Introduction
Endometrial carcinoma (EC), also known as
endometrial cancer, refers to a group of epithelial malignant
tumors originating in the endometrium, and it is the most common
gynecological malignant tumor in the United States in terms of
incidence (1). EC ranks second
among gynecological malignant tumors in China in terms of
incidence, and its mortality rate is increasing year by year
(2). EC accounts for ~85% of newly
diagnosed cases, serous carcinoma accounts for 3–10%, clear cell
carcinoma accounts for <5% and carcinosarcomas are relatively
rare (3). The International
Federation of Gynecology and Obstetrics staging system (4) is used for EC, which is divided into
stages I–IV, and the survival rate decreases as the stage
progresses (4). At present, the
main screening methods for EC are hysteroscopy, segmented curettage
and endometrial biopsy, and surgery combined with chemotherapy is
the most common treatment for EC (5). The procedures involved include pelvic
lymph node dissection, bilateral salpingo-oophorectomy and
hysterectomy (6), and carboplatin
combined with paclitaxel provides the means of first line
chemotherapy (7). However,
although therapies, including surgery, radiotherapy and
chemotherapy, have provided good results (8), there is still an urgent need to
identify novel molecular therapies to target the disease.
Circular RNAs (circRNAs/circs) are a class of
endogenous non-coding RNA molecules that do not have 5′-end caps
and 3′-end poly(A) tails (9), and
form a circular structure with covalent bonds. They are abundant
molecules that are less easily degraded by RNase R and are more
stable than linear RNA (10).
circRNAs are frequently ectopically expressed in a variety of
malignancies, including bladder (11), colorectal (12), breast (13) and lung (14) cancer. circRNAs fulfill important
phenotypic roles, including roles in proliferation, invasion,
metastasis and drug resistance, in various types of malignancies,
including bladder, colorectal, breast and lung cancer (11–14).
circRNAs are also associated with tumor size, lymph node metastasis
and cancer stage (15,16). Furthermore, certain circRNAs have
been identified as molecular biomarkers for the diagnosis,
treatment and prognosis of various types of human cancer (15).
In the present review, the biogenesis,
classification and functions of circRNAs are summarized.
Furthermore, the expression and biological functions of circRNAs
that are involved in EC are reviewed.
Study design
In the present review, relevant literature was
searched for using the PubMed (https://pubmed.ncbi.nlm.nih.gov/) and Web of Science
(http://wokinfo.com) databases, mainly focusing on
the past 5 years (2017–2022), while a small number of studies
identified were published >5 years ago. The search terms used
were ‘endometrial carcinoma’ and ‘circRNAs’. The language limit was
English. Two authors searched relevant literature according to the
topic of this review. The inclusion criteria were: Inclusion of
endometrial carcinoma and circRNAs or association with both. The
exclusion criteria were: No association with endometrial carcinoma
or circRNAs. Articles including the key words ‘endometrial
carcinoma’ and ‘circRNAs’ were searched and cited. Two authors
approved the final list of included studies.
Formation and classification of
circRNAs
Numerous types of circRNA have been found to be
expressed in a cell type-specific or tissue-specific manner
(17,18), suggesting that they may fulfill
biological functions. In the majority of eukaryotes, circRNAs,
transcribed by RNA polymerase II and produced by spliceosomes, are
covalently closed single-stranded RNA molecules that are formed
through the reverse splicing of precursor mRNA (19,20).
The precursor mRNA could be generated either through efficient
canonical splicing to generate linear RNA, or by inefficient
reverse splicing to generate circRNAs (9). circRNAs lack 5′-3′ ends and poly(A)
tails, which render them less susceptible to RNase R degradation
compared with linear RNAs, and thus, they are more stable than
linear RNAs (10). There are three
known mechanisms through which circRNAs may be formed, i.e.,
intron-pairing, lariat-driven circularization and RNA binding
protein (RBP)-driven circularization (21). In the intron-pairing model,
circularization via back-splicing is associated with base-pairing
between different introns, particularly between repetitive
sequences such as ALU repeats, whereas lariat-driven
circularization occurs by adding splice sites of the exons that are
skipped during linear RNA formation (22,23).
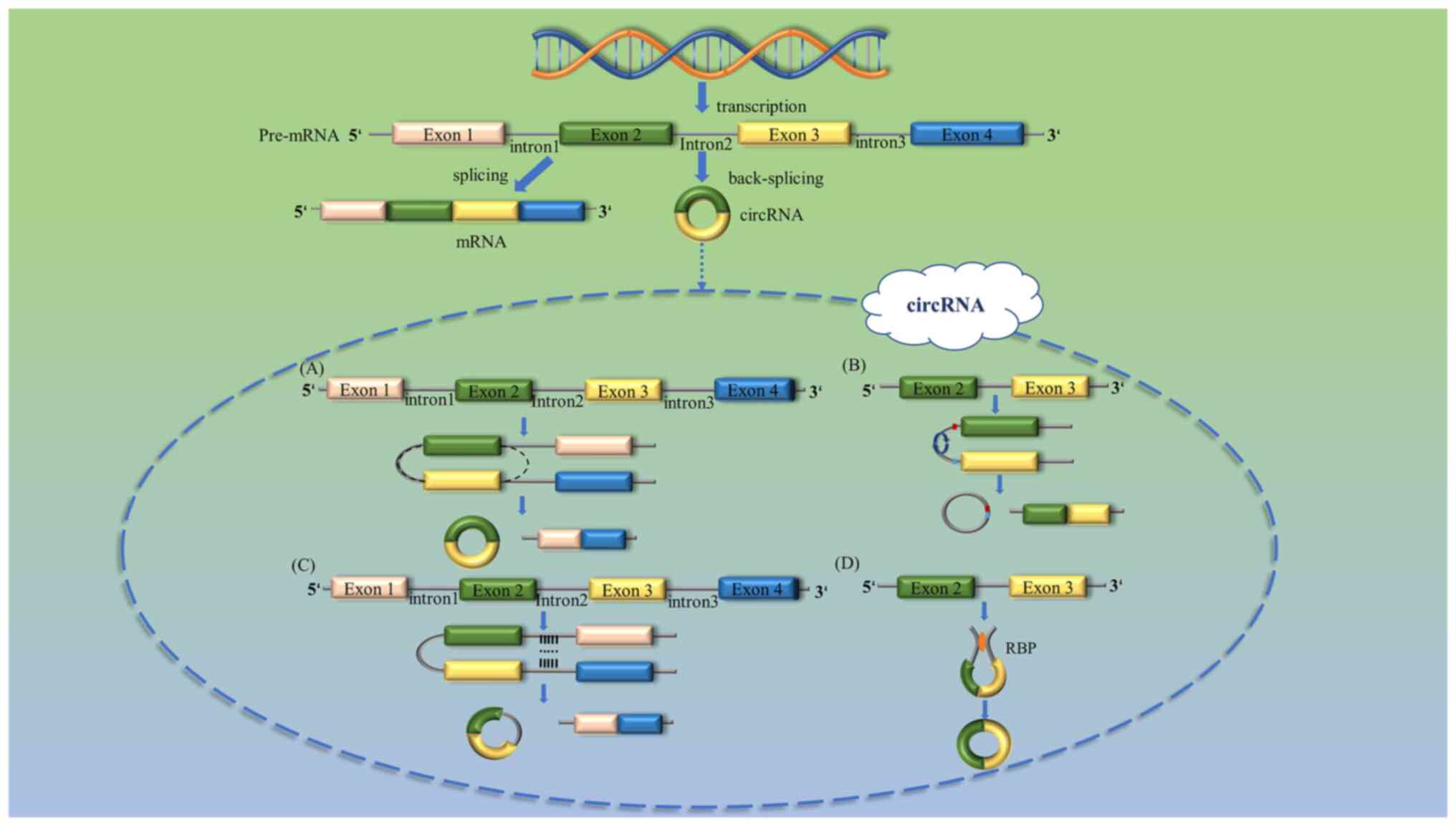
According to the method of splicing, circRNAs may be
classified into four different types: Exonic circRNAs (Fig. 1A), intergenic circRNAs (Fig. 1B), intronic circRNAs (Fig. 1C) and exonic-intron circRNAs
(Fig. 1D) (24,25).
circRNA biogenesis is also regulated by trans-acting factors, such
as RBPs (26). circRNAs are stable
and specifically expressed both intracellularly and in
extracellular fluid (27).
Although circRNAs are generally expressed at low levels (27), increasing evidence has demonstrated
that circRNAs are involved in EC (28–30).
circRNAs carry multiple microRNA (miRNA/miR) binding sites, and
regulate the activity of target miRNAs through their competitive
binding, thereby inhibiting the transcription of downstream
products (31). miRNAs are small
non-coding single-stranded RNAs that bind to the 3′-untranslated
region of mRNAs, thereby inducing mRNA degradation or inhibiting
protein translation (32,33), and thus, the functions of miRNAs
are closely associated with tumor cell development, proliferation,
metastasis, invasion and amino acid transport (34,35).
Targeted therapies that affect tumor cell proliferation, signal
transduction, apoptosis, receptor activation and epigenetic
modification offer major breakthroughs in terms of the treatment of
human malignant tumors (36,37).
For example, as a molecular target of neural tumors, the
Bombesin-receptor family can achieve good therapeutic effect
(38), and as a molecular target
of ovarian cancer, AKT is essential for its treatment (37). circRNAs are becoming increasingly
important in this field due to their relevance in the progression
of malignant tumors (39).
Biological functions of circRNAs
circRNAs act as sponges of miRNAs
miRNAs fulfill an important role in EC development.
circRNAs act as natural ‘sponges’ for miRNA, thereby further
affecting the functions of miRNAs (40). In 2013, Memczak et al
(41) and Hansen et al
(42) demonstrated that antisense
circRNAs derived from cerebellum degeneration-related antigen 1
transcript (CDR1as) were able to act as miRNA sponges for mir-7 to
regulate midbrain development in zebrafish. They also found that
these circRNAs were located in the cytoplasm, and formed
ribonucleoprotein complexes with miR-7 and argonaute proteins
(41,42). These circRNAs contain >70
conserved binding sites for miR-7, enabling them to effectively
inhibit its activity as a molecular sponge (41,42).
Two circRNAs, CDR1as (ciRS-7) and circSry, were found to bind to,
and possibly regulate, specific miRNAs (41,42).
A growing body of evidence suggests that circRNAs may act as
sponges of miRNAs by regulating transcription (43), translation or epigenetic changes of
target genes by inhibiting miRNAs by enveloping the binding sites
of RBPs (44). They have been
demonstrated to be involved in the regulation of various biological
processes and participate in the occurrence and development of
different types of malignant tumor by enriching various forms of
epigenetic modification (45–47).
For example, circRNAs modulate biological processes such as
invasion, proliferation and metastasis through methylation in
bladder cancer (47) and gastric
cancer (48). The potential use of
circRNAs as therapeutic targets and biomarkers for human cancer
types has been previously highlighted (14,30).
circRNAs and RBPs
RBPs are a class of proteins that are widely
involved in gene transcription and translation (44). circRNAs partly exert their
functions via interaction with RBPs in processes such as target
gene genesis, translation, transcriptional regulation and
extracellular transport (44).
circRNAs are able to bind to different RBPs to form specific
circRNPs (44), which are
complexes formed by circRNAs and RBPs (44). The biological function of circRNAs
can be regulated by RBPs through cycles driven by RBPs (21). Certain circRNAs may function as
mRNAs, since they contain a large number of m6A modifications that
are sufficient to drive protein translation in a catabolite
activator protein-independent manner, as opposed to linear mRNAs,
which initiate translation by ordinary ribosomal scanning (49). It has been demonstrated that RBPs
are also involved in the translation of circRNAs (44). Exosomes can act as circRNA-loaded
carriers to transfer genetic information between cells, and
transport circRNAs into the extracellular environment (50). Various types of RBPs, including RNA
binding motif protein 5, RNA binding motif protein 6 and putative
RNA exonuclease NEF-sp, have been reported to act as intracellular
inducers loaded with circRNAs in receptor exosomes to promote
circRNA propagation in the mother cells (51). In order to explore the relationship
between RBPs and circRNAs in more detail, it would be helpful to
further explore the role of circRNAs in the pathophysiology of
malignant tumors.
Translation of circRNAs
In 1995, researchers found that circRNAs, when
translated, could yield repeated polypeptide chains based on the
presence of a continuous open reading frame (ORF), but only when
they contained internal ribosomal entry sites (IRESs) (52). Due to the lack of a 5′-cap in
circRNAs, their translation needs to be initiated by a mechanism
that does not rely on a cap structure (53,54).
Recent studies have demonstrated that the translation of circRNAs
can be initiated in two ways (53,54).
The majority of circRNAs contain IRESs that are able to directly
initiate translation (55).
Downstream of the IRES is the ORF, and different circRNAs have
their own unique ORFs, a feature that is also different from mRNA
(55). At present, three different
types of ORFs have been identified in circRNAs (55). The roles of circRNA-encoding
peptides and proteins depend on the characteristics of overlapping
amino acid sequences (56). The
first ORF starts with the mRNA initiation codon ATG and ends with
the termination codon TGA (57). A
special sequence in this ORF is formed by a series of 5′-ends and
3′-ends in the reverse splicing that occurs after circRNA splicing
(57). When ribosomes translate
this sequence, a novel peptide is generated (57). The second ORF generates a new
sequence, ‘UGAUGA’ or ‘UAAUGA’, from the 5′-ends and 3′-ends. This
is called the overlapping start-stop codon, which encodes a new
polypeptide when the ribosome reads the initiation codon AUG
(56). The third ORF begins at the
initiation codon (AUG) of the mRNA and is similar to the first ORF,
with the exception that the final termination codon is formed by
reverse splicing (58). Infinitely
translatable circRNAs have also been created, with an infinite ORF,
and no IRES, stop codon (59),
5′-cap or 3-poly(A) tail, but with the same ribosomal bound
Shine-Dalgarno sequence and the AUG start codon (60).
circRNAs regulate parental gene
transcription
Since circRNAs are regarded as splicing isomers,
gene transcription may be regulated at various regulatory levels
via translation into functional proteins, or by reducing the number
of canonical splicing transcripts that can be translated into
functional proteins (61).
Muscleblind (MBL) is a post-splicing trans-splicing element, which
is important in tissue-specific alternative splicing (61). The binding of splicing factor MBL
protein to MBL motif located in the flanking intron may promote the
formation of circRNAs via the MBL gene (61). A previous study revealed that
certain exon loops that retain intron segments between exons were
located mainly within the nucleus, and they regulated their
parental genes through cis-modulation via specific RNA-RNA
interactions (62). Several
intron-retained circRNAs, such as circNCOR1, have been demonstrated
to be associated with human RNA polymerase II and are localized
within the nucleus, suggesting that they may regulate gene
expression (62). Further research
has indicated that these circRNAs may interact with U1 small
nuclear ribonucleoprotein, thereby upregulating the expression of
their parental genes (62).
Although, during splicing, circRNAs and their corresponding
colinear forms may compete with each other during biogenesis
(19), the expression of circRNAs
and mRNAs may be promoted simultaneously. circRNAs containing the
initiation codon AUG may affect the expression of linear genes,
whereas the linear transcripts used to splice circRNAs are forced
to use another initiation codon for translation, resulting in a
negative association between the transcription of certain circRNAs
and their parent mRNAs (63),
suggesting that circRNAs may inhibit the expression of their parent
genes. This process also serves an important role in the occurrence
and treatment of tumors. For example, circRNAs can regulate the
transcription of parental genes through RNA-RNA interaction with U1
snRNA (62).
Differential expression of circRNAs in
EC
With the progression of the second generation of
high-throughput sequencing techniques, an ever-increasing number of
circRNAs are being identified (52). Accumulating evidence has indicated
the differential expression and diverse functions of circRNAs in EC
(52,64,65).
As shown in Table I, the
upregulation and downregulation of circRNAs have been shown to
participate in the pathological processes of EC, promoting or
inhibiting EC.
 | Table I.Dysregulated circRNAs in EC. |
Table I.
Dysregulated circRNAs in EC.
| First author,
year | Gene symbol
(circBase ID) | Function in EC | Expression | Downstream
genes | PMID | (Refs.) |
|---|
| Liu et al,
2020 | circIFT80
(hsa_circ_0067835) | Promotes EC cell
proliferation, migration and invasion | Upregulated |
miR-324-5p/HMGA1 | 33169939 | (71) |
| Liu et al,
2020 | circWHSC1 | Promotes the
proliferation, migration and invasion of EC cells, and decreases
apoptosis | Upregulated | miR-646/NPM1 | 32378344 | (28) |
| Hu et al,
2021 | circSLC6A6 | Promotes EC cell
proliferation, migration and invasion | Upregulated | miR-497-5p | 34258297 | (29) |
| Liu et al,
2022 | circATP2C1
(hsa_circ_0005797) | Promotes EC cell
proliferation and invasion | Upregulated | miR-298 | 34852711 | (30) |
| Fang et al,
2021 | circPOLA2 | Promotes cancer
cell proliferation | Upregulated | miR-31 | 34539866 | (70) |
| Shu et al,
2021 | circZNF124 | Promotes cell
proliferation, leucine uptake, migration and invasion in EC
cells | Upregulated |
miR-199b-5p/SLC7A5 | 34145797 | (67) |
| Yang et al,
2021 | circATAD1 | Suppresses EC cell
invasion and migration | Downregulated | miR-10a | 34406448 | (74) |
| Wang et al,
2020 | circWDR26
(hsa_circ_0002577) | Promotes the
proliferation, invasion and metastasis of EC cells | Upregulated | miR-625-5P | 32847606 | (76) |
| Li et al,
2021; | circCORO1C | Inhibits
angiogenesis, proliferation, | Downregulated | / | 34534547 | (78) |
| Lee et al,
2016 |
(hsa_circ_0000437) | migration and
differentiation of EC cells |
|
|
|
|
| Jia et al,
2020 | circESYT2
(hsa_circ_0001776) | Decreases cell
proliferation and glycolysis, and enhances cell apoptosis | Downregulated | miR-182/LRIG2 | 32863771 | (69) |
| Yuan et al,
2021 | circZCCHC7
(hsa_circ_0001860) | Reduces EC
resistance to MPA | Downregulated | miR-520h/Smad7 | 34912799 | (85) |
| Gu et al,
2021 | circTNFRSF21
(hsa_circ_0001610) | Promotes radiation
resistance of EC cells | Upregulated | miR-139-5p | 34462422 | (89) |
| Shi et al,
2020 | circZNF700
(hsa_circ_0109046) | Promotes the
proliferation, migration, invasion and epithelial-mesenchymal
transition of EC cells | Upregulated | miR-136/HMGA2 | 33173333 | (92) |
| Zong et al,
2020 | circPUM1
(hsa_circ_0000043) | Promotes EC cell
proliferation, migration and invasion, and reduces apoptosis | Upregulated |
miR-1271-5p/CTNND1 | 33128584 | (93) |
| Liu et al,
2020 | circTNFRSF21
(hsa_circ_0001610) | Promotes cell
apoptosis and inhibits cell proliferation | Upregulated |
miR-1227-MAPK13/ATF2 | 32299063 | (95) |
| Wu et al,
2021 | circFAT1 | Increases cell
stemness | Upregulated | miR-21 | 34314629 | (96) |
| Shi et al,
2022 | circESRP1 | Promotes EC cell
proliferation, migration, invasion and tumor growth | Upregulated | miR-874-3p | 35317822 | (97) |
Through high-throughput sequencing analysis, Ye
et al (64) analyzed the
differential expression of circRNAs in two grade (G)3 EC samples
and adjacent non-cancerous endometrial tissues, and revealed that
the levels of a total of 62,167 circRNAs were altered in G3 EC and
para-carcinoma endometrial tissues. Among these, the levels of
25,735 circRNAs were elevated, whereas those of 36,432 were lowered
(64).
Chen et al (65) found that, compared with in normal
endometrial tissue, the total expression of circRNAs in EC tissues
was lower, and mainly derived from exon transcription. By comparing
six EC tissues and normal endometrial tissues, they found that 120
circRNAs were differentially expressed. Among the 120
differentially expressed circRNAs, a total of 22 circRNAs were
upregulated in EC, whereas 98 were downregulated. Among the 120
differentially expressed circRNAs, 75 circRNAs were consistently
expressed in EC and normal endometrial tissues, that is, they were
either all upregulated or all downregulated in EC compared with
normal endometrial tissues. The other 45 circRNAs were
inconsistently expressed in all six EC samples.
In a clinical study, Xu et al (66) collected serum samples from 10
patients with EC and 10 matched (location-, age- and sex-matched)
healthy patients. A total of 275 circRNAs were found to be
differentially expressed in EC, including 209 circRNAs that were
upregulated [the first two (hsa_circ_0002577 and hsa_circ_0109046)
were screened from all the expression profiles]. Among the 66
circRNAs that were downregulated, the first two (hsa_circ_0109271
and hsa_circ_0098110) were screened from all expression
profiles.
circRNAs in EC progression
The development of EC is a complex process, and cell
proliferation, invasion and metastasis, resistance to treatment
with drugs and radiation, glycolysis and angiogenesis are all
involved in EC (67). Numerous
tumor phenotypes, including cell proliferation, invasion and
metastasis, resistance to treatment with drugs and radiation,
glycolysis, and angiogenesis, are regulated by circRNAs (67–69).
circRNAs regulate cell proliferation
and motility in EC
Proliferation, invasion and metastasis are the basic
features of malignant tumors, involving a series of complex
multi-step interactions among tumor cells and the host (70). For example, bladder cancer, gastric
cancer (47,48) and EC all have the potential of
proliferation, invasion and metastasis (70,71).
circRNAs are closely involved in the regulation of
cell proliferation via multiple pathways. At present, several
studies have demonstrated that circRNAs adsorb miRNAs through
numerous pathways and fulfill a role in EC development (28,29,71).
For example, circIFT80 [circBase ID (http://www.circbase.org/), hsa_circ_0067835],
circWHSC1, circSLC6A6, circATP2C1 (circBase ID, hsa_circ_0005797)
and circPOLA2 participate in the invasion, proliferation and
metastasis of EC through the adsorption of miR-324-5p (71), miR-646 (28), miR-497-5p (29), miR-298 (30) and miR-31 (70), respectively. Notably, it has been
reported that activation of these pathways depends on exogenous
amino acids for energy (72).
Shu et al (67) reported that knockout of circZNF124
resulted in a marked reduction in the number of EC cells, decreased
cell viability and colony formation, and decreased rates of
invasion and leucine uptake. In EC, circZNF124 was demonstrated to
be directly associated with miR-199b-5p, and miR-199b-5p was
negatively regulated by circZNF124 (67). Solute carrier family 7 member 5
(SLC7A5), also known as L-type amino acid transporter 1, belongs to
the family of L system transporters that transport amino acids,
such as leucine and valine, into cells (73). SLC7A5 promotes cell proliferation,
metastasis and leucine uptake, and its overexpression can reverse
circZNF124 silencing-mediated EC progression (67). These results indicate that
circZNF124 may regulate the proliferation, migration and invasion
of EC cells and their uptake of leucine via the miR-199b-5p/SLC7A5
signaling pathway, thereby promoting the development of EC
(67).
Yang et al (74) analyzed the effects of circATAD1
overexpression on miR-10a expression and methylation through
reverse transcription-quantitative PCR and methylation-specific
PCR. Their study indicated that overexpression of circATAD1 led to
a reduction in miR-10a expression and an increase in the
methylation of the miR-10a gene, which indicated that circATAD1 may
downregulate miR-10a through methylation to inhibit the invasion
and migration of EC cells.
circRNAs are involved in the
angiogenesis of EC
The formation of new capillaries in the existing
vascular network, both into and inside the tumor, is essential for
the growth and metastasis of EC (75). Therefore, inhibition of tumor
angiogenesis is a strategy that may be employed to inhibit EC
growth.
Previous studies have demonstrated that the
proliferative ability of EC cells was enhanced following the
upregulation of the expression of circWDR26 (circBase ID,
hsa_circ_0002577), whereas knocking down its expression effectively
inhibited the proliferation, invasion and metastasis of EC cells
(76,77). Another key to tumor development is
angiogenesis, the creation of a network of new capillaries that
carry oxygen and nutrients needed for tumor tissue to grow
(75). circWDR26 may act as a
miRNA sponge to competitively bind with mir-625-5p and regulate its
expression and function in cells (76). Insulin-like growth factor 1
receptor (IGF1R) has been identified as a downstream target of
miR-625-5P in EC cells (77).
IGF1R is a transmembrane tyrosine kinase receptor involved in a
variety of intracellular signaling pathways associated with tumors,
including the PI3K/Akt signaling pathway (77). Studies have demonstrated that
circWDR26 is able to upregulate IGF1R, and activate the PI3K/Akt
signaling pathway (77) by
competitively binding miR-625-5p through sponging, thereby
adversely affecting EC progression both in vitro and in
vivo (Fig. 2) (76,77).
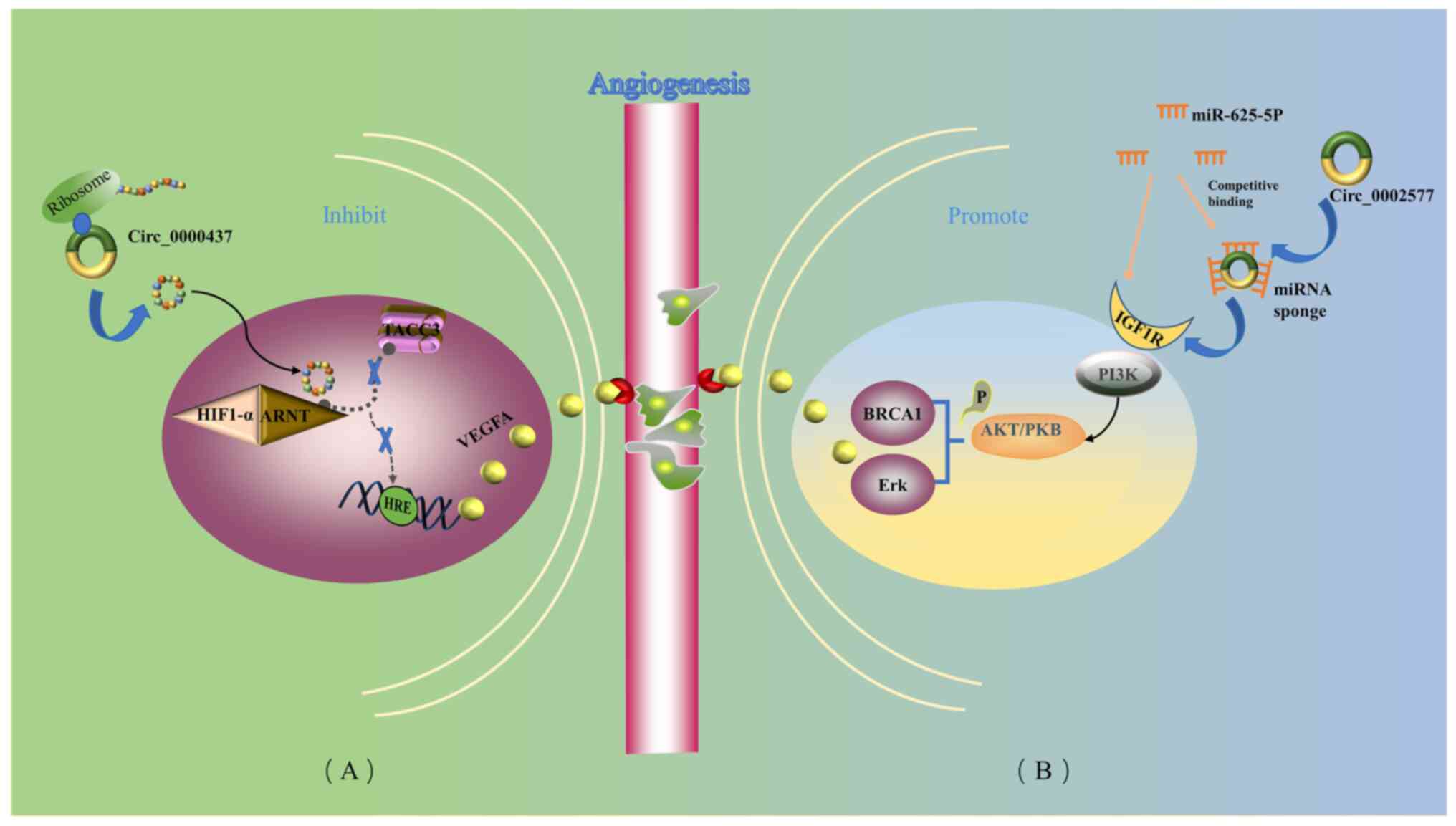 | Figure 2.Formation of new capillaries in the
existing vascular network into and inside the tumor is essential
for the growth and metastasis of EC. circRNAs are involved in
angiogenesis in EC. (A) circ_0000437 reduces VEGFA expression by
blocking the association between ARNT and TACC3, which negatively
regulates tumor angiogenesis and leads to reduced angiogenesis. (B)
circ_002577 competitively binds to miR-625-5p, resulting in the
upregulation of IGF1R and subsequent activation of the PI3K/Akt
signaling pathway, which eventually leads to promotion of the
formation of tumor blood vessels. Thick blue arrows indicate how
circRNAs act. Blue lines indicate the product of the next step.
Black arrows indicate causality. Dashed arrows indicate blocking.
Yellow arrows indicate circRNAs acting as miRNA sponges. ARNT, aryl
hydrocarbon receptor nuclear translocator; circRNA/circ, circular
RNA; EC, endometrial carcinoma; HIF1-α, hypoxia-inducible factor
1-α; HRE, hypoxia response element; IGF1R, insulin-like growth
factor 1 receptor; miRNA/miR, microRNA; P, phosphate group; PKB,
protein kinase B; TACC3, transforming acidic coiled-coil containing
protein 3; VEGFA, vascular endothelial growth factor A. |
Li et al (78) demonstrated that circCORO1C
(circBase ID, hsa_circ_0000437) contains an ORF that encodes a
functional peptide, which competitively binds to aryl hydrocarbon
receptor nuclear translocator and inhibits the coactivatory effect
of transforming acidic coiled-coil containing protein 3 on the
vascular endothelial growth factor (VEGF) promoter, thereby
suppressing the expression of VEGF (Fig. 2). CORO1C-47aa, which serves a role
in the initiation of angiogenesis, not only inhibits angiogenesis,
but also inhibits the proliferation, migration and differentiation
of endothelial cells (78).
circCORO1C expression was found to be downregulated in EC tissues
compared with matched paracancerous non-cancer tissues (68). circCORO1C may serve an
anti-angiogenesis role, and it is anticipated that it may have
potential use in the treatment of EC (68).
circRNAs are involved in the
glycolysis of EC
EC cells mainly obtain their energy through
glycolysis to promote their proliferation (79). Therefore, inhibition of glycolysis
is a further means through which EC may be treated by restraining
the proliferation of EC cells, and effectively killing them.
Jia et al (69) reported that circESYT2 (circBase ID,
hsa_circ_0001776) expression was downregulated in EC tissues
compared with normal tissues, and in G3 EC tissues compared G1 + G2
EC tissues. Another study reported that the metabolic process of
glycolysis fulfills a key role in the development of human
malignant tumors (80), and the
activation of glycolysis, with a consequent increase in lactic acid
production, has been observed in a variety of different types of
cancer cells, leading to changes in energy metabolism that are
associated with the prognosis of patients with malignant tumors
(81). By detecting the
extracellular acidification rate and lactate 2 production, it is
possible to confirm that circESYT2 can regulate glycolysis in EC
cells (82). Leucine rich repeats
and immunoglobulin like domains 2 (LRIG2) are mainly expressed in
the ovary and uterus and serve an important role in the female
reproductive system (82).
circESYT2 acts as a sponge for miR-182 to regulate LRIG2 expression
and inhibit tumor growth in vivo (69). circESYT2 is also able to promote
cell apoptosis, inhibit cell proliferation and glycolysis, and
inhibit the progression of EC via the miR-182/LRIG2 signaling axis
(69).
circRNAs are involved in drug and
radiation resistance of EC
As aforementioned, surgery combined with
radiotherapy and chemotherapy is the most commonly used treatment
for EC, whereas resistance to drugs and radiation poses a major
difficulty in terms of the treatment of EC (5).
To protect the fertility of young patients with EC,
the preferred drugs for maintenance therapy are medroxyprogesterone
acetate (MPA) and megestrol acetate (83). Although the therapy initially
yields benefits, numerous patients with EC eventually develop
resistance to progesterone; 63% of the patients do not respond when
they receive MPA treatment again (84). In a study by Yuan et al
(85), the differential expression
of circZCCHC7 (circBase ID hsa_circ_0001860) in MPA-sensitive EC
cells and MPA-resistant EC cells and tissues was first verified.
circZCCHC7 was revealed to be downregulated in MPA-resistant EC
cells and tissues. circZCCHC7 downregulation enhanced EC resistance
to MPA via the miR-520h/Smad7 axis. It was subsequently
demonstrated that circZCCHC7 acted as a sponge for miRNAs, acting
as a tumor suppressor and fulfilling an important regulatory role
in MPA-resistant and invasive EC (Fig.
3).
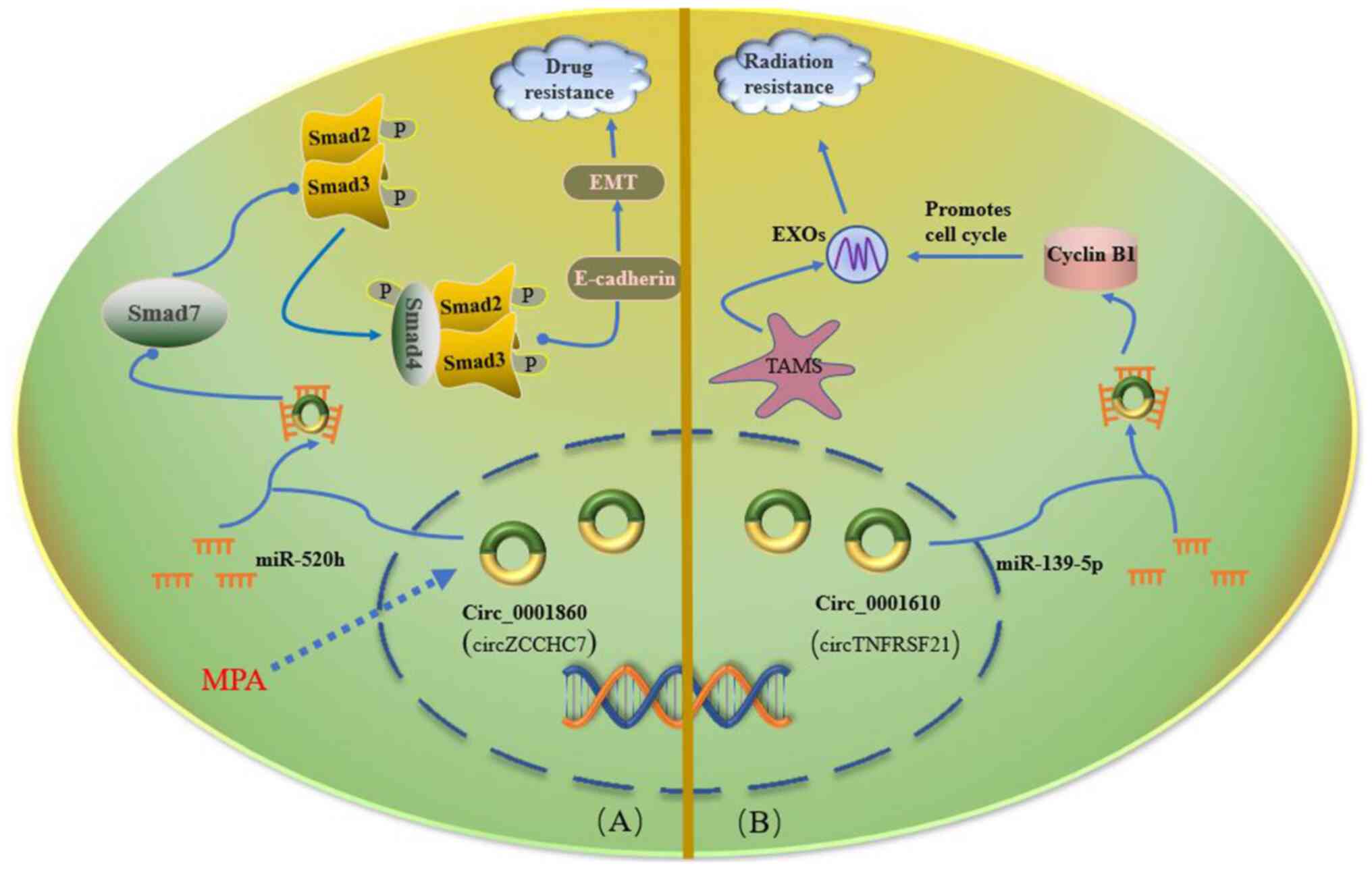 | Figure 3.Surgery combined with radiotherapy
and chemotherapy is the most commonly used treatment for EC, even
though development of resistance to the drugs and radiation is an
obstacle encountered in the treatment of EC. (A) circ_0001860
expression is upregulated in EC cells following MPA treatment, and
circ_0001860 subsequently binds to miR-520h and regulates the
expression and activity of Smad7, thereby inhibiting
phosphorylation of Smad2/3, leading to the formation of the
Smad2/3/4 complex, nuclear translocation of this complex and
enhanced E-cadherin expression. Subsequently, increased E-cadherin
expression can inhibit the progression of EC via EMT. (B)
circ_0001610 can upregulate cyclin B1 expression by endogenously
competing with miR-139-5p binding. Cyclin B1 is a vital promoter of
radiation resistance in EC by regulating the cell cycle. Solid
arrows indicate that the next step is the result of the previous
step. The dashed arrow indicates external intervention. The solid
round head indicates adjustment. Blue lines indicate circRNAs
acting as miRNA sponges. circ, circular RNA; EC, endometrial
carcinoma; EMT, epithelial-mesenchymal transition; EXOs, exosomes;
miR, microRNA; MPA, medroxyprogesterone acetate; P, phosphate
group; TAM, tumor-associated macrophage. |
Radiotherapy is one of the adjuvant treatments for
early-stage EC (86). This therapy
induces apoptosis of the cancer cells by disrupting DNA double
strands and inhibiting cell cycle checkpoint activation (86), thereby leading to improved survival
rates of patients with early EC, and reductions in the local
recurrence of cancer (87).
However, radiation resistance affects the clinical progression of
EC, and can lead to tumor recurrence (88). Compared with normal endometrial
tissue, EC tissue has more evident infiltration of tumor-associated
macrophages (TAMs) (89). There is
evidence that TAMs are able to regulate cancer progression and are
involved in the radiation resistance of EC cells (89). Gu et al (89) reported that circTNFRSF21 (circBase
ID, hsa_circ_0001610) was abundant in M2-polarized
macrophage-derived exosomes, which reduced the radiosensitivity of
EC cells. circTNFRSF21 was also found to promote radiation
resistance of EC cells (Fig.
3).
Other circRNAs in EC
Some researchers (90,91)
have found that the circRNAs heparan sulfate proteoglycan 2 (HSPG2)
and RP11255H23.4 are only expressed in normal endometrial tissues
and are missing in EC tissues (90). They serve an important role in the
pathogenesis of EC through competitive adsorption of corresponding
miRNAs (90). Of the two circRNAs,
HSPG2 is mainly involved in encoding the perlecan protein, a
multidomain heparan sulfate proteoglycan (90), which affects endothelial growth or
regeneration through binding with growth factors on the basement
membrane, thereby inhibiting the progression of EC (91). circZNF700 (hsa_circ_0109046)
promotes EC by acting as a competing endogenous RNA and indirectly
upregulating high mobility group AT-hook 2 expression (92). In addition, downregulation of
circZNF700 has been demonstrated to inhibit the proliferation,
migration, invasion and epithelial-mesenchymal transition (EMT) of
EC cells (92). Furthermore,
circPUM1 serves the role of being a miRNA sponge, as it has been
reported to bind to miR-136, promoting the proliferation, migration
and invasion, and inhibiting the apoptosis, of EC cells (93). Destruction of the structure of
circPUM1 led to a reduction in the tumorigenicity of EC cells in
nude mice (93). A recent genome
sequencing study has revealed that circTNFRSF21 (circBase ID, hsa_
circ_0001610) is upregulated in G3 EC compared with the adjacent
non-cancerous tissues (64). A
previous study on cancer stem-like cells (CSCs) has revealed that
the tumorigenic ability of CSCs is greatly reduced following the
knockdown of MAPK13, suggesting that MAPK13 may serve a role in
tumor progression (94). MAPK13 is
the target of miR-1227 in EC cells (95). Liu et al (95) revealed for the first time that the
novel circRNA circTNFRSF21 could regulate MAPK13 expression via a
competitive interaction with miR-1227, thereby activating the
MAPK13/activating transcription factor 2 signaling pathway and
promoting the formation of EC. circFAT1 has been revealed to be
upregulated in EC, which leads to an increase in the cell stemness
viability of cancer cells via upregulation of miR-21 (96). The cytoplasmic polyadenylation
element-binding protein (CPEB) family is expressed differently in
different types of tumors, thereby exerting an important role in
the occurrence and development of tumors. circESRP1 has been
demonstrated to act as a sponge for miR-874-3p to promote the EMT
process through CPEB4, thereby promoting the proliferation,
invasion and metastasis of EC cells both in vitro and in
vivo, and promoting tumor growth in vivo (97).
Exosomal circRNAs in EC
Extracellular vesicles (EVs) are membranous vesicles
secreted by a variety of cells, facilitating intercellular
communication by transporting intracellular substances (91). Different RNAs can be transferred
between cells through EVs (91,98).
A previous study demonstrated that circRNAs are abundant and stable
in exosomes (91). Secresome is a
key factor in the formation of the tumor microenvironment (98). circRNAs are abundant and expressed
stably in exosomes, and exosomes can be detected in body fluids
(91). In the serum of patients
with EC, the number of upregulated circRNAs has been reported to
exceed the number of downregulated circRNAs (66). circRNAs regulate angiogenesis, drug
resistance, immunity and metabolism via a range of different
mechanisms (66). Tumor
angiogenesis leads to a hypoxic microenvironment, which leads to
acidification of the tumor microenvironment, affecting immune cell
recognition and the response to tumor cells, subsequently leading
to immunosuppression (99). In
addition, tumor cells consume a large amount of glucose, limiting
the use of glucose by T cells, an effect that both weakens the
ability of T cells to kill cancer cells and maintains tumor growth
and metastasis (99). Overall,
circRNAs are stable, conserved and have specific expression
patterns in cells and tissues, suggesting the possibility that
circRNAs may function as molecular diagnostic and prognostic
markers (100,101).
Conclusion and perspectives
It has been widely recognized that circRNAs fulfill
important roles in the development and progression of EC (100,101). EC is a common malignant tumor of
the female reproductive system (102). The incidence of EC has tended to
increase in younger patients between 2006 and 2011 (102). Therefore, it is urgent to find
regulatory factors of EC and to explore novel methods for the
diagnosis, treatment and prognosis of EC. There is increasing
evidence to suggest that circRNAs are closely associated with the
regulation of the biological processes of human cancer, including
EC (16,100,101). At present, circZNF124, circWDR26,
circTNFRSF21, circESYT2 and circCORO1C have all been used as EC
biomarkers (67,76). The present review discusses the
roles of circRNAs in various phenotypes in EC, such as cell
invasion and metastasis, drug and radiation resistance, glycolysis
and angiogenesis, and elaborates the roles of circRNAs in EC in
more detail. To the best of our knowledge, this was the first
review that dealt with this subject. Compared with previous
studies, such as the studies by Shi et al (101) and Takenaka et al (103), the innovation of this review lies
in the first use of the phenotypic classification method to explain
the topic. With the steady progress of RNA technology and its role
in EC, it may be expected that research on the role of circRNAs in
EC will have the potential to undergo great advances in the future.
At present, circRNAs have been found to be abnormally expressed in
numerous types of tumors; however, the abnormal expression of
circRNAs has been reported to vary from disease to disease. Due to
the complexity of tumor pathogenesis, the specific functions of
circRNAs in EC are not yet fully clear, and circRNAs as biomarkers
for EC disease diagnosis and prognosis require further study.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from The National
Natural Science Foundation of China (grant no. 81972522) and
Shenyang Medical College postgraduate Science and Technology
Innovation Fund Project (grant no. Y20210520).
Availability of data and materials
Not applicable.
Authors' contributions
YLi and YW contributed to the conception and design
of the paper. SG and TZ searched relevant literature according to
the topic of this review. The first draft of the manuscript was
written by SG. FM and YLu critically revised the manuscript for
important intellectual content. YW and YLi approved the final list
of included studies. Data authentication is not applicable. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bell DW and Ellenson LH: Molecular
genetics of endometrial carcinoma. Annu Rev Pathol. 14:339–367.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jiang X, Tang H and Chen T: Epidemiology
of gynecologic cancers in China. J Gynecol Oncol. 29:e72018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brooks RA, Fleming GF, Lastra RR, Lee NK,
Moroney JW, Son CH, Tatebe K and Veneris JL: Current
recommendations and recent progress in endometrial cancer. CA
Cancer J Clin. 69:258–279. 2019.PubMed/NCBI
|
|
4
|
Pecorelli S: Revised FIGO staging for
carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol
Obstet. 105:103–104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee YC, Lheureux S and Oza AM: Treatment
strategies for endometrial cancer: Current practice and
perspective. Curr Opin Obstet Gynecol. 29:47–58. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sonoda Y: Surgical treatment for apparent
early stage endometrial cancer. Obstet Gynecol Sci. 57:1–10. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Colombo N, Creutzberg C, Amant F, Bosse T,
González-Martín A, Ledermann J, Marth C, Nout R, Querleu D, Mirza
MR, et al: ESMO-ESGO-ESTRO consensus conference on endometrial
cancer: Diagnosis, treatment and follow-up. Ann Oncol. 27:16–41.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aoki Y, Kanao H, Wang X, Yunokawa M,
Omatsu K, Fusegi A and Takeshima N: Adjuvant treatment of
endometrial cancer today. Jpn J Clin Oncol. 50:753–765. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Suzuki H and Tsukahara T: A view of
pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci.
15:9331–9342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen
W, Jiang B, Qin H, Guo X, Liu M, et al: Circular RNA circSLC8A1
acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer
progression via regulating PTEN. Mol Cancer. 18:1112019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ,
Ma XD, Han K, Chen JW, Judde JG, Deas O, et al:
N6-methyladenosine modification of circNSUN2 facilitates
cytoplasmic export and stabilizes HMGA2 to promote colorectal liver
metastasis. Nat Commun. 10:46952019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang G, Ling Y, Mehrpour M, Saw PE, Liu
Z, Tan W, Tian Z, Zhong W, Lin W, Luo Q, et al:
Autophagy-associated circRNA circCDYL augments autophagy and
promotes breast cancer progression. Mol Cancer. 19:652020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li B, Zhu L, Lu C, Wang C, Wang H, Jin H,
Ma X, Cheng Z, Yu C, Wang S, et al: circNDUFB2 inhibits non-small
cell lung cancer progression via destabilizing IGF2BPs and
activating anti-tumor immunity. Nat Commun. 12:2952021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma
JZ, Sun SH, Yang F and Zhou WP: Circular RNA cSMARCA5 inhibits
growth and metastasis in hepatocellular carcinoma. J Hepatol.
68:1214–1227. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rong Z, Xu J, Shi S, Tan Z, Meng Q, Hua J,
Liu J, Zhang B, Wang W, Yu X and Liang C: Circular RNA in
pancreatic cancer: A novel avenue for the roles of diagnosis and
treatment. Theranostics. 11:2755–2769. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
You X, Vlatkovic I, Babic A, Will T,
Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al:
Neural circular RNAs are derived from synaptic genes and regulated
by development and plasticity. Nat Neurosci. 18:603–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun X, Wang L, Ding J, Wang Y, Wang J,
Zhang X, Che Y, Liu Z, Zhang X, Ye J, et al: Integrative analysis
of Arabidopsis thaliana transcriptomics reveals intuitive splicing
mechanism for circular RNA. FEBS Lett. 590:3510–3516. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lyu D and Huang S: The emerging role and
clinical implication of human exonic circular RNA. RNA Biol.
14:1000–1006. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang F, Nazarali AJ and Ji S: Circular
RNAs as potential biomarkers for cancer diagnosis and therapy. Am J
Cancer Res. 6:1167–1176. 2016.PubMed/NCBI
|
|
25
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lasda E and Parker R: Circular RNAs
co-precipitate with extracellular vesicles: A possible mechanism
for circRNA clearance. PLoS One. 11:e01484072016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu Y, Chen S, Zong ZH, Guan X and Zhao Y:
CircRNA WHSC1 targets the miR-646/NPM1 pathway to promote the
development of endometrial cancer. J Cell Mol Med. 24:6898–6907.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu J, Peng X, Du W, Huang Y, Zhang C and
Zhang X: circSLC6A6 sponges miR-497-5p to promote endometrial
cancer progression via the PI4KB/hedgehog axis. J Immunol Res.
2021:55123912021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Y, Yuan H and He T: Downregulated
circular RNA hsa_circ_0005797 inhibits endometrial cancer by
modulating microRNA-298/catenin delta 1 signaling. Bioengineered.
13:4634–4645. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mitra A, Pfeifer K and Park KS: Circular
RNAs and competing endogenous RNA (ceRNA) networks. Transl Cancer
Res. 7 (Suppl 5):S624–S628. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Quévillon Huberdeau M and Simard MJ: A
guide to microRNA-mediated gene silencing. FEBS J. 286:642–652.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu L, Deng H, Hu J, Huang S, Xiong J and
Deng J: The promising role of miR-296 in human cancer. Pathol Res
Pract. 214:1915–1922. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang X, Zhu X, Yu Y, Zhu W, Jin L, Zhang
X, Li S, Zou P, Xie C and Cui R: Dissecting miRNA signature in
colorectal cancer progression and metastasis. Cancer Lett.
501:66–82. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Padma VV: An overview of targeted cancer
therapy. Biomedicine (Taipei). 5:192015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shariati M and Meric-Bernstam F: Targeting
AKT for cancer therapy. Expert Opin Investig Drugs. 28:977–988.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Moody TW, Lee L, Ramos-Alvarez I,
Iordanskaia T, Mantey SA and Jensen RT: Bombesin receptor family
activation and CNS/neural tumors: Review of evidence supporting
possible role for novel targeted therapy. Front Endocrinol
(Lausanne). 12:7280882021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kristensen LS, Jakobsen T, Hager H and
Kjems J: The emerging roles of circRNAs in cancer and oncology. Nat
Rev Clin Oncol. 19:188–206. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Panda AC: Circular RNAs Act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang
Y, Li X, Wu Z, Yang D, Zhou Y, et al: Circular RNAs in cancer:
Emerging functions in hallmarks, stemness, resistance and roles as
potential biomarkers. Mol Cancer. 18:902019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zang J, Lu D and Xu A: The interaction of
circRNAs and RNA binding proteins: An important part of circRNA
maintenance and function. J Neurosci Res. 98:87–97. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang Y, Mo Y, Gong Z, Yang X, Yang M,
Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, et al: Circular RNAs in
human cancer. Mol Cancer. 16:252017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q,
Wang Q, Xie R, Su Y, Yang M, et al: Circular RNA ACVR2A suppresses
bladder cancer cells proliferation and metastasis through
miR-626/EYA4 axis. Mol Cancer. 18:952019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhu Z, Rong Z, Luo Z, Yu Z, Zhang J, Qiu Z
and Huang C: Circular RNA circNHSL1 promotes gastric cancer
progression through the miR-1306-3p/SIX1/vimentin axis. Mol Cancer.
18:1262019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Weingarten-Gabbay S, Elias-Kirma S, Nir R,
Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A and Segal
E: Comparative genetics. Systematic discovery of cap-independent
translation sequences in human and viral genomes. Science.
351:aad49392016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Janas T, Janas MM, Sapoń K and Janas T:
Mechanisms of RNA loading into exosomes. FEBS Lett. 589:1391–1398.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen CY and Sarnow P: Initiation of
protein synthesis by the eukaryotic translational apparatus on
circular RNAs. Science. 268:415–417. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xia X, Li X, Li F, Wu X, Zhang M, Zhou H,
Huang N, Yang X, Xiao F, Liu D, et al: A novel tumor suppressor
protein encoded by circular AKT3 RNA inhibits glioblastoma
tumorigenicity by competing with active phosphoinositide-dependent
kinase-1. Mol Cancer. 18:1312019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liang WC, Wong CW, Liang PP, Shi M, Cao Y,
Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM and Zhang JF: Translation
of the circular RNA circβ-catenin promotes liver cancer cell growth
through activation of the Wnt pathway. Genome Biol. 20:842019.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hellen CU and Sarnow P: Internal ribosome
entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593–1612.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z,
Xu B, Wu C, Zhou Q, Hu W, Wu C and Jiang J: A novel protein encoded
by a circular RNA circPPP1R12A promotes tumor pathogenesis and
metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer.
18:472019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Abe N, Hiroshima M, Maruyama H, Nakashima
Y, Nakano Y, Matsuda A, Sako Y, Ito Y and Abe H: Rolling circle
amplification in a prokaryotic translation system using small
circular RNA. Angew Chem Int Ed Engl. 52:7004–7008. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Abe N, Matsumoto K, Nishihara M, Nakano Y,
Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y and Abe
H: Rolling circle translation of circular RNA in living human
cells. Sci Rep. 5:164352015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Konieczny P, Stepniak-Konieczna E and
Sobczak K: MBNL proteins and their target RNAs, interaction and
splicing regulation. Nucleic Acids Res. 42:10873–10887. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Smid M, Wilting SM, Uhr K,
Rodríguez-González FG, de Weerd V, Prager-Van der Smissen WJ, van
der Vlugt-Daane M, van Galen A, Nik-Zainal S, Butler A, et al: The
circular RNome of primary breast cancer. Genome Res. 29:356–366.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ye F, Tang QL, Ma F, Cai L, Chen M, Ran
XX, Wang XY and Jiang XF: Analysis of the circular RNA
transcriptome in the grade 3 endometrial cancer. Cancer Manag Res.
11:6215–6227. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen BJ, Byrne FL, Takenaka K, Modesitt
SC, Olzomer EM, Mills JD, Farrell R, Hoehn KL and Janitz M:
Analysis of the circular RNA transcriptome in endometrial cancer.
Oncotarget. 9:5786–5796. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Xu H, Gong Z, Shen Y, Fang Y and Zhong S:
Circular RNA expression in extracellular vesicles isolated from
serum of patients with endometrial cancer. Epigenomics. 10:187–197.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shu L, Peng Y, Zhong L, Feng X, Qiao L and
Yi Y: CircZNF124 regulates cell proliferation, leucine uptake,
migration and invasion by miR-199b-5p/SLC7A5 pathway in endometrial
cancer. Immun Inflamm Dis. 9:1291–1305. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lee II, Maniar K, Lydon JP and Kim JJ: Akt
regulates progesterone receptor B-dependent transcription and
angiogenesis in endometrial cancer cells. Oncogene. 35:5191–5201.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Jia Y, Liu M and Wang S: CircRNA
hsa_circRNA_0001776 inhibits proliferation and promotes apoptosis
in endometrial cancer via downregulating LRIG2 by sponging miR-182.
Cancer Cell Int. 20:4122020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Fang X, Wang J, Chen L and Zhang X:
circRNA circ_POLA2 increases microRNA-31 methylation to promote
endometrial cancer cell proliferation. Oncol Lett. 22:7622021.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liu Y, Chang Y and Cai Y: Circ_0067835
sponges miR-324-5p to induce HMGA1 expression in endometrial
carcinoma cells. J Cell Mol Med. 24:13927–13937. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Hosios AM, Hecht VC, Danai LV, Johnson MO,
Rathmell JC, Steinhauser ML, Manalis SR and Vander Heiden MG: Amino
acids rather than glucose account for the majority of cell mass in
proliferating mammalian cells. Dev Cell. 36:540–549. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang Q and Holst J: L-type amino acid
transport and cancer: Targeting the mTORC1 pathway to inhibit
neoplasia. Am J Cancer Res. 5:1281–1294. 2015.PubMed/NCBI
|
|
74
|
Yang P, Yun K and Zhang R: CircRNA
circ-ATAD1 is downregulated in endometrial cancer and suppresses
cell invasion and migration by downregulating miR-10a through
methylation. Mamm Genome. 32:488–494. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ziyad S and Iruela-Arispe ML: Molecular
mechanisms of tumor angiogenesis. Genes Cancer. 2:1085–1096. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang Y, Yin L and Sun X: CircRNA
hsa_circ_0002577 accelerates endometrial cancer progression through
activating IGF1R/PI3K/Akt pathway. J Exp Clin Cancer Res.
39:1692020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yuan J, Yin Z, Tao K, Wang G and Gao J:
Function of insulin-like growth factor 1 receptor in cancer
resistance to chemotherapy. Oncol Lett. 15:41–47. 2018.PubMed/NCBI
|
|
78
|
Li F, Cai Y, Deng S, Yang L, Liu N, Chang
X, Jing L, Zhou Y and Li H: A peptide CORO1C-47aa encoded by the
circular noncoding RNA circ-0000437 functions as a negative
regulator in endometrium tumor angiogenesis. J Biol Chem.
297:1011822021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wu Q, Zhang W, Liu Y, Huang Y, Wu H and Ma
C: Histone deacetylase 1 facilitates aerobic glycolysis and growth
of endometrial cancer. Oncol Lett. 22:7212021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Helmlinger G, Sckell A, Dellian M, Forbes
NS and Jain RK: Acid production in glycolysis-impaired tumors
provides new insights into tumor metabolism. Clin Cancer Res.
8:1284–1291. 2002.PubMed/NCBI
|
|
82
|
Holmlund C, Nilsson J, Guo D, Starefeldt
A, Golovleva I, Henriksson R and Hedman H: Characterization and
tissue-specific expression of human LRIG2. Gene. 332:35–43. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Rodolakis A, Biliatis I, Morice P, Reed N,
Mangler M, Kesic V and Denschlag D: European society of
gynecological oncology task force for fertility preservation:
clinical recommendations for fertility-sparing management in young
endometrial cancer patients. Int J Gynecol Cancer. 25:1258–1265.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Chen M, Jin Y, Li Y, Bi Y, Shan Y and Pan
L: Oncologic and reproductive outcomes after fertility-sparing
management with oral progestin for women with complex endometrial
hyperplasia and endometrial cancer. Int J Gynaecol Obstet.
132:34–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yuan S, Zheng P, Sun X, Zeng J, Cao W, Gao
W, Wang Y and Wang L: Hsa_Circ_0001860 promotes Smad7 to enhance
MPA resistance in endometrial cancer via miR-520h. Front Cell Dev
Biol. 9:7381892021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Pawlik TM and Keyomarsi K: Role of cell
cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol
Biol Phys. 59:928–942. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Shinde A, Li R, Amini A, Chen YJ, Cristea
M, Dellinger T, Wang W, Wakabayashi M, Beriwal S and Glaser S:
Improved survival with adjuvant brachytherapy in stage IA
endometrial cancer of unfavorable histology. Gynecol Oncol.
151:82–90. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Sorolla MA, Parisi E and Sorolla A:
Determinants of sensitivity to radiotherapy in endometrial cancer.
Cancers (Basel). 12:19062020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Gu X, Shi Y, Dong M, Jiang L, Yang J and
Liu Z: Exosomal transfer of tumor-associated macrophage-derived
hsa_circ_0001610 reduces radiosensitivity in endometrial cancer.
Cell Death Dis. 12:8182021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Hasengaowa, Kodama J, Kusumoto T, Shinyo
Y, Seki N, Nakamura K, Hongo A and Hiramatsu Y: Loss of basement
membrane heparan sulfate expression is associated with tumor
progression in endometrial cancer. Eur J Gynaecol Oncol.
26:403–406. 2005.PubMed/NCBI
|
|
91
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Shi Y, Jia L and Wen H: Circ_0109046
promotes the progression of endometrial cancer via regulating
miR-136/HMGA2 axis. Cancer Manag Res. 12:10993–11003. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zong ZH, Liu Y, Chen S and Zhao Y:
Circ_PUM1 promotes the development of endometrial cancer by
targeting the miR-136/NOTCH3 pathway. J Cell Mol Med. 24:4127–4135.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yasuda K, Hirohashi Y, Kuroda T, Takaya A,
Kubo T, Kanaseki T, Tsukahara T, Hasegawa T, Saito T, Sato N and
Torigoe T: MAPK13 is preferentially expressed in gynecological
cancer stem cells and has a role in the tumor-initiation. Biochem
Biophys Res Commun. 472:643–647. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Liu Y, Chang Y and Cai Y: circTNFRSF21, a
newly identified circular RNA promotes endometrial carcinoma
pathogenesis through regulating miR-1227-MAPK13/ATF2 axis. Aging
(Albany NY). 12:6774–6792. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wu W, Zhou J, Wu Y, Tang X and Zhu W:
Overexpression of circRNA circFAT1 in endometrial cancer cells
increases their stemness by upregulating miR-21 through
methylation. Cancer Biother Radiopharm. Jul 27–2021.(Epub ahead of
print). View Article : Google Scholar
|
|
97
|
Shi R, Zhang W, Zhang J, Yu Z, An L, Zhao
R, Zhou X, Wang Z, Wei S and Wang H: CircESRP1 enhances metastasis
and epithelial-mesenchymal transition in endometrial cancer via the
miR-874-3p/CPEB4 axis. J Transl Med. 20:1392022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Tan S, Xia L, Yi P, Han Y, Tang L, Pan Q,
Tian Y, Rao S, Oyang L, Liang J, et al: Exosomal miRNAs in tumor
microenvironment. J Exp Clin Cancer Res. 39:672020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lyssiotis CA and Kimmelman AC: Metabolic
interactions in the tumor microenvironment. Trends Cell Biol.
27:863–875. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Guo J, Tong J and Zheng J: Circular RNAs:
A promising biomarker for endometrial cancer. Cancer Manag Res.
13:1651–1665. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Shi Y, He R, Yang Y, He Y, Shao K, Zhan
and Wei B: Circular RNAs: Novel biomarkers for cervical, ovarian
and endometrial cancer (Review). Oncol Rep. 44:1787–1798.
2020.PubMed/NCBI
|
|
102
|
Zheng R, Zeng H, Zhang S, Chen T and Chen
W: National estimates of cancer prevalence in China, 2011. Cancer
Lett. 370:33–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Takenaka K, Chen BJ, Modesitt SC, Byrne
FL, Hoehn KL and Janitz M: The emerging role of long non-coding
RNAs in endometrial cancer. Cancer Genet. 209:445–455. 2016.
View Article : Google Scholar : PubMed/NCBI
|