase Src and belongs to the signal transduction and
activation of RNA family of RNA-binding proteins (8,9).
These proteins contain a GRP33/SAM68/GLD-1 domain formed by a
single heterogenous nuclear ribonucleoprotein particle K homology
domain and two flanking regions for RNA-binding (8–10).
Although Sam68 is predominantly localized in the nucleus, it can
shuttle between the nucleus and the cytoplasm, depending on its
functions (9–12). Sam68 has been implicated in RNA
metabolism, including transcription, alternative splicing,
transport, and translation (12–15).
Sam68 is involved in multiple biological events, including
cell-cycle regulation, apoptosis, response to conditions of
external stress, bone metabolism, neural functions, and viral
infection (10,12,13,16,17).
A series of previous studies has also shown a
pro-oncogenic role of Sam68 via the regulation of the signal
transduction pathway, transcription of cancer-related genes,
alternative splicing events, and noncoding RNAs (13,14,18,19).
Researchers have reported that in different types of human cancers,
the dysregulation of Sam68 is implicated in tumorigenesis, tumor
progression, and patient prognosis (20–26).
Nonetheless, it remains unclear whether Sam68 has clinical and
biological significance in OSCC.
Therefore, the aim of the present study was to
investigate the correlation between Sam68 expression and
clinicopathological features in OSCC via immunohistochemical
analysis and to elucidate the biological roles of Sam68 in OSCC
cells.
Materials and methods
Patients and tissue specimens
A total of 77 patients diagnosed with pathologically
confirmed SCC of the tongue, who underwent radical surgery at the
Department of Oral Maxillofacial Surgery at Tokyo Medical and
Dental University Hospital (Tokyo, Japan), between 2014 and 2017,
were enrolled in this retrospective study. Patients who had
previously received any treatment for tongue cancer were excluded
from the study. The surgical specimens were obtained from
formalin-fixed paraffin-embedded tissues. Clinical data on patient
age, sex, TNM classification, pathological differentiation of the
primary tumor, the mode of invasion according to Yamamoto-Kohama
(YK) classification (27),
perineural invasion, lymphovascular invasion, surgical margins,
pathological cervical lymph node metastases, and treatment outcomes
were obtained from the medical records of patients. No patients
received radiotherapy or chemotherapy before radical surgery.
Clinical staging was based on the TNM staging system of the
American Joint Committee on Cancer, seventh edition (28).
This study complied with the Declaration of Helsinki
and was approved (approval no. D2020-025) by the Institutional
Review Board of Tokyo Medical and Dental University. Opt-out
informed consent from patients was obtained by exhibiting the
research information on the official website of the hospital (Tokyo
Medical and Dental University Hospital, Tokyo, Japan). The authors
guarantee the opportunity for refusal by document, call or e-mail
whenever possible. Patients who rejected participation in this
study were excluded.
Immunohistochemistry
The surgical specimens were fixed with 10% formalin
for 24 h at room temperature and then embedded with paraffin. The
paraffin-embedded tissue blocks of the specimens were cut into 4-µm
serial sections and examined immunohistochemically. The sections
were deparaffinized with xylene and rehydrated through a series of
graded alcohol concentrations, followed by transfer and rinse with
phosphate-buffered saline (PBS). The epitopes were then retrieved
by autoclave in 0.01 M citrate buffer at 121°C for 15 min. After
cooling, endogenous peroxidase activity was blocked by 3% hydrogen
peroxide in methanol at room temperature for 15 min and treated
with 10% goat serum (Nichirei Biosciences, Inc.) at room
temperature for 15 min to prevent nonspecific binding. The slides
were washed three times with PBS and incubated overnight at 4°C
with monoclonal rabbit anti-SAM68 (dilution 1:100; cat. no.
ab76471; Abcam) or polyclonal rabbit anti-vimentin (dilution
1:3,000, cat. no. 10366-1-AP; ProteinTech Group, Inc.). Following
additional washing with PBS three times, the sections were
incubated with a biotin-conjugated anti rabbit secondary antibody,
which was included in a Histofine SAB-PO(R) kit (cat. no. 424031;
Nichirei Biosciences, Inc.) and used in undiluted form, at room
temperature for 10 min. Reactive products were detected using this
kit, followed by visualization using the Histofine DAB-3S kit
(Nichirei Biosciences, Inc.) and counterstain with hematoxylin.
Two independent observers who were blinded to the
clinical data analyzed the sections. Doubtful cases were
reassessed, and discrepancies were settled by consensus. The
expression of Sam68 was determined in the nucleus and cytoplasm by
semiquantitative assessment of the percentage of positively stained
tumor cells and staining intensity under a light microscope in
three representative fields on each slide (Olympus System
Microscope Model BX43; Olympus Corporation). The percentage of
positive cells was scored as follows: 1 (0–50% positive cells) and
2 (51–100% positive cells). Staining intensity was scored as
follows: 0, no signal; 1, weak; 2, moderate; and 3, strong. The
staining index was calculated as the production of the score for
the percentage of positive cells and staining intensity. The
nuclear and cytoplasmic indices were calculated (scores of 0, 1, 2,
3, 4, and 6, respectively). The total index was determined as Sam68
expression by summing the nuclear and cytoplasmic indices (scores
of 0–12). The validity of the optimal cutoff value for each
categorical variable was determined using receiver-operating
characteristic curve analysis. A total index ≥8 was considered high
expression in the OSCC tissues. Staining for vimentin was
classified as negative and positive if cytoplasmic immunostaining
occurred in <10% and ≥10% of epithelial tumor cells,
respectively (29,30).
Cell lines
HO-1-N-1 (a human buccal mucosal SCC cell line),
HSC-2 (a human oral SCC cell line), HSC-3 (a human tongue SCC cell
line), and HOC-313 (a human floor-of-the mouth SCC cell line) were
cultured at 37°C in a 5% CO2 atmosphere in Dulbecco's
modified Eagle's medium (DMEM) (Nacalai Tesque, Inc.) containing
10% fetal bovine serum (FBS; Nichirei Biosciences, Inc.). HO-1-N-1
was purchased from the Japanese Collection of Research Bioresources
(cat. no. JCRB0831). HSC-2, HSC-3, and HOC-313 were established
from surgical resected tumors at the Department of Oral and
Maxillofacial Surgery, Tokyo Medical and Dental University
(31,32).
Western blot analysis
Whole-cell lysates were prepared using
radioimmunoprecipitation lysis buffer (Santa Cruz Biotechnology,
Inc.) containing protease inhibitors. Protein content of the
samples was measured using a Qubit Fluorimeter and Qubit Protein
Assay Kits (Thermo Fisher Scientific, Inc.). Next, 20 µg of each
sample was separated by 10% sodium dodecyl sulfate-polyacrylamide
gel electrophoresis and transferred onto nitrocellulose membranes
(BioRad Laboratories, Inc.). Membranes were blocked with 5% non-fat
milk (TPBT) at room temperature for 60 min, then incubated with
monoclonal rabbit anti-SAM68 (dilution 1:500; cat. no. ab76471;
Abcam) or β-actin (1:4,000; cat. no. ab76471; ProteinTech Group,
Inc.) at room temperature for 60 min. The total protein content was
confirmed by β-actin staining. After washing with PBS, membranes
were incubated with a horseradish peroxidase (HRP)-conjugated anti
rabbit secondary antibody dilution 1:4,000; cat. no. 7074; Cell
Signaling Technology) at room temperature for 60 min.
Immunoreactive bands were visualized with chemiluminescence using
Pierce ECL Western Blotting Substrate or SuperSignal West Dura
Extended Duration Substrate (both from Thermo Fisher Scientific,
Inc.).
Sam68 knockdown
Sam68 small interfering RNA (siRNA)#1
(5′-GCAAAGUUGUUACUGAUUU-3′; MISSION predesigned siRNA), Sam68
siRNA#2 (5′-GAGCAAAGUUGUUACUGAU-3′; MISSION predesigned siRNA; both
from Sigma-Aldrich: Merck KGaA), and control siRNA (Silencer
Negative Control No. 1 siRNA; cat. no. AM4611; Ambion; Thermo
Fisher Scientific, Inc.) were transfected into cells (60–70%
confluence) using HilyMax (Dojindo Laboratories, Inc.) transfection
reagent at 37°C according to the manufacturer's instructions. The
concentration of each siRNA was 210 nM. The medium was replaced
with fresh serum-containing medium at 4 h after the transfection.
To evaluate the efficacy of gene silencing, Sam68 protein was
assessed by western blotting at 48 and 144 h after the
transfection.
RNA extraction and quantitative
real-time reverse transcription polymerase chain reaction
(RT-qPCR)
Total cellular RNA was isolated using the RNeasy
Mini Kit (Qiagen GmbH), according to the manufacturer's protocol.
Total RNA (2 µg) was reverse-transcribed using ReverTra Ace qPCR RT
Master Mix (TOYOBO, Inc.). The ABI 7500 real-time polymerase chain
reaction (PCR) system and SYBR Green Real-time PCR Master Mix Plus
(TOYOBO, Inc.) were used for quantitative reverse transcription PCR
(RT-qPCR). The PCR amplification profile was as follows: Initial
denaturation at 95°C for 5 min, followed by 40 cycles of
denaturation at 95°C for 15 sec and annealing and extension at 60°C
for 60 sec. All expression data were normalized to the expression
of β-actin. The primers used in this assay are listed in Table SI. The 2−ΔΔCq method
was used for relative quantification of gene expression levels
(33).
mRNA sequencing, differential
expression analysis, and gene ontology (GO) enrichment
analysis
At 48 h after transfection, total cellular RNA was
obtained from the sample of HO-1-N-1 cells with Sam68 siRNA#1 or
control siRNA using the RNeasy Mini Kit (Qiagen GmbH) according to
the manufacturer's instructions. Samples were prepared for each
group to perform three independent assays. mRNA sequencing was
performed using Illumina NextSeq500 (Illumina, Inc.) at the
Department of Sports Medicine Analysis under the Open Facility
Network Office, University of Tsukuba, in accordance with the
standard protocol (34).
Sequencing was conducted with paired-end reads of 36 bases. After
the sequencing run, the FASTQ files were exported and the basic
information of the NGS run data was verified using CLC Genomics
Workbench 20.0.3 software (Qiagen GmbH). Quality assessment of the
reads confirmed a PHRED score (quality value) of >20 for 99.68%
of all reads, indicating the success of the run. The read number
was approximately 35.4-37.5 million per sample as paired-end reads.
The RNA sequencing data as FASTQ files and expression browser as
table data are deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under
accession number GSE202136. The Metascape tool (https://metascape.org/gp/index.htmls/main/step1) was
used to perform GO enrichment analysis of differentially expressed
genes (35).
Wound healing assay
After 48 h of Sam68 siRNA or control siRNA
transfection, 1.0×106 HO-1-N-1 cells were cultured in
24-well plates until confluent. A wound was made through the
confluent monolayer with a 200-µl pipette tip. There were confluent
cells on either side of the wound. The wells were rinsed with PBS
to remove detached cells. The remaining cells were incubated with
DMEM containing 0.5% FBS. At 72 h after wound creation, the wound
areas were evaluated using a phase-contrast microscope (Olympus
System Microscope Model CKX53; Olympus Corporation). All
experiments were performed in triplicate.
Chamber migration assay
At 48 h after Sam68 siRNA or control siRNA
transfection, HO-1-N-1 cells were detached with TrypLE express
(Thermo Fisher Scientific, Inc.) and resuspended in serum-free
DMEM. A total of 5×105 cells were seeded into the upper
chamber of a Falcon Cell Culture Insert with an 8-µm pore filter
(Corning, Inc.). DMEM with 10% FBS was added to the lower chamber.
Cells were allowed to migrate for 48 h. The filters were then fixed
with 100% ice-cold methanol at room temperature for 15 min and
stained with Carrazzi's hematoxylin (Muto Pure Chemicals Co., Ltd.)
at room temperature for 15 min, followed by removal of nonmigrated
cells with a cotton swab. The filters were mounted on glass slides,
and migrated cells adhering to the bottom side of the chamber were
counted using a light microscope at a magnification of ×40. All
experiments were performed in triplicate.
Statistical analyses
Mann-Whitney U test was used for continuous
variables between the two groups. The associations between Sam68
expression and categorical variables of clinicopathological
features were assessed using Fisher's exact test or Pearson's
chi-square test. To examine the correlation between Sam68
expression and clinicopathological features, multivariate logistic
regression analysis was used. The results are expressed as the mean
± standard deviation (SD) in the wound healing assay, chamber
migration assay, and RT-qPCR analysis. Statistical analyses of
multiple comparisons were performed using one-way analysis of
variance (ANOVA) with a post hoc Dunnett's test of all samples to
control. P<0.05 was considered to indicate a statistically
significant difference. All statistical analyses were performed
using JMP14 (SAS Institute, Inc.).
Results
Sam68 expression in OSCC and adjacent
healthy mucosa
To determine the expression of Sam68 in OSCC and
healthy oral mucosa, immunohistochemical analysis was performed
using the tissue samples of OSCC. In adjacent healthy mucosal
epithelium, Sam68 expression was detected predominantly in the
nucleus with moderate (52%) to strong (35%) intensity, whereas
cytoplasmic Sam68 expression was mostly negative (72%),
occasionally observed with weak intensity (23%), or rarely observed
with moderate intensity (5%) (Fig.
1A). No case was confirmed to have cytoplasmic Sam68 expression
with strong intensity in the adjacent healthy mucosal epithelium.
By contrast, nuclear Sam68 expression with strong intensity was
commonly observed (75%) in OSCC cells. Cytoplasmic Sam68 expression
with moderate (25%) or strong intensity (16%) was observed in OSCC
cells (Fig. 1B-H). As revealed in
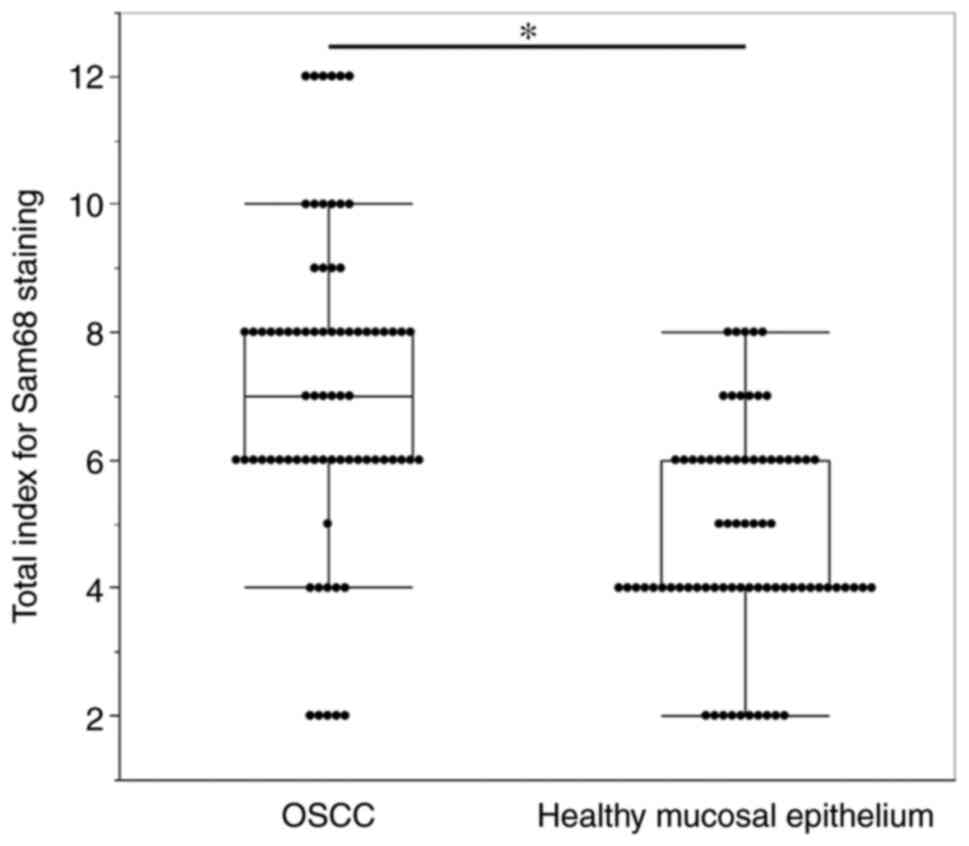
Fig. 2, the total staining index
was significantly higher in OSCC (median index 7, interquartile
range 6–8) than that of adjacent healthy mucosal epithelium (median
index 4, interquartile range 4–6; P<0.01). These findings
indicated that the expression of Sam68 was increased in OSCC
compared with that of adjacent healthy epithelial cells.
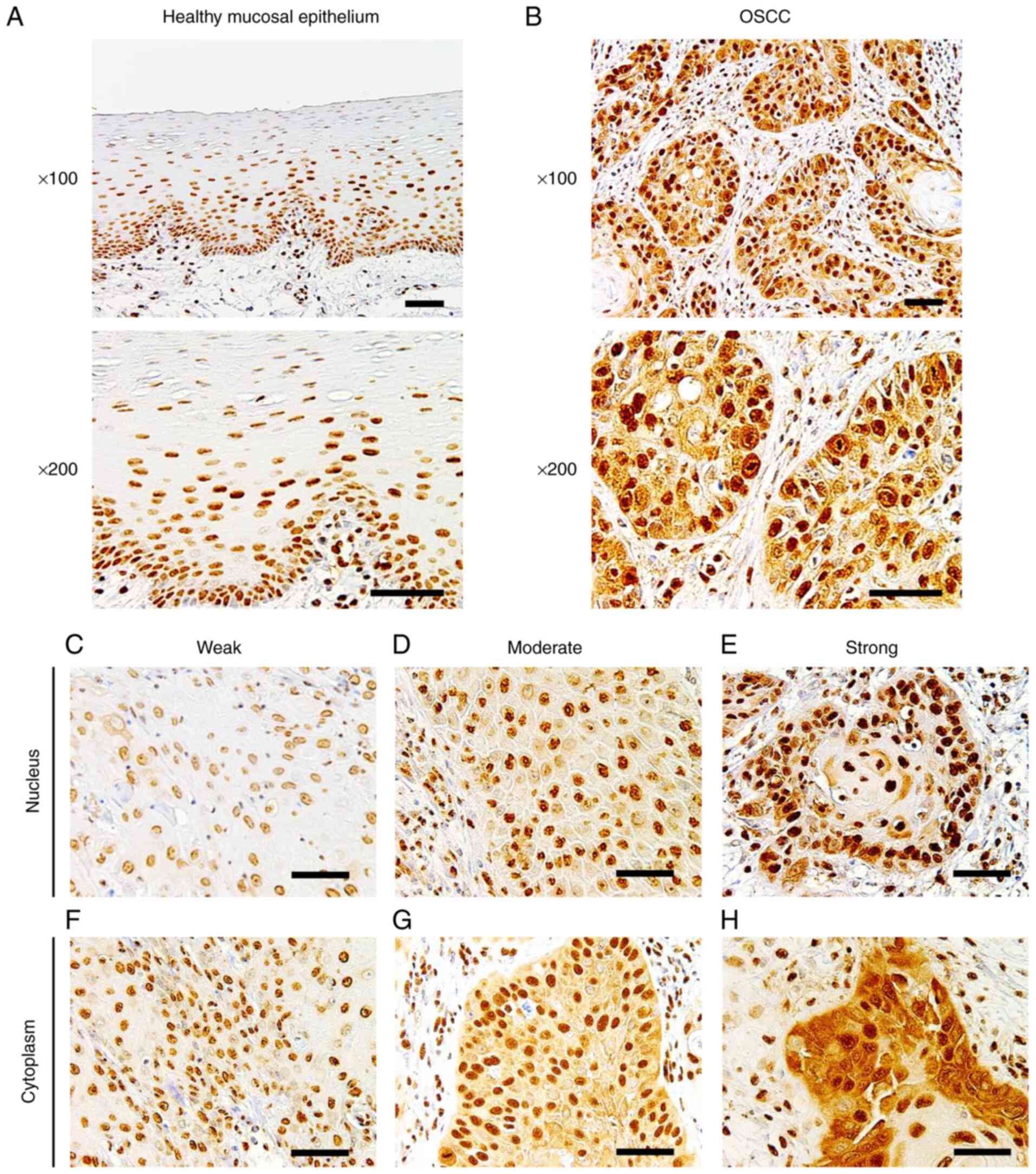 | Figure 1.Immunohistochemistry of Sam68. (A)
Representative images of healthy mucosal epithelium adjacent to
OSCC, in which Sam68 expression was predominantly detected in the
nucleus and negatively observed in the cytoplasm (magnification,
×100 and ×200; scale bar, 50 µm). (B) Representative image of OSCC,
in which Sam68 expression was detected both in the nucleus and
cytoplasm (magnification, ×100 and ×200; scale bar, 50 µm). (C-E)
Nuclear expression of Sam68 with (C) weak, (D) moderate, and (E)
strong staining intensity in OSCC cells (magnification, ×200; scale
bar, 50 µm). (F-H) Cytoplasmic expression of Sam68 with (C) weak,
(D) moderate, and (E) strong staining intensity in OSCC tissue
(magnification, ×200; scale bar, 50 µm). Sam68, Src-associated in
mitosis 68 kDa; OSCC, oral squamous cell carcinoma. |
Correlation between Sam68 expression
and clinicopathological features in patients with OSCC
To investigate the clinical significance of Sam68
expression, the association between the total staining index of
Sam68 and the clinicopathological features of patients with OSCC
was analyzed. The results of the univariate analysis indicated that
a high expression of Sam68 was significantly correlated with
advanced pathological T stage (P=0.01), positive lymphovascular
invasion (P=0.01), and pathological cervical lymph node metastasis
(P<0.01; Table I).
 | Table I.Univariate analyses of the
correlation between Sam68 expression and clinicopathological
features in patients with OSCC. |
Table I.
Univariate analyses of the
correlation between Sam68 expression and clinicopathological
features in patients with OSCC.
|
| Sam68
expression |
|
|---|
|
|
|
|
|---|
| Clinicopathological
features | Low (n=39) | High (n=38) | P-value |
|---|
| Age, years |
|
| 0.56 |
|
<65 | 25 | 16 |
|
|
>65 | 14 | 22 |
|
| Sex |
|
| 0.05 |
|
Male | 29 | 26 |
|
|
Female | 10 | 12 |
|
| pT |
|
| 0.01a |
| T1 +
T2 | 38 | 30 |
|
| T3 +
T4 | 1 | 8 |
|
| Pathological
differentiation |
|
| 0.51 |
|
Well | 20 | 15 |
|
|
Moderate | 16 | 18 |
|
|
Poor | 3 | 5 |
|
| YK
classification |
|
| 0.43 |
| YK1 +
YK2 + YK3 | 26 | 22 |
|
| YK4C +
YK4D | 13 | 16 |
|
| Lymphovascular
invasion |
|
| <0.01 |
|
Negative | 25 | 13 |
|
|
Positive | 14 | 25 |
|
| Perineural
invasion |
|
| 0.19a |
|
Negative | 36 | 31 |
|
|
Positive | 3 | 7 |
|
| Pathological
CLNM |
|
| <0.01 |
|
Negative | 33 | 19 |
|
|
Positive | 6 | 19 |
|
| Primary
recurrence |
|
| 0.99a |
|
Negative | 37 | 36 |
|
|
Positive | 2 | 2 |
|
Cervical lymph node metastasis is the most important
prognostic factor in patients with OSCC; thus, additional
statistical analyses were performed to assess whether Sam68
expression was a critical factor in pathological cervical lymph
node metastasis. A significant correlation between pathological
cervical lymph node metastasis and positive lymphovascular invasion
(P=0.01) and the high expression of Sam68 (P<0.01; Table II) was observed. Moreover,
multivariate logistic regression analysis revealed that a high
expression of Sam68 was an independent factor for cervical lymph
node metastasis [odds ratio (OR): 4.39; 95% confidence interval
(CI): 1.49-14.23; P<0.01; Table
III]. These results indicated that a high expression of Sam68
contributed to tumor progression, particularly cervical lymph node
metastasis, in OSCC.
 | Table II.Univariate analysis of the
correlation between clinicopathological features and lymph node
metastasis. |
Table II.
Univariate analysis of the
correlation between clinicopathological features and lymph node
metastasis.
|
| Pathological
CLNM |
|
|---|
|
|
|
|
|---|
| Clinicopathological
features | Negative
(n=52) | Positive
(n=25) | P-value |
|---|
| Age, years |
|
| 0.11 |
|
<65 | 27 | 14 |
|
|
>65 | 25 | 11 |
|
| Sex |
|
| 0.54 |
|
Male | 36 | 19 |
|
|
Female | 16 | 6 |
|
| pT |
|
| 0.06a |
| T1 +
T2 | 49 | 19 |
|
| T3 +
T4 | 3 | 6 |
|
| Pathological
differentiation |
|
| 0.51 |
|
Well | 26 | 9 |
|
|
Moderate | 21 | 13 |
|
|
Poor | 5 | 3 |
|
| YK
classification |
|
| 0.43 |
| YK1 +
YK2 + YK3 | 34 | 14 |
|
| YK4C +
YK4D | 18 | 11 |
|
| Lymphovascular
invasion |
|
| 0.01 |
|
Negative | 31 | 7 |
|
|
Positive | 21 | 18 |
|
| Perineural
invasion |
|
| 0.21 |
|
Negative | 47 | 20 |
|
|
Positive | 5 | 5 |
|
| Sam68
expression |
|
| <0.01 |
|
High | 33 | 6 |
|
|
Low | 19 | 19 |
|
 | Table III.Multivariate analysis of the
correlation between clinicopathological features and lymph node
metastasis. |
Table III.
Multivariate analysis of the
correlation between clinicopathological features and lymph node
metastasis.
| Clinicopathological
features | Odds ratio | 95%CI | P-value |
|---|
| Lymphovascular
invasion (positive vs. negative) | 2.75 | 0.93-8.59 | 0.07 |
| Sam68 expression
(high vs. low) | 4.39 | 1.49-14.23 | <0.01 |
Sam68 expression and its knockdown in
OSCC cells
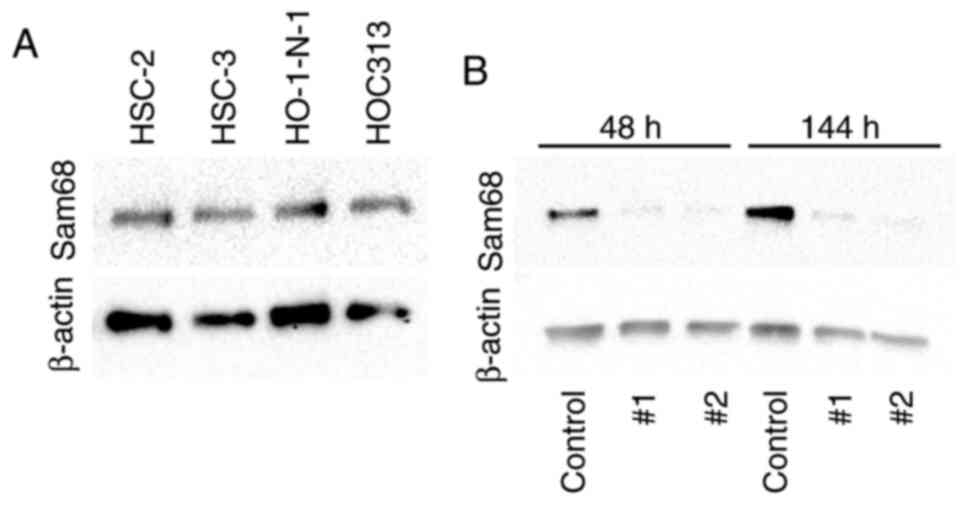
For further investigation, the expression of Sam68
protein in OSCC cells was examined by western blotting. The
expression of Sam68 was detected in all OSCC cell lines (Fig. 3A). To examine the effect of Sam68,
Sam68 knockdown in HO-1-N-1 cells was produced. From 48 to 144 h
after the transfection, the expression of Sam68 was significantly
decreased in Sam68 siRNA-transfected cells compared with that in
control siRNA-transfected cells (Fig.
3B). These cells were subsequently used for the following
experiments to assess the biological roles of Sam68 in OSCC.
Sam68 knockdown affects the phenotype
of OSCC cells
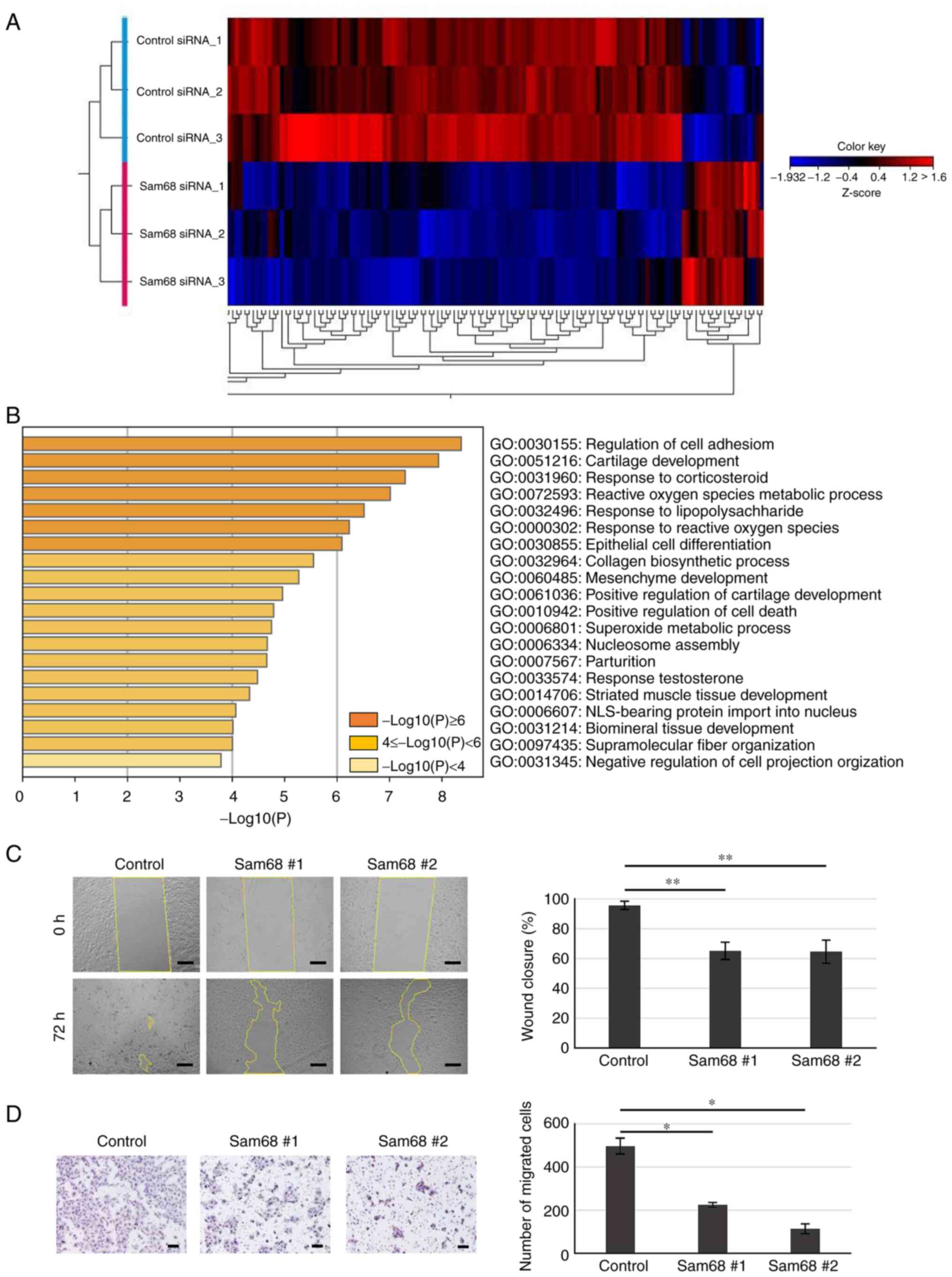
To elucidate the potential mechanism by which Sam68
affects the phenotypes associated with metastasis in OSCC, mRNA
sequencing was used to assess changes in the transcriptome between
Sam68 siRNA- or control siRNA-transfected HO-1-N-1 cells.
Differential expression analysis was performed and it was
determined that genes with a false discovery rate of a P-value of
<0.05 and a cutoff of |fold change|>2 were differentially
expressed between the two conditions. A total of 150 differentially
expressed genes (DEGs) were obtained including 20 upregulated and
130 downregulated genes (Fig. 4A).
To explore the biological process significantly associated with
these DEGs, GO enrichment analysis was further conducted. The
results revealed that enriched GO terms were not detected in the
upregulated genes; in addition, the downregulated genes were
significantly concentrated in various biological processes
(Fig. 4B). In the top 10 enriched
processes, there were some processes related to the
epithelial-mesenchymal transition (EMT): regulation of cell
adhesion, epithelial cell differentiation, and mesenchyme
development. Accumulating evidence has demonstrated that EMT, the
transdifferentiation of epithelial cells into mesenchymal cells, is
integral in cancer progression (36). Namely, epithelial cells
downregulate their epithelial properties, lose their cell-cell
adhesion, and acquire mesenchymal properties, which increase cell
motility (36,37). Wound healing and chamber migration
assays were used to confirm the effect of Sam68 on cell motility.
As revealed in Fig. 4C,
Sam68-knockdown cells demonstrated significantly slower wound
closure compared with control cells. In addition, Sam68 knockdown
markedly decreased the number of migrated cells as determined by
the chamber migration assay (Fig.
4D). These data indicated that Sam68 may be associated with the
EMT process and regulate the motility of OSCC cells.
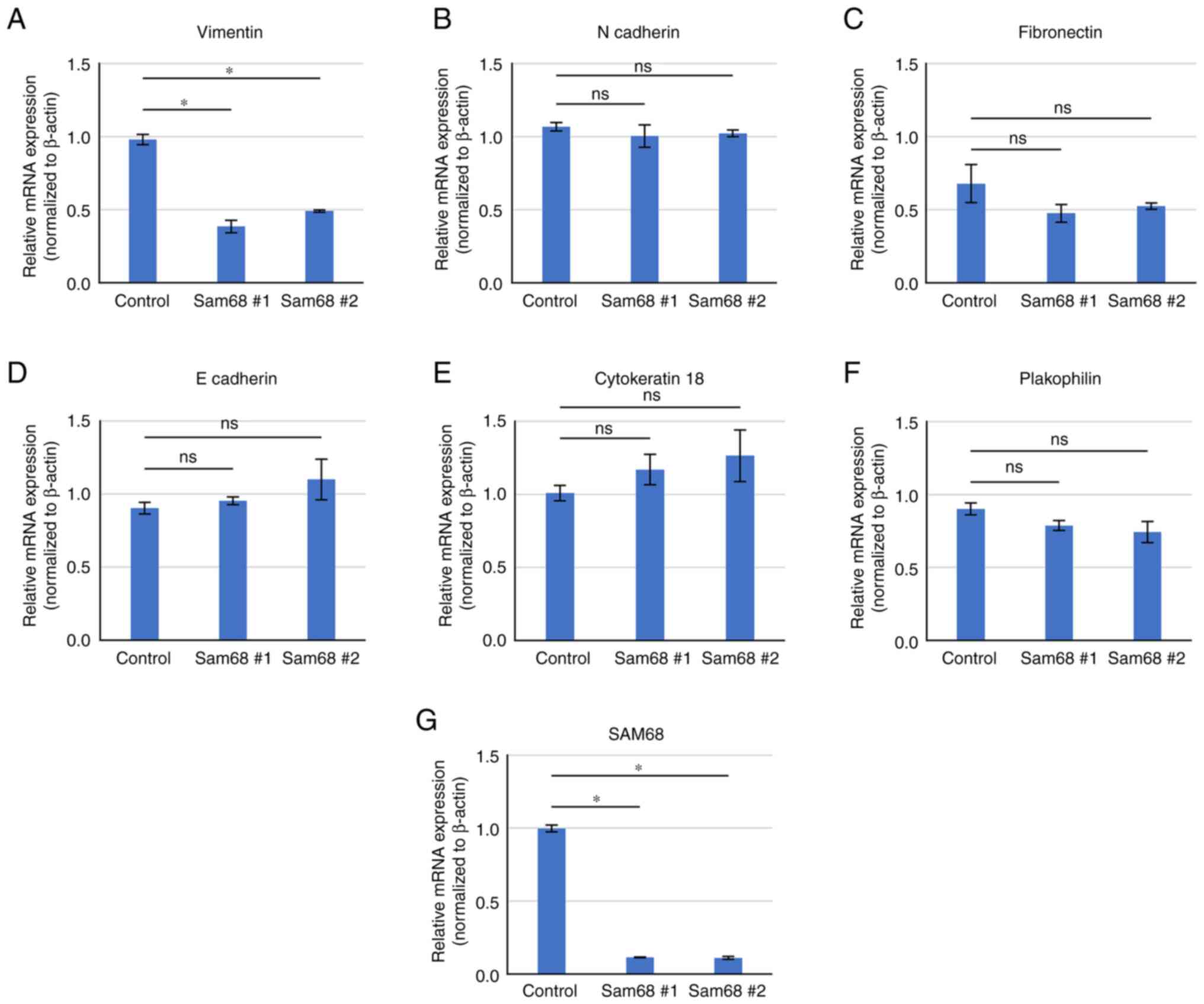
To provide further validation, a literature search
on the 36 downregulated DEGs enriched in the above 3 biological
processes was conducted and a candidate gene associated with EMT
and cell motility was selected. Vimentin is a classical mesenchymal
marker that has been revealed to be elevated in EMT progression and
related to motile activities (38). The differential expression of
vimentin and other EMT markers were verified by RT-qPCR. As
revealed in Fig. 5, a decreased
mRNA level of vimentin was confirmed by qPCR in Sam68-knockdown
cells. The mRNA level of epithelial (E-cadherin, cytokeratin 18,
and plakophilin) and mesenchymal markers (N-cadherin and
fibronectin) were not significantly altered by the reduction of
Sam68 (Fig. 5). These data were
consistent with the results of mRNA sequencing, demonstrating that
vimentin expression was specifically downregulated in
Sam68-knockdown HO-1N-1 cells. The effect of Sam68 knockdown on
vimentin expression was also assessed using RT-qPCR in other OSCC
cell lines, and the mRNA expression of vimentin was confirmed to
have been reduced via Sam68 knockdown in HSC-2 and HOC313 cells
(Fig. S1). Collectively, these
results indicated that Sam68 regulated vimentin expression and the
motile mesenchymal phenotype of OSCC cells.
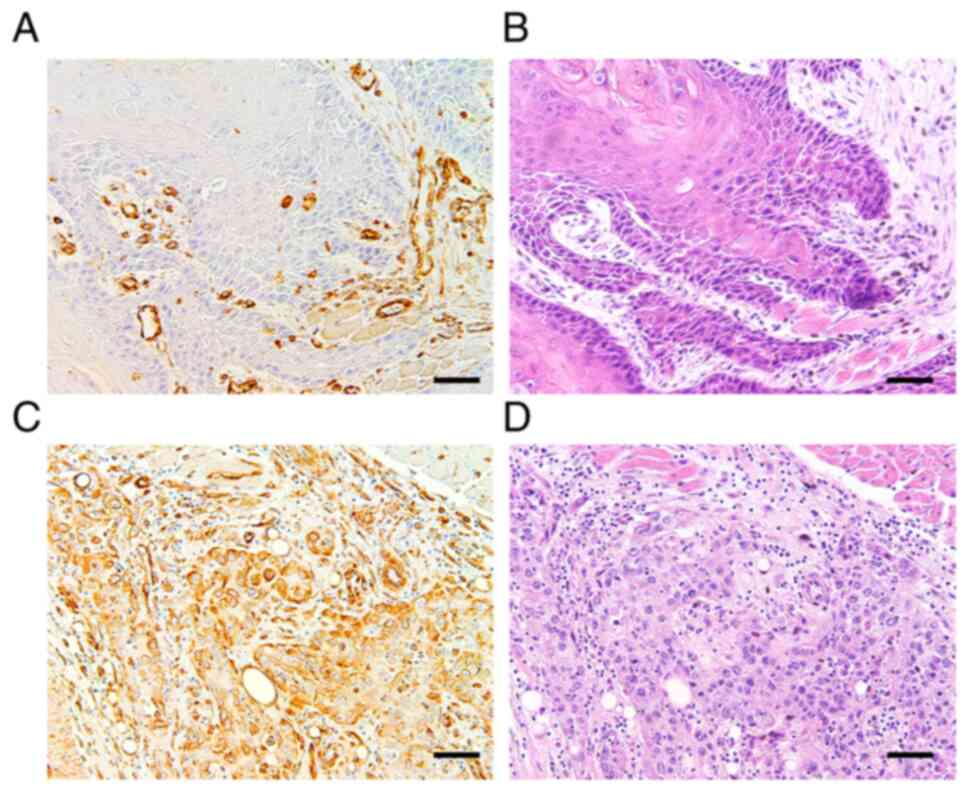
Association between Sam68 and vimentin
expression on tissue samples of OSCC
Next, to clarify whether the expression of vimentin
is correlated with that of Sam68 and cervical lymph node metastasis
in OSCC, an immunohistochemical study of vimentin on 77 tissue
samples was performed. Accordingly, 21 (27%) tumors were found to
exhibit positive vimentin expression (Fig. 6). Although no statistical
significance was observed, the proportion of tumors with Sam68 high
expression was higher in the tumors exhibiting positive vimentin
expression (62%) than those tumors exhibiting negative vimentin
expression (38%) (P=0.17; Table
IV). A similar finding was confirmed in pT1 + T2 tumors (56 and
44%; P=0.26) or pT3 + 4 tumors (80 and 20%; P=0.34), respectively
(data not shown). In addition, the univariate analysis showed that
a positive expression of vimentin was significantly correlated with
pathological cervical lymph node metastasis (P<0.01; Table IV).
 | Table IV.Univariate analysis of the
correlation between vimentin expression and Sam68 expression or
cervical lymph node metastasis. |
Table IV.
Univariate analysis of the
correlation between vimentin expression and Sam68 expression or
cervical lymph node metastasis.
|
|
|
| Variables |
|
|
|---|
|
| Sam68
expression |
| Pathological
CLNM |
|---|
|
|
|
|
|
|---|
|
| Low (n=39) | High (n=38) |
| Negative
(n=52) | Positive
(n=25) |
|---|
| Vimentin
expression |
|
|
|
|
|
|
Negative | 31 | 25 |
| 44 | 12 |
|
Positive | 8 | 13 |
| 8 | 13 |
|
P-value | 0.177 |
| <0.01 |
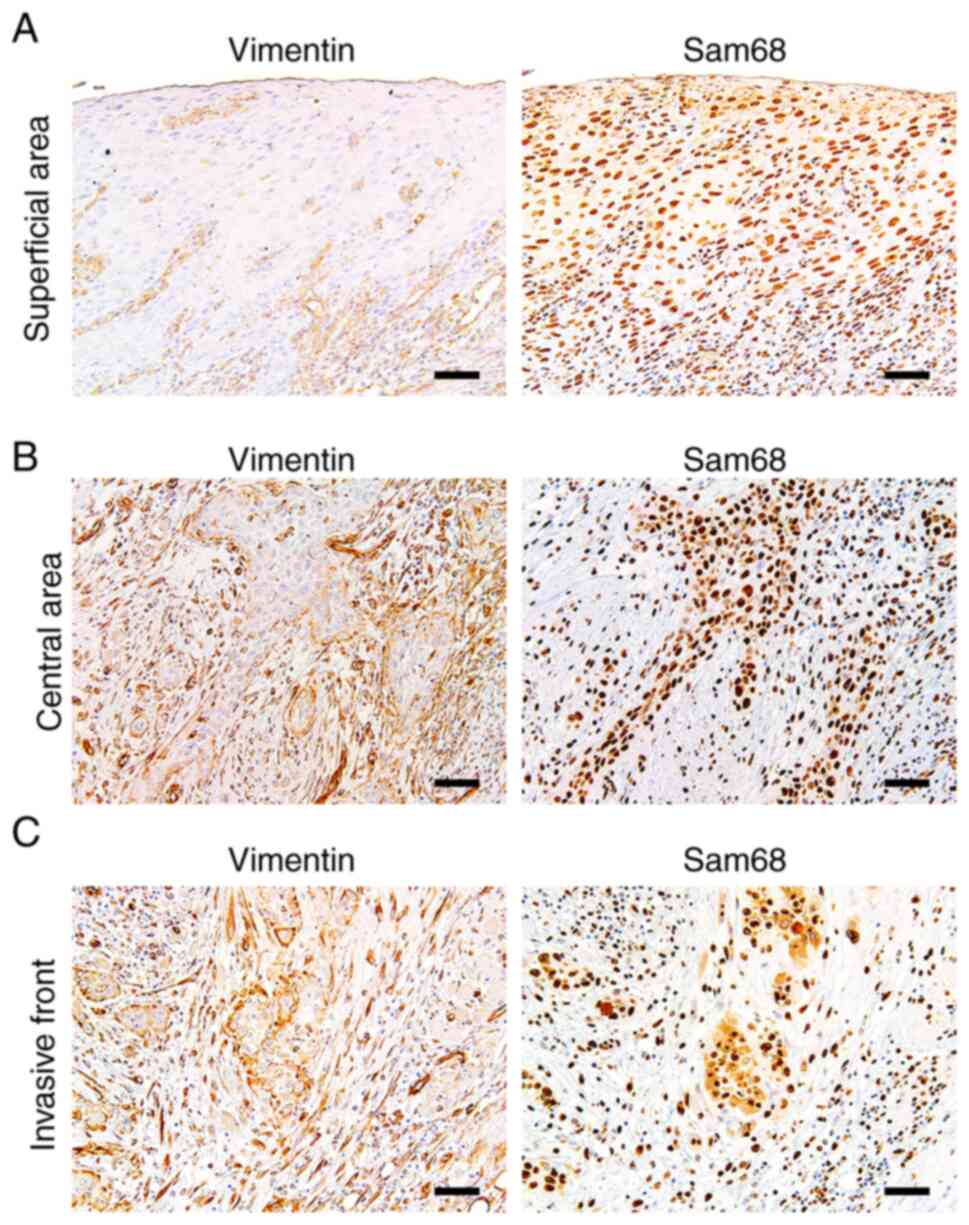
Vimentin expression was reported to be upregulated
at the invasive front in OSCC, which may be related to EMT
(39). Thus, to obtain more
insights into the association between expression of vimentin and
Sam68, the distribution of positive vimentin and the pattern of
Sam68 expression in the tumors exhibiting positive vimentin
expression were investigated microscopically (n=21). Of the 21
tumors, six (28%) broadly exhibited positive vimentin expression at
the invasive front, the central area, and the superficial area of
the tumor. Similarly, the immunoreactivity of Sam68 was uniformly
observed at almost all areas in these tumors. In 10/21 (48%)
tumors, a positive vimentin expression was mostly observed at the
deep area (the invasive front and the central area of the tumor)
but not at the superficial areas of the tumors. Notably, in these
tumors, the tumor cells with vimentin expression at the deep area
were also observed to be accompanied with higher Sam68
immunoreactivity than those without vimentin expression at the
superficial areas of the tumor (Fig.
7). In 3/21 (14%) tumors, positive vimentin expression was
mostly observed at the deep area, but the immunoreactivity of Sam68
was uniformly observed at all areas in these tumors. In addition,
in 2/21 (10%) tumors, the distribution of positive vimentin was
broad, but Sam68 immunoreactivity was found to be higher at the
deep area than those at the superficial area. These microscopic
findings suggest that in the most tumors (16/21; 76%), the
distribution of positive vimentin may be related to the
distribution or degree of Sam68 expression. Collectively, these
results support the association between vimentin expression and
Sam68 expression as well as cervical lymph node metastasis in
OSCC.
Discussion
In the present study, the clinical implication of
Sam68 in OSCC was demonstrated. The immunohistochemical results
demonstrated that advanced OSCC exhibited significantly higher
expression of Sam68. In particular, multivariate analysis
demonstrated that a high expression of Sam68 was an independent
factor associated with cervical lymph node metastasis in OSCC.
Next, to elucidate the biological role of Sam68 in OSCC cells, mRNA
sequencing was performed to measure the changes in the
transcriptome via Sam68 knockdown. The GO analysis revealed that
downregulated DEGs were enriched in the biological process related
to EMT in Sam68-knockdown OSCC cells. It was established that
vimentin expression was specifically downregulated in these cells.
It was also confirmed that the migration activity of OSCC cells was
significantly reduced by Sam68 knockdown. Furthermore, the
immunohistochemical analyses of vimentin demonstrated the
association between vimentin expression and Sam68 expression as
well as cervical lymph node metastasis. These results indicated
that Sam68 may contribute to metastasis by regulating the
expression of vimentin and the cell motility in OSCC.
Multiple studies have shown that Sam68 is associated
with tumor progression, metastasis, and survival of patients in
various cancers, including breast, prostate, lung, gastric, renal,
and cervical cancers (9,18,21–26).
However, few studies have investigated whether Sam68 has clinical
and biological significance in OSCC. Chen et al conducted a
univariate analysis and reported a correlation between higher Sam68
expression and T stage, N stage, nodal status, recurrence, and
survival of patients with tongue cancer; however, these data were
limited in patients with T1-3 classification and clinically
negative nodal diseases (20). To
the best of our knowledge, the present study is the first to assess
the expression profile and evaluate the clinical implication of
Sam68 in patients at all stages of OSCC. In this study, univariate
analysis indicated that pathological T stage, lymphovascular
invasion, and cervical lymph node metastasis were correlated with a
high expression of Sam68 in OSCC. Moreover, multivariate analysis
revealed that a high expression of Sam68 was an independent
predictor of cervical lymph node metastasis. These results suggest
that a high expression of Sam68 is closely related to tumor
progression and metastasis to cervical lymph nodes in OSCC.
To evaluate the association between the invasive
aspect of OSCC on histopathology and Sam68 expression, the
histological grading was assessed by differentiation (2) and mode of invasion according to YK
classification (27). In the
results, those histological parameters were not significantly
related to cervical lymph node metastasis. In addition, there were
no statistically significant differences in those histological
parameters between two groups of Sam68 expression. Conventional
histological grading by differentiation was reported to have a
limited predictive value for metastasis and prognosis (2,40,41),
which is consistent with the result of the present study. In
addition, other histological parameters based on the invasive
pattern of tumors have been recognized to be associated with
cervical lymph node metastasis (27,40).
Conversely, similar to the result of the present study, some
studies have shown no significant association between invasive
pattern and metastasis in OSCC (41,42).
A firm conclusion concerning the association between those
histologic parameters and Sam68 expression cannot be obtained in
the present study alone. Further studies are necessary to clarify
the association between them.
A previous study revealed the association between
Sam68 and resistance to anticancer agents in tongue SCC cells
(20); however, there are no data
in the literature regarding the malignant phenotypes affected by
dysregulation of Sam68 in OSCC. To obtain new insights into the
biological significance of Sam68 in OSCC, mRNA sequencing was
performed to investigate changes in the gene expression profile
through Sam68 knockdown. This result revealed that 36 downregulated
DEGs were statistically enriched in the biological process of GO
related to EMT, including regulation of cell adhesion, epithelial
cell differentiation, and mesenchyme development (36,37,43).
EMT plays a critical role in tumor progression and metastasis
(36–38,44).
During this process, epithelial cells lose cell-cell adhesion and
apicobasal polarity to exhibit migratory mesenchymal properties
(36). It was verified that Sam68
knockdown significantly reduced motile behavior and vimentin
expression in OSCC cells. The immunohistochemical analysis of
vimentin also indicated the association between positive vimentin
and Sam68 expression. In addition, a positive vimentin expression
was found to be significantly correlated with cervical lymph node
metastasis in this study. Recent lines of evidence have
demonstrated that vimentin expression is upregulated in EMT
progression, resulting in a more motile phenotype of tumor cells
(37,38). During the progression of EMT,
epithelial markers such as keratin, E-cadherin, and plakophilin are
decreased and mesenchymal markers such as vimentin, N-cadherin, and
fibronectin are increased (36).
However, the expression profile of EMT markers has been reported to
be widely varied depending on the cancer type, cell lines, and
signaling pathways of EMT (45–47).
Saito et al demonstrated that various OSCC cell lines
exhibited each specific expression of EMT markers (46). In addition, the cell status in
which epithelial properties are retained, known as partial EMT, has
been recognized to possess a higher metastatic ability as compared
with complete EMT, with loss of all epithelial features and
complete acquisition of mesenchymal morphology (48–50).
In this study, vimentin was specifically regulated by Sam68 in OSCC
cells; by contrast, other epithelial and mesenchymal markers were
not significantly altered in these cells. Taken together, the
results indicated that Sam68 may regulate the expression of
vimentin to induce partial EMT and promote the motile mesenchymal
behavior of OSCC.
It has been recognized that changes at the RNA
level, including alternative splicing and noncoding RNA-mediated
control, regulate EMT (19,36).
As a result of alternative splicing, extensive isoforms are
generated in various proteins that regulate EMT. Valacca et
al demonstrated that Sam68 contributes to the regulation of EMT
by changing the splicing profile and overall transcription levels
of serine/arginine-rich splicing factor 1 (SRSF1), which produces a
constitutively active splicing variant of Recepteur d'Origine
Nantais (Ron) proto-oncogene in colon adenocarcinoma cells
(19). In the present study, an
analysis of alternative splicing variants using RNA-sequencing data
was also performed. The results did not reveal a significant
difference in SRSF1 or Ron splicing variants between
Sam68-knockdown OSCC cells and control cells (data not shown). In
addition, recent research has demonstrated that
O-GlcNAcylation of Sam68 may enhance the migratory and
invasive abilities of lung cancer cells (51). O-GlcNAcylation is a
post-translational protein modification catalyzed by
O-GlcNAc transferase (OGT) and has been linked to biological
properties of cancer, including cell proliferation, survival,
invasion, and metastasis (51,52).
It has also been reported that the EMT process is associated with
modulation of O-GlcNAcylation in lung cancer cells (52). In the present study, the
RNA-sequencing data showed that the mRNA level of OGT was not
significantly altered via Sam68-knockdown in OSCC (data not shown).
Collectively, these findings suggest that Sam68 may have the
potential to promote EMT in a different manner, depending on the
type of cancer.
The present study has some limitations. First, is
the relatively small number of patients included in the study.
Second, the sample size between the two groups of pT stage,
perineural invasion, and primary recurrence were biased. Therefore,
the statistical power to draw firm conclusions was
insufficient.
Collectively, the findings of the present study
indicated that Sam68 contributes to metastasis by regulating the
expression of vimentin, which may be associated with partial EMT
and the cell motility of OSCC. Although the detailed pathways by
which Sam68 induces vimentin remain to be identified, it is
concluded that Sam68 may be a promising predictor of cervical lymph
node metastasis and have the potential to be a novel therapeutic
target in oral cancer.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Professor Jun Aida
(Department of Oral Health Promotion, Graduate School of Medical
and Dental Sciences, Tokyo Medical and Dental University, Tokyo,
Japan) for his valuable advice on statistical analysis. We are
grateful to Associate Professor Kei-ichi Morita (Department of
Maxillofacial Surgery, Graduate School of Medical and Dental
Sciences, Tokyo Medical and Dental University, Tokyo, Japan) for
his technical support.
Funding
The present study was supported by JSPS KAKENHI (grant no.
20K10133).
Availability of data and materials
The data sets used and/or analyzed during the
present study are available from the corresponding author upon
reasonable request. The RNA sequencing data as FASTQ files and
expression browser as table data are deposited in the Gene
Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession
number: GSE202136.
Authors' contributions
TKu conceived the present study. TKu and FH designed
the experiments. TKo, YI, HHi, FT, YM, and KK collected the
clinical and pathological data of patients. TKo, TS, and SF
performed the experiments and analyzed these data. TKu and TKo
analyzed statistical data. TKu and HHa confirm the authenticity of
all the raw data. TKu and TKo wrote the manuscript. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The study complied with the standards of the
Declaration of Helsinki and was approved (approval no. D2020-025)
by the Institutional Review Board of Tokyo Medical and Dental
University (Tokyo, Japan).
Patient consent for publication
Opt-out informed consent from patients was obtained
by exhibiting the research information on the official website of
our hospital (Tokyo Medical and Dental University Hospital, Tokyo,
Japan). The authors guarantee the opportunity for refusal by
document, call or e-mail whenever possible. Patients who rejected
participation in this study were excluded.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
DEG
|
differentially expressed genes
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
EMT
|
epithelial-mesenchymal transition
|
|
FBS
|
fetal bovine serum
|
|
GO
|
Gene Ontology
|
|
OSCC
|
oral squamous cell carcinoma
|
|
PBS
|
phosphate-buffered saline
|
|
PCR
|
polymerase chain reaction
|
|
Sam68
|
Src-associated in mitosis 68 kDa
|
|
SCC
|
squamous cell carcinoma
|
|
YK
|
Yamamoto-Kohama
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Naggar AK, Chan JKC, Grandis JR, Takata
T and Slootweg PJ: WHO classification of head and neck tumours. 4th
edition. IARC Press; Lyon: 2017
|
|
3
|
Sato J, Kitagawa Y, Watanabe S, Asaka T,
Ohga N, Hirata K, Shiga T, Satoh A and Tamaki N: Hypoxic volume
evaluated by 18F-fluoromisonidazole positron emission
tomography (FMISO-PET) may be a prognostic factor in patients with
oral squamous cell carcinoma: Preliminary analyses. Int J Oral
Maxillofac Surg. 47:553–560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Larsen SR, Johansen J, Sørensen JA and
Krogdahl A: The prognostic significance of histological features in
oral squamous cell carcinoma. J Oral Pathol Med. 38:657–662. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oikawa Y, Kugimoto T, Kashima Y, Okuyama
K, Ohsako T, Kuroshima T, Hirai H, Tomioka H, Shimamoto H, Michi Y
and Harada H: Surgical treatment for oral tongue squamous cell
carcinoma: A retrospective study of 432 patients. Glob Health Med.
3:157–162. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tomioka H, Yamagata Y, Oikawa Y, Ohsako T,
Kugimoto T, Kuroshima T, Hirai H, Shimamoto H and Harada H: Risk
factors for distant metastasis in locoregionally controlled oral
squamous cell carcinoma: A retrospective study. Sci Rep.
11:52132021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kuroshima T, Onozato Y, Oikawa Y, Ohsako
T, Kugimoto T, Hirai H, Tomioka H, Michi Y, Miura M, Yoshimura R
and Harada H: Prognostic impact of lingual lymph node metastasis in
patients with squamous cell carcinoma of the tongue: A
retrospective study. Sci Rep. 11:205352021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vernet C and Artzt K: STAR, a gene family
involved in signal transduction and activation of RNA. Trends
Genet. 13:479–484. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bielli P, Busà R, Paronetto MP and Sette
C: The RNA-binding protein Sam68 is a multifunctional player in
human cancer. Endocr Relat Cancer. 18:R91–R102. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Henao-Mejia J and He JJ: Sam68
relocalization into stress granules in response to oxidative stress
through complexing with TIA-1. Exp Cell Res. 315:3381–3395. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu J, Zhou L, Tonissen K, Tee R and Artzt
K: The quaking I-5 protein (QKI-5) has a novel nuclear localization
signal and shuttles between the nucleus and the cytoplasm. J Biol
Chem. 274:29202–29210. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Liu Y, Kim BO and He JJ: Direct
participation of Sam68, the 68-kilodalton Src-associated protein in
mitosis, in the CRM1-mediated Rev nuclear export pathway. J Virol.
76:8374–8382. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Babic I, Cherry E and Fujita DJ: SUMO
modification of Sam68 enhances its ability to repress cyclin D1
expression and inhibits its ability to induce apoptosis. Oncogene.
25:4955–4964. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Paronetto MP, Achsel T, Massiello A,
Chalfant CE and Sette C: The RNA-binding protein Sam68 modulates
the alternative splicing of Bcl-x. J Cell Biol. 176:929–939. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hong W, Resnick RJ, Rakowski C, Shalloway
D, Taylor SJ and Blobel GA: Physical and functional interaction
between the transcriptional cofactor CBP and the KH domain protein
Sam68. Mol Cancer Res. 1:48–55. 2002.PubMed/NCBI
|
|
16
|
Richard S, Torabi N, Franco GV, Tremblay
GA, Chen T, Vogel G, Morel M, Cléroux P, Forget-Richard A, Komarova
S, et al: Ablation of the Sam68 RNA binding protein protects mice
from age-related bone loss. PLoS Genet. 1:e742005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Iijima T, Wu K, Witte H, Hanno-Iijima Y,
Glatter T, Richard S and Scheiffele P: SAM68 regulates neuronal
activity-dependent alternative splicing of neurexin-1. Cell.
147:1601–1614. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Frisone P, Pradella D, Di Matteo A,
Belloni E, Ghigna C and Paronetto MP: SAM68: Signal transduction
and RNA metabolism in human cancer. Biomed Res Int.
2015:5289542015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Valacca C, Bonomi S, Buratti E, Pedrotti
S, Baralle FE, Sette C, Ghigna C and Biamonti G: Sam68 regulates
EMT through alternative splicing-activated nonsense-mediated mRNA
decay of the SF2/ASF proto-oncogene. J Cell Biol. 191:87–99. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen S, Li H, Zhuang S, Zhang J, Gao F,
Wang X, Chen W and Song M: Sam68 reduces cisplatin-induced
apoptosis in tongue carcinoma. J Exp Clin Cancer Res. 35:1232016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiao J, Wang Q, Yang Q, Wang H, Qiang F,
He S, Cai J, Yang L and Wang Y: Clinical significance and effect of
Sam68 expression in gastric cancer. Oncol Lett. 15:4745–4752.
2018.PubMed/NCBI
|
|
22
|
Li X, Zhou X, Hua F, Fan Y, Zu L, Wang Y,
Shen W, Pan H and Zhou Q: The RNA-binding protein Sam68 is critical
for non-small cell lung cancer cell proliferation by regulating
Wnt/β-catenin pathway. Int J Clin Exp Pathol. 10:8281–8291.
2017.PubMed/NCBI
|
|
23
|
Li Z, Yu CP, Zhong Y, Liu TJ, Huang QD,
Zhao XH, Huang H, Tu H, Jiang S, Zhang Y, et al: Sam68 expression
and cytoplasmic localization is correlated with lymph node
metastasis as well as prognosis in patients with early-stage
cervical cancer. Ann Oncol. 23:638–646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Z, Li J, Zheng H, Yu C, Chen J, Liu
Z, Li M, Zeng M, Zhou F and Song L: Expression and cytoplasmic
localization of SAM68 is a significant and independent prognostic
marker for renal cell carcinoma. Cancer Epidemiol Biomarkers Prev.
18:2685–2693. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sumithra B, Saxena U and Das AB: A
comprehensive study on genome-wide coexpression network of
KHDRBS1/Sam68 reveals its cancer and patient-specific association.
Sci Rep. 9:110832019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song L, Wang L, Li Y, Xiong H, Wu J, Li J
and Li M: Sam68 up-regulation correlates with, and its
down-regulation inhibits, proliferation and tumourigenicity of
breast cancer cells. J Pathol. 222:227–237. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yamamoto E, Kohama G, Sunakawa H, Iwai M
and Hiratsuka H: Mode of invasion, bleomycin sensitivity, and
clinical course in squamous cell carcinoma of the oral cavity.
Cancer. 51:2175–2180. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti AA: American Joint Committee on Cancer: AJCC
cancer staging manual. 7th edition. Springer-Verlag; New York:
2009
|
|
29
|
Costa LC, Leite CF, Cardoso SV, Loyola AM,
Faria PR, Souza PE and Horta MC: Expression of
epithelial-mesenchymal transition markers at the invasive front of
oral squamous cell carcinoma. J Appl Oral Sci. 23:169–178. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tran CM, Kuroshima T, Oikawa Y, Michi Y,
Kayamori K and Harada H: Clinicopathological and
immunohistochemical characteristics of pigmented oral squamous cell
carcinoma. Oncol Lett. 21:3392021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Momose F, Araida T, Negishi A, Ichijo H,
Shioda S and Sasaki S: Variant sublines with different metastatic
potentials selected in nude mice from human oral squamous cell
carcinomas. J Oral Pathol Med. 18:391–395. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tadokoro K, Ueda M, Ohshima T, Fujita K,
Rikimaru K, Takahashi N, Enomoto S and Tsuchida N: Activation of
oncogenes in human oral cancer cells: A novel codon 13 mutation of
c-H-ras-1 and concurrent amplifications of c-erbB-1 and c-myc.
Oncogene. 4:499–505. 1989.PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tamai S, Fujita SI, Komine R, Kanki Y,
Aoki K, Watanabe K, Takekoshi K and Sugasawa T: Acute cold stress
induces transient MuRF1 upregulation in the skeletal muscle of
zebrafish. Biochem Biophys Res Commun. 608:59–65. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Le Bras GF, Taubenslag KJ and Andl CD: The
regulation of cell-cell adhesion during epithelial-mesenchymal
transition, motility and tumor progression. Cell Adh Migr.
6:365–373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun BO, Fang Y, Li Z, Chen Z and Xiang J:
Role of cellular cytoskeleton in epithelial-mesenchymal transition
process during cancer progression. Biomed Rep. 3:603–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kurihara K, Isobe T, Yamamoto G, Tanaka Y,
Katakura A and Tachikawa T: Expression of BMI1 and ZEB1 in
epithelial-mesenchymal transition of tongue squamous cell
carcinoma. Oncol Rep. 34:771–778. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu B, Salama AM, Valero C, Yuan A, Khimraj
A, Saliba M, Zanoni DK, Ganly I, Patel SG, Katabi N and Ghossein R:
The prognostic role of histologic grade, worst pattern of invasion,
and tumor budding in early oral tongue squamous cell carcinoma: A
comparative study. Virchows Arch. 479:597–606. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kane SV, Gupta M, Kakade AC and D' Cruz A:
Depth of invasion is the most significant histological predictor of
subclinical cervical lymph node metastasis in early squamous
carcinomas of the oral cavity. Eur J Surg Oncol. 32:795–803. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kolokythas A, Park S, Schlieve T, Pytynia
K and Cox D: Squamous cell carcinoma of the oral tongue:
Histopathological parameters associated with outcome. Int J Oral
Maxillofac Surg. 44:1069–1074. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guan Y, Xu F, Wang Y, Tian J, Wan Z, Wang
Z and Chong T: Identification of key genes and functions of
circulating tumor cells in multiple cancers through bioinformatic
analysis. BMC Med Genomics. 13:1402020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Han Y, Yamada SI, Kawamoto M, Gibo T,
Hashidume M, Otagiri H, Tanaka H, Takizawa A, Kondo E, Sakai H, et
al: Immunohistochemical investigation of biomarkers for predicting
adipose tissue invasion in oral squamous cell carcinoma. J Oral
Maxillofac Surg Med Pathol. 34:507–513. 2022. View Article : Google Scholar
|
|
45
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Saito D, Kyakumoto S, Chosa N, Ibi M,
Takahashi N, Okubo N, Sawada S, Ishisaki A and Kamo M: Transforming
growth factor-β1 induces epithelial-mesenchymal transition and
integrin α3β1-mediated cell migration of HSC-4 human squamous cell
carcinoma cells through Slug. J Biochem. 153:303–315. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zeisberg M and Neilson EG: Biomarkers for
epithelial-mesenchymal transitions. J Clin Invest. 119:1429–1437.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jolly MK, Somarelli JA, Sheth M, Biddle A,
Tripathi SC, Armstrong AJ, Hanash SM, Bapat SA, Rangarajan A and
Levine H: Hybrid epithelial/mesenchymal phenotypes promote
metastasis and therapy resistance across carcinomas. Pharmacol
Ther. 194:161–184. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Saitoh M: Involvement of partial EMT in
cancer progression. J Biochem. 164:257–264. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sakakitani S, Podyma-Inoue KA, Takayama R,
Takahashi K, Ishigami-Yuasa M, Kagechika H, Harada H and Watabe T:
Activation of β2-adrenergic receptor signals suppresses mesenchymal
phenotypes of oral squamous cell carcinoma cells. Cancer Sci.
112:155–167. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lin CH, Liao CC, Wang SY, Peng CY, Yeh YC,
Chen MY and Chou TY: Comparative O-GlcNAc proteomic analysis
reveals a role of O-GlcNAcylated SAM68 in lung cancer
aggressiveness. Cancers (Basel). 14:2432022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lucena MC, Carvalho-Cruz P, Donadio JL,
Oliveira IA, de Queiroz RM, Marinho-Carvalho MM, Sola-Penna M, de
Paula IF, Gondim KC, McComb ME, et al: Epithelial mesenchymal
transition induces aberrant glycosylation through hexosamine
biosynthetic pathway activation. J Biol Chem. 291:12917–12929.
2016. View Article : Google Scholar : PubMed/NCBI
|