Introduction
Cervical cancer is the fourth leading cancer in
women with estimated 342,000 cancer-related deaths per year
globally (1). In the past decades,
with the development of cervical cancer prevention and screening
measures, the survival rate of patients with cervical cancer has
been improved. However, the prognosis of advanced cervical cancer
is far from satisfactory. So far, the main treatment methods for
advanced cervical cancer include radical surgery, radiotherapy,
chemotherapy and immunotherapy. However, the curative effect is
limited and the potential adverse reactions are also very serious.
At present, targeted therapy is gradually emerging in cervical
cancer (2). Therefore,
identification of novel molecular targets may help to improve the
survival of patients with cervical cancer.
Protein disulfide isomerase (PDI) is a
redox-dependent protein with both chaperone and oxidoreductase
activities, and is originally discovered in the endoplasmic
reticulum (ER) and participates in protein folding (3). Protein disulfide isomerase family A
member 4 (PDIA4) is one of the PDI family members, and has been
revealed to exert functions in the pathogenesis of various diseases
(4). Similar to other PDI members,
PDIA4 can enhance thrombus formation and initiate coagulation
through a series of cascade reactions (4). Aberrant expression of PDIA4 has been
proved as a self-protection response (5). In human malignancies, PDIA4 was found
to be overexpressed in esophageal squamous cell carcinoma (6). High expression of PDIA4 was
associated with poor survival rate in glioma (7). Modulating the expression of PDIA4 in
cancer cells reveled that PDIA4 facilitated cell growth via
regulating activity of caspases 3 and 7 (8). PDIA4 inhibited prostate cancer cell
apoptosis and drove docetaxel resistance via activating the
Akt-signaling pathway (9), whereas
PDIA4 inactivation could restore a classical mitochondrial
apoptosis pathway (10).
Additionally, PDIA4 also mediated miR-378a-3p activating PI3K/AKT
signaling to promote the growth of ovarian cancer cells (11). However, the role of PDIA4 in
cervical cancer remains unknown.
In the present study, the expression pattern of
PDIA4 in cervical cancer was explored through bioinformatics and
immunohistochemical analysis. Proliferation and migration assays
were performed to reveal the effect of PDIA4 on cervical cancer
cells. Furthermore, Gene set enrichment analysis (GSEA) and
protein-protein interaction network showed that numerous cancer
related pathways may be associated with PDIA4.
Materials and methods
Immunohistochemical (IHC)
analysis
The commercial cervical tissue microarray, including
10 normal cervix samples, 27 cervical intraepithelial neoplasia
(CIN) I samples, 55 CIN II–III samples and 10 cervical squamous
cell cancer samples, was purchased from Shanghai Landian
Biotechnology (Shanghai, China). The clinicopathological data of
these samples are presented in Table
SI. The IHC staining of PDIA4 was performed according to
standard protocols. In brief, the paraffin-embedded tissues (5-µm)
were deparaffinized with xylene and rehydrated with descending
ethanol series. The microarray was microwave-treated with citrate
buffer (pH=6.0) for 30 min. The endogenous peroxidase activity was
blocked at room temperature using 3% H2O2 in
methanol for 15 min, and the slide was blocked with 5% skimmed milk
at room temperature for 15 min. Then the microarray was incubated
with PDIA4 polyclonal antibody (1:100; cat. no. 14712-1-AP;
ProteinTech Group, Inc.) at 4°C overnight. Subsequently, the
microarrays were incubated with goat anti-rabbit secondary antibody
(1:500; cat. no. ab6721; Abcam) for 1 h at room temperature. The
IHC staining was interpreted by two professional pathologists by
using a light microscope (Olympus Corporation). The
immunoreactivity score was used to identify the IHC staining
according to the following criteria: staining extent × staining
intensity. The staining intensity was evaluated as 0=negative,
1=weak, 2=moderate and 3=strong. The staining extent was evaluated
as 0=no positive cells, 1<10%, 2=10–33%, 3=34–66% and 4≥67%. The
present study was approved (approval no. 2021KN81) by the
Institutional Ethics Committee of Shanghai Tenth People's Hospital
(Shanghai, China).
Cell lines, small interfering (siRNA)
and reverse transcription-quantitative (RT-q) PCR
Cervical cancer cell lines SiHa (cat. no. TCHu113),
Me180 (cat. no. TCHu177) and HeLa (cat. no. TCHu187) were purchased
from the Type Culture Collection of the Chinese Academy of Science
(Shanghai, China). All cells were maintained in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% fetal bovine serum
(FBS; both from Invitrogen; Thermo Fisher Scientific, Inc.) and 1%
streptomycin/penicillin, in a humidified atmosphere incubator
containing 5% CO2 at 37°C.
The following siRNA oligonucleotide sequences
against PDIA4 were used in the present study: siRNA against
PDIA4-1, 5′-CCTGAGAGAAGATTACAAATT-3′; PDIA4-2,
5′-GCAAGGTGTCAAACGATGCTA-3′; and control siRNA,
5′-AAGAACAACACAAAAGGACAG-3′. Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect
cervical cancer cells with siRNA in a 12-well plate according to
the manufacturer's protocol. The cells were co-cultured with
lipofectamine/siRNA at room temperature for 30 min, and then
cultured in the incubator at 37°C for 6 h. Final siRNA
concentrations were 6 nM. Following a 48-h siRNA transfection, the
cells were used for subsequent experiments.
RT-qPCR was used to identify the knockdown
efficiency of siPDIA4. Total RNA was isolated from transfected
SiHa, ME180 or HeLa cells using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The cDNA was synthesized by using the
PrimeScript™ Reverse Transcriptase (cat. no. 2690S; Takara
Biotechnology Co., Ltd.) according to the manufacturer's protocol.
SYBR qPCR Mix (cat. no. Q712-02; Nanjing Vazyme Biotech Co., Ltd.)
was used to perform the RT-qPCR. The primer sequences used in the
present study were as follows: PDIA4 forward,
5′-CCACCGCAGAAACAGACCT-3′ and reverse, 5′-GGGCCGTTGTAGTCATAAGGC-3′;
CCND1 forward, 5′-GCTGCGAAGTGGAAACCATC-3′ and reverse,
5′-CCTCCTTCTGCACACATTTGAA-3′; PCNA forward,
5′-ACACTAAGGGCCGAAGATAACG-3′ and reverse,
5′-ACAGCATCTCCAATATGGCTGA-3′; CDH1 forward,
5′-CGAGAGCTACACGTTCACGG-3′ and reverse,
5′-GGGTGTCGAGGGAAAAATAGG-3′; VIM forward,
5′-GACGCCATCAACACCGAGTT-3′ and reverse,
5′-CTTTGTCGTTGGTTAGCTGGT-3′; and GAPDH forward, 5′-
CTGGGCTACACTGAGCACC-3′ and reverse,
5′-AAGTGGTCGTTGAGGGCAATG-3′.
The RT-qPCR thermocycling conditions were as
follows: Initial denaturation at 95°C for 5 min, 40 cycles of 95°C
for 15 sec and 58°C for 30 sec, followed by a melting curve ranging
from 95°C for 15 sec, 60°C for 1 min, to 95°C for 15 min. Relative
quantitation was performed using the comparative 2−∆∆Cq
method (12).
Western blotting
Total protein was extracted by using RIPA lysis
buffer (cat. no. P0013B; Beyotime Institute of Biotechnology).
After using the bicinchoninic acid method to determine the protein
concentration, 4× loading buffer was used to prepare the protein
samples. Equal amounts of proteins (20 µg) were separated on 10%
SDS-PAGE gel and transferred onto a nitrocellulose membrane at 300
mA for 90 min. The membranes were blocked with 5% bovine serum
albumin (BSA; cat. no. E661003; Sangon Biotech, Co., Ltd.) for 1 h
at room temperature, and then incubated with the following primary
antibodies at 4°C overnight: PDIA4 polyclonal antibody (1:500; cat.
no. 14712-1-AP; ProteinTech Group, Inc.), cyclin D1 (1:1,000; cat.
no. 55506S), PCNA (1:1,000; cat. no. 13110), E-cadherin (1:1,000;
cat. no. 14472) and Vimentin (1:1,000; cat. no. 5741; all from Cell
Signaling Technology, Inc.). Next day, the membranes were incubated
with Tween-20 (TBST) for 45 min at room temperature and then
incubated with secondary antibody (HRP-conjugated Affinipure goat
anti-rabbit IgG (H+L); 1:5,000; cat. no. SA00001-2; ProteinTech
Group, Inc.) for 1 h at room temperature. The proteins were
detected by the electrochemiluminescence imaging system (Tanon
Science and Technology Co.). Immunoreactive bands were quantified
using ImageJ software (Version 1.8.0.172; National Institutes of
Health).
Cell Counting Kit-8 (CCK-8) assay
CCK-8 (cat. no. C0037; Beyotime Institute of
Biotechnology) was used to assess the proliferation of SiHa, ME180
or HeLa cells. The transfected cells were placed on a 96-well plate
at a density of 1,000 cells per well. At 72 h after incubation at
37°C, 10 µl of 5 mg/ml CCK-8 reagent was added to the plate well.
The culture was terminated 1 h after adding CCK-8 reagent, and the
optical density value was detected by a microplate reader at 450
nm. The experiments were repeated in triplicate independently.
Transwell assay
Transwell assay was used to determine the migration
of cervical cancer cells. Cells (1×105) in serum-free
medium were added to the top of Transwell chamber (cat. no. 3422;
Corning, Inc.). The lower chamber was filled with DMEM medium
containing 10% FBS as a chemoattractant. After 24 h of incubation
at 37°C, migratory cells were fixed with methanol for 20 min and
stained with 5% crystal violet for 10 min at room temperature. The
number of migratory cells was counted from five randomly selected
fields by using a light microscope.
Bioinformatics analysis
UALCAN platform (http://ualcan.path.uab.edu/) was used to identify the
expression of PDIA4 in The Cancer Genome Atlas (TCGA) cancer
samples (13). Gene Expression
Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) was used to reveal
the expression of PDIA4 in different cervical tissues. The data of
GSE7803 was obtained from the study of Zhai et al (14), which contained 10 normal squamous
cervical epithelial samples and 21 invasive squamous cell
carcinomas of the cervix. The data of GSE9750 was obtained from the
study of Scotto et al (15), which
contained 24 normal cervical epithelium and 33 cervical cancer
samples. The data of GSE7410 was obtained from the study of
Biewenga et al (16), which
contained 5 non-cervical carcinoma samples and 35 cervical cancer
samples.
The Kaplan-Meier plotter (http://kmplot.com/analysis/) was used to explore the
prognostic value of PDIA4, PLOD3, GALNT10, GLB1, MOGS, POFUT1 and
SEC63 in patients with cervical cancer (17). It divided all the samples into
lower and upper expression groups based on the auto cut-off plot.
The multivariate Cox regression analysis was performed to reveal
the association between PDIA4 expression and overall survival (OS)
in different clinical features by using TCGA data.
LinkedOmics website (http://linkedomics.org/login.php) was used to perform
GSEA based on TCGA data to demonstrate the association between the
expression of PDIA4 and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathways, Panther pathways, Gene Ontology biological process
and Gene Ontology molecular function (18). False discovery rate (q-value) was
shown, which was the probability estimation of the possible false
positive results of the standardized enrichment score.
The TIMER website (https://cistrome.shinyapps.io/timer/) was used to show
the association between the expression of PDIA4 and glycan
biosynthesis-related genes, glycosaminoglycan-related genes or
protein export-related genes (19).
The GeneMANIA website (http://genemania.org/) was used to validate the gene
interaction network (20). The
BioGRID database (https://thebiogrid.org/) was used to show the
biomedical interaction network (21).
The GEPIA2021 website (http://gepia2021.cancer-pku.cn/) was used to visualize
the expression of PDIA4 in each immune cell type available in
TCGA/GTEx sub-datasets (22).
Statistical analysis
In the present study, GraphPad Prism (Version 8.0;
GraphPad Software, Inc.) was used to perform statistical analyses.
Paired student's t-test was used to compare the difference between
two different groups. Welch's ANOVA test was used to analyze the
difference among IHC scores of multiple groups. Kaplan-Meier
analysis was used to show the survival rate of patients with
cervical cancer. The R package survival was applied in multivariate
Cox regression analysis. Data were presented as the mean ± SD.
P<0.05 was considered to indicate a statistically significant
difference.
Results
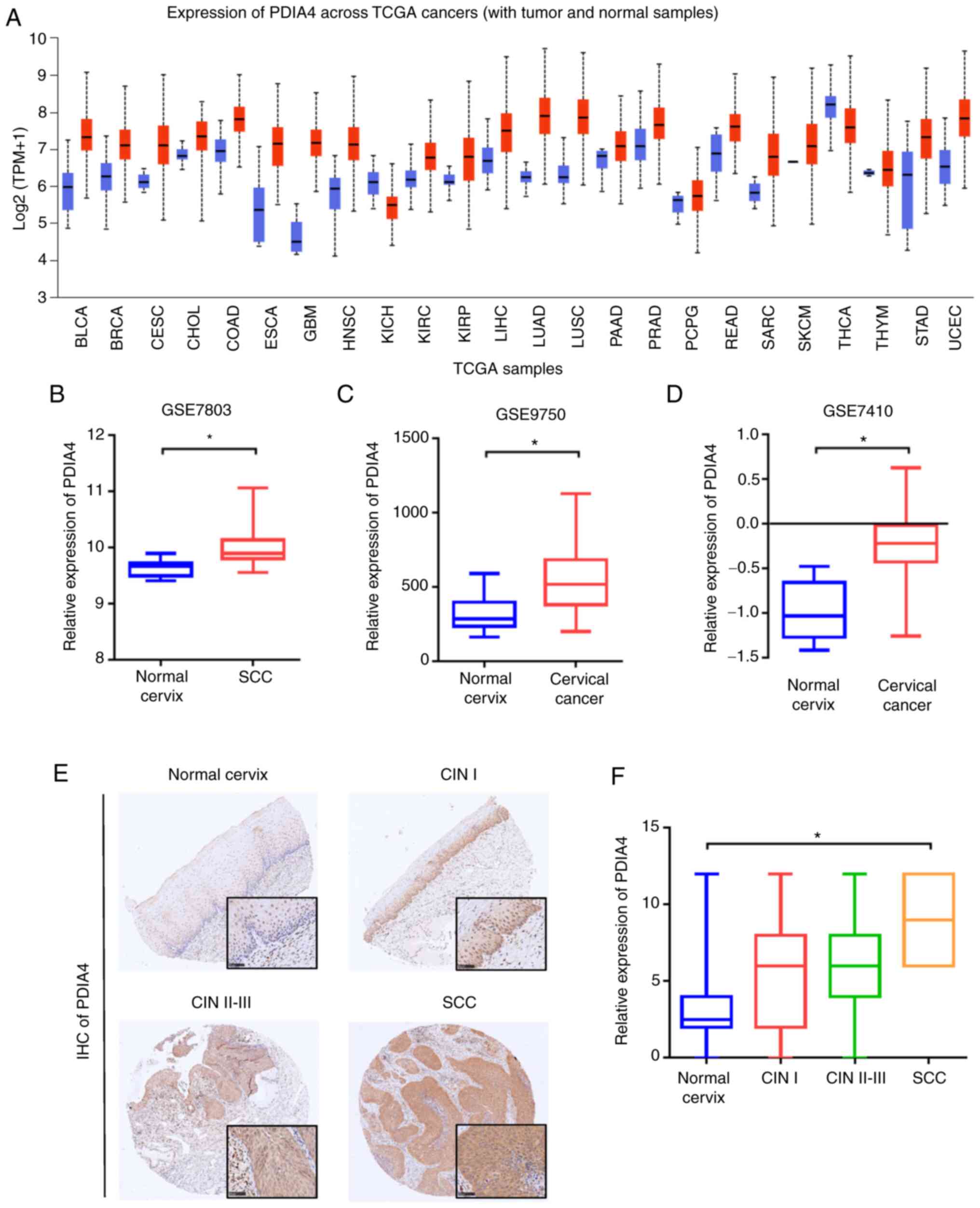
The expression of PDIA4 is upregulated
in cervical cancer
To determine the mRNA expression of PDIA4 in
cervical cancer, the UALCAN platform was first used to study the
expression of PDIA4 between normal and cancer samples. As revealed
in Fig. 1A, PDIA4 was not only
upregulated in cervical cancer, but also highly expressed in other
types of cancer, such as bladder urothelial carcinoma (BLCA),
breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), head
and neck squamous cell carcinoma (HNSC), lung adenocarcinoma (LUAD)
and uterine corpus endometrial carcinoma (UCEC). GEO database
showed that the mRNA expression levels of PDIA4 were increased in
cervical cancer tissues compared with normal cervix tissues
(GSE7803, GSE9750 and GSE7410; Fig.
1B-D). Then, IHC analysis was used to demonstrate the protein
level of PDIA4 in cervical samples. Similar to mRNA expression
pattern, the protein level of PDIA4 was elevated in CIN and
cervical cancer tissues (Fig. 1E and
F). In addition, the mRNA expression of PDIA4 was also analyzed
in different immune cells. The results revealed that the expression
of PDIA4 in CD4+ T cells and CD8+ T cells was
higher than that in B cells and natural killer cells (Fig. S1).
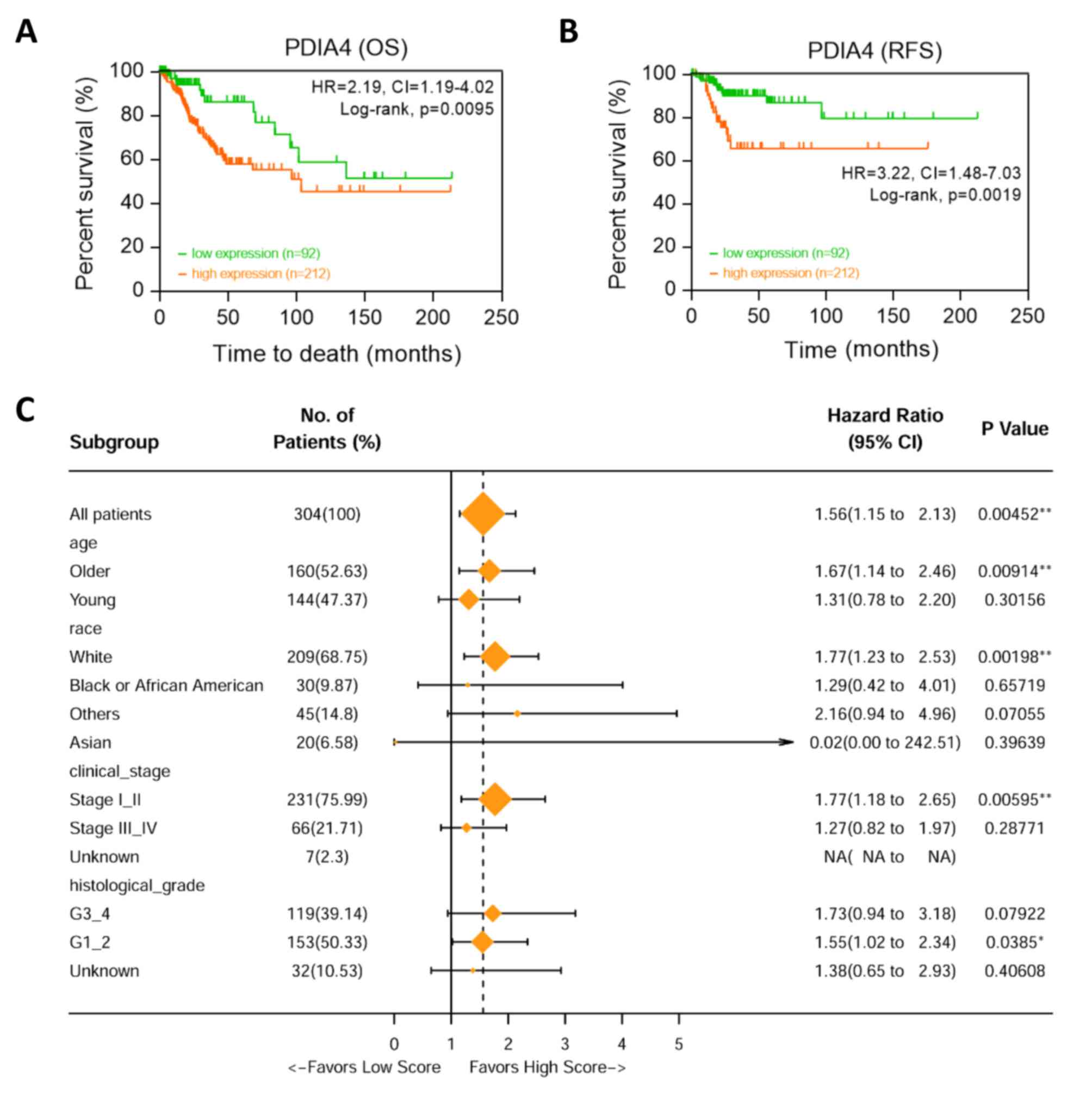
High expression of PDIA4 predicts
worse survival in patients with cervical cancer
Next, the predictive value of PDIA4 in cervical
cancer was analyzed. Kaplan-Meier survival curves revealed that
high mRNA expression of PDIA4 was associated with worse OS
[P=0.0095; hazard ratio (HR)=2.19; 95% confidence interval
(CI)=1.19-4.02] and relapse-free survival (P=0.0019; HR=3.22; 95%
CI=1.48-7.03) of patients with cervical cancer (Fig. 2A and B) based on the auto cut-off
plot. Then the prognostic value of PDIA4 was examined by using
subgroup analysis. The association of PDIA4 mRNA expression with OS
in different clinical features (such as age of patient, human race,
clinical stage and histological grade) was examined via univariate
Cox analysis. High expression of PDIA4 indicated poor clinical
outcome (Fig. 2C).
PDIA4 promotes malignant behavior of
cervical cancer cells
Then, the effect of PDIA4 on the biological activity
of cervical cancer cells was analyzed. CCK-8 assay demonstrated
that silencing of PDIA4 inhibited proliferation of Siha, ME180 and
Hela cell lines (Figs. 3A and
S2A). Furthermore, knockdown of
PDIA4 also reduced the migration of cervical cancer cells as
revealed by Transwell assay (Fig.
3B). Additionally, the expression of proliferation-related
molecules (cyclin D1 and PCNA) and migration-related molecules
(E-cadherin and Vimentin) were detected. The mRNA and protein
expression levels of cyclin D1, PCNA and Vimentin were decreased in
both SiHa and ME180 cells after PDIA4 silencing, whereas the mRNA
expression and protein level of E-cadherin were increased after
knockdown of PDIA4 (Figs. 4C-G and
S2B). These results suggested
that PDIA4 functions as an oncogene in cervical cancer cells.
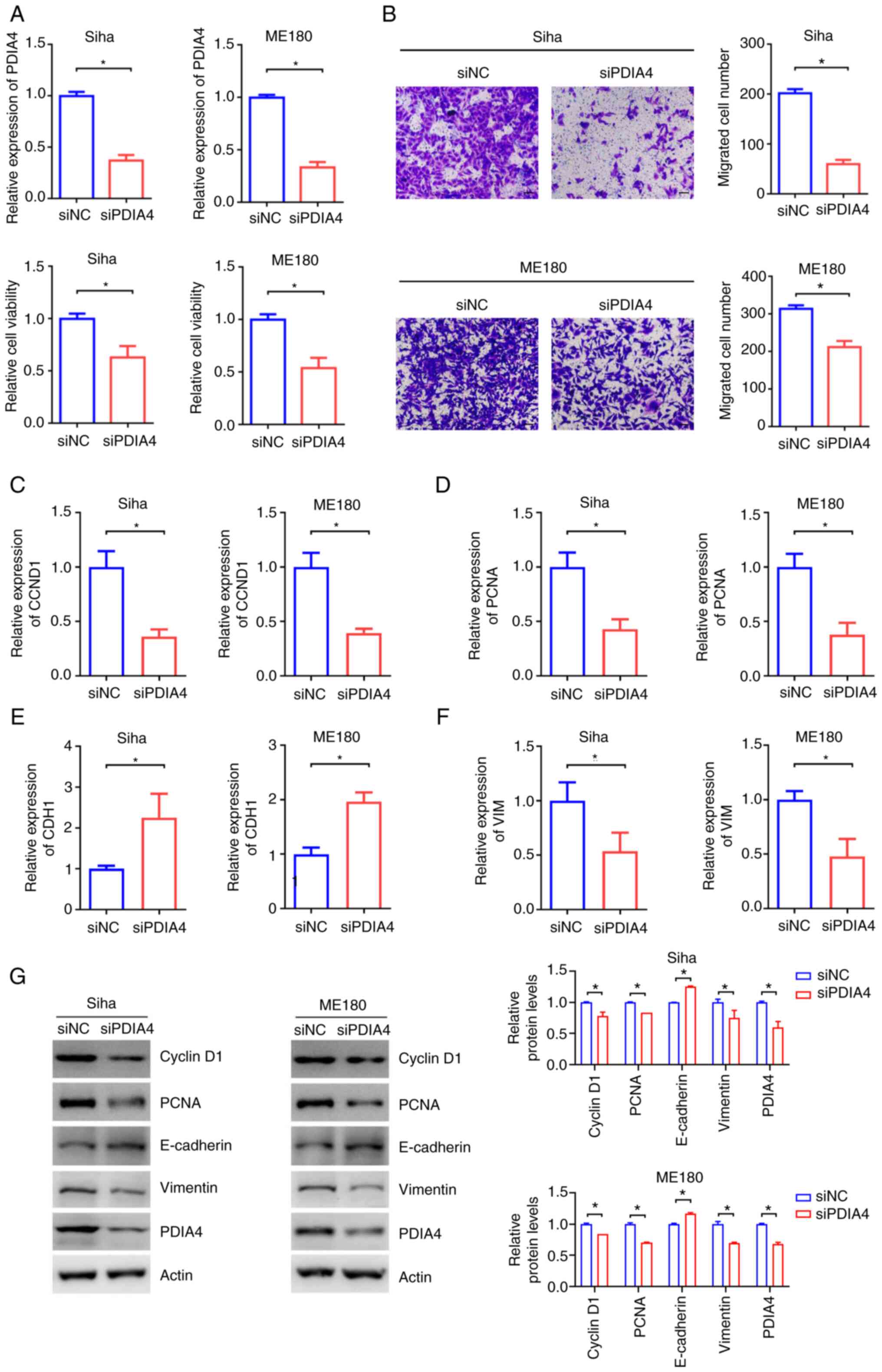 | Figure 3.PDIA4 promotes proliferation and
migration of cervical cancer cells. (A) RT-qPCR and Cell Counting
Kit-8 assay detected PDIA4 mRNA expression and cell growth of Siha
and Me180 cells after knockdown of PDIA4. (B) Transwell assay after
knockdown of PDIA4 in Siha and Me180 cells (Scale bar=50 µm). These
experiments were performed three times. (C-F) The mRNA expression
of (C) CCND1, (D) PCNA (E) CDH1 and (F) VIM was detected using
RT-qPCR after treatment of control or PDIA4 siRNA in Siha and ME180
cells. (G) The protein levels of CCND1, PCNA, E-cadherin and
Vimentin were detected using western blotting after silencing of
PDIA4 in Siha and ME180 cells. These experiments were performed
three times. *P<0.05. PDIA4, protein disulfide isomerase family
A member 4; RT-qPCR, reverse transcription-quantitative PCR; si-,
small interfering; CCND1, cyclin D1; PCNA, proliferating cell
nuclear antigen; CDH1, E-cadherin; VIM, vimentin; NC, negative
control. |
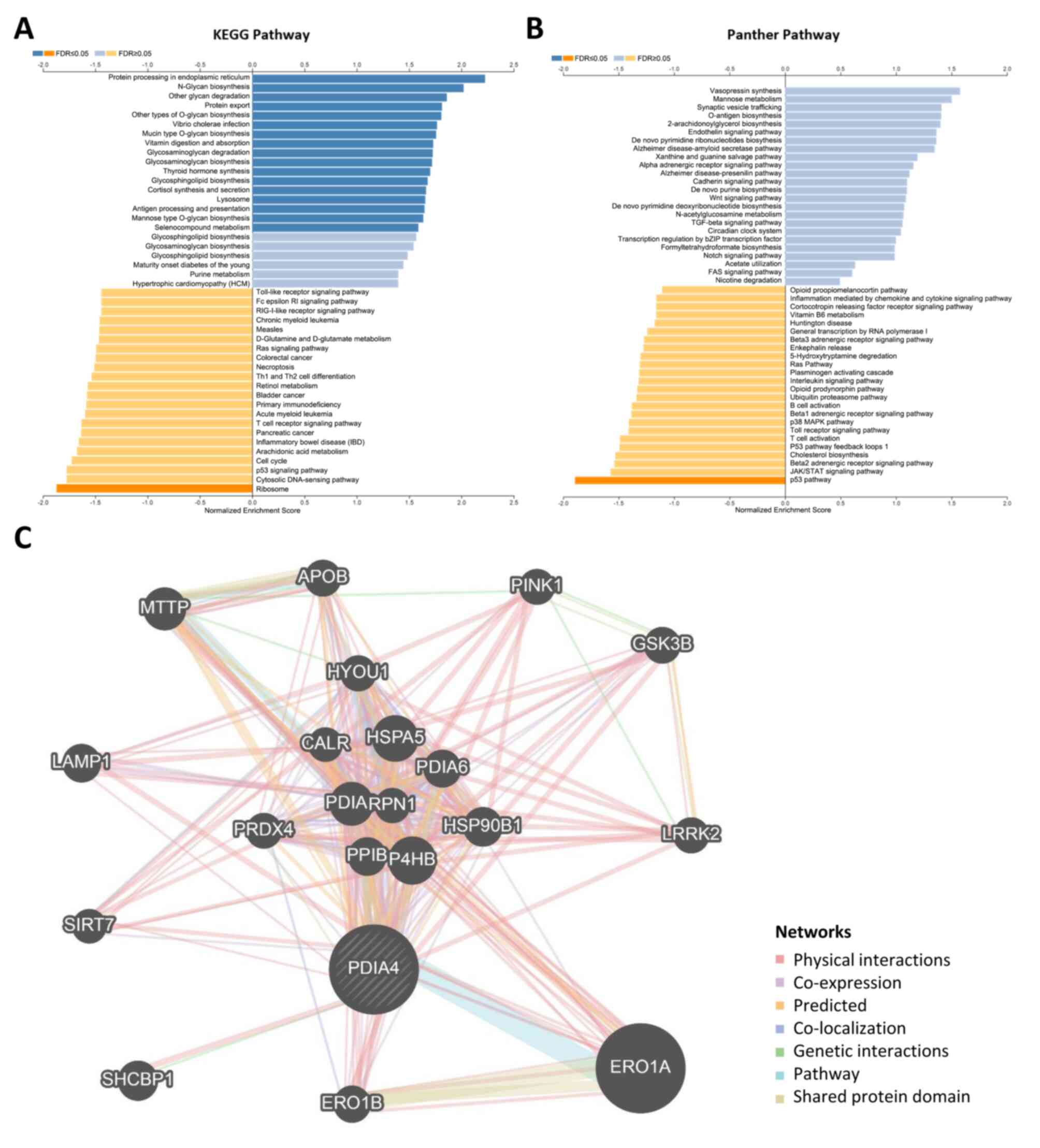
Potential biological functions
regulated by PDIA4 in cervical cancer
As aforementioned, PDIA4 was overexpressed in
cervical cancer and predicted worse survival. Then it was attempted
to study biological functions which were potentially regulated by
PDIA4. GSEA analysis showed that PDIA4 may be involved in several
KEGG pathways, such as glycan biosynthesis, glycosaminoglycan
degradation, protein processing in ER and protein export (Fig. 4A). Similarly, a variety of panther
pathways (such as antigen biosynthesis, the Wnt signaling pathway,
purine biosynthesis and the TGF-β signaling pathway) were
associated with the mRNA expression of PDIA4 (Fig. 4B). GO biological process and
molecular function associated with PDIA4 in cervical cancer were
also explored. As demonstrated in Fig. S3, the mRNA expression of PDIA4 was
associated with glycosylation, response to ER stress, glycoprotein
metabolic process, protein folding and protein hydroxylation.
Furthermore, GeneMANIA platform was used to construct a
PDIA4-interaction network. Several PDIA4-associated genes were
demonstrated in the interaction network, including SIRT7, LAMP1,
ERO1A, PRDX4, LRRK2 and SHCBP1 (Fig.
4C). In addition, the BioGRID database also revealed a series
of proteins that bind to PDIA4 (Fig.
S4).
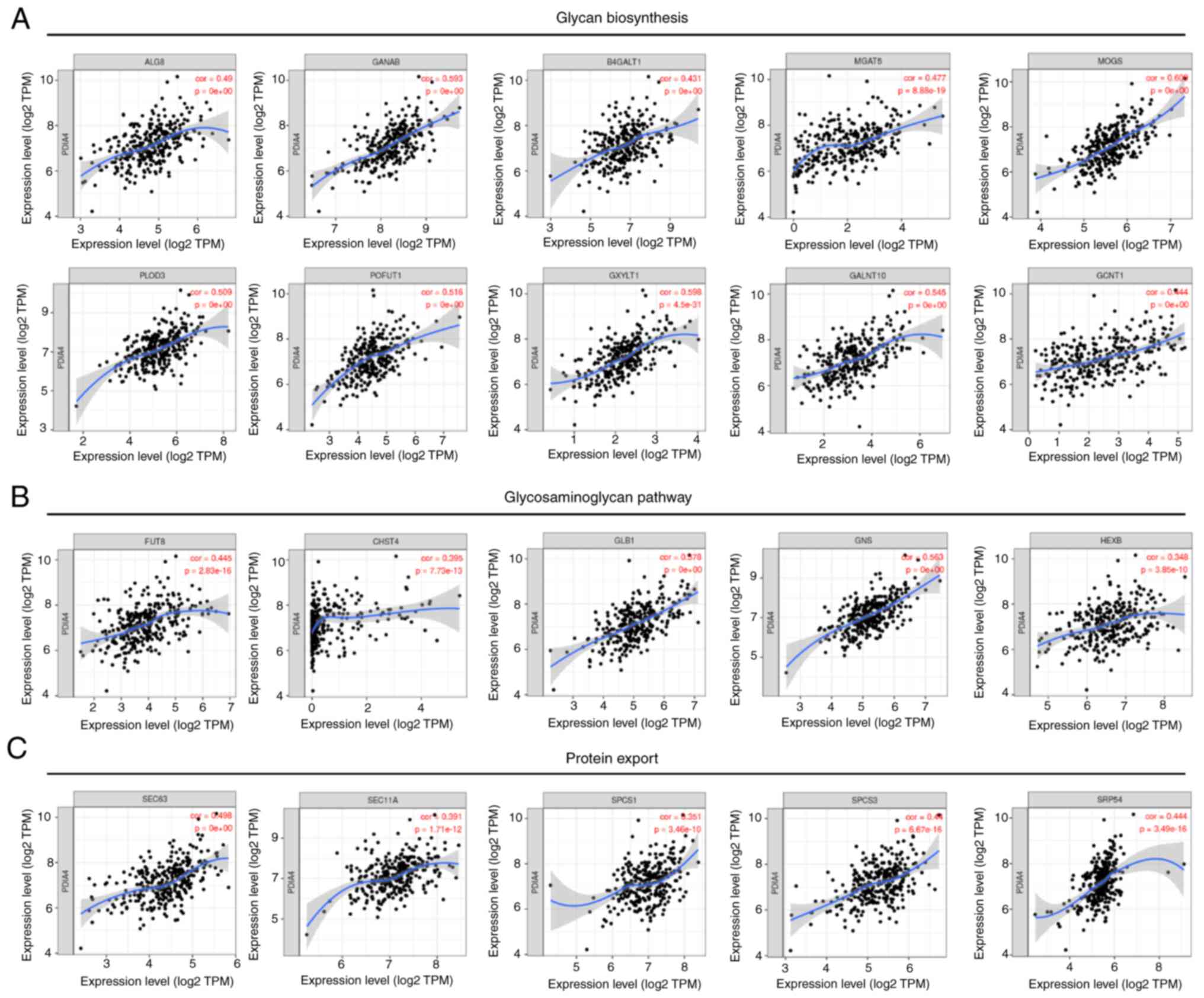
To verify these results, it was further showed that
the mRNA expression of PDIA4 was associated with the expression
levels of glycan biosynthesis-related genes (including ALG8, GANAB,
B4GALT1, MGAT5, PLOD3, POFUT1 and GALNT10),
glycosaminoglycan-related genes (such as FUT8, CHST4, GLB1 and GNS)
and protein export-related genes (such as SCE63, SEC11A, SPCS1 and
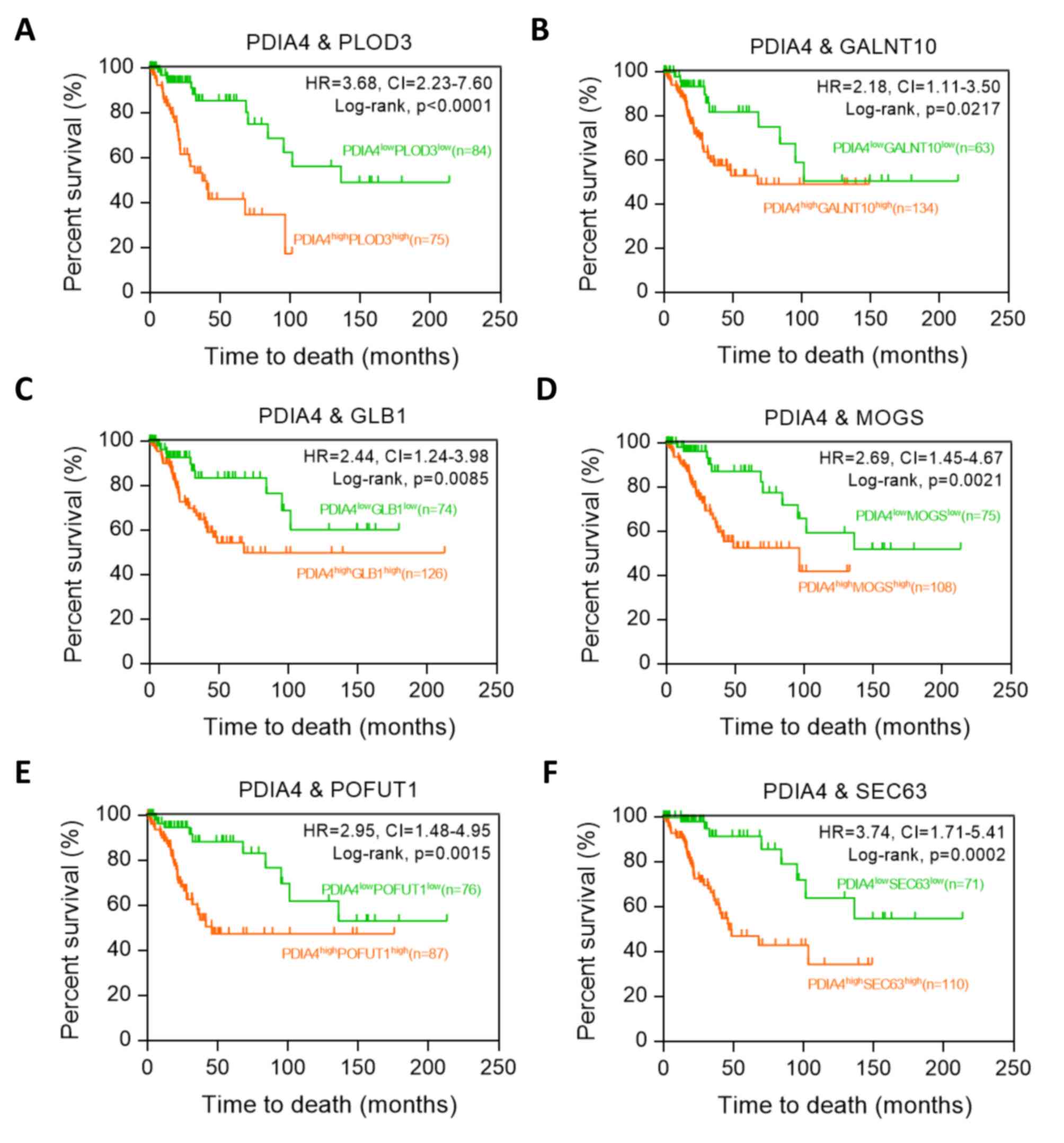
SPCS3) (Fig. 5). Moreover,
Kaplan-Meier analysis revealed that
PDIA4highPLOD3high group was associated with
poor prognosis, compared with
PDIA4lowPLOD3low group in patients with
cervical cancer (P<0.0001; HR=3.68; 95% CI, 2.23-7.60) (Fig. 6A). Similar results were also
observed in the co-expression of PDIA4 combined with GALNT10, GLB1,
MOGS, POFUT1 or SEC63 (Fig.
6B-F).
Discussion
PDI family member PDIA4 has been shown to play roles
in the pathogenesis of various diseases (4). Previous studies have uncovered the
overexpression and cancer-promoting activity of PDIA4 in several
types of cancer (6,7,9). In
the present study, the expression pattern and biological roles of
PDIA4 in cervical cancer were investigated.
The PDI gene family, which is a group of
multifunctional ER enzymes, plays key roles in the correct folding
of polypeptide chains in the ER and the maintenance of several
cellular functions including gluconeogenesis, lipogenesis and
organelle biogenesis (23,24). Gene Expression Atlas datasets have
showed that PDIA1, PDIA3, PDIA4 and PDIA6 are increased in various
types of cancer compared with normal tissue (25). Moreover, the expression of PDIA1 is
significantly higher in the metastatic lymph node breast tumor than
in primary breast tumors (23),
and PDI family genes can also activate membrane proteins such as
matrix metallopeptidases (26),
indicating that PDI is correlated with the promotion of metastasis.
Additionally, several studies have shown that small molecule
irreversible PDI inhibitor propionic acid carbamoyl methyl amides
could play a cytotoxic role in a wide range of cancer cells
(27,28). These results suggested that
targeting key PDI proteins may provide more effective and
personalized treatment strategies in a specific type of cancer.
Previous studies revealed that PDIA4 was abnormally
expressed in several types of cancer. For instance, PDIA4
overexpression was reported in a variety of cancer cell lines, lung
adenocarcinoma tissues and esophageal squamous cell carcinoma
samples (6,8). PDIA4 exhibited a dramatic
upregulation in docetaxel-resistant prostate cancer cells (9). Moreover, PDIA4 was an independent
risk factor for disease-free survival and OS in ovarian cancer
(29) and was associated with poor
prognosis in patients with glioma (7,30).
Recently, Wang et al (31)
reported that PDIA4 could promote glioblastoma progression via the
PI3K/AKT/mTOR pathway. Consistently, the present results showed
that both public database and IHC analysis in cervical samples
revealed the aberrant overexpression of PDIA4 in cervical cancer
tissues. Moreover, Kaplan-Meier survival analysis demonstrated that
cervical cancer patients with higher PDIA4 expression had a poorer
clinical outcome. In addition, high expression of PDIA4 expression
predicted worse survival of patients in other types of cancer.
Functionally, knockdown of PDIA4 significantly impaired cervical
cancer cell proliferation and migration. Moreover, PDIA4 also
affected the expression of proliferation-related genes and
migration-related genes. Therefore, it was indicated that PDIA4 may
be used as an independently prognostic biomarker of cervical
cancer.
The potential biological roles associated with PDIA4
in cervical cancer were revealed by GSEA analysis. Similar to its
role in ER and protein folding, cluster analysis of sequencing data
of TCGA cervical cancer samples revealed that high expression of
PDIA4 was closely related to protein processing in ER, glycoprotein
metabolic process, protein export, folding and hydroxylation. It is
worth noting that the expression of PDIA4 was associated with the
expression levels of glycan biosynthesis-related genes,
glycosaminoglycan-related genes and protein export-related genes.
Additionally, several PDIA4-interacting proteins were also
demonstrated. These results indicated the role of PDIA4 in cervical
cancer. However, the specific molecular mechanism of PDIA4 on
cervical tumorigenesis remains to be explored. Moreover, there are
available effective pan-inhibitors of the PDI family. In future
studies, it is also necessary to carry out relevant in vivo
and in vitro experiments to confirm the roles of PDI in the
treatment of cervical cancer.
Taken together, the present study revealed the
expression features and prognostic values of PDIA4 in cervical
cancers. Silencing PDIA4 could also inhibit the proliferation and
migration of cervical cancer cells. Moreover, potential biological
functions regulated by PDIA4 were demonstrated by functional
cluster analysis and protein-protein interaction map. These
observations provided an understanding of the pathological role of
PDIA4 in cervical cancer, which may help to represent a novel
anti-tumor therapeutic option for cervical cancer.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81874104).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZPC and ZJS developed the project and wrote the
manuscript. FX and ZJS performed the experiments and collected
data. FX performed data analysis and contributed to manuscript
writing. FX and ZJS confirm the authenticity of all the raw data.
The authors are accountable for all aspects of the work in ensuring
that questions related to the accuracy or integrity of any part of
the work are appropriately investigated and resolved. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
2021KN81) by the Institutional Ethics Committee of Shanghai Tenth
People's Hospital (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patrono MG, Calvo MF, Franco JV, Garrote V
and Vietto V: A systematic review and meta-analysis of the
prevalence of therapeutic targets in cervical cancer.
Ecancermedicalscience. 15:12002021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Matsusaki M, Kanemura S, Kinoshita M, Lee
YH, Inaba K and Okumura M: The protein disulfide isomerase family:
From proteostasis to pathogenesis. Biochim Biophys Acta Gen Subj.
1864:1293382020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Z, Zhang H and Cheng Q: PDIA4: The
basic characteristics, functions and its potential connection with
cancer. Biomed Pharmacother. 122:1096882020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Winship AL, Sorby K, Correia J, Rainczuk
A, Yap J and Dimitriadis E: Interleukin-11 up-regulates endoplasmic
reticulum stress induced target, PDIA4 in human first trimester
placenta and in vivo in mice. Placenta. 53:92–100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pawar H, Kashyap MK, Sahasrabuddhe NA,
Renuse S, Harsha HC, Kumar P, Sharma J, Kandasamy K, Marimuthu A,
Nair B, et al: Quantitative tissue proteomics of esophageal
squamous cell carcinoma for novel biomarker discovery. Cancer Biol
Ther. 12:510–522. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li H, Liu Q, Xiao K, He Z, Wu C, Sun J,
Chen X, Chen S, Yang J, Ma Q and Su J: PDIA4 Correlates with poor
prognosis and is a potential biomarker in glioma. Onco Targets
Ther. 14:125–138. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kuo TF, Chen TY, Jiang ST, Chen KW, Chiang
YM, Hsu YJ, Liu YJ, Chen HM, Yokoyama KK, Tsai KC, et al: Protein
disulfide isomerase a4 acts as a novel regulator of cancer growth
through the procaspase pathway. Oncogene. 36:5484–5496. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qian S, Zhang S, Wu Y, Ding Y, Shen and Li
X: Protein disulfide isomerase 4 drives docetaxel resistance in
prostate cancer. Chemotherapy. 65:125–133. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tufo G, Jones AW, Wang Z, Hamelin J,
Tajeddine N, Esposti DD, Martel C, Boursier C, Gallerne C, Migdal
C, et al: The protein disulfide isomerases PDIA4 and PDIA6 mediate
resistance to cisplatin-induced cell death in lung adenocarcinoma.
Cell Death Differ. 21:685–695. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chanjiao Y, Chunyan C, Xiaoxin Q and
Youjian H: MicroRNA-378a-3p contributes to ovarian cancer
progression through downregulating PDIA4. Immun Inflamm Dis.
9:108–119. 2021. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chandrashekar DS, Bashel B, Balasubramanya
SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV and Varambally
S: UALCAN: A portal for facilitating tumor subgroup gene expression
and survival analyses. Neoplasia. 19:649–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ,
Trimble CL, Fearon ER and Cho KR: Gene expression analysis of
preinvasive and invasive cervical squamous cell carcinomas
identifies HOXC10 as a key mediator of invasion. Cancer Res.
67:10163–10172. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M and Murty VV: Identification of copy
number gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Biewenga P, Buist MR, Moerland PD, Ver
Loren van Themaat E, van Kampen AH, ten Kate FJ and Baas F: Gene
expression in early stage cervical cancer. Gynecol Oncol.
108:520–526. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nagy Á, Munkácsy G and Győrffy B:
Pancancer survival analysis of cancer hallmark genes. Sci Rep.
11:60472021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Franz M, Rodriguez H, Lopes C, Zuberi K,
Montojo J, Bader GD and Morris Q: GeneMANIA update 2018. Nucleic
Acids Res. 46:W60–W64. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oughtred R, Rust J, Chang C, Breitkreutz
BJ, Stark C, Willems A, Boucher L, Leung G, Kolas N, Zhang F, et
al: The BioGRID database: A comprehensive biomedical resource of
curated protein, genetic, and chemical interactions. Protein Sci.
30:187–200. 2021. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li C, Tang Z, Zhang W, Ye Z and Liu F:
GEPIA2021: Integrating multiple deconvolution-based analysis into
GEPIA. Nucleic Acids Res. 49:W242–W246. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Galligan JJ and Petersen DR: The human
protein disulfide isomerase gene family. Hum Genomics. 6:62012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hetz C: The unfolded protein response:
Controlling cell fate decisions under ER stress and beyond. Nat Rev
Mol Cell Biol. 13:89–102. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Powell LE and Foster PA: Protein
disulphide isomerase inhibition as a potential cancer therapeutic
strategy. Cancer Med. 10:2812–2825. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu S, Sankar S and Neamati N: Protein
disulfide isomerase: A promising target for cancer therapy. Drug
Discov Today. 19:222–240. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yamada R, Cao X, Butkevich AN, Millard M,
Odde S, Mordwinkin N, Gundla R, Zandi E, Louie SG, Petasis NA and
Neamati N: Discovery and preclinical evaluation of a novel class of
cytotoxic propynoic acid carbamoyl methyl amides (PACMAs). J Med
Chem. 54:2902–2914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu S, Butkevich AN, Yamada R, Zhou Y,
Debnath B, Duncan R, Zandi E, Petasis NA and Neamati N: Discovery
of an orally active small-molecule irreversible inhibitor of
protein disulfide isomerase for ovarian cancer treatment. Proc Natl
Acad Sci USA. 109:16348–16353. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yin F, Yi S, Wei L, Zhao B, Li J, Cai X,
Dong C and Liu X: Microarray-based identification of genes
associated with prognosis and drug resistance in ovarian cancer. J
Cell Biochem. 120:6057–6070. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peng Z, Chen Y, Cao H, Zou H, Wan X, Zeng
W, Liu Y, Hu J, Zhang N, Xia Z, et al: Protein disulfide isomerases
are promising targets for predicting the survival and tumor
progression in glioma patients. Aging (Albany NY). 12:2347–2372.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang M, Zhang W, Liu Y, Ma Z, Xiang W, Wen
Y, Zhang D, Li Y, Li Y, Li T, et al: PDIA4 promotes glioblastoma
progression via the PI3K/AKT/m-TOR pathway. Biochem Biophys Res
Commun. 597:83–90. 2022. View Article : Google Scholar : PubMed/NCBI
|