Introduction
Multiple myeloma (MM) is a heterogenous
hematological malignancy that arises from the clonal proliferation
of plasma cells in the bone marrow, which is frequently accompanied
by the secretion of monoclonal immunoglobulin in serum and/or urine
(1). Clinically, patients with MM
experience hypercalcemia, renal insufficiency, anemia and bony
lesions (2). Despite the current
lack of a definitive cure for MM, the introduction of
immunomodulatory drugs and proteasome inhibitors, as well as the
implementation of hematopoietic stem cell transplantation in
suitable candidates, have substantially improved both
progressive-free survival (PFS) and overall survival (OS) in
patients with MM (1,2). However, the vast majority of patients
with MM inevitably relapse after one or more treatment regimens, or
become refractory to the treatments due to drug resistance and
treatment-related toxicities (3,4).
Advancements in next-generation sequencing technologies and
bioinformatics tools have facilitated the research of the
pathophysiology underlying MM and the identification of novel
treatment targets, as well as potential biomarkers for patients
with MM.
Circular RNAs (circRNAs/circs), a group of
single-stranded non-coding RNAs, are covalently closed ring
structures that lack 5′ end caps and 3′ polyadenylated tails, are
highly stable and have cell- and tissue-specific features (5,6). In
recent years, numerous studies have uncovered the circRNA
expression pattern, regulatory network and potential function [such
as the regulation of important cellular events by sponging
microRNAs (miRNAs/miRs), proteins or DNA] in hematological
malignancies, including MM (7–12).
In MM, only two previous studies have explored the circRNA profiles
of patients with MM and analyzed the clinical implications of
certain dysregulated circRNAs in patients with MM (11,12).
One study revealed that 382 circRNAs were dysregulated in patients
with MM, compared with healthy participates. Further reverse
transcription-quantitative PCR (RT-qPCR) validation showed that 10
circRNAs (including circ-PTK2, circ-RNF217, circ-RERE, circ-NAGPA,
circ-KCNQ5, circ-AFF2, circ-WWC3, circ-DNAJC5, circ-KLHL2,
circ-IQGAP1 and circ-AL137655) were dysregulated in patients with
MM, compared with healthy participants (12). Another study reported that 147
circRNAs were dysregulated in patients with MM compared with
patients with iron deficiency anemia (IDA), and circRNA_101237 was
confirmed to be increased in patients with MM compared with
patients with IDA (11). However,
research regarding the comprehensive evaluation of circRNA profiles
in the prognosis (such as treatment response to induction treatment
and survival benefits) of patients with MM is still lacking.
In the present study, microarray and bioinformatics
analyses were initially performed to assess the circRNA profiles in
eight complete response (CR) and eight non-response (NR) MM cases.
Subsequently, 10 candidate circRNAs (top five upregulated and top
five downregulated circRNAs in CR compared with NR MM cases) were
screened using microarray to validate their prognostic value via
RT-qPCR in 60 patients with MM. Finally, since circ_0026652 was
clinically observed to be a key prognostic marker in MM, the
molecular mechanism of circ_0026652 knockdown on regulating MM
chemosensitivity was further assessed.
Materials and methods
Study patients
A total of 60 newly diagnosed symptomatic patients
with MM from Changzheng Hospital, Second Military Medical
University (Shanghai, China) between January 2017 and December 2019
were enrolled in the present study. Patients were eligible for
inclusion if they had a confirmed diagnosis of symptomatic MM, in
accordance with the International Myeloma Working Group (IMWG)
criteria (13), were >18 years
old and agreed to pre-treatment bone marrow sample collection.
Patients were excluded from the study if they had any other
hematologic malignancies, bone marrow diseases or cancer, had
previously undergone hematopoietic stem cell transplantation, or
had a history of radiotherapy or chemotherapy. The present study
was approved by the Institutional Review Board of Changzheng
Hospital, Second Military Medical University (approval no.
2017SL002), and all patients provided written informed consent.
Sampling and data collection
Bone marrow samples were collected from the patients
prior to the initiation of induction therapy. Plasma cells were
separated from the bone marrow samples using CD138+
Plasma Cell Isolation Kit (Miltenyi Biotec GmbH) and were stored in
liquid nitrogen for further detection. The clinical data of
patients, including age, sex, immunoglobulin subtype, disease
features, laboratory findings, Durie-Salmon stage (14), International Staging System (ISS)
(14) and cytogenetics
abnormalities, were recorded following the initial examinations.
All patients were treated with 3–4 cycles of bortezomib-based
induction treatment (bortezomib/lenalidomide/dexamethasone and
bortezomib/cyclophosphamide/dexamethasone; usually four cycles),
according to the IMWG Guidelines (15). Treatment response was evaluated
using the IMWG uniform response criteria (16,17)
and classified as CR, very good partial response (VGPR), partial
response (PR), stable disease (SD) and progressive disease (PD).
The objective response rate (ORR) was defined as CR + VGPR + PR. NR
was defined as SD + PD. Patients who died during induction therapy
or whose response to induction therapy was not recorded for any
reason were excluded from the final analysis. In addition, all
patients were continuously followed up in accordance with the IMWG
Guidelines (15). PFS and OS were
then evaluated based on the follow-up records (the date of the last
follow-up was Jan 31, 2020). The PFS was defined as the time
elapsed between the start of the treatment and disease progression
or death, whichever came first. OS was defined as the time elapsed
between the start of the treatment and death.
Study design
Plasma cell samples from eight CR cases and eight NR
cases (age- and sex-matched) were selected for circRNA microarray
analysis aimed at identifying the dysregulated circRNAs. Next, the
top five upregulated and top five downregulated circRNAs were
screened out from the dysregulated circRNAs by ranking the absolute
value of Log2 fold-change (FC). Detailed information of
the 10 candidate circRNAs is listed in Table I. Subsequently, the expression of
these 10 circRNAs was further determined via RT-qPCR in plasma cell
samples from 60 patients with MM to verify its association with
treatment response and survival profiles. Following clinical data
analysis, circ_0026652 was found to be associated with the
treatment response and survival of patients with MM. Next, in
vitro experiments were carried out to study the role and
potential mechanisms of circ_0026652 in regulating chemosensitivity
to bortezomib.
 | Table I.Top 10 dysregulated (five upregulated
and five downregulated) circRNAs in microarray assay. |
Table I.
Top 10 dysregulated (five upregulated
and five downregulated) circRNAs in microarray assay.
| CircRNAs | Probe ID | Type | Chrom | Strand | Start | End |
Log2FC | P-value | Padj
value | Trend |
|---|
| Circ_0026652 | ASCRP001444 | Exonic | chr12 | + | 53856276 | 53862616 | −3.41 |
5.33×10−7 |
1.87×10−4 | DOWN |
| Circ_0068708 | ASCRP003866 | Exonic | chr3 | - | 197009549 | 197009716 | −3.08 |
8.18×10−5 |
3.32×10−3 | DOWN |
| Circ_0088128 | ASCRP005132 | Exonic | chr9 | + | 116018393 | 116019465 | −3.01 |
4.26×10−7 |
1.62×10−4 | DOWN |
| Circ_0001566 | ASCRP004311 | Exonic | chr5 | - | 179688683 | 179707608 | −2.90 |
9.33×10−8 |
6.28×10−5 | DOWN |
| Circ_0005327 | ASCRP001893 | Exonic | chr15 | - | 60674540 | 60678285 | −2.76 |
1.45×10−8 |
3.31×10−5 | DOWN |
| Circ_0031113 | ASCRP001671 | Exonic | chr14 | - | 20863608 | 20864108 | 3.06 |
3.21×10−5 |
2.00×10−3 | UP |
| Circ_0083587 | ASCRP004820 | Exonic | chr8 | + | 21958950 | 21959041 | 2.72 |
4.15×10−3 |
4.27×10−2 | UP |
| Circ_0005552 | ASCRP003051 | Exonic | chr2 | + | 63206322 | 63223901 | 2.55 |
6.30×10−5 |
2.82×10−3 | UP |
| Circ_0007171 | ASCRP002789 | Exonic | chr19 | + | 17264776 | 17267848 | 2.24 |
4.73×10−5 |
2.35×10−3 | UP |
| Circ_0007521 | ASCRP004957 | Exonic | chr8 | - | 1.42E+08 | 1.42E+08 | 2.00 |
4.47×10−6 |
6.39×10−4 | UP |
Total RNA extraction and
purification
Total RNA was extracted from the plasma cell samples
using RNeasy Protect Mini Kit (Qiagen GmbH), according to the
manufacturer's instructions. The purity and concentration of total
RNA were then determined using a NanoDrop ND-1000 spectrophotometer
(Thermo Fisher Scientific, Inc.), and the integrity of the RNA was
measured using 1.2% denaturing agarose gel electrophoresis. Next,
the RNA was processed using RNase-Free DNase Set (Qiagen GmbH) for
purification, followed by linear RNA digestion using RNase R
(Epicentre; Illumina, Inc.).
CircRNA microarray analysis
For microarray analysis, purified RNA samples from
eight CR and eight NR cases were subjected to amplification and
transcribed into fluorescent cRNA using Arraystar Super RNA
Labeling Kit (Arraystar, Inc.), and the labeled cRNAs were purified
using RNeasy Mini Kit (Qiagen GmbH). Subsequently, the samples were
hybridized using Arraystar Human CircRNA Array v1 (AS-S-CR-H-V2.0;
Arraystar.) and Gene Expression Microarray Hybridization Kit
(Agilent Technologies, Inc.), according to the manufacturer's
instructions in an Agilent Hybridization Oven (Agilent
Technologies, Inc.). Following hybridization, the hybridized arrays
were scanned using an Agilent Microarray Scanner (Agilent
Technologies, Inc.) and resulting images were analyzed in Agilent
Feature Extraction software (Version 11.0.1.1; Agilent
Technologies, Inc.) to obtain raw data.
Bioinformatics analysis
Quantile normalization and low-intensity filtering
were performed using R software package (R Version 3.1.2) (18). Next, the circRNAs detected in
>50% samples were subjected to further analysis. The principal
component analysis (PCA) plots of circRNA profiles were constructed
using Factoextra package (Version 3.3.3) (19), and a heatmap of circRNA profiles
was plotted using Pheatmap package (Version 1.0.8) (20). The FC of circRNA expression was
calculated, and the circRNA expression difference between CR and NR
cases was determined using an unpaired t-test. The P-value was
corrected using the Benjamini-Hochberg method, which was marked as
Padj-value. The dysregulated circRNAs were defined as
the circRNAs with a FC ≥2.0 (or FC≤0.5) and
Padj<0.05, as shown by volcano plots with R (Version
3.1.2). Gene Ontology (GO) annotation of dysregulated circRNAs
based on located genes was obtained from the GO database
(http://www.geneontology.org/), and
pathway annotation of dysregulated circRNAs based on located genes
was obtained from Kyoko Encyclopedia of Genes and Genomes (KEGG)
database (https://www.kegg.jp/). The target miRNAs
of dysregulated circRNAs were predicted using miRNA target
prediction software miRanda (Version 1.0b) (http://www.microrna.org/). Annotation of the target
miRNAs was performed using GO resource, KEGG, Human Phenotype
Ontology (HP; http://hpo.jax.org/) and Disease
Ontology Identification (DOID; http://www.disease-ontology.org/) databases.
Enrichment analysis was performed using Fisher's exact test. In
addition, a circRNA-miRNA network of the top five upregulated and
top five downregulated circRNAs was established through miRanda
database.
Cell culture
Human U266 and RPMI-8226 MM cell lines were
purchased from the American Type Culture Collection and cultured in
RPMI-1640 medium (Thermo Fisher Scientific, Inc.) containing 10%
FBS (Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100
µg/ml streptomycin (Thermo Fisher Scientific, Inc.) at 37°C with 5%
CO2.
Transfection
Negative control (NC) small interfering RNA (siRNA;
Guangzhou RiboBio Co., Ltd.) (50 pM) and circ_0026652 siRNA
(Guangzhou RiboBio Co., Ltd.) (50 pM) were transfected into
2×105 U266 and RPMI-8226 cells using
Lipofectamine® 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) at 37°C for 6 h. Following
transfection, the cells were divided into si-NC and si-circ cells.
The U266 and RPMI-8226 cells without transfection were used as the
control cells. A total of 24 h after transfection, the expression
of circ_0026652 in control, NC and circ(−) cells were evaluated
using RT-qPCR. The sequences were as follows: circ_0026652 siRNA
sense, 5′-ACUCACCAUUCCAAACGAUUU-3′ and antisense,
5′-UUUGAGUGGUAAGGUUUGCUA-3′; and NC siRNA sense,
5′-GAAUUAAUUAAAGAUGGCCCGUUGUACU-3′ and antisense,
5′-UCAUCGAAGUUAUAGGGAUACAUUACGUGAUC-3′.
Chemosensitivity detection
Following transfection, cells were cultured with
different concentrations of bortezomib (Selleck Chemicals) for 48 h
(the concentration range of U266 and RPMI-8226 cells were 0–16 and
0–4 nM, respectively) at 37°C. Next, a Cell Counting Kit-8 (CCK-8;
Dojindo Molecular Technologies, Inc.) assay was used to evaluate
cell viability, according to the manufacturer's instructions.
Relative cell viability was calculated as previously described
(21). Furthermore, 4 nM
bortezomib was used to treat U266 cells for 24, 48 and 72 h, and 1
nM bortezomib was used to treat RPMI-8226 cells for 24, 48 and 72 h
at 37°C, followed by relative cell viability detection.
Cell apoptosis assessment
Following transfection, cells were incubated with
bortezomib (Selleck Chemicals; 4 nM for U266 cells, 1 nM for
RPMI-8226 cells) for 48 h. The bortezomib concentration for cell
incubation was selected using the IC50, as shown in the
‘Chemosensitivity detection’ subsection. Following incubation,
Annexin V-FITC Apoptosis Detection Kit (MilliporeSigma) was used to
determine cell apoptosis, according to the manufacturer's
instructions. The data were collected by FACSCalibur flow cytometer
(Becton, Dickinson and Company) and analyzed by Flowjo 7.6 (Becton,
Dickinson and Company).
Target miRNA assessment
Using miRanda, miR-608, miR-762 and miR-939 were
identified as the potential targets of circ_0026652. RT-qPCR was
performed to assess the expression of miR-608, miR-762 and miR-939
at 24 h after transfection.
Rescue experiment
The control, si-NC and si-circ cells were
constructed as described in the ‘Transfection’ subsection. Next, 50
pM NC inhibitor (Guangzhou RiboBio Co., Ltd.) and 50 pM miR-608
inhibitor (Guangzhou RiboBio Co., Ltd.) were transfected into si-NC
or si-circ cells using Lipofectamine 2000 for 6 h at 37°C. The
cells were termed as si-NC + NC-inhibitor, si-NC + miR-inhibitor,
si-circ + NC-inhibitor and si-circ + miR-inhibitor cells. At 24 h
after transfection, the expression of circ_0026652 and miR-608 in
the control, si-NC + NC-inhibitor, si-NC + miR-inhibitor, si-circ +
NC-inhibitor and si-circ + miR-inhibitor cells was determined via
RT-qPCR. Following transfection (24 h), chemosensitivity was
detected as described in the ‘Chemosensitivity detection’
subsection, and cell apoptosis was evaluated with the method
described in ‘Cell apoptosis assessment’ subsection. Furthermore,
to detect the transfection efficiency, 50 pM NC inhibitor alone and
50 pM miR-608 inhibitor alone were transfected into
2×105 U266 and RPMI-8226 cells using Lipofectamine 2000
transfection reagent for 6 h at 37°C, followed by detection of
miR-608 expression via RT-qPCR. The sequences were as follows:
miR-608 inhibitor, 5′-ACGGAGCUGUCCCAACACCACCCCU-3′; and NC
inhibitor, 5′-CAGUACUUUUGUGUAGUACAA-3′.
Pathway assessment
In previous studies, miR-608 was reported to
regulate the Wnt/β-catenin pathway in carcinomas (22). In other studies, the Wnt/β-catenin
pathway was reported to play a major role in the regulation of the
chemosensitivity to bortezomib (23–25).
Hence, the expression of Wnt family member 3A (WNT3A) and β-catenin
in control, si-NC + NC-inhibitor, si-NC + miR-inhibitor, si-circ +
NC-inhibitor and si-circ + miR-inhibitor cells was assessed via
western blot analysis at 24 h after transfection.
RT-qPCR
Briefly, total RNA in cells was extracted using
RNeasy Protect Mini Kit (Qiagen GmbH). The purity (A260/A280) range
was 1.90-2.10 and the RNA integrity number was 7.5-9.0. For the
detection of circRNAs, the linear RNA in isolated RNA was digested
using RNase R (Illumina, Inc.) prior to cDNA synthesis. For the
detection of miRNAs, GAPDH and U6, linear RNA digestion was not
performed. Next, cDNA synthesis was carried out using PrimeScript™
RT reagent kit according to the manufacturer's protocol (Takara
Bio, Inc.), followed by cDNA amplification using TB Green™ Fast
qPCR Mix (Takara Bio, Inc.). The thermocycling conditions were as
follows: Initial denaturation at 95°C for 30 seconds (sec); 40
cycles of 95°C for 5 sec and 61°C for 10 sec. Finally, the relative
expression of circRNAs and miRNAs was calculated using the
2−∆∆Cq method (26);
GAPDH, which is a well-known and commonly used internal reference
for circRNA detection, served as an internal reference for
circRNAs, and U6 served as an internal reference for miRNAs
(27–29). The primers used are listed in
Table SI.
Western blot analysis
Protein in cells was extracted using RIPA Lysis and
Extraction Buffer (Thermo Fisher Scientific, Inc.), and protein
quantification was performed using Pierce™ BCA Protein Assay Kit
(Thermo Fisher Scientific, Inc.). Next, the 20 µg protein was
separated on 4–20% TruPAGE™ Precast Gels (MilliporeSigma) for
electrophoresis and then transferred to a PVDF membrane (Pall Life
Sciences). The membrane was blocked by 5% BSA (MilliporeSigma) at
37°C for 1 h, incubated with primary antibodies at 4°C overnight
and then a secondary antibody for 90 min at room temperature.
Finally, the protein was detected by chemiluminescence and
visualized using Immobilon ECL Ultra Western HRP Substrate
(MilliporeSigma). The antibodies used in western blot analysis are
listed in Table SII.
Statistical analysis
Data are presented as the mean ± SD, median
interquartile range or number with percentage [n, (%)]. For the
analysis of clinical data, Student's unpaired t-test or Wilcoxon
rank-sum test was used to determine the difference in clinical
features or the difference in circRNA expression between two
groups. Kaplan-Meier curve was used to evaluate PFS and OS, and
log-rank test was used to determine the difference in PFS and OS
between two groups. For the analysis of the experimental data,
one-way ANOVA followed by Tukey's multiple comparisons was used for
multiple comparisons between groups. Clinical data analysis was
carried out using SPSS 21.0 statistical software (IBM Corp.) and
experimental data analysis was performed using GraphPad Prism 7.02
(GraphPad Software Inc.). P<0.05 was considered to indicate a
statistically significant difference. The experiments were carried
out in triplicate.
Results
Clinical characteristics of patients
with MM
Of the 60 patients with MM, 26 (43.3%) were women
and 34 (56.7%) were men, with a mean age of 54.1±8.3 years
(Table II). Among these patients
with MM, 32 (53.3%), 14 (23.3%) and 14 (23.3%) cases secreted IgG,
IgA or other IgG types, respectively. Furthermore, 33 (55.0%) and
28 (46.7%) cases exhibited bone lesion and renal impairment,
respectively. In addition, seven (11.7%) and 53 (88.3%) cases
presented with Durie-Salmon stage II and III, respectively.
Finally, 14 (23.3%), 14 (23.3%) and 32 (53.3%) cases had ISS stage
I, II and III, respectively. Detailed information on laboratory
findings and cytogenetic abnormalities is displayed in Table II.
 | Table II.Clinical features of patients with MM
(n=60). |
Table II.
Clinical features of patients with MM
(n=60).
| Clinical
characteristics | Patients |
|---|
| Age, years, mean ±
SD | 54.1±8.3 |
| Sex, n (%) |
|
|
Female | 26 (43.3) |
|
Male | 34 (56.7) |
| Immunoglobulin
subtype, n (%) |
|
|
IgG | 32 (53.4) |
|
IgA | 14 (23.3) |
| Other
types | 14 (23.3) |
| Disease features, n
(%) |
|
| Bone
lesion | 33 (55.0) |
| Renal
impairment | 28 (46.7) |
| Laboratory
findings |
|
| Hb,
g/l, mean ± SD | 99.8±23.4 |
|
Calcium, mg/dl, mean ± SD | 9.7±2.2 |
| Scr,
mg/dl, median (IQR) | 1.9 (1.6-2.2) |
|
Albumin, g/l, median
(IQR) | 34.0
(29.0-37.0) |
| β2-MG,
mg/l, median (IQR) | 6.1 (3.3-10.1) |
| LDH,
U/l, median (IQR) | 214.8
(175.4-244.3) |
| Durie
Salmon stage, n (%) |
|
| Stage
II | 7 (11.7) |
| Stage
III | 53 (88.3) |
| ISS
stage, n (%) |
|
| Stage
I | 14 (23.3) |
| Stage
II | 14 (23.3) |
| Stage
III | 32 (53.4) |
| Cytogenetics
abnormalities, n (%) |
|
| t (4;
14) | 7 (11.7) |
| t (14;
16) | 4 (6.7) |
| Del
(17p) | 9 (15.0) |
circRNA profiles in eight CR and eight
NR cases
A total of eight CR cases were randomly selected
from 16 patients who achieved CR using the random number method. A
total of eight NR cases were randomly selected from 17 patients
with NR using the random number method. These eight CR and eight NR
cases were then subjected to circRNA microarray and bioinformatics
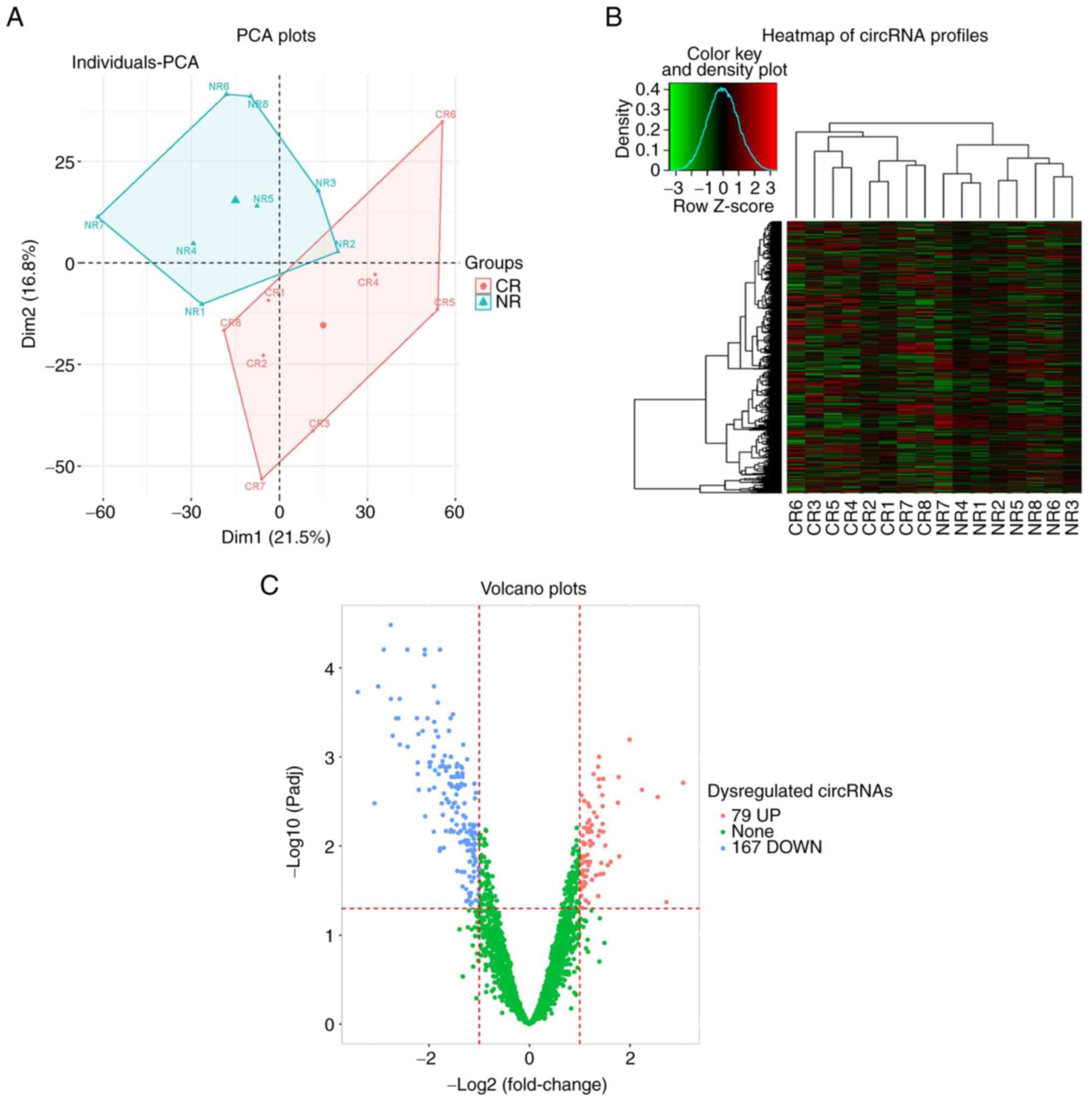
analysis. A clear distinction in circRNA profiles between eight CR
and eight NR cases was revealed by PCA plots, which suggested that
CR cases could be distinguished from NR cases based on their
circRNA profiles (Fig. 1A).
Heatmap analysis of circRNA profiles revealed a relatively good
consistency of circRNA profiles in CR and NR cases (Fig. 1B). Furthermore, volcano plots
showed that 79 circRNAs were upregulated and 167 circRNAs were
downregulated in patients with CR, compared with NR cases [FC ≥2.0
(or FC≤0.5) and Padj<0.05; Fig. 1C].
Enrichment analyses
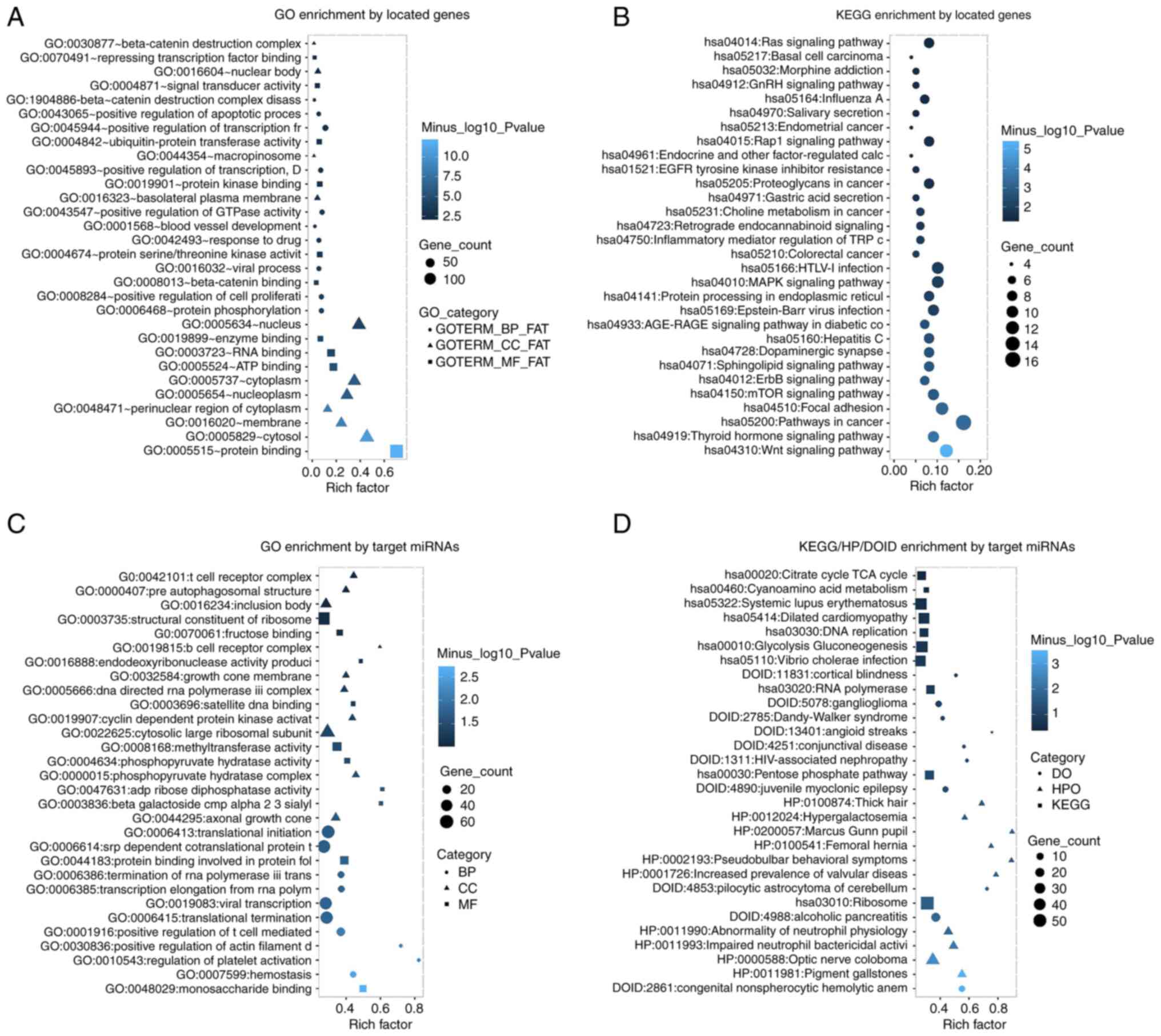
GO enrichment analysis of the located genes revealed
that dysregulated circRNAs were enriched in various biological
processes (such as ‘positive regulation of cell proliferation’,
‘positive regulation of GTPase activity’ and ‘positive regulation
of transcription, DNA-templated’), cellular components (such as
‘cytosol’, ‘membrane’ and ‘perinuclear region of cytoplasm’) and
molecular functions (such as ‘protein binding’, ‘β-catenin binding’
and ‘protein kinase binding’; Fig.
2A). KEGG enrichment analysis of the located genes showed that
dysregulated circRNAs were mainly enriched in the ‘Wnt signaling
pathway’, ‘mTOR signaling pathway’ and ‘MAPK signaling pathway’
(Fig. 2B). Furthermore, GO
enrichment analysis of target miRNAs showed that dysregulated
circRNAs were enriched in multiple biological processes (such as
‘positive regulation of T cell-mediated cytotoxicity’,
‘translational termination’ and ‘translational initiation’),
cellular components (such as ‘cytosolic large ribosomal subunit’,
‘B cell receptor complex’ and ‘T cell receptor complex’) and
molecular function (such as ‘protein binding involved in protein
folding’, ‘methyltransferase activity’ and ‘satellite DNA binding’;
Fig. 2C). KEGG/HP/DOID enrichment
analysis of the target miRNAs revealed that the dysregulated
circRNAs in acute myeloid leukemia were enriched in disease
ontology (such as ‘congenital nonspherocytic hemolytic anemia’,
‘alcoholic pancreatitis’ and ‘pilocytic astrocytoma of
cerebellum’), HP (such as ‘impaired neutrophil bactericidal
activity’, ‘abnormality of neutrophil physiology’ and ‘increased
prevalence of valvular diseases’) and cellular signaling pathways
(such as ‘ribosome’, ‘RNA polymerase’ and ‘DNA replication’;
Fig. 2D). So as to the detailed
information of enrichment analyses, they are exhibited in Table SIII, Table SIV, Table SV Table SVI.
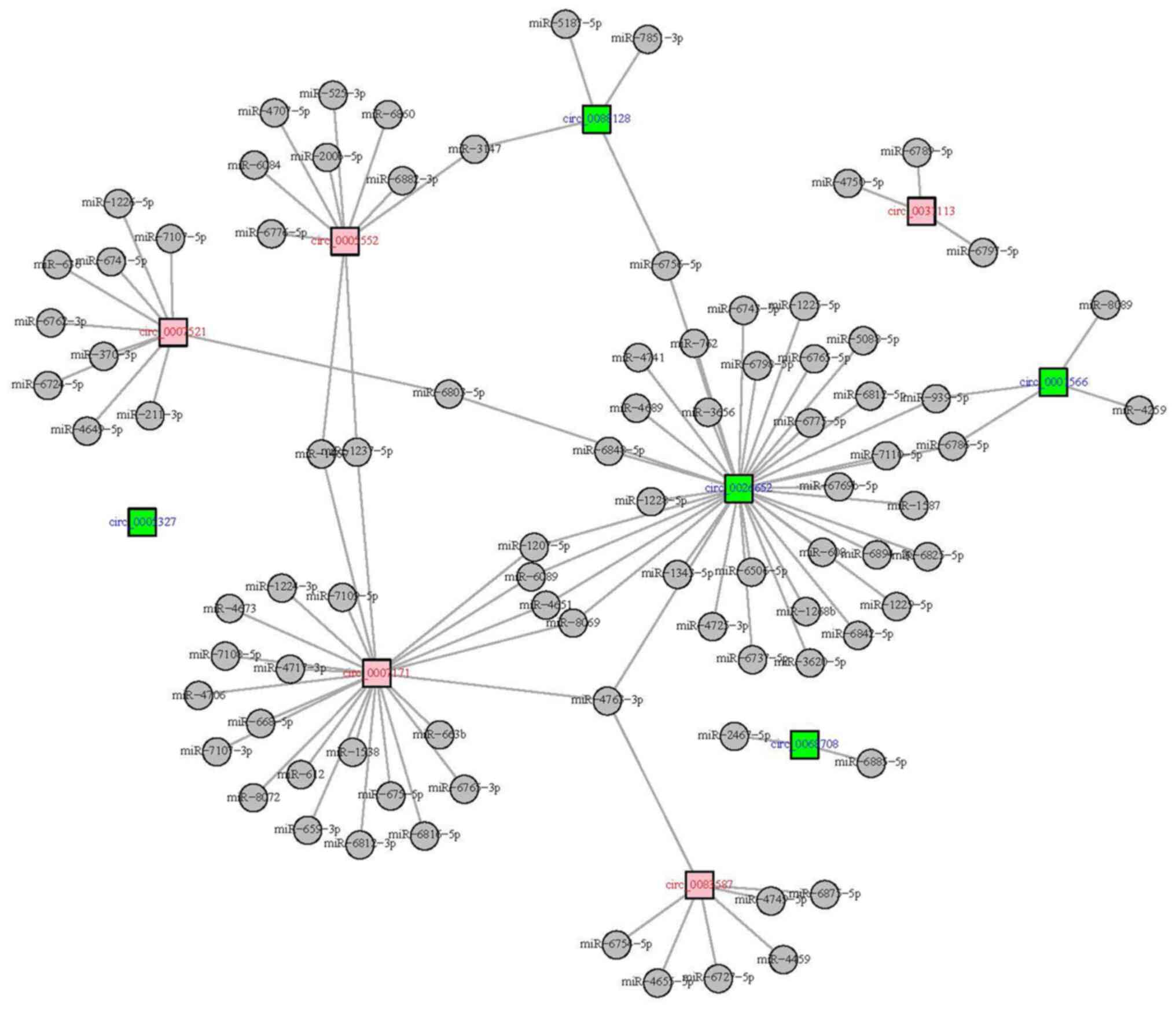
CircRNA-miRNA network of top five
upregulated and top five downregulated circRNAs
Among the dysregulated circRNAs, 10 candidate
circRNAs (top five upregulated and top five downregulated) were
further screened out by ranking the absolute value of
Log2FC. Then, circRNA-miRNA network of top five
upregulated and top five downregulated circRNAs was constructed
using the miRanda database. As shown in the circRNA-miRNA network,
half of the dysregulated circRNAs (including circ_0026652,
circ_0007171, circ_0007521, circ_0005552 and circ_0083587) had a
large number of target miRNAs, while circ_0001566 and circ_0088128
had four target miRNAs; Circ_0031113 had three target miRNAs,
circ_0068708 had two target miRNAs and circ_0005327 did not have
any (Fig. 3).
Validation of association between 10
candidate circRNAs and treatment response via RT-qPCR
To validate the association between 10 candidate
circRNAs (screened out using microarray and bioinformatics
analysis) and treatment response in 60 patients with MM, their
expression was further assessed via RT-qPCR. A total of 16 cases
(26.7%) achieved CR, 43 cases (71.7%) achieving ORR and 17 (28.3%)
had NR (data not shown).
It was found that the expression levels of
circ_0026652 (P<0.001; Fig.
4A), circ_0068708 (P=0.017; Fig.
4B), circ_0088128 (P=0.009; Fig.
4C) and circ_0001566 (P=0.034; Fig. 4D) were decreased, that of
circ_0031113 (P=0.001; Fig. 4F),
circ_0083587 (P=0.011; Fig. 4G),
circ_0005552 (P=0.028; Fig. 4H)
and circ_0007171 (P=0.019; Fig.
4I) were increased, and that of circ_0005327 (P=0.068; Fig. 4E) and circ_0007521 (P=0.127;
Fig. 4J) showed no change in
patients with CR compared with NR cases.
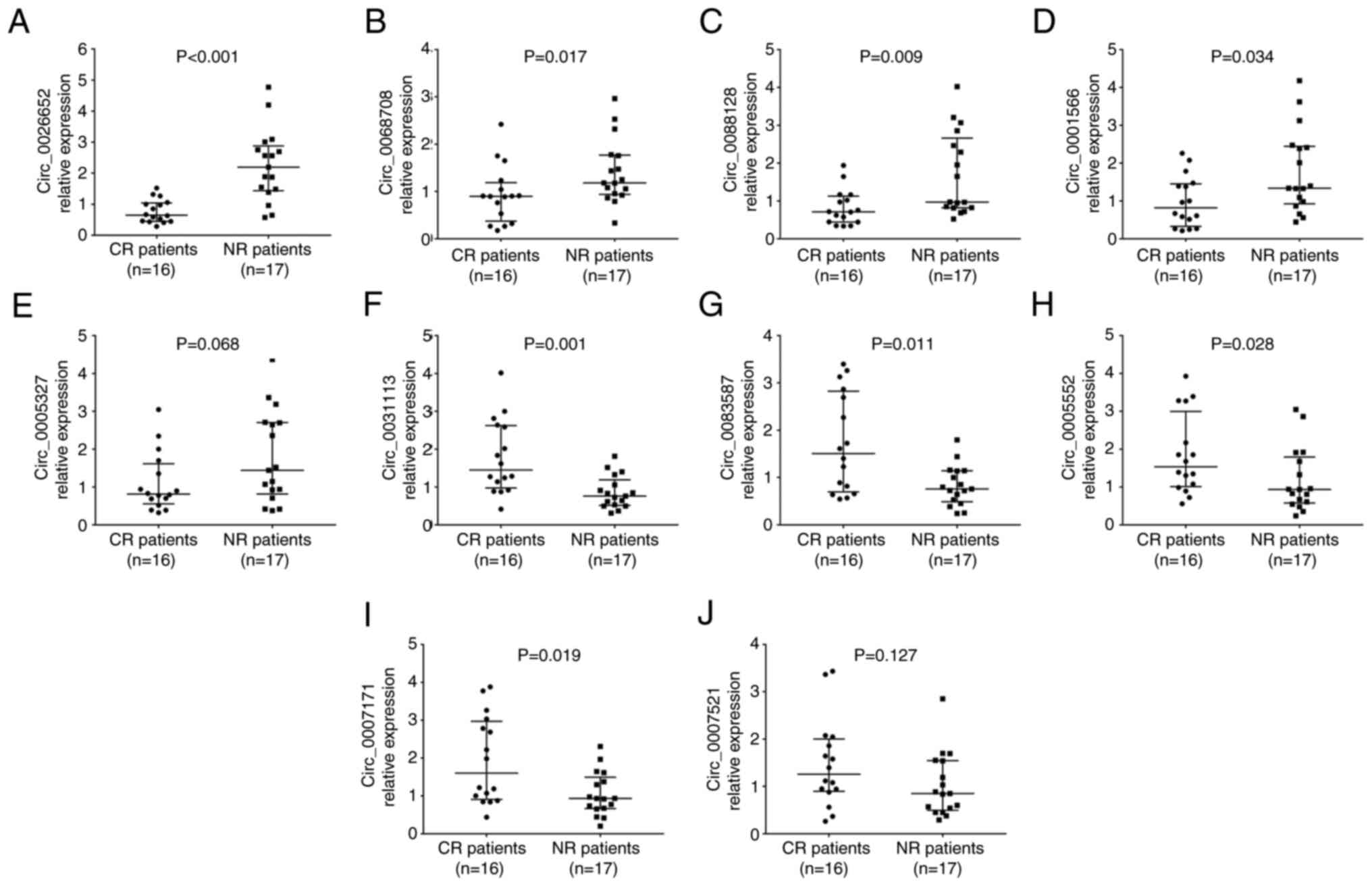 | Figure 4.Ten candidate circRNAs between CR
patients and NR patients. Comparisons of (A) circ_0026652, (B)
circ_0068708, (C) circ_0088128, (D) circ_0001566, (E) circ_0005327,
(F) circ_0031113, (G) circ_0083587, (H) circ_0005552, (I)
circ_0007171 and (J) circ_0007521 expression levels between CR
patients (n=16) and NR patients (n=17). CircRNA/circ, circular RNA;
CR, complete response; NR, no response. |
The expression levels of circ_0026652 (P=0.002;
Fig. 5A), circ_0068708 (P=0.045;
Fig. 5B) and circ_0088128
(P=0.001; Fig. 5C) were decreased,
that of circ_0031113 (P=0.007; Fig.
5F), circ_0083587 (P=0.041; Fig.
5G), circ_0005552 (P=0.010; Fig.
5H) and circ_0007171 (P=0.009; Fig. 5I) were increased, and that of
circ_0001566 (P=0.159; Fig. 5D),
circ_0005327 (P=0.093; Fig. 5E)
and circ_0007521 (P=0.240; Fig.
5J) showed no change in patients with CR compared with non-CR
cases.
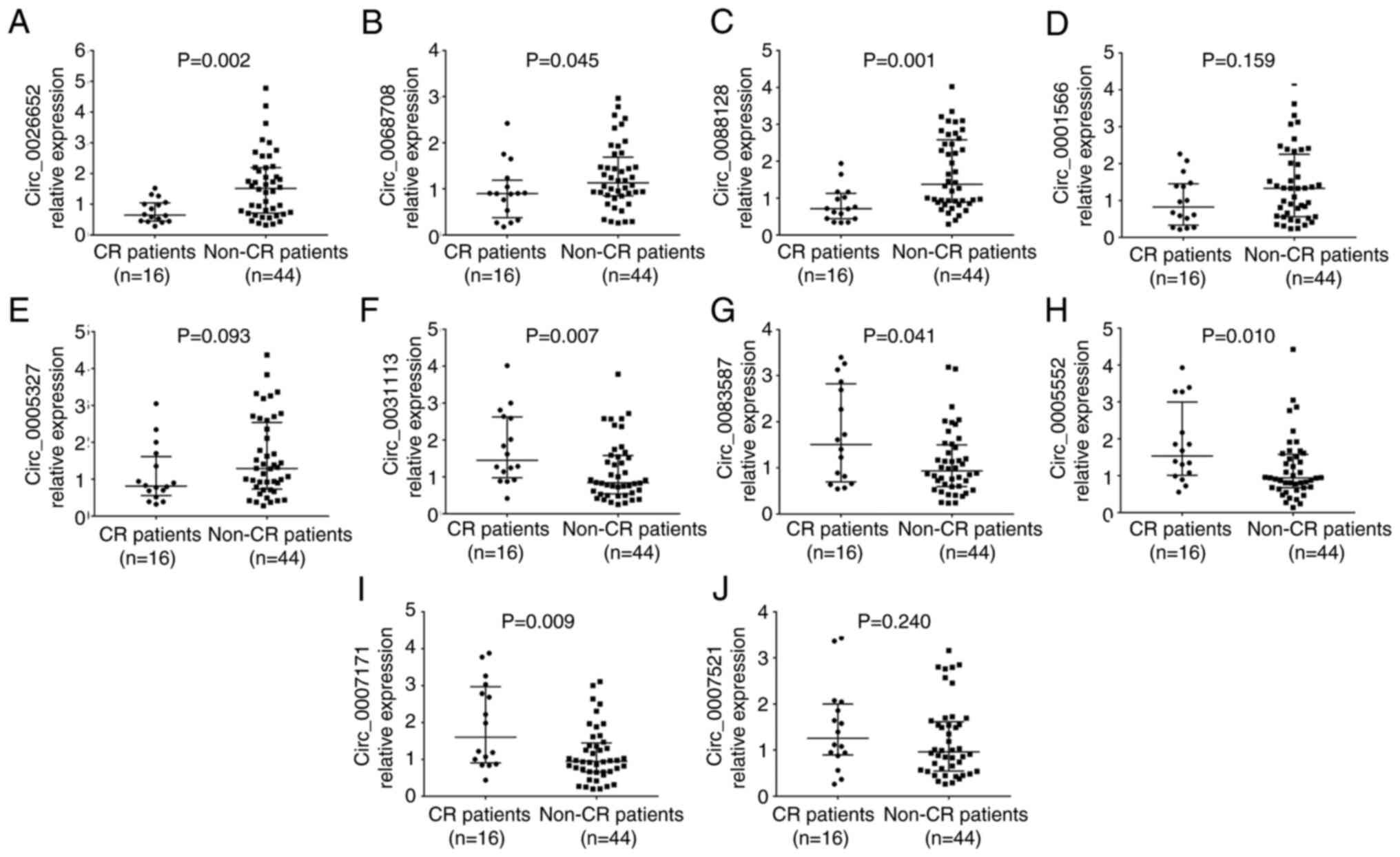 | Figure 5.Ten candidate circRNAs between CR
patients and non-CR patients. Comparisons of (A) circ_0026652, (B)
circ_0068708, (C) circ_0088128, (D) circ_0001566, (E) circ_0005327,
(F) circ_0031113, (G) circ_0083587, (H) circ_0005552, (I)
circ_0007171 and (J) circ_0007521 expression levels between CR
patients (n=16) and non-CR (n=44) patients. CircRNA/circ, circular
RNA; CR, complete response. |
In addition, the expression levels of circ_0026652
(P<0.001; Fig. 6A) and
circ_0001566 (P=0.027; Fig. 6D)
were decreased, that of circ_0031113 (P=0.015; Fig. 6F) and circ_0083587 (P=0.012;
Fig. 6G) were increased, and that
of circ_0068708 (P=0.063; Fig.
6B), circ_0088128 (P=0.326; Fig.
6C), circ_0005327 (P=0.175; Fig.
6E), circ_0005552 (P=0.262; Fig.
6H), circ_0007171 (P=0.216; Fig.
6I) and circ_0007521 (P=0.171; Fig. 6J) showed no change in patients with
ORR compared with non-ORR cases.
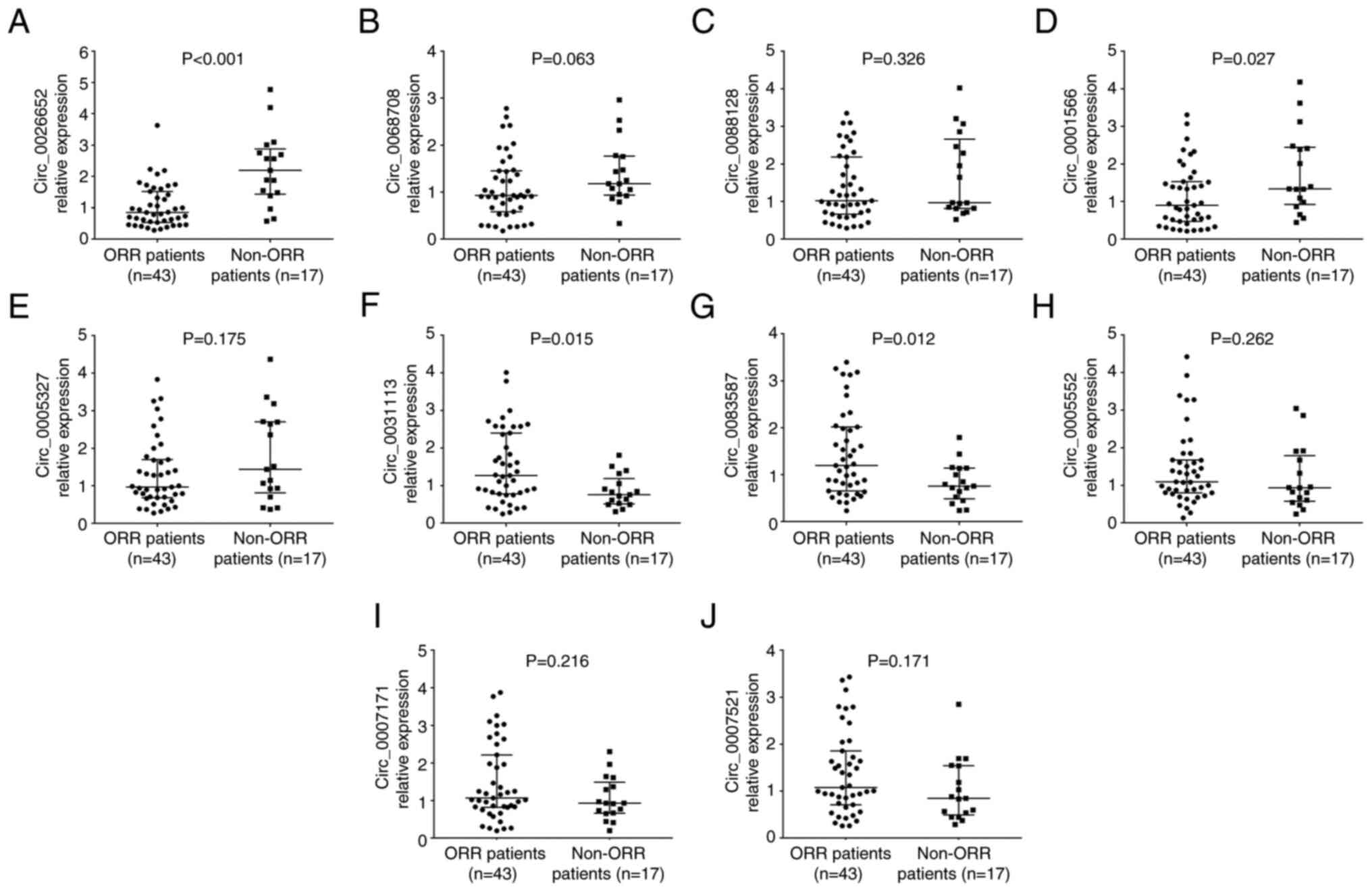 | Figure 6.Ten candidate circRNAs between ORR
patients and non-ORR patients. Comparisons of (A) circ_0026652, (B)
circ_0068708, (C) circ_0088128, (D) circ_0001566, (E) circ_0005327,
(F) circ_0031113, (G) circ_0083587, (H) circ_0005552, (I)
circ_0007171 and (J) circ_0007521 expression levels between ORR
patients (n=43) and non-ORR (n=17) patients. CircRNA/circ, circular
RNA; ORR, objective response rate. |
Furthermore, it was also noted that these ten
candidate circRNAs were of no difference between BLD regimen
treated patients and BCD regimen treated patients (Fig. S1A-J). Besides, the treatment
outcomes, such as response, PFS and OS, were of no difference
between BLD regimen treated patients and BCD regimen treated
patients (data not shown).
Validation of the association between
10 candidate circRNAs and survival via RT-qPCR
circ_0026652 high (P=0.017; Fig. 7A) and circ_0068708 high (P=0.040;
Fig. 7B) expression were
associated with a shorter PFS, circ_0031113 high (P=0.019; Fig. 7F) and circ_0005552 high (P=0.014;
Fig. 7H) expression were
associated with a long PFS, and no association of circ_0088128 high
(P=0.180; Fig. 7C), circ_0001566
high (P=0.057; Fig. 7D),
circ_0005327 high (P=0.245; Fig.
7E), circ_0083587 high (P=0.250; Fig. 7G), circ_0007171 high (P=0.092;
Fig. 7I) and circ_0007521 high
(P=0.242; Fig. 7J) expression with
PFS was observed.
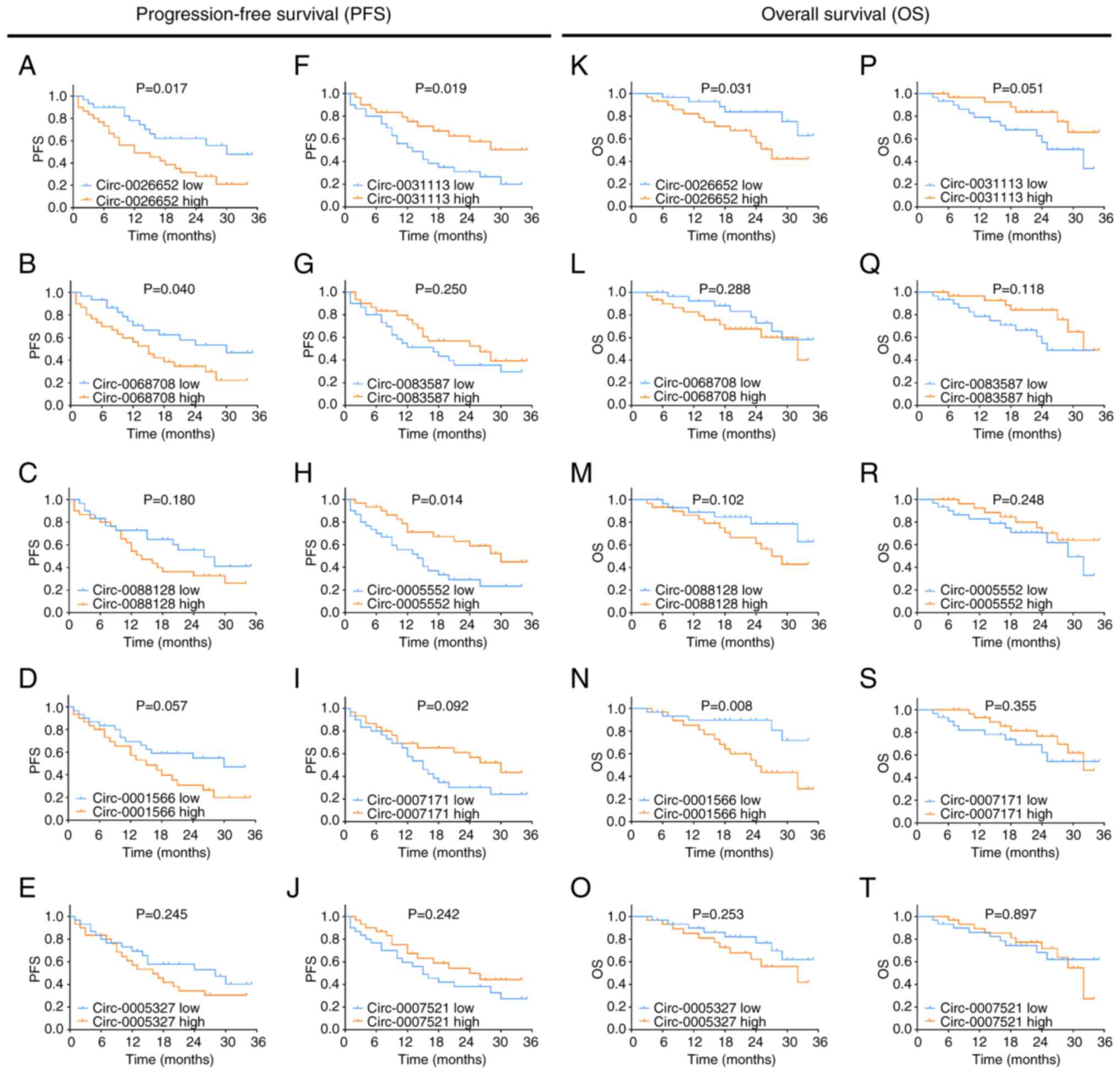 | Figure 7.Associations between candidate
circRNAs and PFS and OS. Associations between circ_0026652,
circ_0068708, circ_0088128, circ_0001566, circ_0005327,
circ_0031113, circ_0083587, circ_0005552, circ_0007171 and
circ_0007521 expression levels and (A-J) PFS and (K-T) OS.
CircRNA/circ, circular RNA; PFS, progressive-free survival; OS,
overall survival. |
circ_0026652 high (P=0.031; Fig. 7K) and circ_0001566 high (P=0.008;
Fig. 7N) expression were
associated with a short OS, while no association of circ_0068708
high (P=0.288; Fig. 7L),
circ_0088128 high (P=0.102; Fig.
7M), circ_0005327 high (P=0.253; Fig. 7O), circ_0031113 high (P=0.051;
Fig. 7P), circ_0083587 high
(P=0.118; Fig. 7Q), circ_0005552
high (P=0.248; Fig. 7R),
circ_0007171 high (P=0.355; Fig.
7S) and circ_0007521 high (P=0.897; Fig. 7T) expression with OS was
observed.
Circ_0026652 knockdown enhances MM
chemosensitivity to bortezomib
Among the 10 selected candidate circRNAs,
circ_0026652 was associated with a lower CR, ORR, PFS and OS in MM
(Table SVII), which indicated
that circ_0026652 was a key prognostic marker in MM; therefore,
further cellular experiments were conducted to investigate the
effect of circ_002665 knockdown on the regulation of
chemosensitivity to bortezomib in U266 and RPMI-8226 cells.
In U266 cells, circ_0026652 expression was reduced
in si-circ compared with the si-NC cells (P<0.001; Fig. 8A). Relative cell viability
following treatment with 2, 4, 8 and 16 nM bortezomib treatment was
decreased in si-circ compared with si-NC cells (all P<0.05;
Fig. 8B). In addition, under 4 nM
bortezomib treatment, relative cell viability was decreased in
si-circ compared with si-NC cells at 24, 48 and 72 h (all
P<0.05; Fig. S2A).
Subsequently, cell apoptosis was assessed following treatment with
4 nM (IC50 value) bortezomib, and the results showed
that cell apoptosis rate was increased in si-circ compared with
si-NC cells (P<0.05; Fig. 8C and
D).
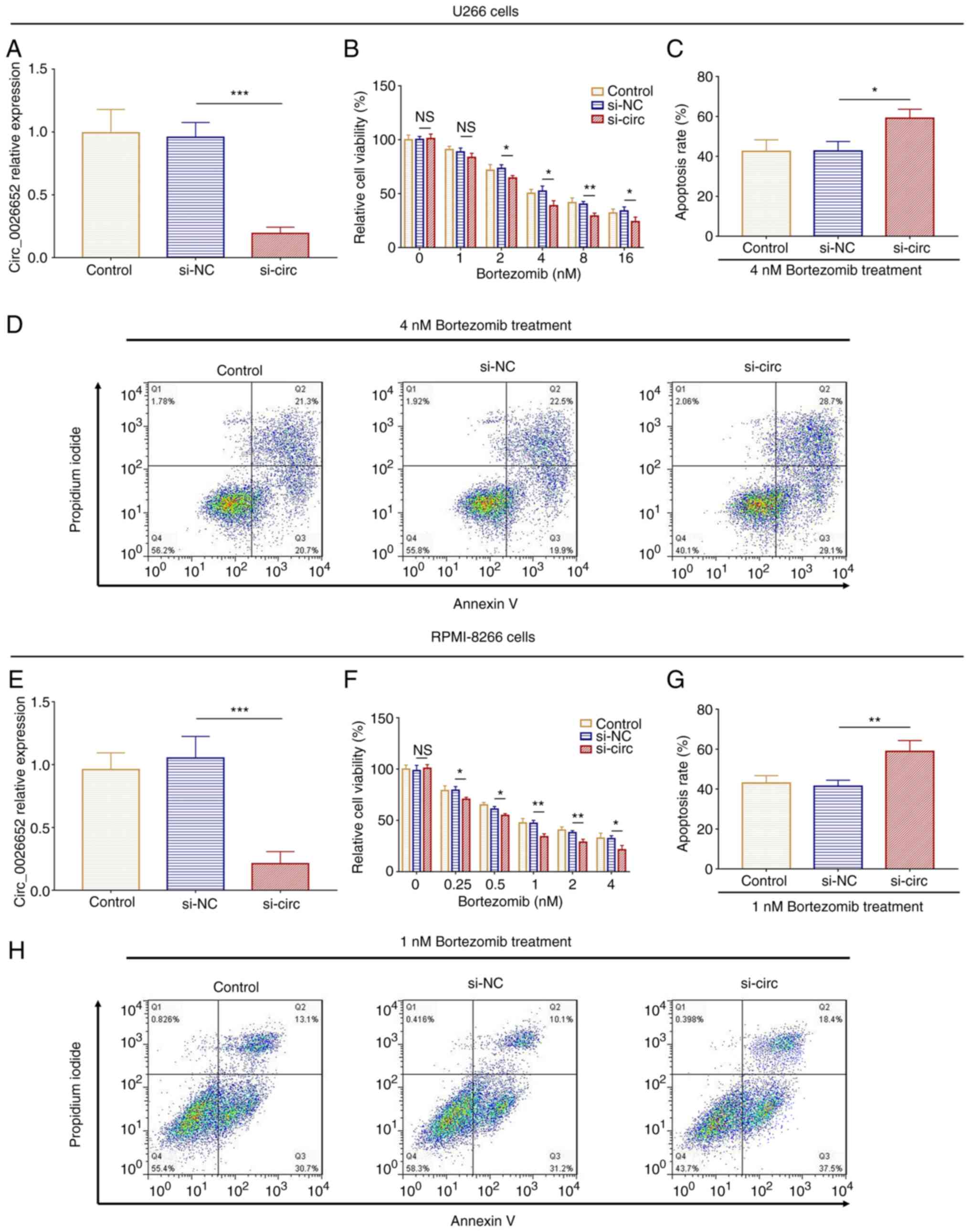 | Figure 8.Effect of circ_0026652 knockdown on
multiple myeloma chemosensitivity to bortezomib. (A) circ_0026652
expression, (B) relative cell viability under different bortezomib
treatment (0, 1, 2, 4, 8 and 16 nM) and (C and D) cell apoptosis
under 4 nM bortezomib treatment in U266 cells transfected with
si-circ and si-NC. (E) circ_0026652 expression, (F) relative cell
viability under different bortezomib concentrations (0, 0.25, 0.5,
1, 2 and 4 nM) and (G and H) cell apoptosis under 1 nM bortezomib
treatment in RPMI-8226 cells transfected with si-circ and si-NC.
*P<0.05, **P<0.01, ***P<0.001. circ, circular
RNA; si, small interfering RNA; NC, negative control. |
In RPMI-8226 cells, circ_0026652 expression was
lower in si-circ than in si-NC cells (P<0.001; Fig. 8E). Relative cell viability
following treatment with 0.25, 0.5, 1, 2 and 4 nm bortezomib was
decreased in si-circ compared with si-NC cells (all P<0.05;
Fig. 8F). In addition, following
treatment with 1 nM bortezomib, relative cell viability was
decreased in si-circ compared with si-NC cells at 24, 48 and 72 h
(all P<0.05; Fig. S2B). Next,
cell apoptosis was detected following treatment with 1 nM
(IC50 value) bortezomib, and the results showed that the
cell apoptosis rate was elevated in si-circ compared with si-NC
cells (P<0.01; Fig. 8G and
H).
Collectively, these data indicated that circ_0026652
knockdown promoted chemosensitivity to bortezomib in MM.
Circ_0026652 knockdown facilitates MM
chemosensitivity to bortezomib by regulating the miR-608-mediated
Wnt/β-catenin pathway
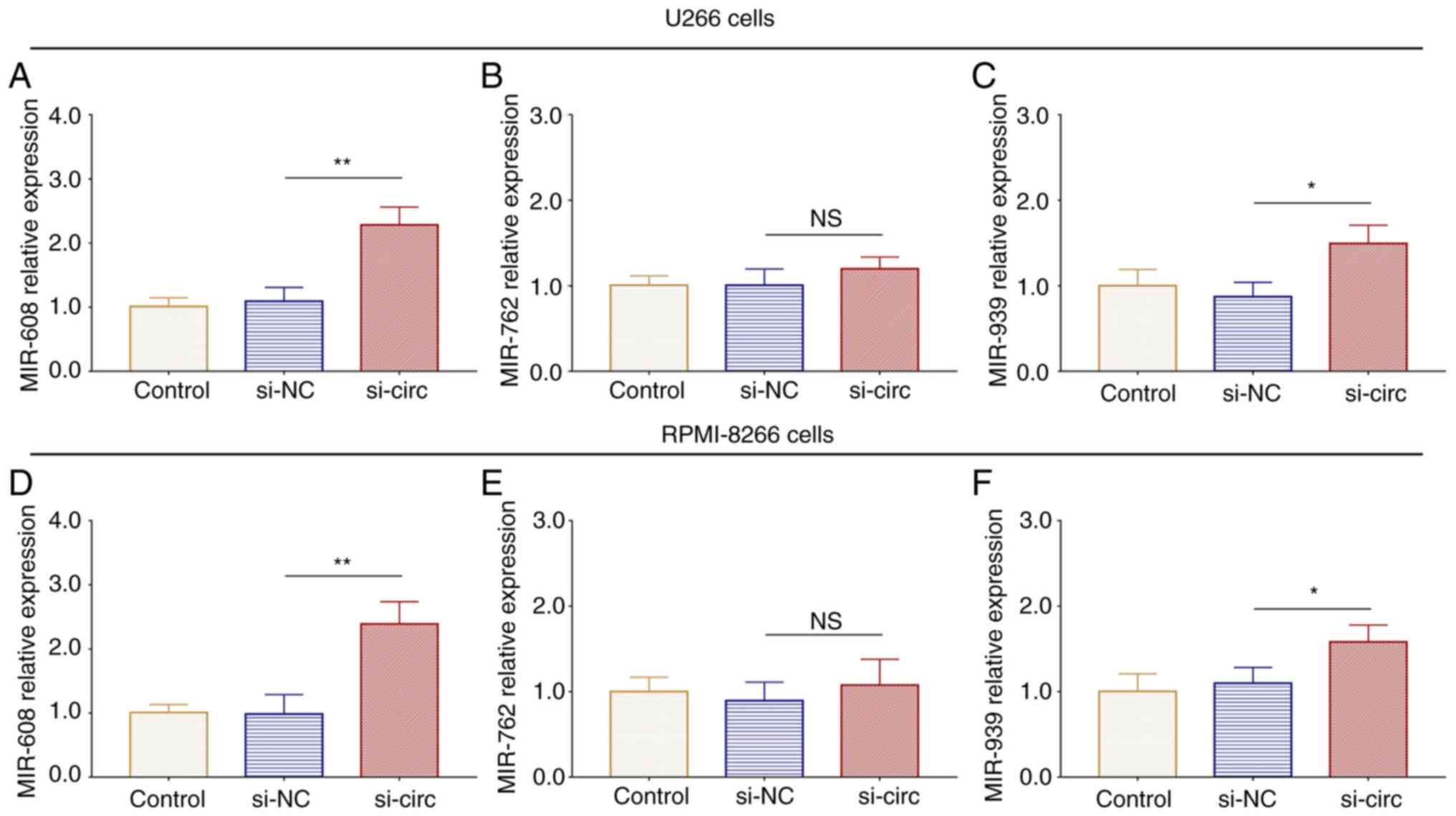
In both U266 (Fig.
9A-C) and RPMI-8226 cells (Fig.
9D-F), miR-608 and miR-939 expression levels were increased
(all P<0.05) and miR-762 expression showed no change (both
P>0.05) in si-circ cells compared with si-NC cells. miR-608 has
been reported to regulate the Wnt/β-catenin pathway, which plays an
essential role in the regulation of chemosensitivity to bortezomib
(22–25), hence, further rescue experiments
were performed to explore how the interaction between circ_0026652
and miR-608 regulated MM chemosensitivity to bortezomib. miR-608
inhibitor transfection efficiency is shown in Fig. S3A and B.
In the si-NC + miR-inhibitor and si-circ +
miR-inhibitor U266 cells (Fig.
10A-E) and si-NC + miR-inhibitor and si-circ + miR-inhibitor
RPMI-8226 cells (Fig. 10F-J),
miR-608 expression was decreased (all P<0.01) and did not affect
circ_0026652 expression (all P>0.05). Furthermore, in the si-NC
+ miR-inhibitor and si-circ + miR-inhibitor groups, relative cell
viability was increased (all P<0.05) and cell apoptosis was
decreased (all P<0.05) following bortezomib treatment, thus
indicating that the miR-608 inhibitor enhanced chemosensitivity to
bortezomib.
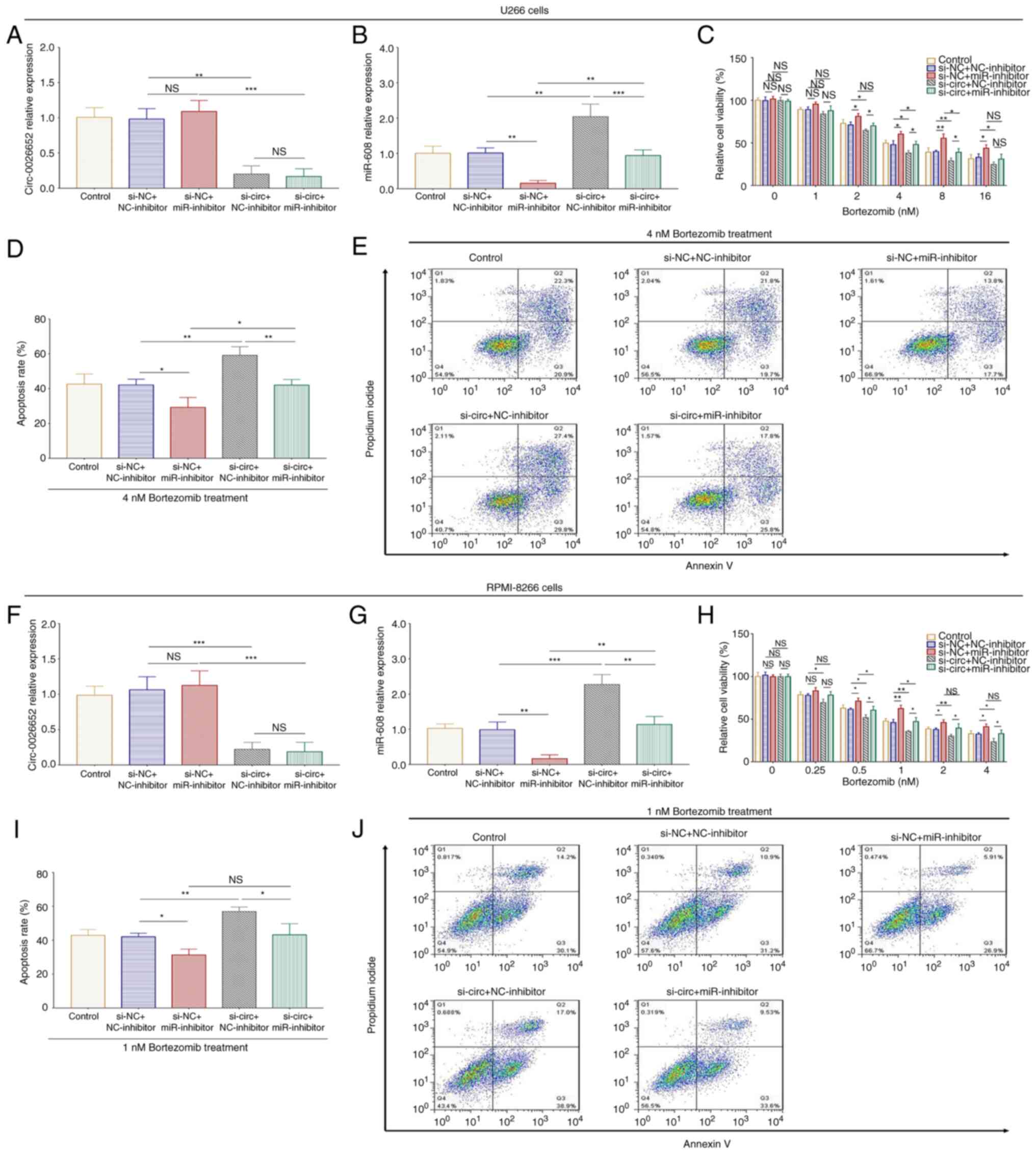 | Figure 10.miR-608 knockdown reverses the
effects of circ_0026652 knockdown on multiple myeloma
chemosensitivity to bortezomib. (A) circ_0026652 expression, (B)
miR-608 expression, (C) relative cell viability under different
bortezomib treatment (0, 1, 2, 4, 8 and 16 nM) and (D and E) cell
apoptosis under 4 nM bortezomib treatment in U266 cells in the
control, si-NC + NC-inhibitor, si-NC + miR-inhibitor, si-circ +
NC-inhibitor and si-circ + miR-inhibitor groups. (F) circ_0026652
expression, (G) miR-608 expression, (H) relative cell viability
under different bortezomib treatment (0, 0.25, 0.5, 1, 2 and 4 nM)
and (I and J) cell apoptosis under 1 nM bortezomib treatment in
RPMI-8226 cells in the control, si-NC + NC-inhibitor, si-NC +
miR-inhibitor, si-circ + NC-inhibitor and si-circ + miR-inhibitor
groups. *P<0.05, **P<0.01, ***P<0.001. miR,
microRNA; circ, circular RNA; si, small interfering RNA; NC,
negative control. |
In addition, the effect of circ_0026652 and miR-608
knockdown on the Wnt/β-catenin pathway was assessed; in both U266
and RPMI-8226 cells, WNT3A and β-catenin protein expression was
decreased in si-circ + NC-inhibitor compared with si-NC +
NC-inhibitor cells and partially increased in si-NC + miR-inhibitor
cells compared with si-NC + NC-inhibitor cells. Further rescue
experiments showed that transfection with the miR-608 inhibitor
compensated the suppressive effect of circ_0026652 knockdown on the
protein expression of WNT3A and β-catenin to some extent (Fig. 11A and B).
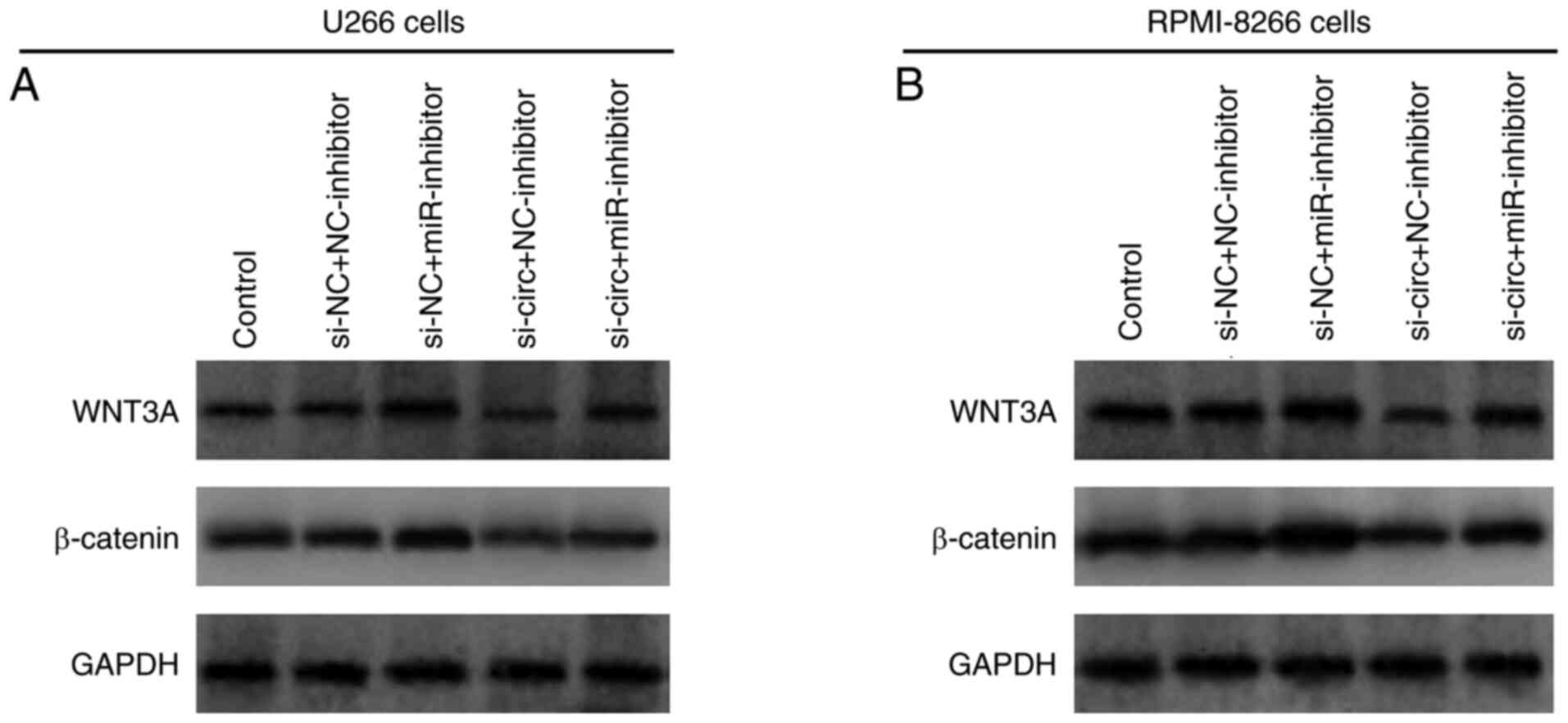 | Figure 11.miR-608 knockdown reverses the
effects of circ_0026652 knockdown on the Wnt/β-catenin pathway. (A)
Protein expression levels of WNT3A and β-catenin in U266 cells in
the control, si-NC + NC-inhibitor, si-NC + miR-inhibitor, si-circ +
NC-inhibitor and si-circ + miR-inhibitor groups. (B) Protein
expression levels of WNT3A and β-catenin in RPMI-8226 cells in the
control, si-NC + NC-inhibitor, si-NC + miR-inhibitor, si-circ +
NC-inhibitor and si-circ + miR-inhibitor groups. miR, microRNA;
circ, circular RNA; si, small interfering RNA; NC, negative
control; WNT3A, Wnt family member 3A. |
Collectively, the results showed that circ_0026652
knockdown enhanced chemosensitivity to bortezomib by regulating the
miR-608-mediated Wnt/β-catenin pathway in MM.
Discussion
Due to their high stability, and cell- and
tissue-specific expression patterns, the study of circRNA is a
research hotspot, and previous studies using next-generation
sequencing technologies and bioinformatics tools have reported that
dysregulated circRNAs are essential for the development and
progression of hematological malignancies (7–12),
including MM (11,12). One study reported 122 upregulated
and 260 downregulated circRNAs in patients with MM compared with
healthy participates. Among these dysregulated circRNAs, circ-PTK2,
circ-RNF217, circ-RERE, circ-NAGPA, circ-KCNQ5, circ-AFF2,
circ-WWC3, circ-DNAJC5, circ-KLHL2, circ-IQGAP1 and circ-AL137655,
were confirmed by RT-qPCR to be dysregulated in MM compared with
the controls (12). Another study
showed that circRNA microarray analysis revealed 40 upregulated and
10 downregulated circRNAs in patients with MM compared with
patients with IDA (11).
Subsequent RT-qPCR validation studies showed that circ_101237 is
upregulated in patients with MM compared with patients with IDA,
and its upregulation is associated with 13q14 deletion, 1q21
amplification, P53 deletion, t(4,14)
and t(14,16) chromosomal abnormalities (11). The role of circRNA expression
profiles in the prognosis of MM has not been comprehensively
explored.
In the present study, microarray assays and
bioinformatics analyses were initially performed in eight CR and
eight NR MM cases to comprehensively characterize the circRNA
profiles in the prognosis of MM. A total of 246 circRNAs were found
to be dysregulated (79 upregulated and 167 downregulated) in CR MM
cases compared with NR MM cases. Subsequent enrichment analyses
revealed that the dysregulated circRNAs were mainly enriched in
MM-related signaling pathways, such as the ‘Wnt signaling pathway’,
‘mTOR signaling pathway’ and ‘MAPK signaling pathway’. Wnt
signaling functions as an important regulator of cell
proliferation, cell fate, migration and cell polarity (30). In MM, aberrant Wnt signaling
pathway activation induces MM cell proliferation, migration and
invasion by modulating the expression of cell cycle genes,
Rho-associated protein kinase and protein kinase C family members,
which cause drug resistance and malignancy progression (31). mTOR (a highly conserved
PI3K-related serine/threonine kinase) is essential in regulating
cell proliferation, survival, invasion, migration and
chemoresistance, which is involved in the pathogenesis of MM
(32,33). The MAPK (a serine-threonine protein
kinase) signaling pathway plays a vital role in modulating MM cell
proliferation, survival and drug resistance (34). In combination, dysregulated
circRNAs appear to play a key role in drug resistance in MM.
To validate the association between the 10 candidate
circRNAs and MM prognosis, their expression was further determined
via RT-qPCR in a larger sample of patients with MM. It was revealed
that 8/10 circRNAs (including circ_0026652, circ_0068708,
circ_0088128, circ_0001566, circ_0031113, circ_0083587,
circ_0005552 and circ_0007171) were dysregulated in CR compared
with NR cases, 7/10 circRNAs (including circ_0026652, circ_0068708,
circ_0088128, circ_0031113, circ_0083587, circ_0005552 and
circ_0007171) were dysregulated in CR compared with non-CR cases,
4/10 circRNAs (including circ_0026652, circ_0001566, circ_0031113
and circ_0083587) were dysregulated in ORR compared with non-ORR
cases, 4/10 circRNAs (including circ_0026652, circ_0068708,
circ_0031113 and circ_0005552) were associated with PFS and 2/10
circRNAs (including circ_0026652 and circ_0001566) were associated
with OS. In combination, these findings indicated that dysregulated
circRNAs were closely linked with prognosis in patients with MM.
The possible explanations were as follows: i) The dysregulated
circRNAs may regulate the transcription of their parental genes
either positively or negatively. For instance, according to a
previous study, circ_0031113 likely enhances the transcription of
its parental gene telomerase protein component 1 (TEP1), and TEP1
acts as a tumor suppressor gene that may inhibit MM cell
proliferation, while promoting apoptosis; therefore, circ_0031113
could ameliorate disease progression and improve prognosis in
patients with MM (35). ii) The
dysregulated circRNAs may regulate the pathogenesis of MM by
sponging their target miRNAs. For instance, through the use of the
miRanda database, it has been found that circ_0083587 may serve as
a sponge for miR-6875-5p to repress the function of miR-6875-5p in
maintaining cell survival and enhancing cell proliferation,
resulting in alleviated disease progression and is associated with
favorable prognosis in patients with MM (36).
Of note, a key prognostic marker, circ_0026652, was
identified in the present study, which was not only associated with
CR and ORR, but also with PFS and OS in patients with MM. The
possible explanations were as follows: i) Subsequent experiments
revealed that circ_0026652 knockdown enhanced chemosensitivity to
bortezomib treatment, and high circ_0026652 expression is likely
associated with poor treatment response and survival in patients
with MM; ii) subsequent experiments revealed that circ_0026652
knockdown suppressed the Wnt/β-catenin pathway; meanwhile, the
Wnt/β-catenin pathway was demonstrated to induce MM cell
proliferation, migration and invasion, and thus high circ_0026652
expression may promote the survival and aggressiveness of MM cells
via the Wnt/β-catenin pathway, leading to a poor prognosis in
patients with MM (31).
To further explore the potential mechanisms of
circ_0026652 in the regulation of chemosensitivity to bortezomib in
MM, cellular experiments were performed. The results showed that
circ_0026652 knockdown promoted MM chemosensitivity to bortezomib
treatment by regulating the miR-608-mediated Wnt/β-catenin pathway.
These results can be explained as follows: i) circ_0026652 may act
as a miRNA sponge to directly suppress the activity of miR-608;
meanwhile, miR-608 has been reported to directly inhibit the
expression of frizzled family receptor 4 that induces the
activation of the oncogenic Wnt/β-catenin pathway (22,37);
in addition, the Wnt//β-catenin pathway is known to mediate
proliferation, migration and drug resistance of MM cells by
modulating the expression of cell cycle genes (such as cyclin D1,
MYC proto-oncogene, BHLH transcription factor and aurora kinase A)
(31). Thereby, circ_0026652
knockdown likely promotes chemosensitivity to bortezomib through
the miR-608-mediated Wnt/β-catenin pathway in MM (22,31,37).
Despite the notable findings, the present study had
certain limitations. First, due to the limited number of patients
with MM in our hospital, only 60 patients with MM were enrolled,
which may have reduced the statistic power of the analyses.
Secondly, only patients with MM who underwent bortezomib-based
induction treatment were enrolled, and thus the findings may not be
applicable to patients with MM who have not been treated with
bortezomib. Finally, although cellular experiments were conducted
to explore the potential mechanism of circ_0026652 in regulating
chemosensitivity to bortezomib in MM, further in vivo
studies are needed for validation.
In conclusion, the present study provided a
comprehensive overview of the role of dysregulated circRNAs in MM
prognosis, and confirmed that circ_0026652, circ_0068708,
circ_0088128, circ_0001566, circ_0031113, circ_0083587,
circ_0005552 and circ_0007171 may serve as potential prognostic
biomarkers for treatment response or survival in patients with MM.
Among these circRNAs, circ_0026652 is a key and promising
prognostic marker of MM. In addition, circ_0026652 knockdown
promoted chemosensitivity to bortezomib by regulating the
miR-608-mediated Wnt/β-catenin pathway in MM. These findings may
lay the foundations for developing potential drug targets, as well
as optimizing the treatment and improving the prognosis of MM.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to institutional
policy prohibits data sharing, but are available from the
corresponding author on reasonable request.
Authors' contributions
WF conceived and designed the present study. WF and
LL confirm the authenticity of all the raw data. LL, JLi and JD
performed the experiments and collected the data. HJ, HH and JLu
analyzed the data. WQ and NH mainly designed the experiments and
wrote the manuscript. PG and YZ are responsible for analysis and
interpretation of data and revising the article. approved the final
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by Institutional Review
Board of Changzheng Hospital, Second Military Medical University
(Shanghai, China), and all patients provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
MM
|
Multiple myeloma
|
|
PFS
|
progressive-free survival
|
|
OS
|
overall survival
|
|
CircRNAs
|
Circular RNAs
|
|
miRNAs
|
microRNAs
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
IDA
|
iron deficiency anemia
|
|
CR
|
complete response
|
|
NR
|
non-response
|
|
IMWG
|
International Myeloma Working
Group
|
|
ISS
|
International Staging System
|
|
VGPR
|
very good partial response
|
|
SD
|
stable disease
|
|
PD
|
progressive disease
|
|
FC
|
fold-change
|
|
PCA
|
principal component analysis
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoko Encyclopedia of Genes and
Genomes
|
|
HP
|
Human Phenotype
|
|
IQR
|
interquartile range
|
References
|
1
|
Röllig C, Knop S and Bornhäuser M:
Multiple myeloma. Lancet. 385:2197–2208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kunacheewa C and Orlowski RZ: New drugs in
multiple myeloma. Annu Rev Med. 70:521–547. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pinto V, Bergantim R, Caires HR, Seca H,
Guimãraes JE and Vasconcelos MH: Multiple myeloma: Available
therapies and causes of drug resistance. Cancers (Basel).
12:4072020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pawlyn C and Davies FE: Toward
personalized treatment in multiple myeloma based on molecular
characteristics. Blood. 133:660–675. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jamal M, Song T, Chen B, Faisal M, Hong Z,
Xie T, Wu Y, Pan S, Yin Q, Shao L and Zhang Q: Recent progress on
circular RNA research in acute myeloid leukemia. Front Oncol.
9:11082019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kristensen LS, Andersen MS, Stagsted LV,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu Z, Sun H, Li J and Jin H: Circular RNAs
in leukemia. Aging (Albany NY). 11:4757–4771. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li W, Zhong C, Jiao J, Li P, Cui B, Ji C
and Ma D: Characterization of hsa_circ_0004277 as a new biomarker
for acute myeloid leukemia via circular RNA profile and
bioinformatics analysis. Int J Mol Sci. 18:5972017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lv C, Sun L, Guo Z, Li H, Kong D, Xu B,
Lin L, Liu T, Guo D, Zhou J and Li Y: Circular RNA regulatory
network reveals cell-cell crosstalk in acute myeloid leukemia
extramedullary infiltration. J Transl Med. 16:3612018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ding Y, Dong Y, Lu H, Luo X, Fu J, Xiu B,
Liang A and Zhang W: Circular RNA profile of acute myeloid
leukaemia indicates circular RNA annexin A2 as a potential
biomarker and therapeutic target for acute myeloid leukaemia. Am J
Transl Res. 12:1683–1699. 2020.PubMed/NCBI
|
|
11
|
Liu X, Tang H, Liu J and Wang X:
hsa_circRNA_101237: A novel diagnostic and prognostic biomarker and
potential therapeutic target for multiple myeloma. Cancer Manag
Res. 12:2109–2118. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou F, Wang D, Wei W, Chen H, Shi H, Zhou
N, Wu L and Peng R: Comprehensive profiling of circular RNA
expressions reveals potential diagnostic and prognostic biomarkers
in multiple myeloma. BMC Cancer. 20:402020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rajkumar SV, Dimopoulos MA, Palumbo A,
Blade J, Merlini G, Mateos MV, Kumar S, Hillengass J, Kastritis E,
Richardson P, et al: International myeloma working group updated
criteria for the diagnosis of multiple myeloma. Lancet Oncol.
15:e538–e548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chinese Hematology Association and Chinese
Society of Hematology, . Zhonghua Nei Ke Za Zhi. 61:480–487.
2022.(Article in Chinese). PubMed/NCBI
|
|
15
|
Ludwig H, Miguel JS, Dimopoulos MA,
Palumbo A, Garcia Sanz R, Powles R, Lentzsch S, Ming Chen W, Hou J,
Jurczyszyn A, et al: International myeloma working group
recommendations for global myeloma care. Leukemia. 28:981–992.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rajkumar SV, Harousseau JL, Durie B,
Anderson KC, Dimopoulos M, Kyle R, Blade J, Richardson P, Orlowski
R, Siegel D, et al: Consensus recommendations for the uniform
reporting of clinical trials: Report of the international myeloma
workshop consensus panel 1. Blood. 117:4691–4695. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Durie BG, Harousseau JL, Miguel JS, Bladé
J, Barlogie B, Anderson K, Gertz M, Dimopoulos M, Westin J,
Sonneveld P, et al: International uniform response criteria for
multiple myeloma. Leukemia. 20:1467–1473. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Skuja E, Butane D, Nakazawa-Miklasevica M,
Daneberga Z, Purkalne G and Miklasevics E: Deletions in metastatic
colorectal cancer with chromothripsis. Exp Oncol. 41:323–327.
2019.PubMed/NCBI
|
|
19
|
Wang L, Liu X, Yue M, Liu Z, Zhang Y, Ma
Y, Luo J, Li W, Bai J, Yao H, et al: Identification of hub genes in
bladder cancer based on weighted gene co-expression network
analysis from TCGA database. Cancer Rep (Hoboken).
e15572021.(Online ahead of print). PubMed/NCBI
|
|
20
|
Cheng Q and Wang L: LncRNA XIST serves as
a ceRNA to regulate the expression of ASF1A, BRWD1M, and PFKFB2 in
kidney transplant acute kidney injury via sponging hsa-miR-212-3p
and hsa-miR-122-5p. Cell Cycle. 19:290–299. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou Y, Liu X, Xue J, Liu L, Liang T, Li
W, Yang X, Hou X and Fang H: Discovery of peptide boronate
derivatives as histone deacetylase and proteasome dual inhibitors
for overcoming bortezomib resistance of multiple myeloma. J Med
Chem. 63:4701–4715. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Y, Zhang W, Wang Y and Wang S:
HOXD-AS1 promotes cell proliferation, migration and invasion
through miR-608/FZD4 axis in ovarian cancer. Am J Cancer Res.
8:170–182. 2018.PubMed/NCBI
|
|
23
|
Jin Y, Xu L, Wu X, Feng J, Shu M, Gu H,
Gao G, Zhang J, Dong B and Chen X: Synergistic efficacy of the
demethylation agent decitabine in combination with the protease
inhibitor bortezomib for treating multiple myeloma through the
Wnt/β-catenin pathway. Oncol Res. 27:729–737. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yao R, Sun X, Xie Y, Liu L, Han D, Yao Y,
Li H, Li Z and Xu K: Lithium chloride inhibits cell survival,
overcomes drug resistance, and triggers apoptosis in multiple
myeloma via activation of the Wnt/β-catenin pathway. Am J Transl
Res. 10:2610–2618. 2018.PubMed/NCBI
|
|
25
|
Savvidou I, Khong T, Cuddihy A, McLean C,
Horrigan S and Spencer A: β-catenin inhibitor BC2059 is efficacious
as monotherapy or in combination with proteasome inhibitor
bortezomib in multiple myeloma. Mol Cancer Ther. 16:1765–1778.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen X, Mao R, Su W, Yang X, Geng Q, Guo
C, Wang Z, Wang J, Kresty LA, Beer DG, et al: Circular RNA
circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα
signaling in STK11 mutant lung cancer. Autophagy. 16:659–671. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fang Y, Wang X, Li W, Han J, Jin J, Su F,
Zhang J, Huang W, Xiao F, Pan Q and Zou L: Screening of circular
RNAs and validation of circANKRD36 associated with inflammation in
patients with type 2 diabetes mellitus. Int J Mol Med.
42:1865–1874. 2018.PubMed/NCBI
|
|
29
|
Zhang M, Wen F and Zhao K: Circular
RNA_0001946 is insufficiently expressed in tumor tissues, while its
higher expression correlates with less lymph node metastasis, lower
TNM stage, and improved prognosis in NSCLC patients. J Clin Lab
Anal. 35:e236252021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gajos-Michniewicz A and Czyz M: WNT
signaling in melanoma. Int J Mol Sci. 21:48522020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
van Andel H, Kocemba KA, Spaargaren M and
Pals ST: Aberrant Wnt signaling in multiple myeloma: Molecular
mechanisms and targeting options. Leukemia. 33:1063–1075. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Peterson TR, Laplante M, Thoreen CC,
Sancak Y, Kang SA, Kuehl WM, Gray NS and Sabatini DM: DEPTOR is an
mTOR inhibitor frequently overexpressed in multiple myeloma cells
and required for their survival. Cell. 137:873–886. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li J, Zhu J, Cao B and Mao X: The mTOR
signaling pathway is an emerging therapeutic target in multiple
myeloma. Curr Pharm Des. 20:125–135. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Podar K, Chauhan D and Anderson KC: Bone
marrow microenvironment and the identification of new targets for
myeloma therapy. Leukemia. 23:10–24. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu TH, Huang CC, Lin PR, Chang HW, Ger LP,
Lin YW, Changchien CS, Lee CM and Tai MH: Expression and prognostic
role of tumor suppressor gene PTEN/MMAC1/TEP1 in hepatocellular
carcinoma. Cancer. 97:1929–1940. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xie Y, Du J, Liu Z, Zhang D, Yao X and
Yang Y: MiR-6875-3p promotes the proliferation, invasion and
metastasis of hepatocellular carcinoma via BTG2/FAK/Akt pathway. J
Exp Clin Cancer Res. 38:72019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gupta S, Iljin K, Sara H, Mpindi JP,
Mirtti T, Vainio P, Rantala J, Alanen K, Nees M and Kallioniemi O:
FZD4 as a mediator of ERG oncogene-induced WNT signaling and
epithelial-to-mesenchymal transition in human prostate cancer
cells. Cancer Res. 70:6735–6745. 2010. View Article : Google Scholar : PubMed/NCBI
|