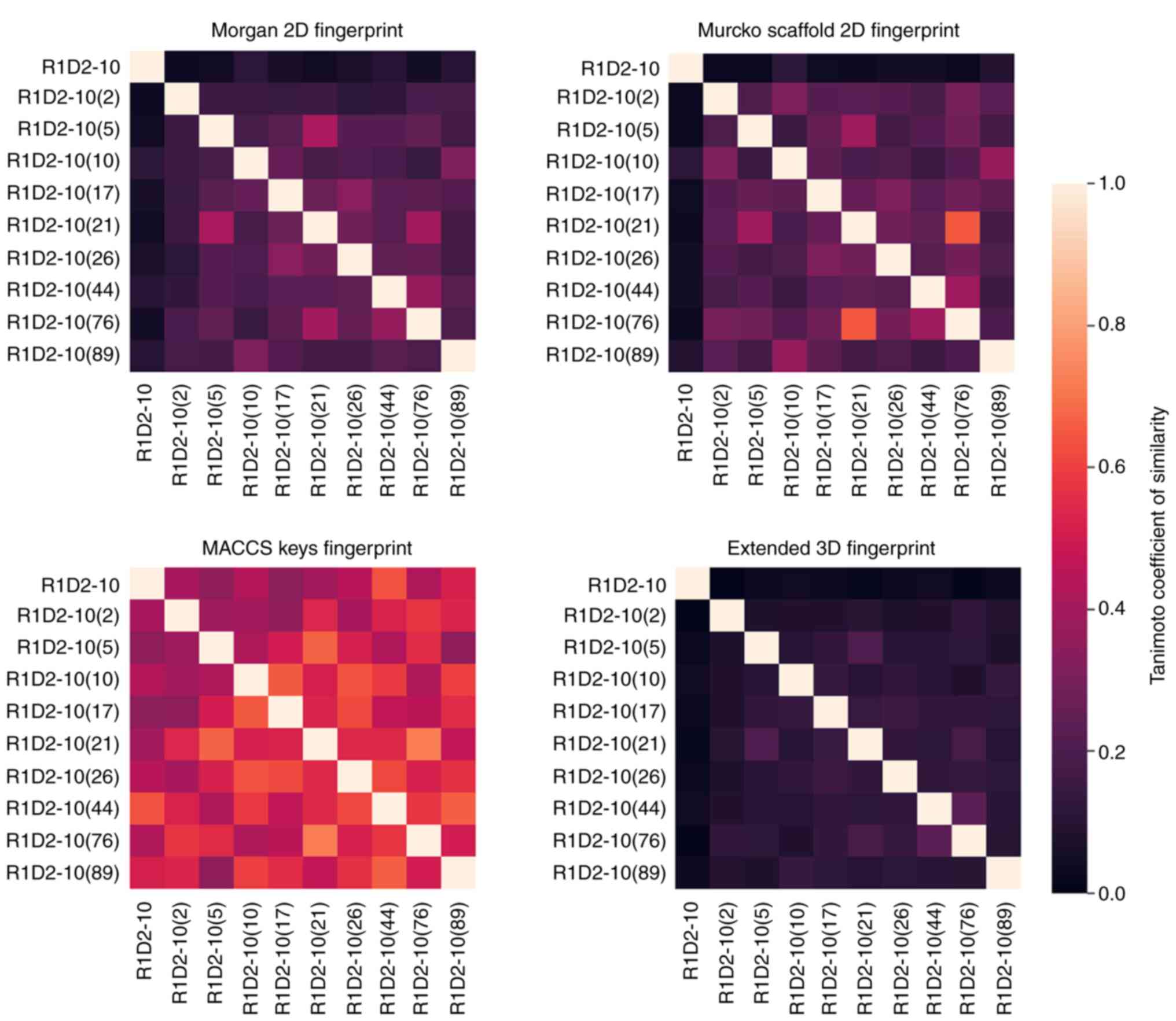
|
1
|
Jafri MA, Ansari SA, Alqahtani MH and Shay
JW: Roles of telomeres and telomerase in cancer, and advances in
telomerase-targeted therapies. Genome Med. 8:692016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Berardinelli F, Coluzzi E, Sgura A and
Antoccia A: Targeting telomerase and telomeres to enhance ionizing
radiation effects in in vitro and in vivo cancer models. Mutat Res
Rev Mutat Res. 773:204–219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lipinska N, Romaniuk A, Paszel-Jaworska A,
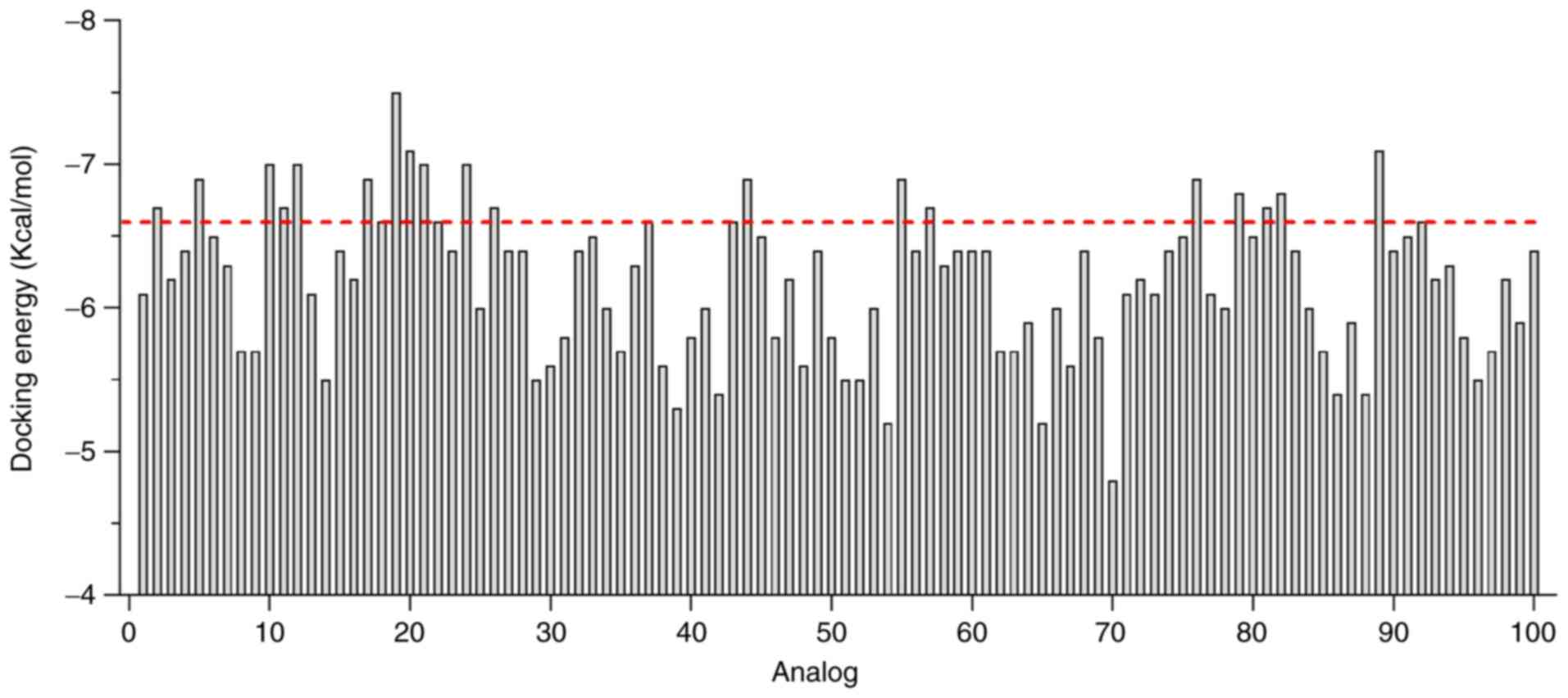
Toton E, Kopczynski P and Rubis B: Telomerase and drug resistance
in cancer. Cell Mol Life Sci. 74:4121–4132. 2017. View Article : Google Scholar
|
|
4
|
Mender I, LaRanger R, Luitel K, Peyton M,
Girard L, Lai TP, Batten K, Cornelius C, Dalvi MP, Ramirez M, et
al: Telomerase-Mediated strategy for overcoming non-small cell lung
cancer targeted therapy and chemotherapy resistance. Neoplasia.
20:826–837. 2018. View Article : Google Scholar
|
|
5
|
Sengupta S, Sobo M, Lee K, Senthil Kumar
S, White AR, Mender I, Fuller C, Chow LML, Fouladi M, Shay JW and
Drissi R: Induced telomere damage to treat telomerase expressing
therapy-resistant pediatric brain tumors. Mol Cancer Ther.
17:1504–1514. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang G, Wu LW, Mender I, Barzily-Rokni M,
Hammond MR, Ope O, Cheng C, Vasilopoulos T, Randell S, Sadek N, et
al: Induction of telomere dysfunction prolongs disease control of
therapy-resistant melanoma. Clin Cancer Res. 24:4771–4784. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu Y, Zhong D, Li Y, Wu H, Xu X, Yang J
and Gu Z: Tumor-Oriented telomerase-terminated nanoplatform as
versatile strategy for multidrug resistance reversal in cancer
treatment. Adv Healthc Mater. 9:e19017392020. View Article : Google Scholar : PubMed/NCBI
|
|
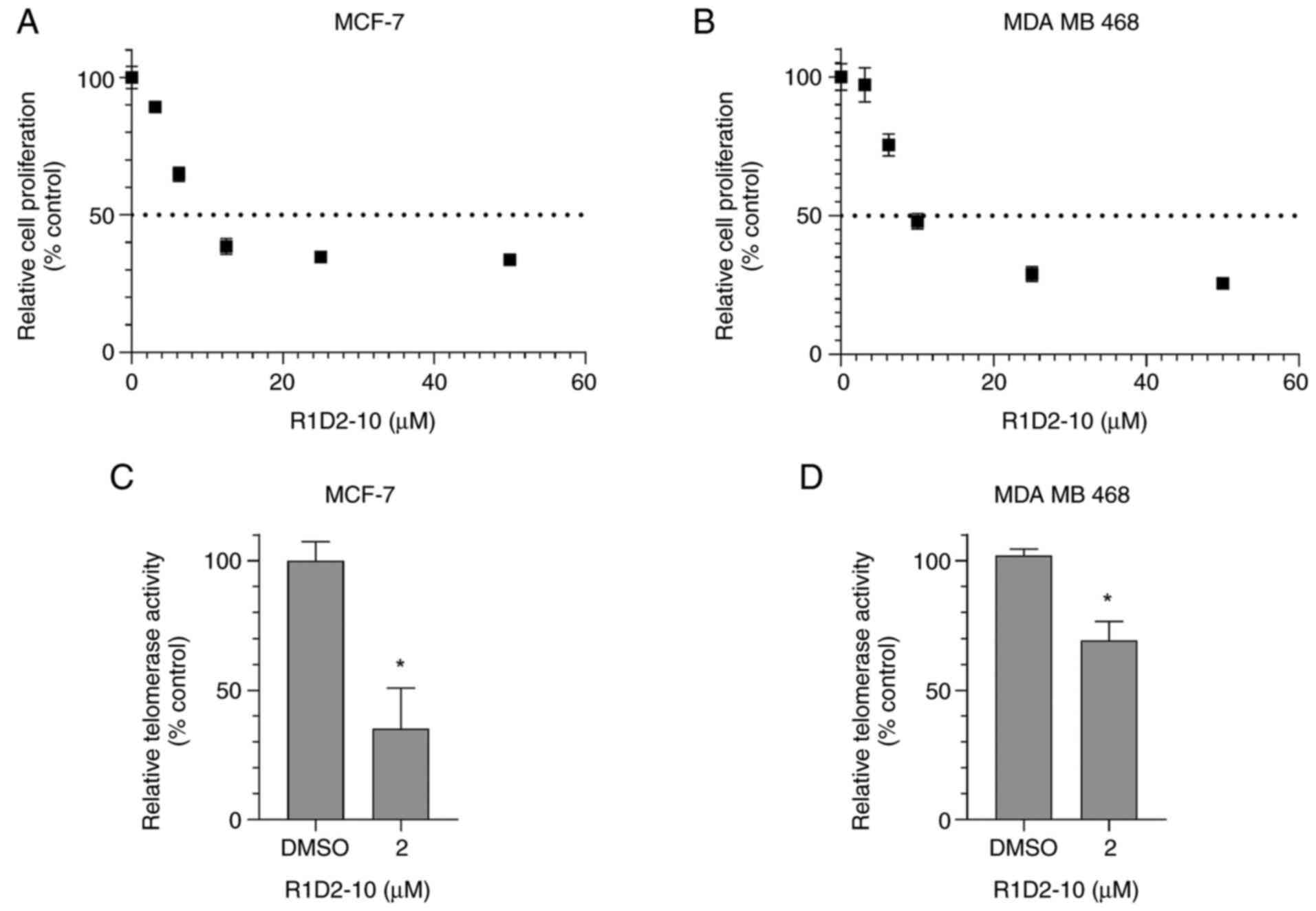
8
|
Gomez DL, Armando RG, Cerrudo CS,
Ghiringhelli PD and Gomez DE: Telomerase as a cancer target.
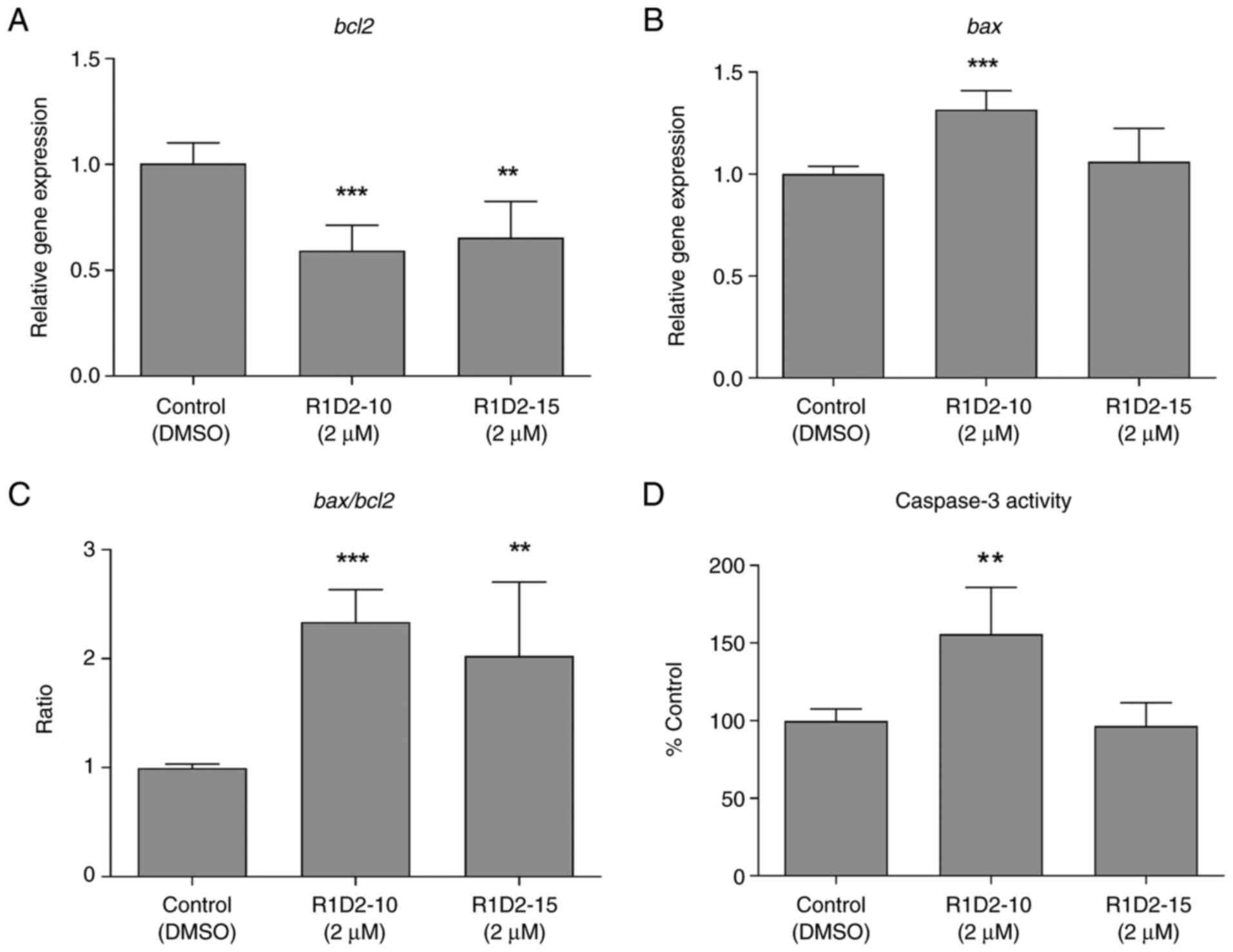
Development of new molecules. Curr Top Med Chem. 16:2432–2440.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
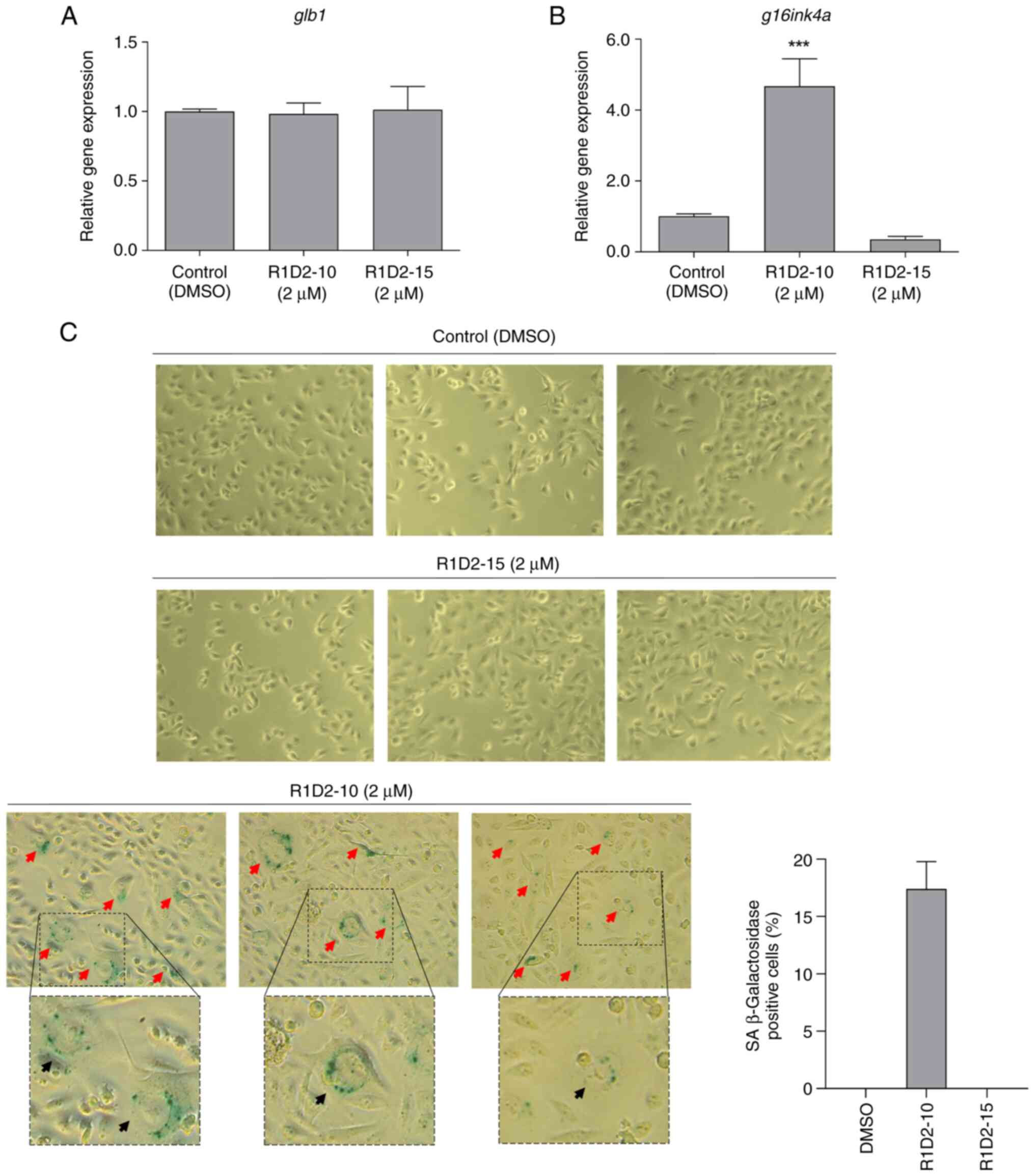
9
|
Guterres AN and Villanueva J: Targeting
telomerase for cancer therapy. Oncogene. 39:5811–5824. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jager K and Walter M: Therapeutic
Targeting of Telomerase. Genes. 7:2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Armando RG, Mengual Gomez DL, Juritz EI,
Lorenzano Menna P and Gomez DE: Homology model and docking-based
virtual screening for ligands of human dyskerin as new inhibitors
of telomerase for cancer treatment. Int J Mol Sci. 19:32162018.
View Article : Google Scholar
|
|
12
|
Jaiswal RK and Yadava PK: Assessment of
telomerase as drug target in breast cancer. J Biosci. 45:722020.
View Article : Google Scholar
|
|
13
|
Holliday DL and Speirs V: Choosing the
right cell line for breast cancer research. Breast Cancer Res.
13:2152011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cawthon RM: Telomere measurement by
quantitative PCR. Nucleic Acids Res. 30:e472002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Skalic M, Jimenez J, Sabbadin D and De
Fabritiis G: Shape-Based generative modeling for de novo drug
design. J Chem Inf Model. 59:1205–1214. 2019. View Article : Google Scholar
|
|
17
|
RDKit, . Open-source cheminformatics.
GitHub and SourceForge, 2021. https://www.rdkit.org/
|
|
18
|
Rappé AK, Casewit CJ, Colwell K, Goddard
WA III and Skiff WM: UFF, a full periodic table force field for
molecular mechanics and molecular dynamics simulations. J Am Chem
Soc. 114:10024–10035. 1992. View Article : Google Scholar
|
|
19
|
Morris GM, Huey R, Lindstrom W, Sanner MF,
Belew RK, Goodsell DS and Olson AJ: AutoDock4 and AutoDockTools4:
Automated docking with selective receptor flexibility. J Comput
Chem. 30:2785–2791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Salentin S, Schreiber S, Haupt VJ, Adasme
MF and Schroeder M: PLIP: Fully automated protein-ligand
interaction profiler. Nucleic Acids Res. 43((W1)): W443–447. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schildge S, Bohrer C, Beck K and
Schachtrup C: Isolation and culture of mouse cortical astrocytes. J
Vis Exp. ((71)): 500792013.
|
|
22
|
Pires DE, Blundell TL and Ascher DB:
pkCSM: Predicting small-molecule pharmacokinetic and toxicity
properties using graph-based signatures. J Med Chem. 58:4066–4072.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Daina A, Michielin O and Zoete V:
SwissTargetPrediction: Updated data and new features for efficient
prediction of protein targets of small molecules. Nucleic Acids
Res. 47((W1)): W357–W364. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sander T, Freyss J, von Korff M and
Rufener C: DataWarrior: An open-source program for chemistry aware
data visualization and analysis. J Chem Inf Model. 55:460–473.
2015. View Article : Google Scholar
|
|
25
|
Gasteiger J and Engel T: Chemoinformatics:
a textbook. John Wiley & Sons, 2006. Chapter 2.9. Volume.
1:92–110. 2006.
|
|
26
|
Leach AR and Gillet VJ: An introduction to
chemoinformatics. Springer, 2007. Chapter 5 - Similiraty Methods.
Volume. 1:99–117. 2007.
|
|
27
|
Sharma A and Lal SP: Tanimoto based
similarity measure for intrusion detection system. J Inf Sec.
2:195–201. 2011.
|
|
28
|
Willett P, Barnard JM and Downs GM:
Chemical similarity searching. J Chem Inf Comput Sci. 38:983–996.
1998. View Article : Google Scholar
|
|
29
|
Bemis GW and Murcko MA: The properties of
known drugs. 1. Molecular frameworks. J Med Chem. 39:2887–2893.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Durant JL, Leland BA, Henry DR and Nourse
JG: Reoptimization of MDL keys for use in drug discovery. J Chem
Inf Comput Sci. 42:1273–1280. 2002. View Article : Google Scholar
|
|
31
|
BIOVIA, . The keys to understanding MDL
keyset technology. Dassault Systemes; Waltham, MA: 2011, https://docplayer.net/64556108-The-keys-to-understanding-mdl-keyset-technology-white-paper.html
|
|
32
|
Axen SD, Huang XP, Caceres EL, Gendelev L,
Roth BL and Keiser MJ: A Simple Representation of three-dimensional
molecular structure. J Med Chem. 60:7393–7409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Deng Y, Chan SS and Chang S: Telomere
dysfunction and tumour suppression: The senescence connection. Nat
Rev Cancer. 8:450–458. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Roake CM and Artandi SE: Control of
cellular aging, tissue function, and cancer by p53 downstream of
telomeres. Cold Spring Harb Perspect Med. 7:a0260882017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin J and Epel E: Stress and telomere
shortening: Insights from cellular mechanisms. Ageing Res Rev.
73:1015072022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han Y, Zhang J, Hu CQ, Zhang X, Ma B and
Zhang P: In silico ADME and toxicity prediction of ceftazidime and
its impurities. Front Pharmacol. 10:4342019. View Article : Google Scholar
|
|
37
|
Hou T, Wang J, Zhang W and Xu X: ADME
evaluation in drug discovery. 7. Prediction of oral absorption by
correlation and classification. J Chem Inf Model. 47:208–218. 2007.
View Article : Google Scholar
|
|
38
|
Holt K, Nagar S and Korzekwa K: Methods to
predict volume of distribution. Curr Pharmacol Rep. 5:391–399.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Muehlbacher M, Spitzer GM, Liedl KR and
Kornhuber J: Qualitative prediction of blood-brain barrier
permeability on a large and refined dataset. J Comput Aided Mol
Des. 25:1095–1106. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Suenderhauf C, Hammann F and Huwyler J:
Computational prediction of blood-brain barrier permeability using
decision tree induction. Molecules. 17:10429–10445. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
McDonnell AM and Dang CH: Basic review of
the cytochrome p450 system. J Adv Pract Oncol. 4:263–268. 2013.
|
|
42
|
Laufkotter O, Sturm N, Bajorath J, Chen H
and Engkvist O: Combining structural and bioactivity-based
fingerprints improves prediction performance and scaffold hopping
capability. J Cheminform. 11:542019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bajusz D, Racz A and Heberger K: Why is
Tanimoto index an appropriate choice for fingerprint-based
similarity calculations? J Cheminform. 7:202015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Snarey M, Terrett NK, Willett P and Wilton
DJ: Comparison of algorithms for dissimilarity-based compound
selection. J Mol Graph Model. 15:372–385. 1997. View Article : Google Scholar
|
|
45
|
Zhang JM and Zou L: Alternative
lengthening of telomeres: From molecular mechanisms to therapeutic
outlooks. Cell Biosci. 10:302020. View Article : Google Scholar
|
|
46
|
Shay JW and Wright WE: Telomeres and
telomerase in normal and cancer stem cells. FEBS Lett.
584:3819–3825. 2010. View Article : Google Scholar
|
|
47
|
Gurung RL, Lim SN, Low GK and Hande MP:
MST-312 alters telomere dynamics, gene expression profiles and
growth in human breast cancer cells. J Nutrigenet Nutrigenomics.
7:283–298. 2014.PubMed/NCBI
|
|
48
|
Kazemi-Lomedasht F, Rami A and Zarghami N:
Comparison of inhibitory effect of curcumin nanoparticles and free
curcumin in human telomerase reverse transcriptase gene expression
in breast cancer. Adv Pharm Bull. 3:127–130. 2013.
|
|
49
|
Wardi L, Alaaeddine N, Raad I, Sarkis R,
Serhal R, Khalil C and Hilal G: Glucose restriction decreases
telomerase activity and enhances its inhibitor response on breast
cancer cells: Possible extra-telomerase role of BIBR 1532. Cancer
Cell Int. 14:602014. View Article : Google Scholar
|
|
50
|
Noureini SK and Wink M: Dose-dependent
cytotoxic effects of boldine in HepG-2 cells-telomerase inhibition
and apoptosis induction. Molecules. 20:3730–3743. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang YL, Huang PH, Chiu HC, Kulp SK and
Chen CS, Kuo CJ, Chen HD and Chen CS: Histone deacetylase inhibitor
AR42 regulates telomerase activity in human glioma cells via an
Akt-dependent mechanism. Biochem Biophys Res Commun. 435:107–112.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bashash D, Ghaffari SH, Mirzaee R,
Alimoghaddam K and Ghavamzadeh A: Telomerase inhibition by
non-nucleosidic compound BIBR1532 causes rapid cell death in pre-B
acute lymphoblastic leukemia cells. Leuk Lymphoma. 54:561–568.
2013. View Article : Google Scholar
|
|
53
|
Chen RJ, Wu PH, Ho CT, Way TD, Pan MH,
Chen HM, Ho YS and Wang YJ: P53-dependent downregulation of hTERT
protein expression and telomerase activity induces senescence in
lung cancer cells as a result of pterostilbene treatment. Cell
Death Dis. 8:e29852017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fang JL and Beland FA: Long-term exposure
to zidovudine delays cell cycle progression, induces apoptosis, and
decreases telomerase activity in human hepatocytes. Toxicol Sci.
111:120–130. 2009. View Article : Google Scholar
|
|
55
|
Hu Y, Bobb D, He J, Hill DA and Dome JS:
The HSP90 inhibitor alvespimycin enhances the potency of telomerase
inhibition by imetelstat in human osteosarcoma. Cancer Biol Ther.
16:949–957. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tejera AM, Alonso DF, Gomez DE and Olivero
OA: Chronic in vitro exposure to 3′-azido-2′, 3′-dideoxythymidine
induces senescence and apoptosis and reduces tumorigenicity of
metastatic mouse mammary tumor cells. Breast Cancer Res Treat.
65:93–99. 2001. View Article : Google Scholar
|
|
57
|
Frink RE, Peyton M, Schiller JH, Gazdar
AF, Shay JW and Minna JD: Telomerase inhibitor imetelstat has
preclinical activity across the spectrum of non-small cell lung
cancer oncogenotypes in a telomere length dependent manner.
Oncotarget. 7:31639–31651. 2016. View Article : Google Scholar
|
|
58
|
Morais KS, Guimaraesb AFR, Ramos DAR,
Silva FP and de Oliveira DM: Long-term exposure to MST-312 leads to
telomerase reverse transcriptase overexpression in MCF-7 breast
cancer cells. Anticancer Drugs. 28:750–756. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mueller S, Hartmann U, Mayer F, Balabanov
S, Hartmann JT, Brummendorf TH and Bokemeyer C: Targeting
telomerase activity by BIBR1532 as a therapeutic approach in germ
cell tumors. Invest New Drugs. 25:519–524. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
van Deursen JM: The role of senescent
cells in ageing. Nature. 509:439–446. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sharpless NE and Sherr CJ: Forging a
signature of in vivo senescence. Nat Rev Cancer. 15:397–408. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhao R, Choi BY, Lee MH, Bode AM and Dong
Z: Implications of genetic and epigenetic alterations of CDKN2A
(p16(INK4a)) in cancer. EBioMedicine. 8:30–39. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liang Y, Liang B, Wu XR, Chen W and Zhao
LZ: Network pharmacology-based systematic analysis of molecular
mechanisms of dingji fumai decoction for ventricular arrhythmia.
Evid Based Complement Alternat Med. 2021:55354802021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bernadotte A, Mikhelson VM and Spivak IM:
Markers of cellular senescence. Telomere shortening as a marker of
cellular senescence. Aging (Albany NY). 8:3–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Burchett KM, Yan Y and Ouellette MM:
Telomerase inhibitor imetelstat (GRN163L) limits the lifespan of
human pancreatic cancer cells. PloS One. 9:e851552014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lee BY, Han JA, Im JS, Morrone A, Johung
K, Goodwin EC, Kleijer WJ, DiMaio D and Hwang ES:
Senescence-associated beta-galactosidase is lysosomal
beta-galactosidase. Aging Cell. 5:187–195. 2006. View Article : Google Scholar
|
|
67
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol
Cancer. 14:482015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Asghari-Kia L, Bashash D, Safaroghli-Azar
A, Momeny M, Hamidpour M and Ghaffari SH: Targeting human
telomerase RNA component using antisense oligonucleotide induces
rapid cell death and increases ATO-induced apoptosis in APL cells.
Eur J Pharmacol. 809:215–223. 2017. View Article : Google Scholar
|
|
69
|
Bashash D, Ghaffari SH, Zaker F, Kazerani
M, Hezave K, Hassani S, Rostami M, Alimoghaddam K and Ghavamzadeh
A: BIBR 1532 increases arsenic trioxide-mediated apoptosis in acute
promyelocytic leukemia cells: Therapeutic potential for APL.
Anticancer Agents Med Chem. 13:1115–1125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Vairano M, Graziani G, Tentori L, Tringali
G, Navarra P and Dello Russo C: Primary cultures of microglial
cells for testing toxicity of anticancer drugs. Toxicol Lett.
148:91–94. 2004. View Article : Google Scholar
|
|
71
|
Cardama GA, Comin MJ, Hornos L, Gonzalez
N, Defelipe L, Turjanski AG, Alonso DF, Gomez DE and Menna PL:
Preclinical development of novel Rac1-GEF signaling inhibitors
using a rational design approach in highly aggressive breast cancer
cell lines. Anticancer Agents Med Chem. 14:840–851. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ramana M, Lokhande R, Bhar S, Ranade P,
Mehta A and Gadre G: In Silico design, synthesis and bioactivity of
N-(2, 4-Dinitrophenyl)-3-oxo-3-phenyl-N-(aryl) phenyl propanamide
derivatives as breast cancer inhibitors. Curr Comput Aided Drug
Des. 13:112–126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Gerbelli BB, Vassiliades SV, Rojas JEU,
Pelin JNBD, Mancini RSN, Pereira WSG, Aguilar AM, Venanzi M,
Cavalieri F, Giuntini F and Alves WA: Hierarchical Self-assembly of
peptides and its applications in bionanotechnology. Bioin Bioba
Mater. 220:19000852019.
|
|
74
|
Smith AJ, Zhang X, Leach AG and Houk KN:
Beyond picomolar affinities: Quantitative aspects of noncovalent
and covalent binding of drugs to proteins. J Med Chem. 52:225–233.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Alqahtani S: In silico ADME-Tox modeling:
Progress and prospects. Expert Opin Drug Metab Toxicol.
13:1147–1158. 2017. View Article : Google Scholar
|
|
76
|
Nandini Asha R, Ravindran Durai Nayagam B
and Bhuvanesh N: Synthesis, molecular docking, and in silico ADMET
studies of 4-benzyl-1-(2,4,6-trimethyl-benzyl)-piperidine:
Potential inhibitor of SARS-CoV2. Bioorg Chem. 112:1049672021.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Almeleebia TM, Shahrani MA, Alshahrani MY,
Ahmad I, Alkahtani AM, Alam MJ, Kausar MA, Saeed A, Saeed M and
Iram S: Identification of new mycobacterium tuberculosis proteasome
inhibitors using a knowledge-based computational screening
approach. Molecules. 26:23262021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Gentile D, Floresta G, Patamia V,
Chiaramonte R, Mauro GL, Rescifina A and Vecchio M: An integrated
pharmacophore/Docking/3D-QSAR approach to screening a large library
of products in search of future botulinum neurotoxin a inhibitors.
Int J Mol Sci. 21:94702020. View Article : Google Scholar
|
|
79
|
Daina A, Michielin O and Zoete V:
SwissADME: A free web tool to evaluate pharmacokinetics,
drug-likeness and medicinal chemistry friendliness of small
molecules. Sci Rep. 7:427172017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Gfeller D, Grosdidier A, Wirth M, Daina A,
Michielin O and Zoete V: SwissTargetPrediction: A web server for
target prediction of bioactive small molecules. Nucleic Acids Res.
42:(Web Server Issue). W32–W38. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Abdelfatah S, Bockers M, Asensio M,
Kadioglu O, Klinger A, Fleischer E and Efferth T: Isopetasin and
S-isopetasin as novel P-glycoprotein inhibitors against
multidrug-resistant cancer cells. Phytomedicine. 86:1531962021.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Greish KF, Salerno L, Al Zahrani R, Amata
E, Modica MN, Romeo G, Marrazzo A, Prezzavento O, Sorrenti V,
Rescifina A, et al: Novel structural insight into inhibitors of
heme oxygenase-1 (HO-1) by new imidazole-based compounds:
Biochemical and in vitro anticancer activity evaluation. Molecules.
23:12092018. View Article : Google Scholar
|
|
83
|
Verma K, Kannan K, V S, R S, V K and K R:
Exploring β-tubulin inhibitors from plant origin using
computational approach. Phytochem Anal. 28:230–241. 2017.
View Article : Google Scholar
|
|
84
|
Lo YC and Torres JZ: Chemical similarity
networks for drug discovery. Special Topics in Drug Discovery. Chen
T: IntechOpen; Volume 1. pp. 53–70. 2016
|
|
85
|
Duran-Frigola M, Pauls E, Guitart-Pla O,
Bertoni M, Alcalde V, Amat D, Juan-Blanco T and Aloy P: Extending
the small-molecule similarity principle to all levels of biology
with the Chemical Checker. Nat Biotechnol. 38:1087–1096. 2020.
View Article : Google Scholar
|
|
86
|
Jasial S, Hu Y, Vogt M and Bajorath J:
Activity-relevant similarity values for fingerprints and
implications for similarity searching. F1000Res. 5:Chem Inf Sci.
–591. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Kropiwnicki E, Evangelista JE, Stein DJ,
Clarke DJB, Lachmann A, Kuleshov MV, Jeon M, Jagodnik KM and
Ma'ayan A: Drugmonizome and Drugmonizome-ML: Integration and
abstraction of small molecule attributes for drug enrichment
analysis and machine learning. Database (Oxford). 2021:baab0172021.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Lambert LJ, Grotegut S, Celeridad M,
Gosalia P, Backer LJ, Bobkov AA, Salaniwal S, Chung TD, Zeng FY,
Pass I, et al: Development of a robust high-throughput screening
platform for inhibitors of the striatal-enriched tyrosine
phosphatase (STEP). Int J Mol Sci. 22:44172021. View Article : Google Scholar
|
|
89
|
Lopez-Lopez E, Cerda-Garcia-Rojas CM and
Medina-Franco JL: Tubulin inhibitors: A chemoinformatic analysis
using cell-based data. Molecules. 26:24832021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Thomas M, Smith RT, O'Boyle NM, de Graaf C
and Bender A: Comparison of structure- and ligand-based scoring
functions for deep generative models: A GPCR case study. J
Cheminform. 13:392021. View Article : Google Scholar : PubMed/NCBI
|