|
1
|
Bell DW and Ellenson LH: Molecular
genetics of endometrial carcinoma. Annu Rev Pathol. 14:339–367.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
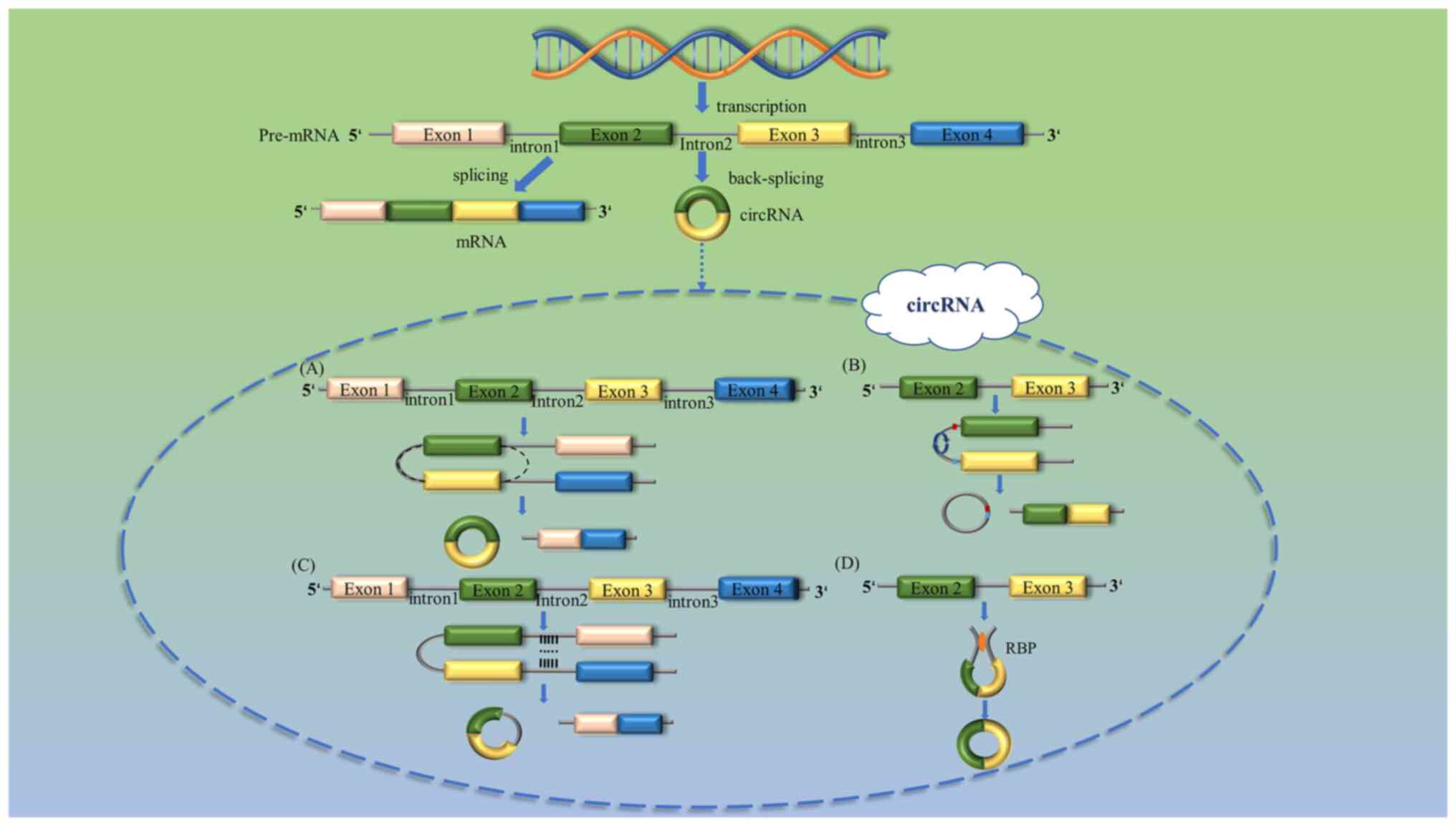
|
Jiang X, Tang H and Chen T: Epidemiology
of gynecologic cancers in China. J Gynecol Oncol. 29:e72018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brooks RA, Fleming GF, Lastra RR, Lee NK,
Moroney JW, Son CH, Tatebe K and Veneris JL: Current
recommendations and recent progress in endometrial cancer. CA
Cancer J Clin. 69:258–279. 2019.PubMed/NCBI
|
|
4
|
Pecorelli S: Revised FIGO staging for
carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol
Obstet. 105:103–104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee YC, Lheureux S and Oza AM: Treatment
strategies for endometrial cancer: Current practice and
perspective. Curr Opin Obstet Gynecol. 29:47–58. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sonoda Y: Surgical treatment for apparent
early stage endometrial cancer. Obstet Gynecol Sci. 57:1–10. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Colombo N, Creutzberg C, Amant F, Bosse T,
González-Martín A, Ledermann J, Marth C, Nout R, Querleu D, Mirza
MR, et al: ESMO-ESGO-ESTRO consensus conference on endometrial
cancer: Diagnosis, treatment and follow-up. Ann Oncol. 27:16–41.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aoki Y, Kanao H, Wang X, Yunokawa M,
Omatsu K, Fusegi A and Takeshima N: Adjuvant treatment of
endometrial cancer today. Jpn J Clin Oncol. 50:753–765. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Suzuki H and Tsukahara T: A view of
pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci.
15:9331–9342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen
W, Jiang B, Qin H, Guo X, Liu M, et al: Circular RNA circSLC8A1
acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer
progression via regulating PTEN. Mol Cancer. 18:1112019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ,
Ma XD, Han K, Chen JW, Judde JG, Deas O, et al:
N6-methyladenosine modification of circNSUN2 facilitates
cytoplasmic export and stabilizes HMGA2 to promote colorectal liver
metastasis. Nat Commun. 10:46952019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang G, Ling Y, Mehrpour M, Saw PE, Liu
Z, Tan W, Tian Z, Zhong W, Lin W, Luo Q, et al:
Autophagy-associated circRNA circCDYL augments autophagy and
promotes breast cancer progression. Mol Cancer. 19:652020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li B, Zhu L, Lu C, Wang C, Wang H, Jin H,
Ma X, Cheng Z, Yu C, Wang S, et al: circNDUFB2 inhibits non-small
cell lung cancer progression via destabilizing IGF2BPs and
activating anti-tumor immunity. Nat Commun. 12:2952021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma
JZ, Sun SH, Yang F and Zhou WP: Circular RNA cSMARCA5 inhibits
growth and metastasis in hepatocellular carcinoma. J Hepatol.
68:1214–1227. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rong Z, Xu J, Shi S, Tan Z, Meng Q, Hua J,
Liu J, Zhang B, Wang W, Yu X and Liang C: Circular RNA in
pancreatic cancer: A novel avenue for the roles of diagnosis and
treatment. Theranostics. 11:2755–2769. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
You X, Vlatkovic I, Babic A, Will T,
Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al:
Neural circular RNAs are derived from synaptic genes and regulated
by development and plasticity. Nat Neurosci. 18:603–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun X, Wang L, Ding J, Wang Y, Wang J,
Zhang X, Che Y, Liu Z, Zhang X, Ye J, et al: Integrative analysis
of Arabidopsis thaliana transcriptomics reveals intuitive splicing
mechanism for circular RNA. FEBS Lett. 590:3510–3516. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lyu D and Huang S: The emerging role and
clinical implication of human exonic circular RNA. RNA Biol.
14:1000–1006. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang F, Nazarali AJ and Ji S: Circular
RNAs as potential biomarkers for cancer diagnosis and therapy. Am J
Cancer Res. 6:1167–1176. 2016.PubMed/NCBI
|
|
25
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lasda E and Parker R: Circular RNAs
co-precipitate with extracellular vesicles: A possible mechanism
for circRNA clearance. PLoS One. 11:e01484072016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu Y, Chen S, Zong ZH, Guan X and Zhao Y:
CircRNA WHSC1 targets the miR-646/NPM1 pathway to promote the
development of endometrial cancer. J Cell Mol Med. 24:6898–6907.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu J, Peng X, Du W, Huang Y, Zhang C and
Zhang X: circSLC6A6 sponges miR-497-5p to promote endometrial
cancer progression via the PI4KB/hedgehog axis. J Immunol Res.
2021:55123912021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Y, Yuan H and He T: Downregulated
circular RNA hsa_circ_0005797 inhibits endometrial cancer by
modulating microRNA-298/catenin delta 1 signaling. Bioengineered.
13:4634–4645. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mitra A, Pfeifer K and Park KS: Circular
RNAs and competing endogenous RNA (ceRNA) networks. Transl Cancer
Res. 7 (Suppl 5):S624–S628. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Quévillon Huberdeau M and Simard MJ: A
guide to microRNA-mediated gene silencing. FEBS J. 286:642–652.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu L, Deng H, Hu J, Huang S, Xiong J and
Deng J: The promising role of miR-296 in human cancer. Pathol Res
Pract. 214:1915–1922. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang X, Zhu X, Yu Y, Zhu W, Jin L, Zhang
X, Li S, Zou P, Xie C and Cui R: Dissecting miRNA signature in
colorectal cancer progression and metastasis. Cancer Lett.
501:66–82. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Padma VV: An overview of targeted cancer
therapy. Biomedicine (Taipei). 5:192015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shariati M and Meric-Bernstam F: Targeting
AKT for cancer therapy. Expert Opin Investig Drugs. 28:977–988.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Moody TW, Lee L, Ramos-Alvarez I,
Iordanskaia T, Mantey SA and Jensen RT: Bombesin receptor family
activation and CNS/neural tumors: Review of evidence supporting
possible role for novel targeted therapy. Front Endocrinol
(Lausanne). 12:7280882021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kristensen LS, Jakobsen T, Hager H and
Kjems J: The emerging roles of circRNAs in cancer and oncology. Nat
Rev Clin Oncol. 19:188–206. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Panda AC: Circular RNAs Act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang
Y, Li X, Wu Z, Yang D, Zhou Y, et al: Circular RNAs in cancer:
Emerging functions in hallmarks, stemness, resistance and roles as
potential biomarkers. Mol Cancer. 18:902019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zang J, Lu D and Xu A: The interaction of
circRNAs and RNA binding proteins: An important part of circRNA
maintenance and function. J Neurosci Res. 98:87–97. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang Y, Mo Y, Gong Z, Yang X, Yang M,
Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, et al: Circular RNAs in
human cancer. Mol Cancer. 16:252017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q,
Wang Q, Xie R, Su Y, Yang M, et al: Circular RNA ACVR2A suppresses
bladder cancer cells proliferation and metastasis through
miR-626/EYA4 axis. Mol Cancer. 18:952019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhu Z, Rong Z, Luo Z, Yu Z, Zhang J, Qiu Z
and Huang C: Circular RNA circNHSL1 promotes gastric cancer
progression through the miR-1306-3p/SIX1/vimentin axis. Mol Cancer.
18:1262019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Weingarten-Gabbay S, Elias-Kirma S, Nir R,
Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A and Segal
E: Comparative genetics. Systematic discovery of cap-independent
translation sequences in human and viral genomes. Science.
351:aad49392016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Janas T, Janas MM, Sapoń K and Janas T:
Mechanisms of RNA loading into exosomes. FEBS Lett. 589:1391–1398.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen CY and Sarnow P: Initiation of
protein synthesis by the eukaryotic translational apparatus on
circular RNAs. Science. 268:415–417. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xia X, Li X, Li F, Wu X, Zhang M, Zhou H,
Huang N, Yang X, Xiao F, Liu D, et al: A novel tumor suppressor
protein encoded by circular AKT3 RNA inhibits glioblastoma
tumorigenicity by competing with active phosphoinositide-dependent
kinase-1. Mol Cancer. 18:1312019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liang WC, Wong CW, Liang PP, Shi M, Cao Y,
Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM and Zhang JF: Translation
of the circular RNA circβ-catenin promotes liver cancer cell growth
through activation of the Wnt pathway. Genome Biol. 20:842019.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hellen CU and Sarnow P: Internal ribosome
entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593–1612.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z,
Xu B, Wu C, Zhou Q, Hu W, Wu C and Jiang J: A novel protein encoded
by a circular RNA circPPP1R12A promotes tumor pathogenesis and
metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer.
18:472019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Abe N, Hiroshima M, Maruyama H, Nakashima
Y, Nakano Y, Matsuda A, Sako Y, Ito Y and Abe H: Rolling circle
amplification in a prokaryotic translation system using small
circular RNA. Angew Chem Int Ed Engl. 52:7004–7008. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Abe N, Matsumoto K, Nishihara M, Nakano Y,
Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y and Abe
H: Rolling circle translation of circular RNA in living human
cells. Sci Rep. 5:164352015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Konieczny P, Stepniak-Konieczna E and
Sobczak K: MBNL proteins and their target RNAs, interaction and
splicing regulation. Nucleic Acids Res. 42:10873–10887. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Smid M, Wilting SM, Uhr K,
Rodríguez-González FG, de Weerd V, Prager-Van der Smissen WJ, van
der Vlugt-Daane M, van Galen A, Nik-Zainal S, Butler A, et al: The
circular RNome of primary breast cancer. Genome Res. 29:356–366.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ye F, Tang QL, Ma F, Cai L, Chen M, Ran
XX, Wang XY and Jiang XF: Analysis of the circular RNA
transcriptome in the grade 3 endometrial cancer. Cancer Manag Res.
11:6215–6227. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen BJ, Byrne FL, Takenaka K, Modesitt
SC, Olzomer EM, Mills JD, Farrell R, Hoehn KL and Janitz M:
Analysis of the circular RNA transcriptome in endometrial cancer.
Oncotarget. 9:5786–5796. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Xu H, Gong Z, Shen Y, Fang Y and Zhong S:
Circular RNA expression in extracellular vesicles isolated from
serum of patients with endometrial cancer. Epigenomics. 10:187–197.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shu L, Peng Y, Zhong L, Feng X, Qiao L and
Yi Y: CircZNF124 regulates cell proliferation, leucine uptake,
migration and invasion by miR-199b-5p/SLC7A5 pathway in endometrial
cancer. Immun Inflamm Dis. 9:1291–1305. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lee II, Maniar K, Lydon JP and Kim JJ: Akt
regulates progesterone receptor B-dependent transcription and
angiogenesis in endometrial cancer cells. Oncogene. 35:5191–5201.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Jia Y, Liu M and Wang S: CircRNA
hsa_circRNA_0001776 inhibits proliferation and promotes apoptosis
in endometrial cancer via downregulating LRIG2 by sponging miR-182.
Cancer Cell Int. 20:4122020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Fang X, Wang J, Chen L and Zhang X:
circRNA circ_POLA2 increases microRNA-31 methylation to promote
endometrial cancer cell proliferation. Oncol Lett. 22:7622021.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liu Y, Chang Y and Cai Y: Circ_0067835
sponges miR-324-5p to induce HMGA1 expression in endometrial
carcinoma cells. J Cell Mol Med. 24:13927–13937. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Hosios AM, Hecht VC, Danai LV, Johnson MO,
Rathmell JC, Steinhauser ML, Manalis SR and Vander Heiden MG: Amino
acids rather than glucose account for the majority of cell mass in
proliferating mammalian cells. Dev Cell. 36:540–549. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang Q and Holst J: L-type amino acid
transport and cancer: Targeting the mTORC1 pathway to inhibit
neoplasia. Am J Cancer Res. 5:1281–1294. 2015.PubMed/NCBI
|
|
74
|
Yang P, Yun K and Zhang R: CircRNA
circ-ATAD1 is downregulated in endometrial cancer and suppresses
cell invasion and migration by downregulating miR-10a through
methylation. Mamm Genome. 32:488–494. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ziyad S and Iruela-Arispe ML: Molecular
mechanisms of tumor angiogenesis. Genes Cancer. 2:1085–1096. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
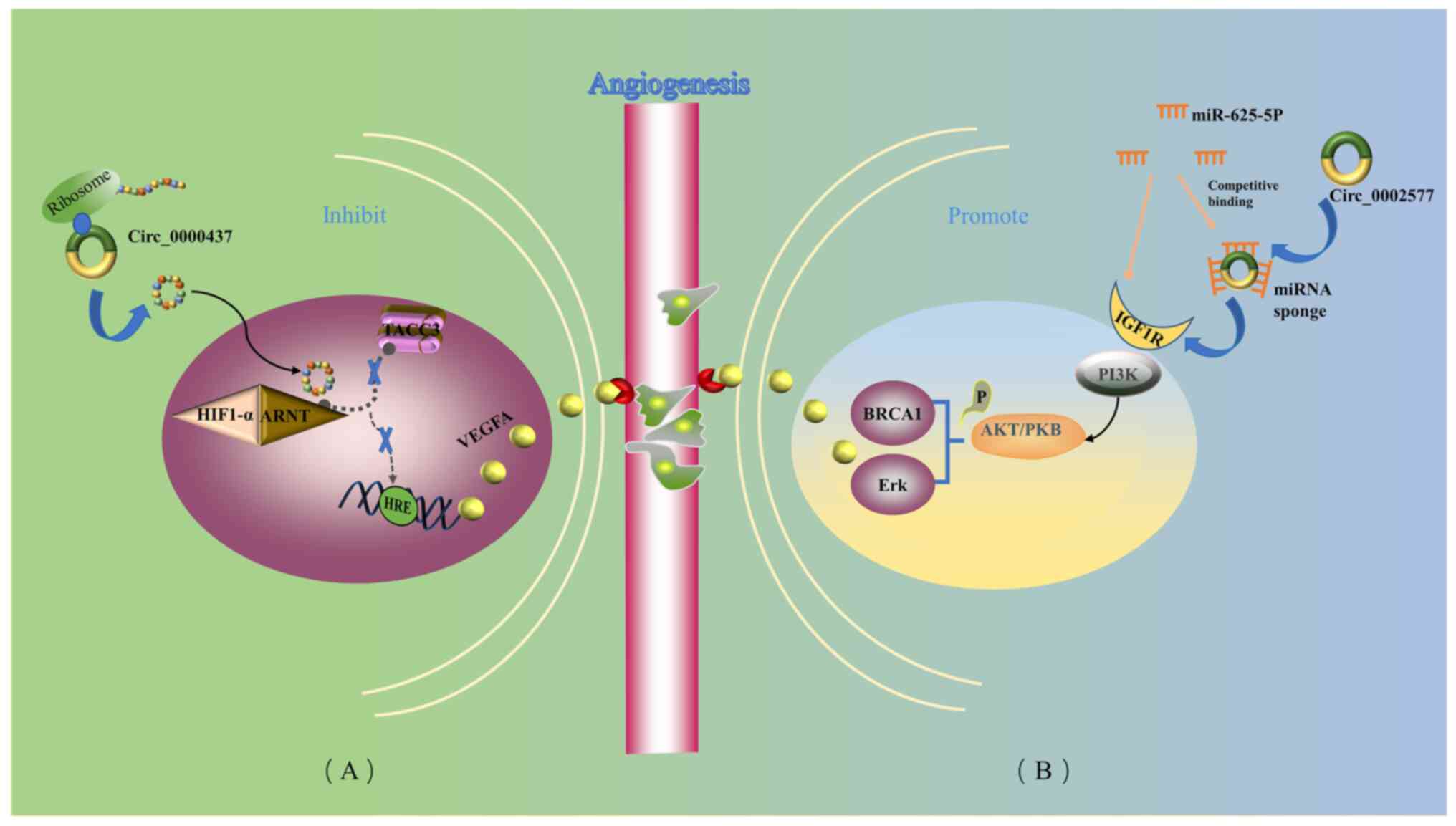
Wang Y, Yin L and Sun X: CircRNA
hsa_circ_0002577 accelerates endometrial cancer progression through
activating IGF1R/PI3K/Akt pathway. J Exp Clin Cancer Res.
39:1692020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yuan J, Yin Z, Tao K, Wang G and Gao J:
Function of insulin-like growth factor 1 receptor in cancer
resistance to chemotherapy. Oncol Lett. 15:41–47. 2018.PubMed/NCBI
|
|
78
|
Li F, Cai Y, Deng S, Yang L, Liu N, Chang
X, Jing L, Zhou Y and Li H: A peptide CORO1C-47aa encoded by the
circular noncoding RNA circ-0000437 functions as a negative
regulator in endometrium tumor angiogenesis. J Biol Chem.
297:1011822021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wu Q, Zhang W, Liu Y, Huang Y, Wu H and Ma
C: Histone deacetylase 1 facilitates aerobic glycolysis and growth
of endometrial cancer. Oncol Lett. 22:7212021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Helmlinger G, Sckell A, Dellian M, Forbes
NS and Jain RK: Acid production in glycolysis-impaired tumors
provides new insights into tumor metabolism. Clin Cancer Res.
8:1284–1291. 2002.PubMed/NCBI
|
|
82
|
Holmlund C, Nilsson J, Guo D, Starefeldt
A, Golovleva I, Henriksson R and Hedman H: Characterization and
tissue-specific expression of human LRIG2. Gene. 332:35–43. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Rodolakis A, Biliatis I, Morice P, Reed N,
Mangler M, Kesic V and Denschlag D: European society of
gynecological oncology task force for fertility preservation:
clinical recommendations for fertility-sparing management in young
endometrial cancer patients. Int J Gynecol Cancer. 25:1258–1265.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Chen M, Jin Y, Li Y, Bi Y, Shan Y and Pan
L: Oncologic and reproductive outcomes after fertility-sparing
management with oral progestin for women with complex endometrial
hyperplasia and endometrial cancer. Int J Gynaecol Obstet.
132:34–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
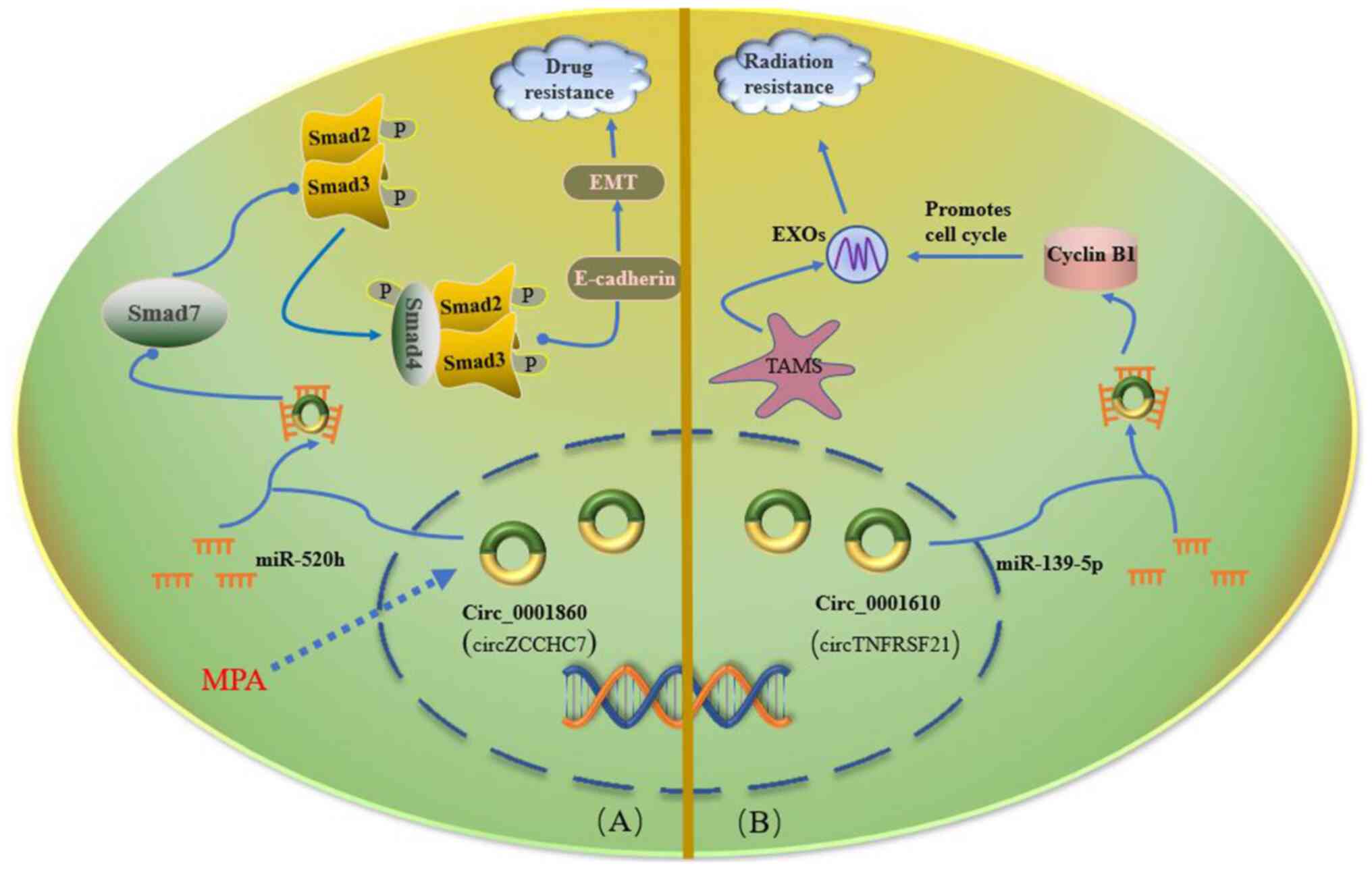
Yuan S, Zheng P, Sun X, Zeng J, Cao W, Gao
W, Wang Y and Wang L: Hsa_Circ_0001860 promotes Smad7 to enhance
MPA resistance in endometrial cancer via miR-520h. Front Cell Dev
Biol. 9:7381892021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Pawlik TM and Keyomarsi K: Role of cell
cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol
Biol Phys. 59:928–942. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Shinde A, Li R, Amini A, Chen YJ, Cristea
M, Dellinger T, Wang W, Wakabayashi M, Beriwal S and Glaser S:
Improved survival with adjuvant brachytherapy in stage IA
endometrial cancer of unfavorable histology. Gynecol Oncol.
151:82–90. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Sorolla MA, Parisi E and Sorolla A:
Determinants of sensitivity to radiotherapy in endometrial cancer.
Cancers (Basel). 12:19062020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Gu X, Shi Y, Dong M, Jiang L, Yang J and
Liu Z: Exosomal transfer of tumor-associated macrophage-derived
hsa_circ_0001610 reduces radiosensitivity in endometrial cancer.
Cell Death Dis. 12:8182021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Hasengaowa, Kodama J, Kusumoto T, Shinyo
Y, Seki N, Nakamura K, Hongo A and Hiramatsu Y: Loss of basement
membrane heparan sulfate expression is associated with tumor
progression in endometrial cancer. Eur J Gynaecol Oncol.
26:403–406. 2005.PubMed/NCBI
|
|
91
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Shi Y, Jia L and Wen H: Circ_0109046
promotes the progression of endometrial cancer via regulating
miR-136/HMGA2 axis. Cancer Manag Res. 12:10993–11003. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zong ZH, Liu Y, Chen S and Zhao Y:
Circ_PUM1 promotes the development of endometrial cancer by
targeting the miR-136/NOTCH3 pathway. J Cell Mol Med. 24:4127–4135.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yasuda K, Hirohashi Y, Kuroda T, Takaya A,
Kubo T, Kanaseki T, Tsukahara T, Hasegawa T, Saito T, Sato N and
Torigoe T: MAPK13 is preferentially expressed in gynecological
cancer stem cells and has a role in the tumor-initiation. Biochem
Biophys Res Commun. 472:643–647. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Liu Y, Chang Y and Cai Y: circTNFRSF21, a
newly identified circular RNA promotes endometrial carcinoma
pathogenesis through regulating miR-1227-MAPK13/ATF2 axis. Aging
(Albany NY). 12:6774–6792. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wu W, Zhou J, Wu Y, Tang X and Zhu W:
Overexpression of circRNA circFAT1 in endometrial cancer cells
increases their stemness by upregulating miR-21 through
methylation. Cancer Biother Radiopharm. Jul 27–2021.(Epub ahead of
print). View Article : Google Scholar
|
|
97
|
Shi R, Zhang W, Zhang J, Yu Z, An L, Zhao
R, Zhou X, Wang Z, Wei S and Wang H: CircESRP1 enhances metastasis
and epithelial-mesenchymal transition in endometrial cancer via the
miR-874-3p/CPEB4 axis. J Transl Med. 20:1392022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Tan S, Xia L, Yi P, Han Y, Tang L, Pan Q,
Tian Y, Rao S, Oyang L, Liang J, et al: Exosomal miRNAs in tumor
microenvironment. J Exp Clin Cancer Res. 39:672020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lyssiotis CA and Kimmelman AC: Metabolic
interactions in the tumor microenvironment. Trends Cell Biol.
27:863–875. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Guo J, Tong J and Zheng J: Circular RNAs:
A promising biomarker for endometrial cancer. Cancer Manag Res.
13:1651–1665. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Shi Y, He R, Yang Y, He Y, Shao K, Zhan
and Wei B: Circular RNAs: Novel biomarkers for cervical, ovarian
and endometrial cancer (Review). Oncol Rep. 44:1787–1798.
2020.PubMed/NCBI
|
|
102
|
Zheng R, Zeng H, Zhang S, Chen T and Chen
W: National estimates of cancer prevalence in China, 2011. Cancer
Lett. 370:33–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Takenaka K, Chen BJ, Modesitt SC, Byrne
FL, Hoehn KL and Janitz M: The emerging role of long non-coding
RNAs in endometrial cancer. Cancer Genet. 209:445–455. 2016.
View Article : Google Scholar : PubMed/NCBI
|