Physical external factors (ultraviolet, ionizing
radiation), chemical drugs or poisons [benzo(a)pyrene, alkylating
agents, platinum compounds, psoralens], as well as endogenous
byproducts (metabolites, free radicals) result in numerous forms of
DNA damage in cells (1). The
primary pathways for DNA damage repair (DDR) include mismatch
repair, nucleotide excision repair (NER) and base excision repair
(BER) for single-strand break (SSB) and double-strand break (DSB)
repair mechanisms (2).
Homologous recombination (HR), classical
non-homologous end joining (NHEJ), alternative end joining and
single strand annealing repair are key repair pathways of DSBs
(3). The HR-based repair pathway,
known as gene transformation pathway, is generally considered to be
the sole error-free pathway that maintain DNA integrity and initial
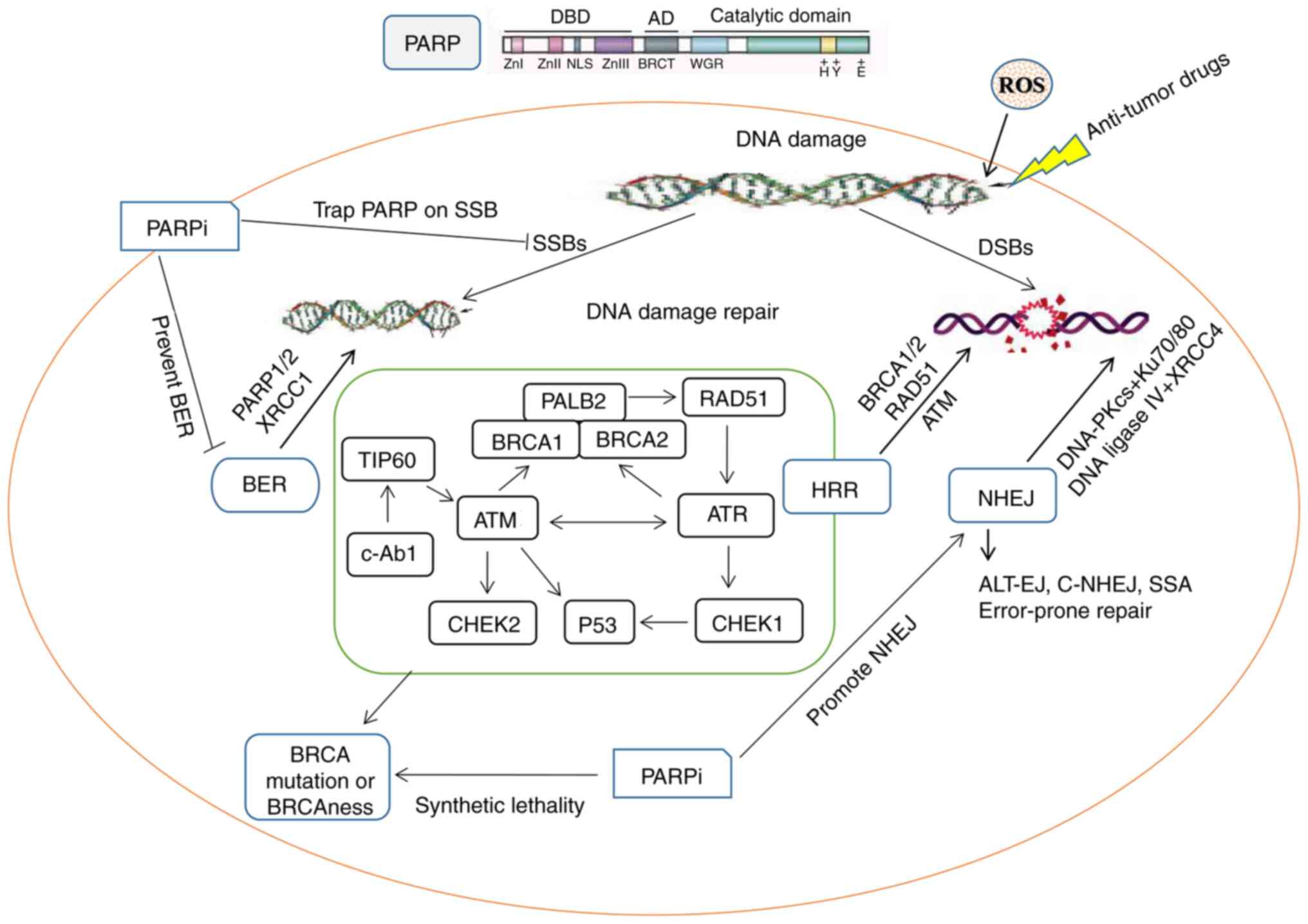
DNA sequence. The poly (ADP-ribose) polymerase (PARP) family
consists of 17 abundant nuclear enzymes that are present in the
majority of eukaryotic cells and promote formation of ADP-ribose
polymer (PAR) (4). DNA damage
recruits and activates PARP-1 and PARP-2, leading to
ADP-ribosylation at multiple sites. PARP-1 mediates several
processes involved in DNA metabolism, such as single-strand damage
repair, NER, DSB repair and regulation of chromatin structure
(5). By identifying endogenous or
exogenous DNA damage, PARP-1 aggregates and bind to the site of DNA
strand breaks to participate in BER. In BER, PARP-1 is catalyzed
and activated by synthesis of PAR polymers by PARylation. A series
of repair proteins, such as X-ray repair cross complementary
combination-1 (XRCC1) and DNA polymerase β, are assembled to repair
the DNA damage sites (6).
PARP inhibitors (PARPis) lead to cell death, notably
in cells with defective DDR function, by trapping PARP-1 on damaged
chromatin (11). Only three of the
17 members of the PARP enzyme family (PARP-1, PARP-2 and PARP-3)
localize to the nucleus in response to early DNA damage and serve a
crucial role in DDR (8). PARP1 has
been implicated in the 10 hallmarks of cancer, while other PARPs
modify certain cancer hallmarks, including PARP2 and PARP5a/5b,
which modify cancer metabolism and replicative immortality,
respectively (12).
Renal cell carcinoma (RCC) is one of the ten most
common types of cancer in developed countries (13) and accounts for 2–3% of all adult
tumors (14). RCC has a 2.2%
incidence and 1.8% mortality rate worldwide (15). RCC is insensitive to traditional
chemotherapy and radiotherapy and readily develops drug resistance;
present treatment methods for localized RCC primarily include
surgery, ablation and surveillance (16). Furthermore, targeted therapies for
metastatic RCC are primarily focused on antiangiogenic therapy,
such as inhibitors sunitinib and pazopanib, which are directed at
the tyrosine kinase domain of vascular endothelial growth factor
receptor (VEGFR) (17). In recent
years, the combination of immunotherapy [programmed cell death
1/programmed cell death ligand (PD-L1) blockade] with tyrosine
kinase inhibitors (TKIs) has increased the overall survival rate
for patients with RCC (18,19).
However, as a type of targeted therapy, the roles of PARPi direct
to DDR in RCC treatment have not been fully explored.
Ongoing studies have been combined PARPi,
antiangiogenic therapy and novel immunomodulators to improve the
outcome of urinary tract tumors (20,21).
In addition, studies have explored the association between DDR and
prostate or bladder cancer progression and treatment (22–24).
To the best of our knowledge, however, there is a lack of
systematic reviews of DDR, PARP and RCC. Although PARPis have been
investigated in prostate cancer treatment, their use in RCC
requires additional investigation and molecular classification
(25). Advanced stages of certain
types of cancer develop resistance against PARPis (26). Therefore, optimization of the use
of PARPis is a challenge. The present review article aimed to
summarize the association between DDR pathway and RCC progression
and treatment.
DDR genes can be useful for predicting progression
and clinical benefits of immunotherapy for ccRCC (27). Multigene tests including DDR gene
provide a more comprehensive risk assessment for patients with
early-onset renal cancer in comparison with the control population
in genome aggregation database (28,29).
DDR gene may predict future prognosis of patients with ccRCC as
well as immunotherapy response (30–32).
Deleterious DDR gene alterations are associated with advanced ccRCC
and may affect outcome of immunotherapy in ccRCC (29).
ccRCC is characterized by chromosomal instability,
which is primarily caused by errors in DDR and affects DSB repair
mechanisms (33). Among DSB repair
mechanisms, NHEJ is an error-prone repair mechanism in which broken
DNA ends are joined compared with HR repair in DNA integrity and
initial DNA sequence (34). Genes
encoding HR proteins include BRCA1, BRCA2, ataxia telangiectasia
mutated (ATM) gene Rad3-associated kinase (ATR), BRCA1-associated
RING domain 1, Bloom's syndrome (BLM) RecQ like helicase and RAD51
recombinase, and HR deficiency is defined by the inability to
repair DNA damage by the normal HR repair pathway (35). Although a single gene mutation
cannot result in cell death, mutations of both genes (i.e., BRCA1/2
and PARP1) lead to cell death and are considered to have a
synthetic lethality effect (26).
Polymerase θ produces synthetic lethal effects with
multiple DNA repair genes, such as BRCA1/2, making it key in
HR-deficient cancer (36).
Although PARP1 binds to DNA damage sites, PARPi may prevent
PARylation, which in turn hinders recruitment of BER enzyme,
impedes DNA repair and leads to DSBs and synthetic lethality in a
HR-deficient mechanism (37). By
contrast, the cytotoxicity of the captured PARP-DNA complexes is
higher than that of the unrepaired SSBs induced by PARP
inactivation, indicating that PARPi can be toxic by trapping the
PARP enzyme on the DNA (38).
PARP1/2 and other repair factors were activated and recruited by
DNA breaks and clinical PARPis are considered to prolong the
presence of PARP1/2 molecules at break sites and chromatin, called
‘trapping’ (39). Furthermore,
PARP mediates DSB and NER damage repair, stability of replication
forks and chromatin regulation of DNA (5).
PARPis, such as olaparib, niraparib, talazoparib and
rucaparib, have been recently approved by the US Food and Drug
Administration for treatment of ovarian and germline BRCA DNA
repair associated mutant breast cancer (40). PARG inhibitors also promote
sensitivity to radiation-induced DNA damage, inhibit the
progression of replication fork and hinder the survival of cancer
cells, further supporting the hypothesis that selective inhibition
of PARG may prevent the survival of cancer cells (41). Therefore, although the action sites
of PARG inhibitors and PARPi are different, PARPi destabilize
replication forks and PARG inhibitors may complement PARPi in
exacerbating the replication deficiencies of tumor cells by causing
DNA damage, hindering DNA repair (42). The roles of PARP1 in DDR and
synthetic lethality are shown in Fig.
1; Briefly, the DNA damage caused by SSBs and DSBs correspond
to different repair pathways, while the simultaneous defects of the
BER and HR repair pathways force transition to NHEJ repair, an
error-prone repair mechanism that leads to cell death (Fig. 1). In addition, the DDR-associated
target molecules are summarized and shown in Table I.
The histone PARylation factor 1 forms a joint active
site with PARP1/2 and enables recognition of the DNA damage site by
PARP1/2. The latter binds to the site of DNA damage and uses
NAD+ to convert receptor protein PAR into the ADP
ribosomal polymer, thus facilitating the separation of PARP1/2 from
the DNA break and subsequent repair (43). Following activation by DNA damage,
PARP participates in genome integrity, tumor formation and stemness
via the PARP1-Krüppel like factor 4 complex, thus regulating the
expression of telomerase in tumor and embryonic stem cells
(44). PARylation is a reversible
post-transcriptional modification that requires PARP1 and PARG. By
activating PARylation of target proteins, PARP regulates numerous
physiological processes, including chromatin remodeling, DNA damage
response, apoptosis and mitosis (45). Inhibition of PARG can also lead to
synthetic lethality along with factors that inhibit DNA
replication, such as checkpoint kinase 1 inhibitors. PARG
inhibitors compensate for the decreased efficacy of PARPis with
inhibition of DNA replication factors [i.e., Checkpoint kinase
1(CHK1) inhibitors] in ovarian cancer treatment (46). In addition to BRCA1/2 mutations,
other HR repair-associated genes or predictive biomarkers are also
currently explored in the early treatment of tumors by adjuvant and
neoadjuvant therapy with PARPis (47).
The rs5751129 polymorphic genotype and mRNA
expression levels of XRCC6 (Ku70) are associated with RCC etiology
and may serve as a marker for increased susceptibility of the
Taiwanese population to RCC (48).
A non-invasive panel comprising circulating tumor cell and urine
cellular polymorphisms of XPC (polymorphic site: rs2228001, A2815C)
and XRCC1 (polymorphic site: rs25487, G1196A) showed high
sensitivity for bladder and prostate cancer and RCC screening
(49). The XRCC1 Arg194 allele and
urinary 8-hydroxy-2 deoxyguanosine levels and total arsenic
concentration are predictive factors for RCC prognosis (50). In addition, dual PARP and RAD51
inhibitor conjugates disrupt resistance mechanisms to olaparib
treatment in breast cancer cells regardless of the mutation status
of BRCA (51). BRCA1-associated
protein 1 (BAP1) encodes a widely expressed deubiquitinase of
histone H2A, resulting in increased sensitivity of bromodomain and
extra-terminal inhibitors to BAP1-deficient cancer (such as
cutaneous and uveal melanoma and ccRCC) (52).
Loss or inactivation of PARP1 delays
starvation-induced autophagy, which plays an important role in
contributing to survival during nutrient starvation conditions to
optimize the usage of limited energy supplies, while autophagy and
PARP1 activation exert a pro-survival effect following nutrient
deprivation (59). In addition to
autophagy and apoptosis, necroptosis caused by tumor necrosis
factor (TNF)-related apoptosis-inducing ligand, which mediates the
receptor-interacting serine/threonine-protein kinase (RIPK)
1/RIPK3-dependent activation of PARP1 pathway, is associated with
liver injury (60). Unlike the
induction of apoptosis, necrosis and other forms of cell death,
parthanatos is a PARP1-dependent and caspase-independent cell-death
pathway (61). During
PARP1-mediated cell death, mitochondrial protein apoptosis-inducing
factor is released and transferred to the nucleus (62). In BRCA wild-type) ovarian cancer,
PARP inhibition promotes ferroptosis by suppressing solute carrier
family 7 member 11 and synergizing with ferroptosis inducers
(63). Antioxidants and the PARP1
inhibitor olaparib rescue the death of RCC cells triggered by
zafirlukast, a cysteinyl leukotriene receptor 1 antagonist,
dependent on that of hypoxia-inducible factor (HIF)-2α (64). Therefore, the understanding of the
association between PARP and forms of cell death as well as its
role in gene stability is key for design of novel chemotherapeutic
drugs for various types of cancer.
Certain PARP family enzymes exhibit poly-catalyzed
enzymatic activity, whereas others are characterized by
mono-catalyzed enzymatic activity (Table II) (65). PARPis competitively bind to the
NAD+ binding sites of the PARP1/2 enzymes and inhibit
PARylation, improving the clinical benefits in BRCA mutant tumors
(Table I). The use of the oral
inhibitor of PARP1/2 olaparib has been approved by FDA. This
compound is primarily used for treatment of patients with ovarian
cancer containing the BRCA1/2 mutation (66). Patients with breast and ovarian
cancer who possess BRCA1/2 mutations or deletions may benefit from
PARP1 and PARP2 inhibitors (67).
Ovarian cancer cells with higher expression of NADP+ are
more sensitive to PARPi (68).
Synthetic lethality enables PARPis to achieve their desired
efficacy in clinical trials (26,69,70).
Presently, the PARPi is used in tumors with BRCA1/2 mutations, such
as those derived from breast and ovarian cancer, and ongoing
research is exploited beyond germline BRCA mutations to identify
suitable biomarkers to predict treatment response (71).
As PARP trapping activity may exceed the inhibitory
potential of PARylation reactions, the clinical exploration
processes for the antitumor activity of PARPis are different
(72). Moreover, the supply domain
of PARP1 binds to the nicotinamide-riboni site of NAD+;
PARPis simulate the nicotinamide structural domain and
competitively bind the Ni binding site of PARP, which leads to
partial binding to the receptor binding site of PARP (73). As NAD+ competitors are
prone to off-target effects, a novel inhibitor of PARP1 that
specifically targets the histone-dependent PARP1 activation pathway
has been developed to overcome the limits of NAD-like PARP1
inhibitors, which exhibit high specificity to PARP1 and a
potentially potent therapeutic effect on urological tumors
(74). In addition, classical
NAD-like PARP1 inhibitors may inhibit the survival of normal kidney
epithelial cells at high concentrations, whereas novel non-NAD-like
PARP1 inhibitors are only active against malignant cells (75). As PARP activity is associated with
various types of cell death, development of PARP activators that
contribute to tumor cell death is being investigated (Table II).
RCCs are classified into three primary
histopathological classifications as follows: ccRCC, pRCC and
chRCC, with a proportion of incidence 70–75%, 10–16% and 5%,
respectively (76). Non-ccRCC
(nccRCC) include the pRCC, chRCC, unclassified, collecting duct,
and translocation carcinoma (77).
As most prevalent subtype of RCC, Approximately 70% of the ccRCC
cases are are linked to the mutation or inactivation of a tumor
suppressor gene, namely von Hippel-Lindau (VHL), resulting in
activation of the HIF/VEGF pathway (78). TKIs targeting VEGFR are key RCC
treatment drugs (18). VEGFR-based
TKIs, such as sunitinib and pazopanib, have been the mainstay of
treatment for patients with advanced RCC. However, RCC cells are
prone to develop drug resistance to VEGFR-TKIs, which limits the
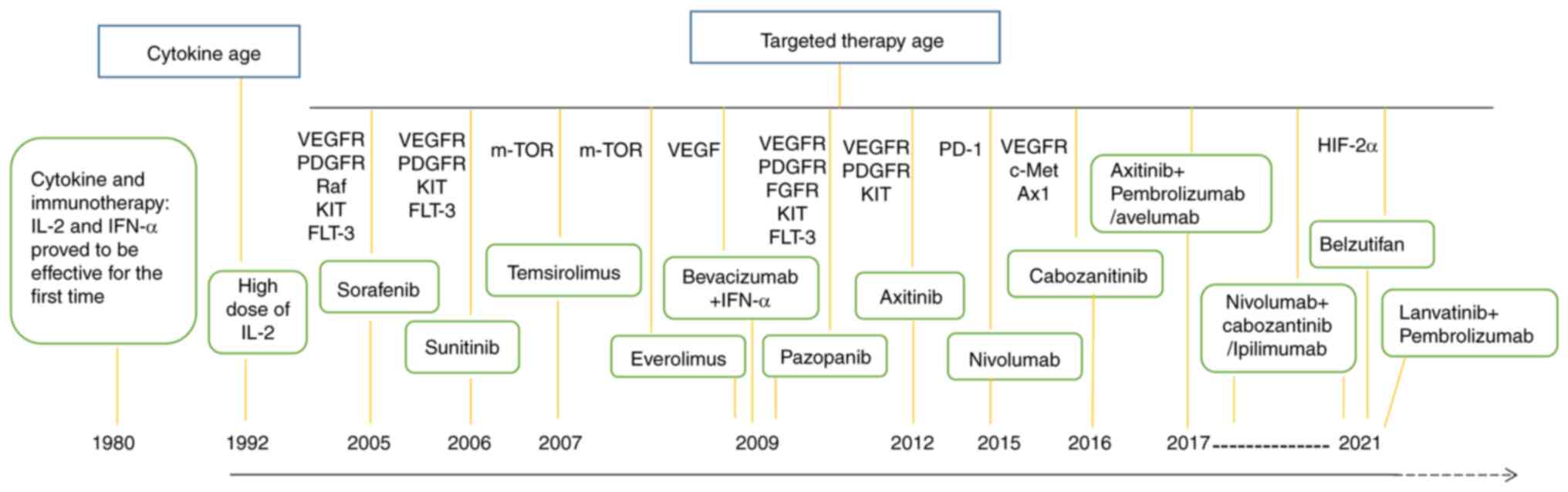
efficacy of targeted therapy (79). Recently, several therapeutic
options have been approved (immunotherapy and immunotherapy/TKIs)
for first-line treatment of metastatic ccRCC, such as axtinib +
pembrolizumab or avelumab, nivolumab + cabozantinib or ipilimumab
and pembrolizumab + lenvatinib (Fig.
2) (19).
To date, various abnormal gene indicators of RCC
have been detected and evaluated. The mutations of polybromo 1
(PBRM1), BRCA1-associated protein 1 and lysine demethylase 5C genes
affect the outcome of targeted therapy with sunitinib in patients
with metastatic ccRCC (80).
Global tumor cell expression profile may be altered by loss of
PBRM1 in ccRCC, thereby influencing the responsiveness to immune
checkpoint therapy (81). The
increase in expression levels of p53 and alteration of targets of
the rapamycin pathway, such as neurofibromin 1 and
phosphatidylinositol-4,5-biphosphate 3-kinase catalytic subunit α,
may be associated with resistance of patients following first-line
VEGF-directed therapy (82).
Cyclin-dependent kinase inhibitor 2A and MET proto-oncogene
receptor tyrosine kinase are the most frequently altered somatic
genes in nccRCC tumors and are treated by cabozantinib (83).
In a patient with chRCC, biallelic TSC complex
subunit 2 mutations have been associated with a notable response to
temsirolimus (84). Atezolizumab
and bevacizumab have been shown to be safe and produce an objective
response in patients with RCC and variant histology or >20%
sarcomatoid differentiation, particularly in patients with
PD-L1-positive tumors (85).
Belzutifan, a HIF-2 inhibitor, has been approved by the FDA for
treatment of patients with VHL syndrome-associated RCC that do not
require immediate surgery (86).
However, in clinical trials, only some ccRCC patients appear to be
benefit from the HIF-2 inhibitors, and using intact p53 pathway
status may be premature to predict sensitivity ccRCC patients to
HIF-2 inhibitors (87). Compared
with normal kidney cells, benzo[4]helicenium shows specific killing
efficiency against RCC, selectively damaging mitochondria and DNA
in RCC cancer cells, providing a potential targeted drug for RCC
precision therapy (88).
Resistance to targeted therapies remains a key
obstacle in clinical treatment of cancer. Sunitinib induces genomic
instability of RCC cells by affecting the interaction of
microtubule-associated protein 1A/1B-light chain 3-II and PARP1
(89). As a PARPi, olaparib
reverses drug resistance to sorafenib in liver cancer (90). A total of 27–32% of RCC tissue
samples have mutations in HR genes, while PARPi agents (such as
niraparib, talazoparib and rucaparib) that target DDR mutations may
be effective treatment options for RCC (91). Due to increased PARP1 expression
and decreased PARG levels, ccRCC is accompanied by increased levels
of poly(ADPribose) (pADPr). The development of ccRCC is associated
with accumulation of pADPr (75).
VHL-deficient RCC is associated with downregulation of DNA repair
induced by hypoxia, conferring increased sensitivity to PARPis
(92). Impaired DNA repair
ability, which is associated with the BRCA1A complex, sensitizes
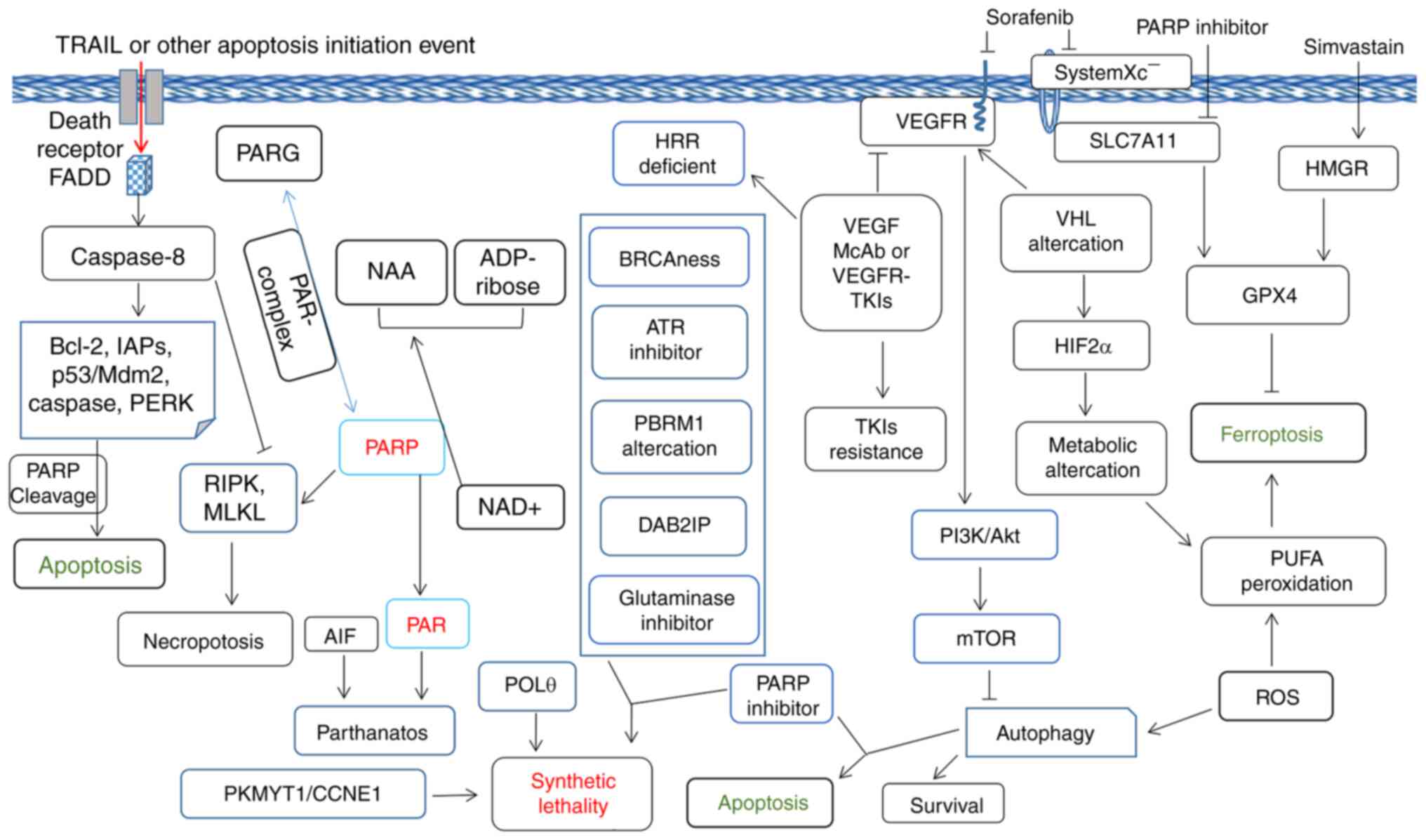
folliculin-deficient RCC cells to olaparib treatment (93). Therefore, it is desirable to
explore the combination effect between VEGFR-TKIs and PARPi in RCC
(Fig. 3). The PARP and PARPi
function in modulation of RCC tumor cell death was shown in
Fig. 3.
In addition to DNA damage, PARPs serve metabolic
regulatory roles and are involved in obesity, which modulates
carbohydrate and lipid metabolism and guides pathologically
metabolic abnormalities (94).
PARPs act as cofactors of nuclear receptors or transcription
factors that are activated in a lipid-responsive manner. PARPs
modulate lipid metabolism and homeostasis, while activation PARP
disrupts lipid metabolism signal (95). As aforementioned, ccRCC is often
associated with VHL mutations or deletions, which affect its
metabolic properties and enhances sensitivity to glutathione
peroxidase 4 inhibitors that induce ferroptosis (96). Glutaminase inhibitors inhibit
pyrimidine synthesis and increase levels of reactive oxygen species
in VHL-deficient RCC cells, leading to DNA replication stress and
suppression of VHL−/− RCC cell proliferation, while
olaparib, a PARPi, inhibits proliferation of these cells by acting
in a synergistic mechanism with glutaminase inhibitors (97). This combined treatment supports the
development of novel treatment strategies that target VHL-deficient
RCC and chemoresistant ovarian cancers (97,98).
By utilizing hypoxia-inducible lipid droplet associated protein,
the VHL/HIF-2α pathway may induce ferroptosis by upregulating lipid
peroxidation levels (99).
The Warburg effect, demonstrated by increased
glycolytic intermediate labeling, decreased pyruvate dehydrogenase
flow and decreased tricarboxylic acid cycle labeling, has been
observed in various primary ccRCC cases (100). By regulating PARP1 expression,
lipid metabolism-associated drugs simvastatin and tanshinone I
inhibit proliferation of melanoma and renal tumour cells (101). Therefore, PARP inhibition may
exert a synergistic inhibitory effect on tumor growth of ccRCC with
other metabolic inhibitory molecules (97). The gene mutations of RCC are
heterozygous both at the primary site and metastatic lesions, but
the distant metastatic lesions may exihibit more significant growth
and invasion phenotype, which may be associated with the function
of the DDR response. Therefore, the primary site, and, particularly
the metastatic lesions, should be considered for the evaluation of
metastatic RCC.
RCC is non-sensitive to chemoradiotherapy; however,
its mechanism remains unknown. The increase of PARP1 in RCC cells
leads to radioresistance and certain PARP1 inhibitors enhance
sensitivity of radiotherapy in human RCC xenograft and head and
neck squamous cell carcinoma cell models (102,103). DAB adaptor protein 2 interactive
protein (DAB2IP) can degrade PARP1 by forming a complex with PARP1
and E3 ligases, causing DAB2IP deficient RCC cells acquire
resistance to ionizing radiation (102). Inhibition of DNA repair by
radiation therapy combined with veliparib accelerates induction of
tumor cell senescence and induces expression of immune stimulators
to activate cytotoxic T lymphocytes and mediate antitumor response
(104). Patients with ccRCC with
high exosome component 1 show poor prognosis; exosome component 1
cleaves single-stranded DNA and sensitizes human ccRCC cells to
PARPis (105).
Increased progression-free survival has been noted
in patients with advanced RCC treated with first-line nivolumab
combined with cabozantinib compared with those treated with
sunitinib (106). PARP1 low
expression protein levels are associated with higher patient
overall survival following treatment with immune checkpoint
inhibitors (ICIs) (107). The
overall survival and progression-free survival of patients with
PBR-mutated 1 (PBRM1) ccRCC treated with nivolumab were
significantly increased in the PARP1-low group compared with that
noted in the PARP1-high expression (107). Inactivation of PBRM1 occurs in
40% of ccRCC cases and contributes to a synthetic lethal effect for
PARP and ATR inhibitors, which provides a basis for evaluating the
efficacy of these inhibitors in treatment of patients with
PBRM1-deficient cancer (108).
DEAD/H-box helicase 11 may be a novel biomarker for patients with
RCC resistance to TKIs and immunotherapy and may predict PARPi
sensitivity in RCC (109).
In collecting duct RCC (CDRCC), expression levels of
baculoviral IAP repeat containing 5, pituitary tumor-transforming
gene 1 regulator of sister chromatid separation, centromere protein
F and cyclin-dependent kinase inhibitor 3 [specific marker genes of
cancer stem cells (CSCs)] are associated with poor, while
inhibitors of PARP, histone deacetylase 2 and fibroblast growth
factor receptor are effective against CSCs and may serve as
potential therapeutic options for CDRCC (110). ATM is a key tumor suppressor gene
found in almost 3% of RCC and is involved in HR repair (Fig. 1). ATM mutation may affect RCC tumor
response to veliparib (a PARPi), which produces months of disease
control, decreased levels of lactate dehydrogenase and improved
performance status, as demonstrated in a case report of pRCC
(111). A case for niraparib to
sorafenib-axitinib-everolimus-resistance metastatic ccRCC with
BAP1-Frame shift mutation has achieved a partial response and
lasted for 5 months (112).
Although has not been formally clinically tested, to the best of
our knowledge, the above cases provide examples for the use of
PARPi in the treatment of certain type of RCC. By producing
fumarate, fumarate hydratase (FH) was defined as DNA repair
required in NHEJ in cells (113).
Inactivation or germline mutations of FH lead to hereditary
leiomyomatosis and RCC (HLRCC), while loss of FH and accumulation
of fumarate lead to decreased G2 checkpoint, which increases the
possibility of endogenous DNA damage (114). Succinate dehydrogenase
(SDH)-related hereditary paraganglioma and pheochromocytoma is
another hereditary cancer syndrome associated with mutations in
SDH. These mutations suppress the HR DNA repair pathway, thus
rendering tumor cells susceptible to synthetic lethality by PARPi
(115). Metabolites associated
with germline mutations of FH and SDH genes (SDHA, SDHB, SDHC and
SDHD) suppress HR repair pathway, conferring sensitivity to PARPis
in vivo experiment or in clinical trials (116,117).
Since PARP is associated with various forms of cell
death, such as apoptosis, autophagy and necroptosis, numerous PARP
activators have been developed that exhibit inhibitory effects on
RCC cells. For example, tumor protein p53-inducible nuclear protein
2 (TP53INP2) activates expression of PARP in patients with ccRCC
and the overexpression of TP53INP2 inhibits ccRCC cell
proliferation, migration and invasion (118). The induction of apoptosis in RCC
cells is accompanied by elevation of reactive oxygen species (ROS)
levels and induction of cleaved-PARP expression (119). PARP activation also plays a key
role in necroptosis induced by glutamate, which is blocked by
necrostatin-1 (120). Oridonin, a
key ingredient of traditional Chinese medicine Rabdosia rubescens,
enhances cytotoxicity of 5-fluorouracil in RCC cells by enhancing
activity of PARP1 and inducing necroptosis (121). By increasing ROS levels,
decreasing pro-PARP and increasing cleaved PARP expression levels,
shikonin, a component of traditional Chinese medicine Comfrey,
triggers programmed death of different types of RCC cell (122).
Inhibition of angiogenesis may be associated with
cardiovascular and kidney toxicity (123,124), as well as liver injury (125). Genetic deletion or suppression of
PARP1 appears to be protective against toxic insult in various
organs (i.e., hemodynamic dysfunction, multiple organ failure in
patients with sepsis) (126,127). Pharmacological suppression or
genetic deletion of PARP1 markedly decreases cisplatin-induced
kidney injury, suggesting that pharmacological inhibition of PARP
may be a promising method for inhibiting nephropathy caused by
cisplatin (128). By attenuating
the intrarenal inflammatory cascade, amelparib, a PARPi, exerts
favorable effects in an mice model of ischemic acute kidney injury
and promotes hypoxic HK-2 cell proliferation (129). Olaparib, a clinically approved
PARPi for the treatment of HR-deficient tumors, improves organ
function, suppresses inflammatory responses and expedites wound
healing in severe burn injury (130). Therefore, genetic deletion or
suppression of PARP1 may have a protective effect in various
organs, suggesting the detoxification and synergism effect of
PARPis on RCC treatment. Although there is no evidence to suggest
that accumulation of renal toxicity is associated with occurrence
of RCC, inhibition of PARP has a key protective effect on the
kidney and its role in the prevention or treatment of RCC is worth
investigation.
Currently, the primary indications for the use of
PARPis include tumors with BRCA1/2 mutation (131). However, identification of
additional predictive biomarkers is key to determine the treatment
options according to the RCC molecular characteristics (132). Drugs that promote HR repair
defects in tumor cells (i.e., PI3K inhibitors, cyclin-dependent
kinase inhibitors) may increase sensitivity to PARPis (133–136). As a specific polymerase θ
inhibitor, novobiocin is combined with PARPi in treatment of
HR-deficient tumors, as well in tumors that have acquired PARPi
resistance (137). In addition,
with the wider clinical use of PARPi, whether long-term inhibition
of PARP activity may lead to mutations or protection in normal
cells, or other unknown negative or positive effects, will more
thoroughly be verified.
In addition to immunotherapy, development of
synthetic lethal associated targets [such as protein kinase,
membrane associated tyrosine/threonine 1)/cyclin E1 and
e-cadherin/ROS1 inhibitor] may offer novel directions for tumor
therapy (141,142). In addition, dual PARP and RAD51
or other DNA repair target inhibitor conjugates have the potential
to overcome resistance mechanisms to PARPi. The positive results of
inhibition of PARP in other DDR-associated genes, such as PALB2
(partner and localized of BRCA2) in prostate cancer, may benefit
further explorations of PARP inhibition in RCC treatment.
Therefore, DDR genes are key tumor targets involved in various
forms of cell death. The application of DDR inhibitors (such as
PARPi) and other targeted, immunotherapy and tumor
metabolism-associated drugs in RCC should be explored in
future.
RCC incidence and mortality rate are high worldwide
and lack useful immunotherapy options. The enzymatic activity or
expression level of DDR genes, particularly PARP1, may serve as
potential tumor therapeutic targets. Moreover, PARP1 may serve as a
key biological marker to predict the therapeutic effect of ICIs and
evaluate prognosis of patients with ccRCC. The role of DDR pathways
in RCC progression may provide potential therapeutic targets for
treatment of certain types of RCC.
Not applicable.
The present study was supported by the National Natural Science
Foundation of China (grant no. 81803576) and Shenzhen Futian
District Public Health Research Project (grant nos. FTWS2021073 and
FTWS2020026).
YCL, FCT, ZCL, HXZ, CHC, CL, YFY and ZHH contributed
to manuscript writing. CHC, CL, YFY and ZHH conceptualized the
study. YCL constructed tables and figures. FCT, ZBL, HZH and YCL
critically revised the manuscript. All authors have read and
approved the final manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Abbotts R and Wilson DR: Coordination of
DNA single strand break repair. Free Radic Biol Med. 107:228–244.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Anand SK, Sharma A, Singh N and Kakkar P:
Entrenching role of cell cycle checkpoints and autophagy for
maintenance of genomic integrity. DNA Repair (Amst). 86:1027482020.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Oh JM and Myung K: Crosstalk between
different DNA repair pathways for DNA double strand break repairs.
Mutat Res Genet Toxicol Environ Mutagen. 873:5034382022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lavrik OI: PARPs' impact on base excision
DNA repair. DNA Repair (Amst). 93:1029112020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ray CA and Nussenzweig A: The multifaceted
roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol
Cell Biol. 18:610–621. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koczor CA, Saville KM, Andrews JF, Clark
J, Fang Q, Li J, Al-Rahahleh RQ, Ibrahim M, McClellan S, Makarov
MV, et al: Temporal dynamics of base Excision/Single-Strand break
repair protein complex assembly/disassembly are modulated by the
PARP/NAD+/SIRT6 axis. Cell Rep. 37:1099172021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Richard IA, Burgess JT, O'Byrne KJ and
Bolderson E: Beyond PARP1: The potential of other members of the
poly (ADP-Ribose) polymerase family in DNA repair and cancer
therapeutics. Front Cell Dev Biol. 9:8012002021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Covarrubias AJ, Perrone R, Grozio A and
Verdin E: NAD+ metabolism and its roles in cellular
processes during ageing. Nat Rev Mol Cell Biol. 22:119–141. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gupte R, Liu Z and Kraus WL: PARPs and
ADP-ribosylation: Recent advances linking molecular functions to
biological outcomes. Genes Dev. 31:101–126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao Y, Li C, Wei L, Teng Y, Nakajima S,
Chen X, Xu J, Leger B, Ma H, Spagnol ST, et al: SSRP1 cooperates
with PARP and XRCC1 to facilitate single-strand DNA break repair by
chromatin priming. Cancer Res. 77:2674–2685. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Prokhorova E, Zobel F, Smith R, Zentout S,
Gibbs-Seymour I, Schutzenhofer K, Peters A, Groslambert J, Zorzini
V, Agnew T, et al: Serine-linked PARP1 auto-modification controls
PARP inhibitor response. Nat Commun. 12:40552021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Demeny MA and Virag L: The PARP enzyme
family and the hallmarks of cancer part 1. Cell intrinsic
hallmarks. Cancers (Basel). 13:20422021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shaw G: The silent disease. Nature. 537
(Suppl):S98–S99. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Linehan WM and Ricketts CJ: The Cancer
Genome Atlas of renal cell carcinoma: Findings and clinical
implications. Nat Rev Urol. 16:539–552. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
Cancers in 185 Countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mao W, Wang K, Wu Z, Xu B and Chen M:
Current status of research on exosomes in general, and for the
diagnosis and treatment of kidney cancer in particular. J Exp Clin
Cancer Res. 40:3052021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lai Y, Zeng T, Liang X and Wu W, Zhong F
and Wu W: Cell death-related molecules and biomarkers for renal
cell carcinoma targeted therapy. Cancer Cell Int. 19:2212019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xiong W, Zhang B, Yu H, Zhu L, Yi L and
Jin X: RRM2 Regulates sensitivity to sunitinib and PD-1 blockade in
renal cancer by stabilizing ANXA1 and activating the AKT pathway.
Adv Sci (Weinh). 8:e21008812021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Popovic M, Matovina-Brko G, Jovic M and
Popovic LS: Immunotherapy: A new standard in the treatment of
metastatic clear cell renal cell carcinoma. World J Clin Oncol.
13:28–38. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Criscuolo D, Morra F, Giannella R,
Visconti R, Cerrato A and Celetti A: New combinatorial strategies
to improve the PARP inhibitors efficacy in the urothelial bladder
Cancer treatment. J Exp Clin Cancer Res. 38:912019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuasa T, Urasaki T and Oki R: Recent
advances in medical therapy for urological cancers. Front Oncol.
12:7469222022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yin M, Grivas P, Wang QE, Mortazavi A,
Emamekhoo H, Holder SL, Drabick JJ, Woo MS, Pal S, Vasekar M, et
al: Prognostic value of DNA damage response genomic alterations in
Relapsed/Advanced urothelial cancer. Oncologist. 25:680–688. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang W, van Gent DC, Incrocci L, van
Weerden WM and Nonnekens J: Role of the DNA damage response in
prostate cancer formation, progression and treatment. Prostate
Cancer Prostatic Dis. 23:24–37. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chakraborty G, Armenia J, Mazzu YZ,
Nandakumar S, Stopsack KH, Atiq MO, Komura K, Jehane L, Hirani R,
Chadalavada K, et al: Significance of BRCA2 and RB1 Co-loss in
aggressive prostate cancer progression. Clin Cancer Res.
26:2047–2064. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rimar KJ, Tran PT, Matulewicz RS, Hussain
M and Meeks JJ: The emerging role of homologous recombination
repair and PARP inhibitors in genitourinary malignancies. Cancer-Am
Cancer Soc. 123:1912–1924. 2017.PubMed/NCBI
|
|
26
|
Lord CJ and Ashworth A: PARP inhibitors:
Synthetic lethality in the clinic. Science. 355:1152–1158. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo E, Wu C, Ming J, Zhang W, Zhang L and
Hu G: The clinical significance of DNA damage repair signatures in
clear cell renal cell carcinoma. Front Genet. 11:5930392020.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hartman TR, Demidova EV, Lesh RW, Hoang L,
Richardson M, Forman A, Kessler L, Speare V, Golemis EA, Hall MJ,
et al: Prevalence of pathogenic variants in DNA damage response and
repair genes in patients undergoing cancer risk assessment and
reporting a personal history of early-onset renal cancer. Sci Rep.
10:135182020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Karczewski KJ, Francioli LC, Tiao G,
Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A,
Birnbaum DP, et al: The mutational constraint spectrum quantified
from variation in 141,456 humans. Nature. 581:434–443. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peng L, Liang J, Wang Q and Chen G: A DNA
Damage repair gene signature associated with immunotherapy response
and clinical prognosis in clear cell renal cell carcinoma. Front
Genet. 13:7988462022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meng H, Jiang X, Cui J, Yin G, Shi B, Liu
Q, Xuan H and Wang Y: Genomic analysis reveals novel specific
metastatic mutations in Chinese clear cell renal cell carcinoma.
Biomed Res Int. 2020:24951572020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ged Y, Chaim JL, DiNatale RG, Knezevic A,
Kotecha RR, Carlo MI, Lee CH, Foster A, Feldman DR, Teo MY, et al:
DNA damage repair pathway alterations in metastatic clear cell
renal cell carcinoma and implications on systemic therapy. J
Immunother Cancer. 8:e0002302020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tapia-Laliena MA, Korzeniewski N,
Pena-Llopis S, Scholl C, Frohling S, Hohenfellner M, Duensing A and
Duensing S: Cullin 5 is a novel candidate tumor suppressor in renal
cell carcinoma involved in the maintenance of genome stability.
Oncogenesis. 8:42019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bhattacharjee S and Nandi S: Choices have
consequences: The nexus between DNA repair pathways and genomic
instability in cancer. Clin Transl Med. 5:452016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang R and Zhou PK: DNA damage repair:
Historical perspectives, mechanistic pathways and clinical
translation for targeted cancer therapy. Signal Transduct Target
Ther. 6:2542021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schrempf A, Slyskova J and Loizou JI:
Targeting the DNA repair enzyme polymerase theta in cancer therapy.
Trends Cancer. 7:98–111. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Horton JK, Stefanick DF, Prasad R, Gassman
NR, Kedar PS and Wilson SH: Base excision repair defects invoke
hypersensitivity to PARP inhibition. Mol Cancer Res. 12:1128–1139.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Murai J, Huang SY, Das BB, Renaud A, Zhang
Y, Doroshow JH, Ji J, Takeda S and Pommier Y: Trapping of PARP1 and
PARP2 by clinical PARP inhibitors. Cancer Res. 72:5588–5599. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shao Z, Lee BJ, Rouleau-Turcotte E,
Langelier MF, Lin X, Estes VM, Pascal JM and Zha S: Clinical PARP
inhibitors do not abrogate PARP1 exchange at DNA damage sites in
vivo. Nucleic Acids Res. 48:9694–9709. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rao PD, Sankrityayan H, Srivastava A,
Kulkarni YA, Mulay SR and Gaikwad AB: ‘PARP'ing fibrosis:
Repurposing poly (ADP ribose) polymerase (PARP) inhibitors. Drug
Discov Today. 25:1253–1261. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Houl JH, Ye Z, Brosey CA,
Balapiti-Modarage L, Namjoshi S, Bacolla A, Laverty D, Walker BL,
Pourfarjam Y, Warden LS, et al: Selective small molecule PARG
inhibitor causes replication fork stalling and cancer cell death.
Nat Commun. 10:56542019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Slade D: PARP and PARG inhibitors in
cancer treatment. Genes Dev. 34:360–394. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Suskiewicz MJ, Zobel F, Ogden T, Fontana
P, Ariza A, Yang JC, Zhu K, Bracken L, Hawthorne WJ, Ahel D, et al:
HPF1 completes the PARP active site for DNA damage-induced
ADP-ribosylation. Nature. 579:598–602. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hsieh MH, Chen YT, Chen YT, Lee YH, Lu J,
Chien CL, Chen HF, Ho HN, Yu CJ, Wang ZQ and Teng SC: PARP1
controls KLF4-mediated telomerase expression in stem cells and
cancer cells. Nucleic Acids Res. 45:10492–10503. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miwa M and Masutani M:
PolyADP-ribosylation and cancer. Cancer Sci. 98:1528–1535. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pillay N, Tighe A, Nelson L, Littler S,
Coulson-Gilmer C, Bah N, Golder A, Bakker B, Spierings D, James DI,
et al: DNA replication vulnerabilities render ovarian cancer cells
sensitive to poly(ADP-Ribose) glycohydrolase inhibitors. Cancer
Cell. 35:519–533. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pilie PG, Gay CM, Byers LA, O'Connor MJ
and Yap TA: PARP inhibitors: Extending benefit beyond BRCA-mutant
cancers. Clin Cancer Res. 25:3759–3771. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chang WS, Ke HL, Tsai CW, Lien CS, Liao
WL, Lin HH, Lee MH, Wu HC, Chang CH, Chen CC, et al: The role of
XRCC6 T-991C functional polymorphism in renal cell carcinoma.
Anticancer Res. 32:3855–3860. 2012.PubMed/NCBI
|
|
49
|
Wu C, Xu C, Wang G, Zhang D and Zhao X:
Noninvasive circulating tumor cell and urine cellular XPC
(rs2228001, A2815C) and XRCC1 (rs25487, G1196A) polymorphism
detection as an effective screening panel for genitourinary system
cancers. Transl Cancer Res. 8:2803–2812. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hsueh YM, Lin YC, Chen WJ, Huang CY, Shiue
HS, Pu YS, Chen CH and Su CT: The polymorphism XRCC1 Arg194Trp and
8-hydroxydeoxyguanosine increased susceptibility to arsenic-related
renal cell carcinoma. Toxicol Appl Pharmacol. 332:1–7. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Malka MM, Eberle J, Niedermayer K, Zlotos
DP and Wiesmuller L: Dual PARP and RAD51 inhibitory drug conjugates
show synergistic and selective effects on breast cancer cells.
Biomolecules. 11:9812021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xu YY, Ren ZL, Liu XL, Zhang GM, Huang SS,
Shi WH, Ye LX, Luo X, Liu SW, Li YL and Yu L: BAP1 loss augments
sensitivity to BET inhibitors in cancer cells. Acta Pharmacol Sin.
43:1803–1815. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li X, Zhang Z, Fan B, Li Y, Song D and Li
GY: PARP-1 Is a potential marker of retinal photooxidation and a
key signal regulator in retinal light injury. Oxid Med Cell Longev.
2022:68813222022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li X and Darzynkiewicz Z: Cleavage of
Poly(ADP-ribose) polymerase measured in situ in individual cells:
Relationship to DNA fragmentation and cell cycle position during
apoptosis. Exp Cell Res. 255:125–132. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Desroches A and Denault JB: Caspase-7 uses
RNA to enhance proteolysis of poly(ADP-ribose) polymerase 1 and
other RNA-binding proteins. Proc Natl Acad Sci USA.
116:21521–21528. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Koh DW, Dawson TM and Dawson VL: Mediation
of cell death by poly(ADP-ribose) polymerase-1. Pharmacol Res.
52:5–14. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
D'Amours D, Sallmann FR, Dixit VM and
Poirier GG: Gain-of-function of poly(ADP-ribose) polymerase-1 upon
cleavage by apoptotic proteases: Implications for apoptosis. J Cell
Sci. 114:3771–3778. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Huang Q and Shen HM: To die or to live:
The dual role of poly(ADP-ribose) polymerase-1 in autophagy and
necrosis under oxidative stress and DNA damage. Autophagy.
5:273–276. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Rodriguez-Vargas JM, Ruiz-Magana MJ,
Ruiz-Ruiz C, Majuelos-Melguizo J, Peralta-Leal A, Rodriguez MI,
Munoz-Gamez JA, de Almodovar MR, Siles E, Rivas AL, et al:
ROS-induced DNA damage and PARP-1 are required for optimal
induction of starvation-induced autophagy. Cell Res. 22:1181–1198.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jouan-Lanhouet S, Arshad MI,
Piquet-Pellorce C, Martin-Chouly C, Le Moigne-Muller G, Van
Herreweghe F, Takahashi N, Sergent O, Lagadic-Gossmann D,
Vandenabeele P, et al: TRAIL induces necroptosis involving
RIPK1/RIPK3-dependent PARP-1 activation. Cell Death Differ.
19:2003–2014. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhou Y, Liu L, Tao S, Yao Y, Wang Y, Wei
Q, Shao A and Deng Y: Parthanatos and its associated components:
Promising therapeutic targets for cancer. Pharmacol Res.
163:1052992021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang Y, Kim NS, Haince JF, Kang HC, David
KK, Andrabi SA, Poirier GG, Dawson VL and Dawson TM:
Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is
critical for PAR polymerase-1-dependent cell death (parthanatos).
Sci Signal. 4:a202011. View Article : Google Scholar
|
|
63
|
Hong T, Lei G, Chen X, Li H, Zhang X, Wu
N, Zhao Y, Zhang Y and Wang J: PARP inhibition promotes ferroptosis
via repressing SLC7A11 and synergizes with ferroptosis inducers in
BRCA-proficient ovarian cancer. Redox Biol. 42:1019282021.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wolf C, Smith S and van Wijk SJL:
Zafirlukast Induces VHL- and HIF-2α-dependent oxidative cell death
in 786-O clear cell renal carcinoma cells. Int J Mol Sci.
23:35672022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Manco G, Lacerra G, Porzio E and Catara G:
ADP-Ribosylation Post-translational modification: An overview with
a focus on RNA biology and new pharmacological perspectives.
Biomolecules. 12:4432022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Deeks ED: Olaparib: First global approval.
Drugs. 75:231–240. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kummar S, Chen A, Parchment RE, Kinders
RJ, Ji J, Tomaszewski JE and Doroshow JH: Advances in using PARP
inhibitors to treat cancer. BMC Med. 10:252012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bian C, Zhang C, Luo T, Vyas A, Chen SH,
Liu C, Kassab MA, Yang Y, Kong M and Yu X: NADP+ is an
endogenous PARP inhibitor in DNA damage response and tumor
suppression. Nat Commun. 10:6932019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Murata S, Zhang C, Finch N, Zhang K, Campo
L and Breuer EK: Predictors and modulators of synthetic lethality:
An update on PARP inhibitors and personalized medicine. Biomed Res
Int. 2016:23465852016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zatreanu D, Robinson H, Alkhatib O,
Boursier M, Finch H, Geo L, Grande D, Grinkevich V, Heald RA,
Langdon S, et al: Polθ inhibitors elicit BRCA-gene synthetic
lethality and target PARP inhibitor resistance. Nat Commun.
12:36362021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Boussios S, Rassy E, Moschetta M, Ghose A,
Adeleke S, Sanchez E, Sheriff M, Chargari C and Pavlidis N: BRCA
mutations in ovarian and prostate cancer: Bench to bedside. Cancers
(Basel). 14:36362021.
|
|
72
|
Min A and Im SA: PARP inhibitors as
therapeutics: Beyond modulation of PARylation. Cancers (Basel).
12:3942020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Kinoshita T, Nakanishi I, Warizaya M,
Iwashita A, Kido Y, Hattori K and Fujii T: Inhibitor-induced
structural change of the active site of human poly(ADP-ribose)
polymerase. Febs Lett. 556:43–46. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Makhov P, Uzzo RG, Tulin AV and Kolenko
VM: Histone-dependent PARP-1 inhibitors: A novel therapeutic
modality for the treatment of prostate and renal cancers. Urol
Oncol. 39:312–315. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Karpova Y, Guo D, Makhov P, Haines AM,
Markov DA, Kolenko V and Tulin AV: Poly(ADP)-Ribosylation
Inhibition: A promising approach for clear cell renal cell
carcinoma therapy. Cancers (Basel). 13:49732021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Shuch B, Amin A, Armstrong AJ, Eble JN,
Ficarra V, Lopez-Beltran A, Martignoni G, Rini BI and Kutikov A:
Understanding pathologic variants of renal cell carcinoma:
Distilling therapeutic opportunities from biologic complexity. Eur
Urol. 67:85–97. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wang X, Lopez R, Luchtel RA, Hafizi S,
Gartrell B and Shenoy N: Immune evasion in renal cell carcinoma:
Biology, clinical translation, future directions. Kidney Int.
99:75–85. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Nguyen-Tran HH, Nguyen TN, Chen CY and Hsu
T: Endothelial reprogramming stimulated by oncostatin m promotes
inflammation and tumorigenesis in VHL-deficient kidney tissue.
Cancer Res. 81:5060–5073. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Sharma R, Kadife E, Myers M, Kannourakis
G, Prithviraj P and Ahmed N: Determinants of resistance to VEGF-TKI
and immune checkpoint inhibitors in metastatic renal cell
carcinoma. J Exp Clin Cancer Res. 40:1862021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Hsieh JJ, Chen D, Wang PI, Marker M,
Redzematovic A, Chen YB, Selcuklu SD, Weinhold N, Bouvier N,
Huberman KH, et al: Genomic biomarkers of a randomized trial
comparing first-line everolimus and sunitinib in patients with
metastatic renal cell carcinoma. Eur Urol. 71:405–414. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Miao D, Margolis CA, Gao W, Voss MH, Li W,
Martini DJ, Norton C, Bosse D, Wankowicz SM, Cullen D, et al:
Genomic correlates of response to immune checkpoint therapies in
clear cell renal cell carcinoma. Science. 359:801–806. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Pal SK, Sonpavde G, Agarwal N, Vogelzang
NJ, Srinivas S, Haas NB, Signoretti S, McGregor BA, Jones J, Lanman
RB, et al: Evolution of circulating tumor DNA profile from
first-line to subsequent therapy in metastatic renal cell
carcinoma. Eur Urol. 72:557–564. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Martinez CN, Xie W, Asim BM, Dzimitrowicz
H, Burkart J, Geynisman DM, Balakrishnan A, Bowman IA, Jain R,
Stadler W, et al: Cabozantinib in advanced non-clear-cell renal
cell carcinoma: A multicentre, retrospective, cohort study. Lancet
Oncol. 20:581–590. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Maroto P, Anguera G, Roldan-Romero JM,
Apellaniz-Ruiz M, Algaba F, Boonman J, Nellist M, Montero-Conde C,
Cascon A, Robledo M and Rodríguez-Antona C: Biallelic TSC2
mutations in a patient with chromophobe renal cell carcinoma
showing extraordinary response to temsirolimus. J Natl Compr Canc
Netw. 16:352–358. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
McGregor BA, McKay RR, Braun DA, Werner L,
Gray K, Flaifel A, Signoretti S, Hirsch MS, Steinharter JA, Bakouny
Z, et al: Results of a multicenter Phase II study of atezolizumab
and bevacizumab for patients with metastatic renal cell carcinoma
with variant histology and/or sarcomatoid features. J Clin Oncol.
38:63–70. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Fallah J, Brave MH, Weinstock C, Mehta GU,
Bradford D, Gittleman H, Bloomquist EW, Charlab R, Hamed SS, Miller
CP, et al: FDA approval summary: Belzutifan for von Hippel-Lindau
disease associated tumors. Clin Cancer Res. Jun 21–2022.(Epub ahead
of print). View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Stransky LA, Vigeant SM, Huang B, West D,
Denize T, Walton E, Signoretti S and Kaelin WJ: Sensitivity of VHL
mutant kidney cancers to HIF2 inhibitors does not require an intact
p53 pathway. Proc Natl Acad Sci USA. 119:e21204031192022.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
He X, Gan F, Zhou Y, Zhang Y, Zhao P, Zhao
B, Tang Q, Ye L, Bu J, Mei J, et al: Nonplanar Helicene
Benzo[4]Helicenium for the precise treatment of renal cell
carcinoma. Small Methods. 5:e21007702021. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yan S, Liu L, Ren F, Gao Q, Xu S, Hou B,
Wang Y, Jiang X and Che Y: Sunitinib induces genomic instability of
renal carcinoma cells through affecting the interaction of LC3-II
and PARP-1. Cell Death Dis. 8:e29882017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yang XD, Kong FE, Qi L, Lin JX, Yan Q,
Loong J, Xi SY, Zhao Y, Zhang Y, Yuan YF, et al: PARP inhibitor
Olaparib overcomes Sorafenib resistance through reshaping the
pluripotent transcriptome in hepatocellular carcinoma. Mol Cancer.
20:202021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Pletcher JP, Bhattacharjee S, Doan JP,
Wynn R, Sindhwani P, Nadiminty N and Petros FG: The Emerging role
of poly (ADP-Ribose) polymerase inhibitors as effective therapeutic
agents in renal cell carcinoma. Front Oncol. 11:6814412021.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Scanlon SE, Hegan DC, Sulkowski PL and
Glazer PM: Suppression of homology-dependent DNA double-strand
break repair induces PARP inhibitor sensitivity in VHL-deficient
human renal cell carcinoma. Oncotarget. 9:4647–4660. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhang Q, Xu Y, Zhang Z, Li J, Xia Q and
Chen Y: Folliculin deficient renal cancer cells exhibit BRCA1 a
complex expression impairment and sensitivity to PARP1 inhibitor
olaparib. Gene. 769:1452432021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Szanto M and Bai P: The role of ADP-ribose
metabolism in metabolic regulation, adipose tissue differentiation,
and metabolism. Genes Dev. 34:321–340. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Szanto M, Gupte R, Kraus WL, Pacher P and
Bai P: PARPs in lipid metabolism and related diseases. Prog Lipid
Res. 84:1011172021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zou Y, Palte MJ, Deik AA, Li H, Eaton JK,
Wang W, Tseng YY, Deasy R, Kost-Alimova M, Dancik V, et al: A
GPX4-dependent cancer cell state underlies the clear-cell
morphology and confers sensitivity to ferroptosis. Nat Commun.
10:16172019. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Okazaki A, Gameiro PA, Christodoulou D,
Laviollette L, Schneider M, Chaves F, Stemmer-Rachamimov A,
Yazinski SA, Lee R, Stephanopoulos G, et al: Glutaminase and
poly(ADP-ribose) polymerase inhibitors suppress pyrimidine
synthesis and VHL-deficient renal cancers. J Clin Invest.
127:1631–1645. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Shen YA, Hong J, Asaka R, Asaka S, Hsu FC,
Suryo RY, Jung JG, Chen YW, Yen TT, Tomaszewski A, et al:
Inhibition of the MYC-Regulated glutaminase metabolic axis is an
effective synthetic lethal approach for treating chemoresistant
ovarian cancers. Cancer Res. 80:4514–4526. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhao S, Li P, Wu W, Wang Q, Qian B, Li X
and Shen M: Roles of ferroptosis in urologic malignancies. Cancer
Cell Int. 21:6762021. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Courtney KD, Bezwada D, Mashimo T,
Pichumani K, Vemireddy V, Funk AM, Wimberly J, McNeil SS, Kapur P,
Lotan Y, et al: Isotope tracing of human clear cell renal cell
carcinomas demonstrates suppressed glucose oxidation in vivo. Cell
Metab. 28:793–800. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhang Y, Huang J, Huang Y, Zhang S and Wu
W, Long H, Duan X, Lai Y and Wu W: Tanshinone I and simvastatin
inhibit melanoma tumour cell growth by regulating poly (ADP ribose)
polymerase 1 expression. Mol Med Rep. 23:402021.PubMed/NCBI
|
|
102
|
Yun EJ, Lin CJ, Dang A, Hernandez E, Guo
J, Chen WM, Allison J, Kim N, Kapur P, Brugarolas J, et al:
Downregulation of human DAB2IP gene expression in renal cell
carcinoma results in resistance to ionizing radiation. Clin Cancer
Res. 25:4542–4551. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhou C, Fabbrizi MR, Hughes JR, Grundy GJ
and Parsons JL: Effectiveness of PARP inhibition in enhancing the
radiosensitivity of 3D spheroids of head and neck squamous cell
carcinoma. Front Oncol. 12:9403772022. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Meng Y, Efimova EV, Hamzeh KW, Darga TE,
Mauceri HJ, Fu YX, Kron SJ and Weichselbaum RR: Radiation-inducible
immunotherapy for cancer: Senescent tumor cells as a cancer
vaccine. Mol Ther. 20:1046–1055. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Liu Q, Xiao Q, Sun Z, Wang B, Wang L, Wang
N, Wang K, Song C and Yang Q: Exosome component 1 cleaves
single-stranded DNA and sensitizes human kidney renal clear cell
carcinoma cells to poly(ADP-ribose) polymerase inhibitor. Elife.
10:e694542021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Cella D, Motzer RJ, Suarez C, Blum SI,
Ejzykowicz F, Hamilton M, Wallace JF, Simsek B, Zhang J, Ivanescu
C, et al: Patient-reported outcomes with first-line nivolumab plus
cabozantinib versus sunitinib in patients with advanced renal cell
carcinoma treated in CheckMate 9ER: An open-label, randomised,
phase 3 trial. Lancet Oncol. 23:292–303. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Hagiwara M, Fushimi A, Matsumoto K and Oya
M: The Significance of PARP1 as a biomarker for predicting the
response to PD-L1 blockade in patients with PBRM1-mutated clear
cell renal cell carcinoma. Eur Urol. 81:145–148. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Chabanon RM, Morel D, Eychenne T,
Colmet-Daage L, Bajrami I, Dorvault N, Garrido M, Meisenberg C,
Lamb A, Ngo C, et al: PBRM1 deficiency confers synthetic lethality
to DNA repair inhibitors in cancer. Cancer Res. 81:2888–2902. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Park JS, Lee ME, Jang WS, Rha KH, Lee SH,
Lee J and Ham WS: The DEAD/DEAH box helicase, DDX11, is essential
for the survival of advanced clear cell renal cell carcinoma and is
a determinant of PARP inhibitor sensitivity. Cancers (Basel).
13:25742021. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Pan XW, Zhang H, Xu D, Chen JX, Chen WJ,
Gan SS, Qu FJ, Chu CM, Cao JW, Fan YH, et al: Identification of a
novel cancer stem cell subpopulation that promotes progression of
human fatal renal cell carcinoma by single-cell RNA-seq analysis.
Int J Biol Sci. 16:3149–3162. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Olson D, Bhalla S, Yang X, Martone B and
Kuzel TM: Novel use of targeted therapy via PARP-Inhibition in a
rare form of papillary renal cell carcinoma: A case report and
literature review. Clin Genitourin Cancer. 14:e445–e448. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Lian BJ, Zhang K, Fang XD, Li F, Dai Z,
Chen WY and Qi XP: Clinical benefit of Niraparib to
TKI/mTORi-resistance metastatic ccRCC with BAP1-frame shift
mutation: Case report and literature review. Front Oncol.
12:9272502022. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Saatchi F and Kirchmaier AL: Tolerance of
DNA replication stress is promoted by fumarate through modulation
of histone demethylation and enhancement of replicative
intermediate processing in saccharomyces cerevisiae. Genetics.
212:631–654. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Johnson TI, Costa A, Ferguson AN and
Frezza C: Fumarate hydratase loss promotes mitotic entry in the
presence of DNA damage after ionising radiation. Cell Death Dis.
9:9132018. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Sulkowski PL, Sundaram RK, Oeck S, Corso
CD, Liu Y, Noorbakhsh S, Niger M, Boeke M, Ueno D, Kalathil AN, et
al: Krebs-cycle-deficient hereditary cancer syndromes are defined
by defects in homologous-recombination DNA repair. Nat Genet.
50:1086–1092. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Sulkowski PL, Oeck S, Dow J, Economos NG,
Mirfakhraie L, Liu Y, Noronha K, Bao X, Li J, Shuch BM, et al:
Oncometabolites suppress DNA repair by disrupting local chromatin
signalling. Nature. 582:586–591. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Ueno D, Vasquez JC, Sule A, Liang J, van
Doorn J, Sundaram R, Friedman S, Caliliw R, Ohtake S, Bao X, et al:
Targeting Krebs-cycle-deficient renal cell carcinoma with Poly
ADP-ribose polymerase inhibitors and low-dose alkylating
chemotherapy. Oncotarget. 13:1054–1067. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Li X, Hu D, Li Y, Luo Y, Liang B, Yu K,
Xiong W and Zuo D: Overexpression of TP53INP2 promotes apoptosis in
clear cell renal cell cancer via caspase-8/TRAF6 signaling pathway.
J Immunol Res. 2022:12604232022.PubMed/NCBI
|
|
119
|
Lee HK, Cha HS, Nam MJ, Park K, Yang YH,
Lee J and Park SH: Broussochalcone A induces apoptosis in human
renal cancer cells via ROS level elevation and activation of FOXO3
signaling pathway. Oxid Med Cell Longev. 2021:28007062021.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Xu X, Chua CC, Zhang M, Geng D, Liu CF,
Hamdy RC and Chua BH: The role of PARP activation in
glutamate-induced necroptosis in HT-22 cells. Brain Res.
1343:206–212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Zheng W, Zhou CY, Zhu XQ, Wang XJ, Li ZY,
Chen XC, Chen F, Che XY and Xie X: Oridonin enhances the
cytotoxicity of 5-FU in renal carcinoma cells by inducting
necroptotic death. Biomed Pharmacother. 106:175–182. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Tsai MF, Chen SM, Ong AZ, Chung YH, Chen
PN, Hsieh YH, Kang YT and Hsu LS: Shikonin induced program cell
death through generation of reactive oxygen species in renal cancer
cells. Antioxidants (Basel). 10:18312021. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Clou E and Luque Y: Angiogenesis
inhibitors: Mechanism of action and nephrotoxicity. Nephrol Ther.
18:1–6. 2022.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Al-Harbi NO, Imam F, Alharbi MM, Khan MR,
Qamar W, Afzal M, Algahtani M, Alobaid S, Alfardan AS, Alshammari
A, et al: Role of rivaroxaban in sunitinib-induced renal injuries
via inhibition of oxidative stress-induced apoptosis and
inflammation through the tissue nacrosis factor-α induced nuclear
factor-κappa B signaling pathway in rats. J Thromb Thrombolysis.
50:361–370. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Studentova H, Volakova J, Spisarova M,
Zemankova A, Aiglova K, Szotkowski T and Melichar B: Severe
tyrosine-kinase inhibitor induced liver injury in metastatic renal
cell carcinoma patients: Two case reports assessed for causality
using the updated RUCAM and review of the literature. BMC
Gastroenterol. 22:492022. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Wang Y, An R, Umanah GK, Park H, Nambiar
K, Eacker SM, Kim B, Bao L, Harraz MM, Chang C, et al: A nuclease
that mediates cell death induced by DNA damage and poly(ADP-ribose)
polymerase-1. Science. 354:aad68722016. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Santos SS, Brunialti M, Soriano FG, Szabo
C and Salomao R: Repurposing of clinically approved
Poly-(ADP-Ribose) polymerase inhibitors for the therapy of sepsis.
Shock. 56:901–909. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Mukhopadhyay P, Horvath B, Kechrid M,
Tanchian G, Rajesh M, Naura AS, Boulares AH and Pacher P:
Poly(ADP-ribose) polymerase-1 is a key mediator of
cisplatin-induced kidney inflammation and injury. Free Radic Biol
Med. 51:1774–1788. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Jang HR, Lee K, Jeon J, Kim JR, Lee JE,
Kwon GY, Kim YG, Kim DJ, Ko JW and Huh W: Poly (ADP-Ribose)
polymerase inhibitor treatment as a novel therapy attenuating renal
ischemia-reperfusion injury. Front Immunol. 11:5642882020.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Ahmad A, Olah G, Herndon DN and Szabo C:
The clinically used PARP inhibitor olaparib improves organ
function, suppresses inflammatory responses and accelerates wound
healing in a murine model of third-degree burn injury. Br J
Pharmacol. 175:232–245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Onji H and Murai J: Reconsidering the
mechanisms of action of PARP inhibitors based on clinical outcomes.
Cancer Sci. 113:2943–2951. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Simonaggio A, Epaillard N, Elaidi R, Sun
CM, Moreira M, Oudard S and Vano YA: Impact of molecular signatures
on the choice of systemic treatment for metastatic kidney cancer.
Bull Cancer. 107 (Suppl):S24–S34. 2020.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Konstantinopoulos PA, Barry WT, Birrer M,
Westin SN, Cadoo KA, Shapiro GI, Mayer EL, O'Cearbhaill RE, Coleman
RL, Kochupurakkal B, et al: Olaparib and α-specific PI3K inhibitor
alpelisib for patients with epithelial ovarian cancer: A
dose-escalation and dose-expansion phase 1b trial. Lancet Oncol.
20:570–580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Abbotts R, Dellomo AJ and Rassool FV:
Pharmacologic induction of BRCAness in BRCA-proficient cancers:
Expanding PARP inhibitor use. Cancers (Basel). 14:26402022.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Nelson LJ, Castro KE, Xu B, Li J, Dinh NB,
Thompson JM, Woytash J, Kipp KR and Razorenova OV: Synthetic
lethality of cyclin-dependent kinase inhibitor Dinaciclib with
VHL-deficiency allows for selective targeting of clear cell renal
cell carcinoma. Cell Cycle. 21:1103–1119. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhao Y, Zhou K, Xia X, Guo Y and Tao L:
Chk1 inhibition-induced BRCAness synergizes with olaparib in
p53-deficient cancer cells. Cell Cycle. 1–13. 2022.doi:
10.1080/15384101.2022.2111769 (Epub ahead of print). View Article : Google Scholar
|
|
137
|
Zhou J, Gelot C, Pantelidou C, Li A, Yucel
H, Davis RE, Farkkila A, Kochupurakkal B, Syed A, Shapiro GI, et
al: A first-in-class polymerase theta inhibitor selectively targets
homologous-recombination-deficient tumors. Nat Cancer. 2:598–610.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Ding L, Chen X, Xu X, Qian Y, Liang G, Yao
F, Yao Z, Wu H, Zhang J, He Q and Yang B: PARP1 suppresses the
transcription of PD-L1 by Poly(ADP-Ribosyl)ating STAT3. Cancer
Immunol Res. 7:136–149. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Jiao S, Xia W, Yamaguchi H, Wei Y, Chen
MK, Hsu JM, Hsu JL, Yu WH, Du Y, Lee HH, et al: PARP inhibitor
upregulates PD-L1 expression and enhances cancer-associated
immunosuppression. Clin Cancer Res. 23:3711–3720. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Higuchi T, Flies DB, Marjon NA,
Mantia-Smaldone G, Ronner L, Gimotty PA and Adams SF: CTLA-4
blockade synergizes therapeutically with PARP inhibition in
BRCA1-deficient ovarian cancer. Cancer Immunol Res. 3:1257–1268.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Gallo D, Young JTF, Fourtounis J, Martino
G, Alvarez-Quilon A, Bernier C, Duffy NM, Papp R, Roulston A,
Stocco R, et al: CCNE1 amplification is synthetic lethal with
PKMYT1 kinase inhibition. Nature. 604:749–756. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Bajrami I, Marlow R, van de Ven M, Brough
R, Pemberton HN, Frankum J, Song F, Rafiq R, Konde A, Krastev DB,
et al: E-Cadherin/ROS1 inhibitor synthetic lethality in breast
cancer. Cancer Discov. 8:498–515. 2018. View Article : Google Scholar : PubMed/NCBI
|