Introduction
Two-dimensional (2D) cell cultures have an important
role in assessing drugs due to their simplicity, robustness,
rapidity and cost-effectiveness. The elimination of several
anticancer drugs during clinical development is often due to the
overestimation of their anticancer activity on a 2D-culture-based
screening platform. Considerable data have suggested the
significance of three-dimensional (3D) cell culture systems over 2D
culture systems, due to their ability to better mimic the actual
tumor microenvironment (TME) (1).
The transcriptional profiling of several genes and limited signal
transduction pathways, including hypoxia, TGF, Wnt and
epithelial-mesenchymal transition pathways, in 3D culture systems
have shown similarities with tumor xenografts and patients with
cancer (2,3). These data indicated that 3D culture
systems may reduce the number of animal models required in drug
screening and the failure rates of clinical trials.
The stroma is considered a critical component of the
TME, which markedly affects numerous hallmarks of cancer (4). The tumor stroma is essential in
various molecular processes that aid cancer progression, metastasis
and tumor cell resistance to therapeutic drugs. An accurate
understanding of tumor-stroma interactions may result in more
effective ameliorative strategies, ultimately improving patient
health outcomes. Cancer therapeutic strategies that do not account
for the stroma are inadequate (4);
this is one of the main reasons for the non-performance of a number
of oncology drugs in clinical trials, irrespective of their high
efficiency in 2D-cultured cell line-based models. Notably, only
6.7% of drugs are approved during their transition from the
preclinical phase to phase I clinical trials (5). In addition, most drugs fail in phase
III clinical trials, which is considered to be the most expensive
phase of clinical trials. It should be noted that the median cost
of phase I clinical trials is ~$3.4 million. By contrast, single
phase II and III clinical trials cost ~$8.6 and $19 million,
respectively (6,7). One of the most challenging oncology
issues is the problem of developing productive drugs in a
time-saving and economical manner. Therefore, there is an urgent
need for the development of cost- and time-efficient platforms for
the screening of therapeutic drugs.
Various 3D innovative technologies are currently
available, which have advanced the screening of antitumor agents,
such as magnetic levitation, gel embedding technologies, 3D
bioprinting and microfluidic cell culture. In the magnetic
levitation procedure, magnetic forces are applied to deliver
magnetic nanoparticles to 2D cells, which help in forming 3D
spheroids by making physiologically relevant extracellular matrix
(ECM) (8). In the gel embedding
technology, a hydrophilic polymer-based gel is used to form a 3D
spheroid-like structure; this 3D structure facilitates
cell-cell/cell-ECM interaction and supports signaling involved in
inducing drug resistance of cancer cells (9). Although 3D culture systems have
received attention over 2D culture systems, they lack the
interactions of tumor cells with other cells of the stroma, such as
fibroblasts or endothelial cells. Therefore, these 3D systems
cannot mimic the TME. Co-culture of tumor cells with other cells of
the stroma could be a partial solution to issues related with the
failure of oncology drugs in clinical trials. Therefore, there is
an urgent need to develop advanced 3D culture platforms for drug
screening (10).
The present study developed a 3D triculture model
using epithelial MCF-7 cells, fibroblast MRC5 cells and human
umbilical vein endothelial cells (HUVECs) embedded in the AXTEX-4D™
platform. Briefly, the AXTEX-4D is a platform composed of a
nonwoven fabric base matrix (polyethylene terephthalate) that
receives and supports the growth of tissueoids. These polymers are
less prone to heat and stress, making them autoclavable and capable
of providing mechanical strength. In addition, the size of pores in
the platform is 65 mm, which is designed to sustain cell adhesion
and cell morphology. The present study also explored the use of the
triculture model in testing the action of chemotherapeutic drugs in
solid cancers compared with 3D monoculture and 2D culture
models.
Materials and methods
Materials
The anti-collagen I (cat. no. AB745) and
anti-laminin antibodies (cat. no. L8271) were purchased from
MilliporeSigma. Anti-Ki67 antibodies were purchased from Abcam
(cat. no. ab15580). Anti-mouse Alexa Fluor®
594-conjugated secondary antibodies (cat. no. A11005; 1:1,000) were
purchased from Thermo Fisher Scientific, Inc., and anti-rabbit
Alexa Fluor 488-conjugated secondary antibodies (cat. no. ab150077;
1:1,000) were obtained from Abcam. CFSE Blue (cat. no. C34574) and
CFSE Green (cat. no. C34554) dyes were from Invitrogen; Thermo
Fisher Scientific, Inc. Dil Red (cat. no. D3911) dye was obtained
from Thermo Fisher Scientific, Inc. CFSE Blue, CFSE Green and Dil
Red were used to stain the tumor cells (MCF-7 and HT-29), HUVECs
and MRC5 cells, respectively.
Cell lines
Human breast cancer MCF-7 cells, colorectal cancer
HT-29 cells, HUVECs (passage 1; CRL-1730™) and human lung
fibroblast MRC-5 cells were obtained from ATCC. EMEM
(MilliporeSigma) was used to culture MCF-7, HT-29 and MRC-5 cells,
whereas HUVECs and Jurkat T cells (TIB-152™; ATCC) were cultured in
EBM2 (Lonza Group Ltd.) and RPMI-1640 (MilliporeSigma),
respectively. All media were supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.) and 2 mM glutamine
(MilliporeSigma). Cells were cultured at 37°C and 8% CO2
was supplied to cells under static conditions. Two tumor cell lines
(MCF-7 and HT-29) were used to show the versality of the AXTEX-4D
platform.
Establishing 3D monocultures and
tricultures
3D monoculture and triculture tissueoids on the
AXTEX-4D
The AXTEX-4D platform (Premas Biotech Pvt Ltd.) was
used to form 3D tissueoids as described previously (7,11,12).
A patent for the platform has been filed (patent no. US 20200326330
A1; application filed, January 29, 2020; publication date, October
15, 2020) by Premas Biotech Pvt Ltd. The platform is in the
exploratory phase and currently has no catalogue number. For 3D
monoculture tissueoids, 5,000 MCF-7 cells were used; for 3D
triculture tissueoids, a suspension of the MCF-7 cell line was
mixed with MRC-5 and HUVEC cell lines in a 1:2:1 ratio (1,250 tumor
cells, 2,500 fibroblast cells and 1,250 endothelial cells). The
cell population was used to make monoculture and triculture
tissueoids on the platform and further used for various
experiments. Briefly, ~0.8×106 cells (monoculture and
triculture) were seeded in 60-mm dish and were allowed to grow
until 70–80% confluence was reached. Cells were then washed and
centrifuged at 120 × g for 5 min at room temperature. Finally, a
cell suspension was generated so that each drop of media contained
5,000 cells. A single drop of media containing 5,000 cells was used
to make 3D tissueoids on the AXTEX-4D platform and underwent
subsequent experiments.
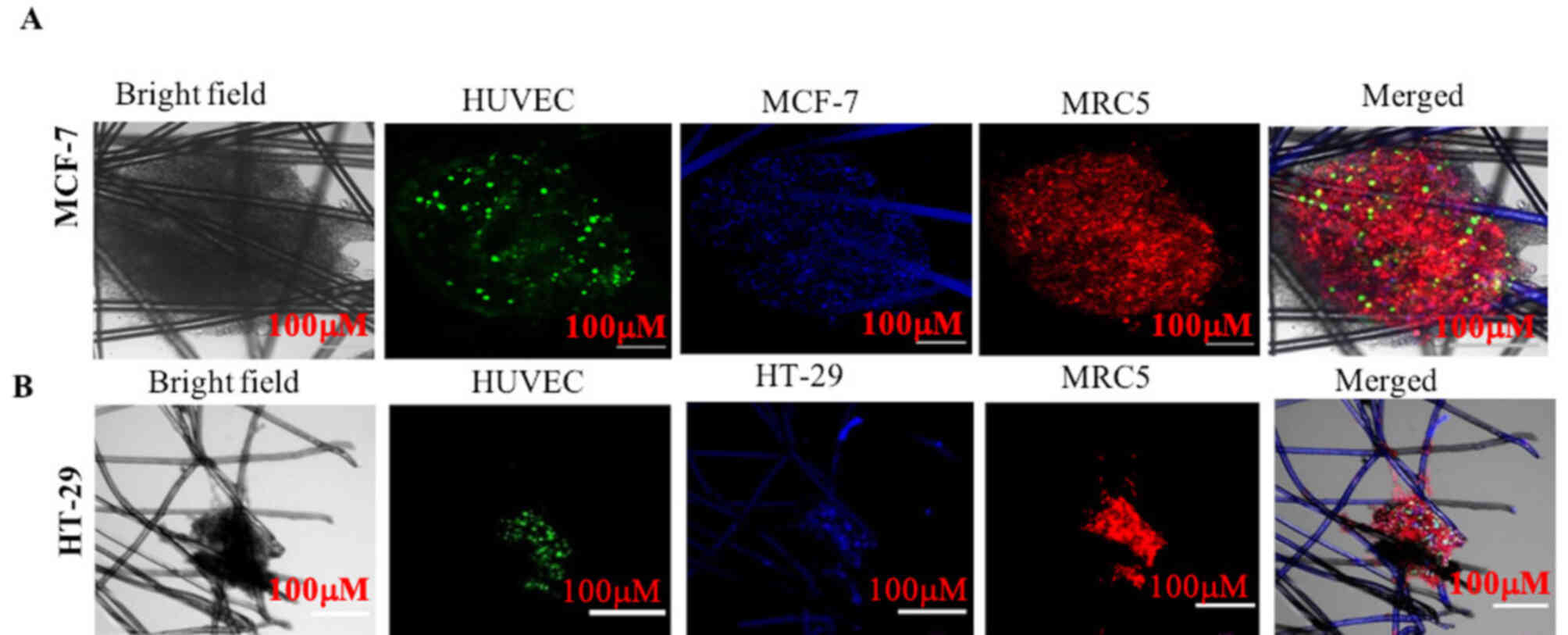
To evaluate interactions among tumor cells,
endothelial cells (HUVECs) and fibroblast cells (MRC5), MCF-7 and
HT-29 tumor cells were grown as a monoculture and as tricultures on
the AXTEX-4D platform for 4 days, since the HUVEC network would not
last beyond the 7-day time point (13). 3D MCF-7 or HT-29 tissueoids were
labelled with a blue fluorescence dye, whereas HUVECs and MRC5
cells were labelled with green and red fluorescence dyes,
respectively. The localization of all of the three cell types was
visualized via bright field or confocal microscopy and were
visually assessed.
Proliferation and ECM formation
3D monoculture and triculture tissueoids were
stained with specific antibodies to assess proliferation and ECM
formation. Briefly, tissueoids were formed on the AXTEX-4D platform
after 24 h of culture and subsequently fixed with 4%
paraformaldehyde for 15 min at room temperature, followed by
washing with 1X PBS. Subsequently, permeabilization was performed
with 0.1% Triton-X for 4 min at room temperature. After
permeabilization, samples were blocked with 1% BSA (MilliporeSigma)
for 1 h at room temperature, and were then incubated with
anti-Ki67, anti-laminin and anti-collagen I antibodies (1:1,000,
1:500 and 1:50, respectively) overnight at 4°C. Finally, tissueoids
were incubated at 37°C for 2 h with Alexa Fluor 488-conjugated
anti-rabbit secondary antibodies (for Ki67 and collagen I; 1:1,000)
and Alexa Fluor 594-conjugated anti-mouse secondary antibodies (for
laminin; 1:1,000), and were washed with PBS. Tissueoids were
counterstained with Hoechst and mounted using Prolong Gold mounting
media. Imaging was then performed using a Leica TCS SP8 confocal
microscope (Leica Microsystems, Inc.).
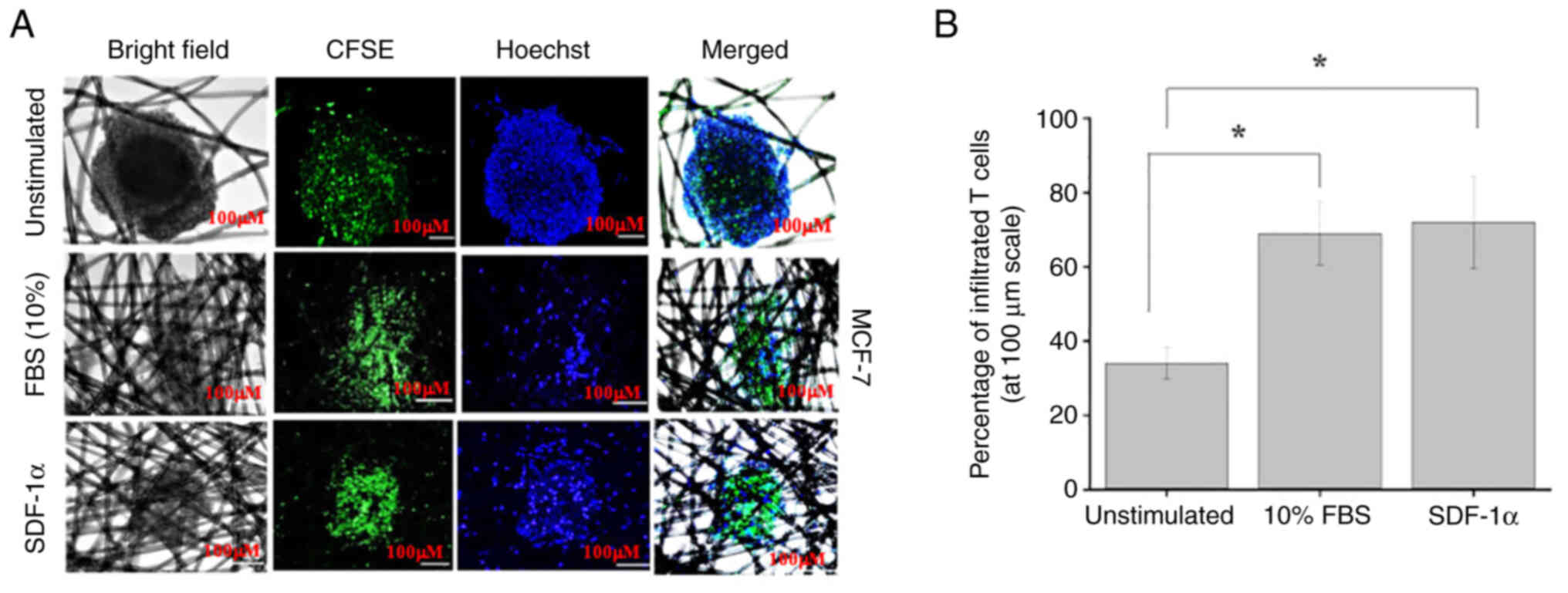
Tumor immune infiltration
Triculture tissueoids of breast cancer cells were
generated after 24 h of culture on the AXTEX-4D platform. Briefly,
100 µl Jurkat cells (5×104 cells) were poured on the
upper chamber of a Transwell system (Corning™ HTS
Transwell®−96 Tissue Culture System; cat. no. 3387;
Corning, Inc.). Jurkat cells were stained with CFSE at 37°C for 20
min and a chemoattractant (SDF1α or 10% FBS; Shenandoah
Biotechnology, Inc.) was added to the lower chamber containing
triculture tissueoids. The infiltration assay was performed as
described previously by our group. The experiment was repeated
three times, and one tissueoid per well was used to count T cells
during each repeat. Images were captured using a Nikon confocal
microscope (A1R HD25; Nikon Corp.) at ×10 objective. The Cell
Counter plug-in (version-2) of ImageJ (National Institutes of
Health) was used to count the infiltrated T cells (14). First, channels were split using
ImageJ. Using the 3D-OC set measurements, the cell measurements
were set with dot and font size of 20. Subsequently, cells present
on the edges were excluded and the background staining was
minimized using a size filter. The number of cells was then
quantified.
Alamar Blue assay
Tissueoids were formed after 24 h of culture on the
AXTEX-4D platform and were subsequently treated with drugs. Briefly
the 3D monoculture, triculture and 2D culture of MCF-7 breast
cancer cells were treated with different concentrations of
raloxifene (1, 10, 50 and 100 µM; cat. no. R0109; Tokyo Chemical
Industry Co., Ltd.) and doxorubicin (2.5, 5 and 10 µM;
MilliporeSigma) for 48 h at 37°C. Subsequently, 20 µl Alamar Blue
solution (cat. no. DAL1025; Invitrogen; Thermo Fisher Scientific,
Inc.) was directly added to 200 µl medium and the cells were
incubated for 4 h at 37°C. The Alamar Blue assay was performed as
described previously (15). An
ELISA plate reader (Spectramax Gemini EM; Molecular Devices, LLC)
was used to determine relative fluorescence units (RFU). The
following formula was used to calculate cell viability (15): Viability (%)=RFU value of cells
treated with raloxifene/RFU value of the control untreated cells
×100.
Statistical analysis
Each experiment was repeated three times and results
are presented as the mean ± SEM. The difference in cell viability
was assessed using one-way analysis of variance followed by
Bonferroni-post hoc test. OriginPro (Version:2020b) (Konark
solutions Bangalore Private Limited) was used for all statistical
analyses. P£0.05 was considered to indicate a statistically
significant difference.
Results
Establishing a triculture tissueoid
model using the AXTEX 4D platform
When two or more cell populations are used together
to develop a co-culture model, they co-exist together (16). However, in the native environment,
several phenotypically distinct cells exist together near to each
other or in a more compact organizational form and exert strong
paracrine effects (17).
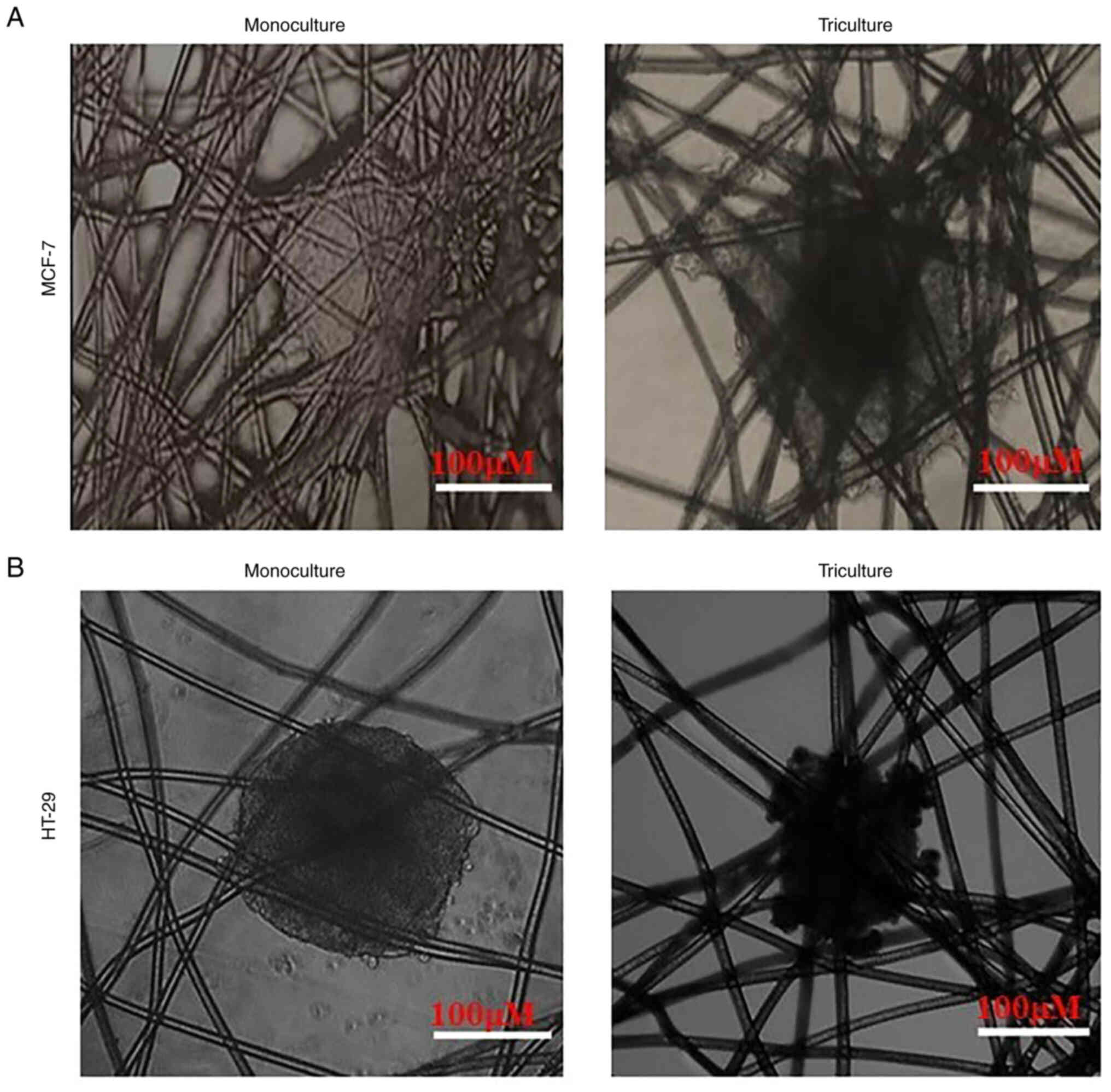
Proliferation of solid cancer cells along with HUVECs and MRC5
cells resulted in the formation of MCF-7 and HT-29 cell triculture
tissueoids (Figs. 1 and 2). It is evident that all three cells
could co-exist together in the triculture model.
Study of cellular proliferation and
ECM interactions in cancer tissueoids
The aim of the present study was to develop a
versatile triculture model that could be used for the screening of
chemotherapeutic drugs with high efficiency against cancer by
evaluating their effects on tumor progression and cell
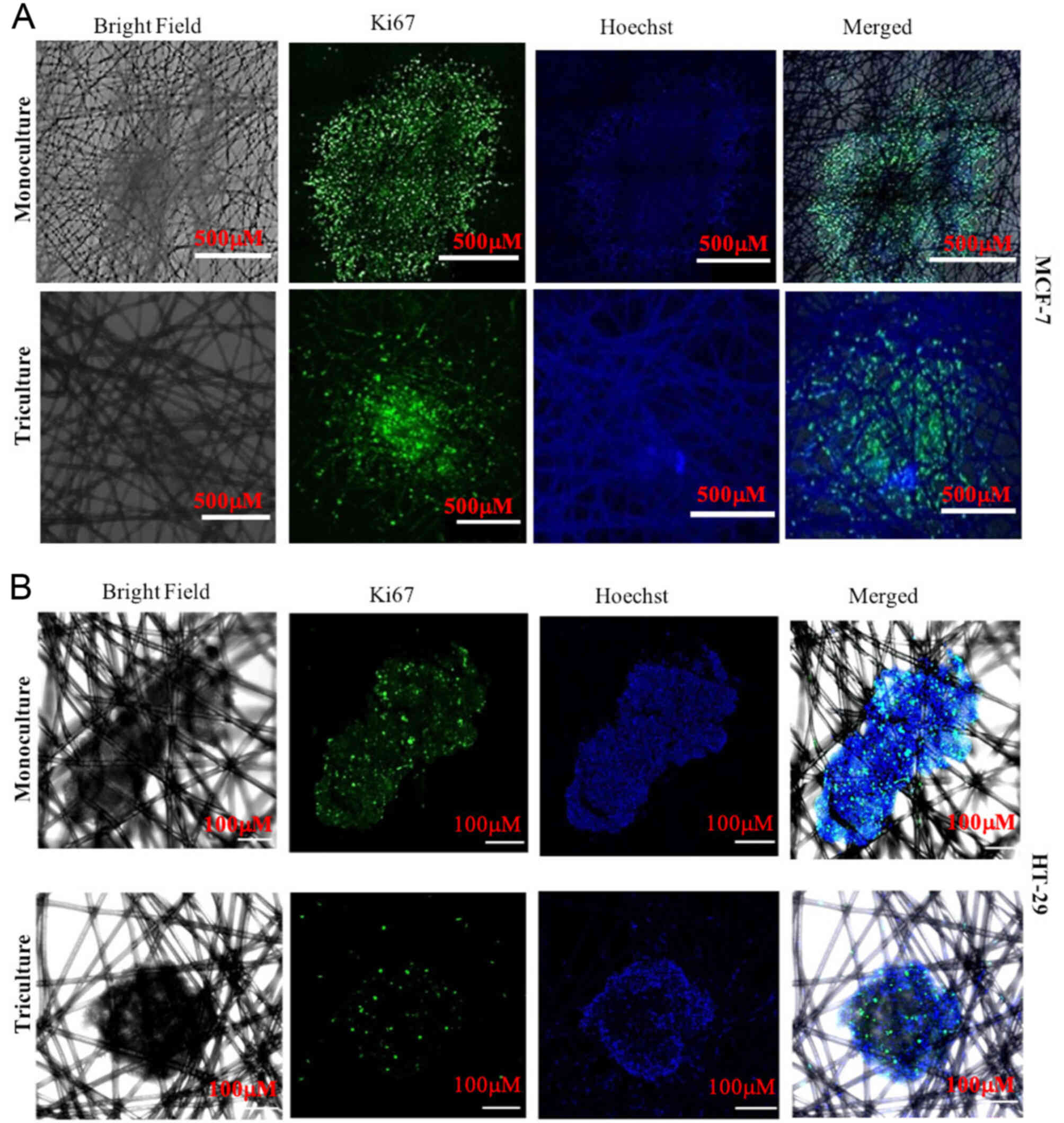
cytotoxicity. The expression of Ki67 (a marker of proliferation)
and ECM formation are strongly associated with tumor cell
proliferation, tumor progression, survival and therapeutic
resistance in the tumor microenvironment. Therefore, they are
widely used in routine screening of chemotherapeutic drugs
(18–21). Ki67 immunostaining (Fig. 3A and B) indicated that MCF-7
triculture tissueoids displayed a homogeneous distribution pattern
of Ki67-positive cells, whereas Ki67 staining was mostly restricted
to the outmost cell layers in MCF-7 monoculture tissueoids
(Fig. 3A). In contrast to MCF-7
cells, HT-29 monoculture and triculture tissueoids displayed
homogenous distribution of Ki67-positive cells, indicating that
different cancer cell types exhibit different cellular organization
in their native environment (Fig.
3B).
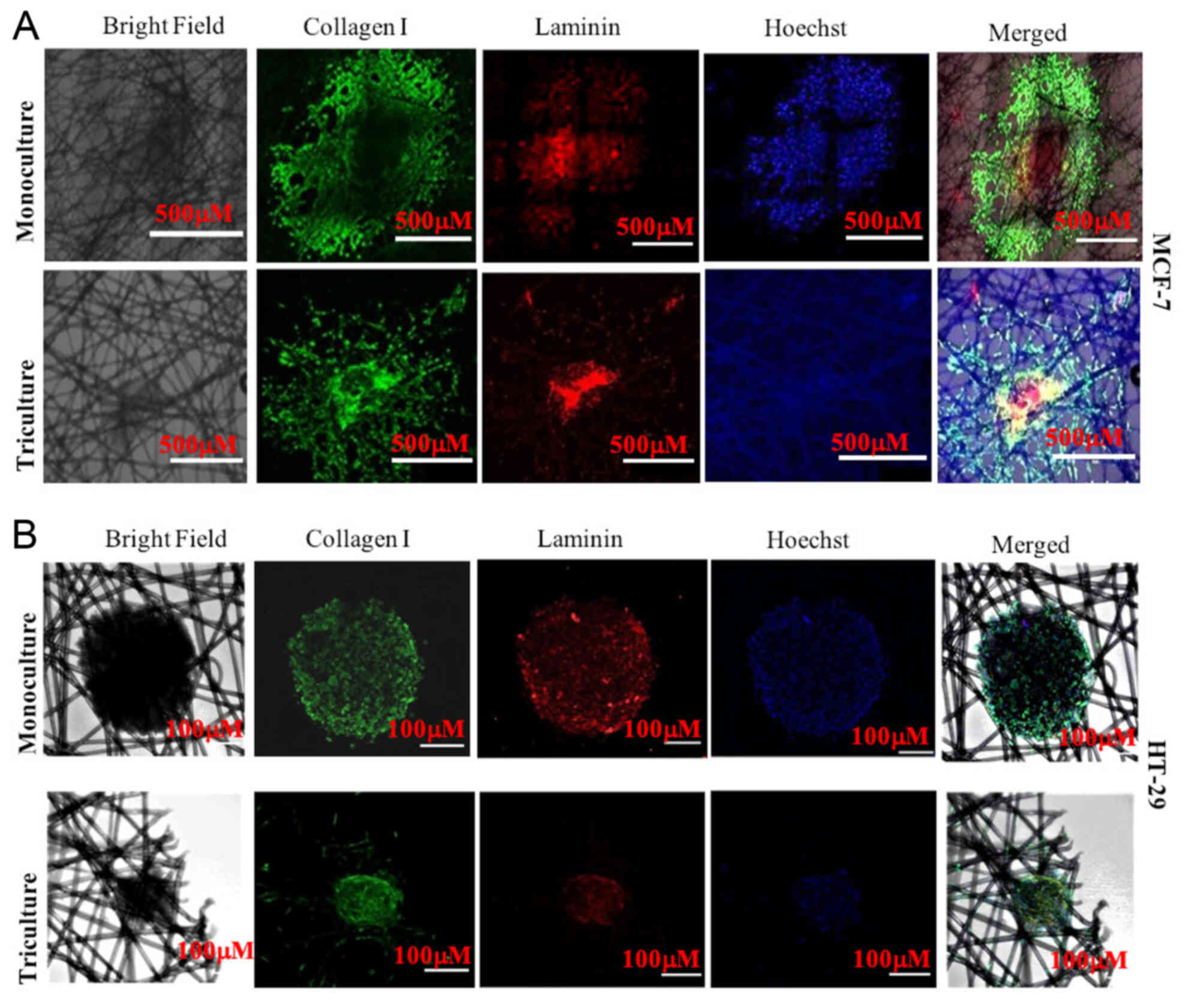
Collagens and laminins are the main component of the
ECM, which provide structural support, and regulate cell
attachment, migration and differentiation. These components promote
tissue repair and modulate cellular behavior (22,23).
The interaction of collagen and laminin is expected to be essential
for ECM formation; however, little work has been done to study the
interactions (24). In the present
study, strong signals were detected for laminin in the innermost
region of MCF-7 monoculture and triculture tissueoids, whereas
collagen was detected in the outermost region in the case of MCF-7
monoculture and triculture tissueoids (Fig. 4A). In HT-29 monoculture and
triculture tissueoids, collagen and laminin signals were detected
throughout the tissueoid (Fig.
4B). These results suggested no visible interconnection between
collagen and laminin in the MCF-7 3D tissueoids; however, it was
visible in both monoculture and triculture 3D tissueoids of HT-29
cells (Fig. 4B), suggesting the
strong structural support of the cells. However, further
investigation is essential to confirm these findings.
Infiltration of Jurkat T cells in
triculture model
The identification of drugs that manipulate the
interaction of immune system cells, fibroblasts and endothelial
cells with tumor cells may lead to novel cancer treatments
(25). Our previous study explored
the infiltration of T cells in a monoculture tissueoid model
(11). Using the same strategy in
the present study, the infiltration of immune T cells was
investigated in the triculture model. Confocal microscopy revealed
the significant infiltration of Jurkat T cells in the 3D MCF-7
triculture tissueoids in the presence of both 10% FBS and SDF-1a in
comparison to unstimulated cells (Fig.
5). However, T-cell infiltration was not obvious in the
unstimulated (0% FBS) 3D triculture tissueoid model. The results
were similar to those detected in the monoculture tissueoids in our
previous study (11).
Drug sensitivity in 2D culture, and 3D
monoculture and triculture
The lack of interactions between tumor cells and
stromal tissues or blood flow through endothelial cells in a 3D
culture model make them unable to completely mimic the TME.
Previous studies have shown greater clinical relevance of 3D
tricultures, due to their enhanced resistance to chemotherapeutics,
compared with 2D cultures (26,27).
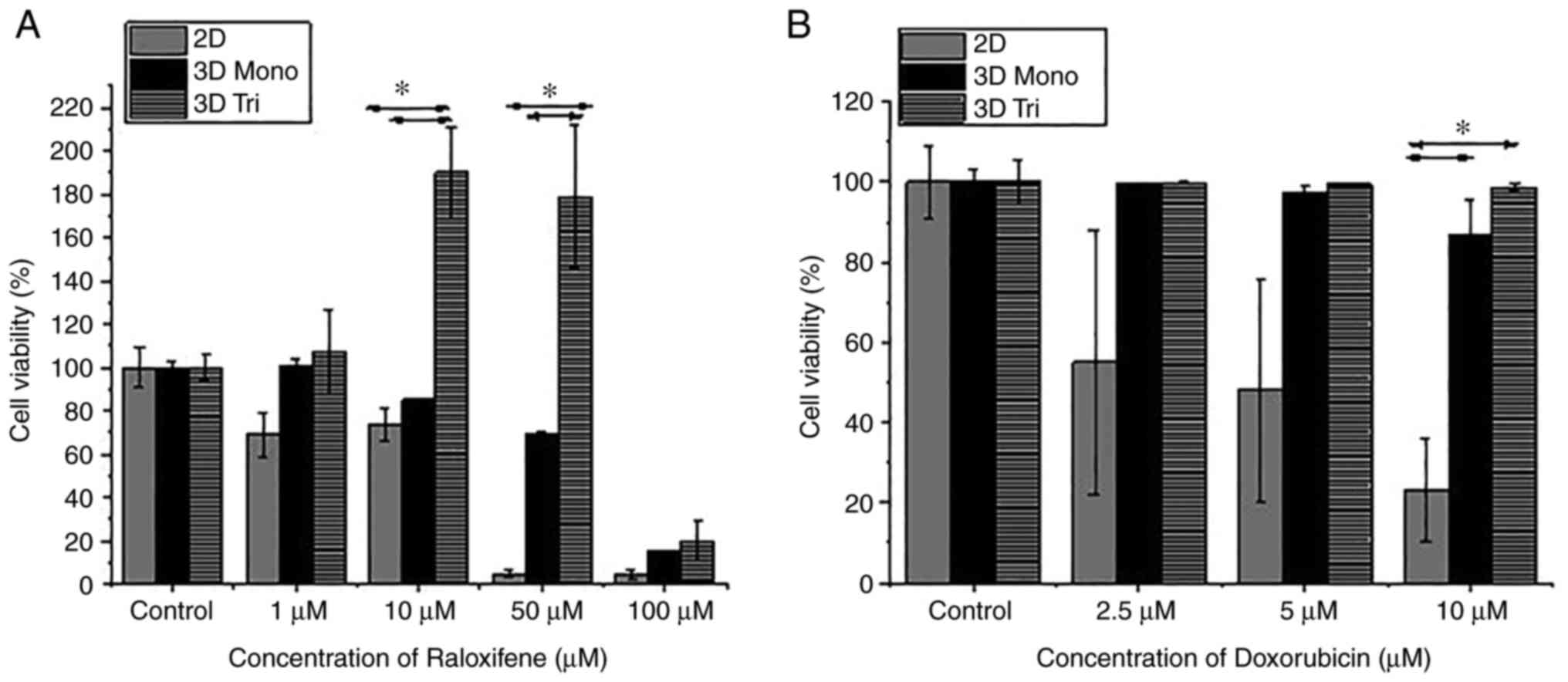
To this end, the present study evaluated drug sensitivity in a 2D
culture, and in 3D monoculture and tricultures tissueoids of MCF-7
cells by Alamar Blue assay to demonstrate drug-induced cytotoxicity
(Fig. 6). Breast cancer 3D
tricultures and monocultures were treated with different doses of
raloxifene (1, 10, 50 and 100 µM) and were compared with 2D
monocultures for 48 h. Compared with the 3D tricultures, MCF-7
cells cultured in 2D and 3D monocultures exhibited reduced cellular
viability following raloxifene treatment. The percentages of cell
viability following raloxifene treatment were as follows: 3D
triculture, 1 µM, 107.01±19.81%; 10 µM, 190.23±21.13%; 50 µM,
179.12±32.62%; 100 µM, 20.31±8.50%. 3D monoculture, 1 µM,
101.05±2.26%; 10 µM, 85.05±0.46%; 50 µM, 69.13±1.05%, 100 µM,
15.19±0.16%. 2D culture, 1 µM, 69.06±10.41%; 10 µM, 73.18±7.73%; 50
µM, 4.71±1.32%; 100 µM, 4.63±1.92% (Fig. 6A).
Similar results were obtained using doxorubicin
(Fig. 6B). The maximum serum
concentration achievable for doxorubicin is 6.73 µM (28); therefore, breast cancer 3D
tricultures and monocultures were treated with different doses of
doxorubicin (2.5, 5 and 10 µM) and compared with 2D monocultures
for 48 h. Compared with the 3D tricultures, both 2D cultures and 3D
monocultures of breast cancer cells were most sensitive to the drug
doses applied. The percentages of cell viability following
doxorubicin treatment were as follows: 3D triculture, 2.5 µM,
99.81±0.18%; 5 µM, 99.45±0.48%; 10 µM, 98.71±0.74%. 3D monoculture,
2.5 µM, 99.75±0.18%; 5 µM, 97.50±1.88%; 10 µM, 86.73±8.88%. 2D
culture, 2.5 µM, 55.03± 33%; 5 µM, 48.12±28%; 10 µM, 23.14±13%
(Fig. 6B). These results suggested
that MCF-7 3D tricultures and monocultures were significantly more
resistant to raloxifene and doxorubicin than 2D cultures, with the
triculture showing maximum drug resistance. Xenograft models of
MCF-7 tumors have been shown to exhibit drug resistance (29–32).
These results suggested that the response to anticancer drugs in
the 3D triculture and monoculture systems was more similar to the
response observed for the same drugs in xenograft models; however,
it is not true in the case of the 2D culture systems. These results
further suggested that the developed 3D monoculture and triculture
models may regulate mechanisms associated with the drug resistance
of tumor cells, such as mechanisms associated with drug
inactivation, multi-drug resistance, cell death inhibition,
DNA-repair and target gene amplification and may be used to assess
mechanisms associated with drug resistance (33).
Discussion
The present study developed a triculture tissueoid
model containing endothelial cells, fibroblasts and tumor cells on
the AXTEX-4D platform, which mimics the TME. The TME in solid
tumors not only contains tumor cells but also contains endothelial
cells, ECM, stromal cells and immune cells, which are an essential
and larger part of the tumor mass. Interactions among tumor,
vascular and other cells promotes cancer growth through cell-cell
and ECM interactions (34–36). Previous studies have also shown the
role of direct interaction of cancer cells with fibroblasts and/or
vascular cells in cancer cell invasion and metastasis (37,38).
The present study demonstrated the utility of the AXTEX-4D platform
in developing a triculture tissueoid model that may be used in
studying tumor, vascular and fibroblast cell interactions, which
serve an essential role in tumor growth, metastasis, and the
evaluation of anti-angiogenic and vascular targeting therapies.
Additionally, the ability of the AXTEX-4D platform in evaluating
the expression of proliferative markers and ECM formation, which
may control the tumor response toward therapy, was assessed.
Although the present study demonstrated the importance of the
developed triculture model in assessing tumor-related
characteristics, further investigations at the molecular level to
measure the difference in the expression levels of tumor markers
are necessary.
The ECM is associated with numerous factors, such as
tissue stiffness, interactions with relevant receptors and tumor
progression (39). The expression
of laminin receptors, laminin, collagen, collagenase and Ki67 have
been shown to be associated with pathological grade and the
clinical behavior of tumors (40).
Consistent with these studies, the present study observed the
expression of Ki67, collagen I and laminin (ECM markers) in 3D
monoculture and triculture tissueoids of MCF-7 and HT-29 cells.
However, flow cytometric analysis of proliferative and ECM markers
is required to confirm the statistical differences, which will be
performed in our future studies. Notably, differences were observed
in the localization of Ki67, collagen and laminin staining between
monocultures and tricultures. As reported previously, the
interaction of tumor cells with other cells in the stroma in the
triculture model affects the image quality of confocal microscopy
and results in a lower resolution, most likely due to altered
compactness or different cell types (41–44)
and could be the reason for the differences in the localization of
Ki67 between the monoculture and triculture models. However, more
experimental data are required to verify this. These results
suggested that the tissueoid model may be used to evaluate and
screen anticancer agents targeting proliferation and ECM formation,
and may help study the therapeutic resistance of tumor cells.
The lack of interactions between tumor cells and
stromal tissues or blood flow through endothelial cells in 3D
culture models make them unable to completely mimic the TME.
Several studies have demonstrated the drug resistance features of
3D models over 2D cultures (45,46).
For example, the 3D culture of paclitaxel-treated ovarian cancer
cells exhibited reduced cell death (40 or 60%) compared with the 2D
culture (80%) (45). These results
indicated that chemotherapeutic drugs have reduced activity in 3D
culture models and may be associated with increased drug
resistance. Limited diffusion through the spheroid, hypoxia and the
presence of stromal cells are other factors that could contribute
to drug resistance (47,48). Similar drug resistance as observed
in 3D spheroids has also been observed in vivo (49,50).
Similar to xenograft models, breast cancer spheroids have been
shown to develop multi-lobular structures, wavy protrusions and
drug resistance (51). However,
the triculture models, compared with 2D culture models, provide a
physiologically relevant model for assessing cancer cell growth,
drug response and therapeutic screening (27,51,52).
The present study demonstrated higher drug resistance to raloxifene
and doxorubicin in the triculture tissueoids compared with in the
3D monoculture and 2D culture models. However, it should be noted
that in contrast to raloxifene, doxorubicin is a DNA topoisomerase
II inhibitor that is not applied for the treatment of ERa-positive
breast cancer, such as MCF-7, which show resistance against
doxorubicin treatment (53).
In the present study, cell cytotoxicity was observed
using clinically achievable concentrations of drugs (28). These findings suggested that the
use of a 3D culture model may avert the overestimation of drug
efficacy. Increased cell viability was detected in response to
every dose of raloxifene and doxorubicin in the triculture model
compared with that in the monoculture model. Nevertheless, the
absence of significant differences in cell viability in 3D
monocultures compared with in 3D tricultures is an unpredicted
observation and needs further refinement.
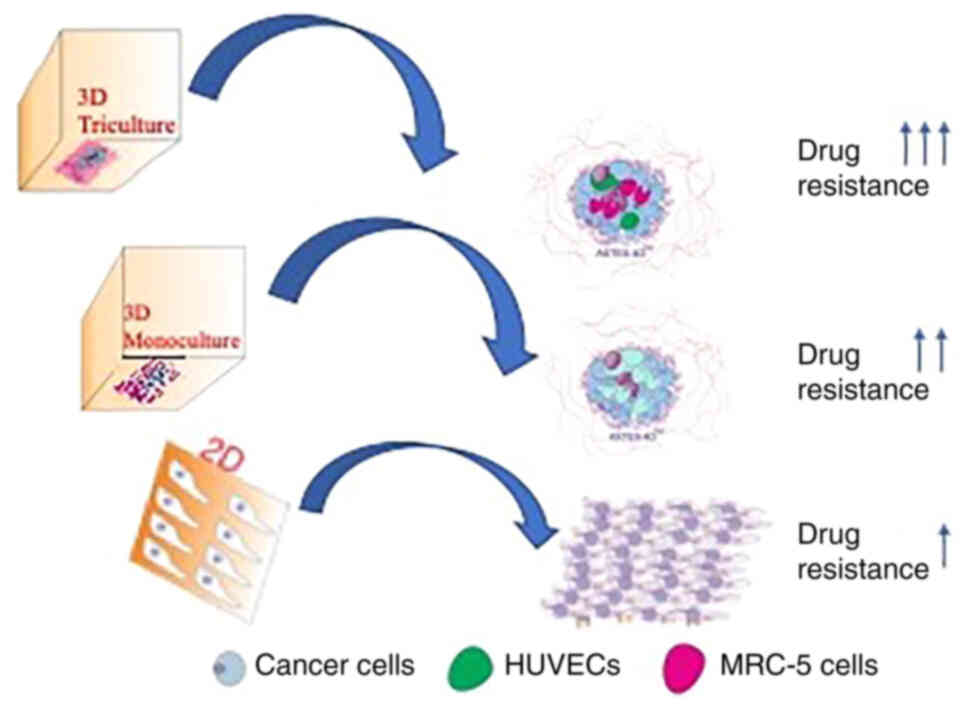
It was hypothesized that the triculture tissueoid
model involving three different cell types will produce more
clinically relevant results than 2D and 3D monocultures (Fig. 7). It should be noted that there are
several advantages and limitations to the use of advanced 3D
approaches in terms of analyzing information-rich biological data.
For example, microfluidic cell culture has the advantage of more
closely mimicking the TME, as in this system a continuous supply of
media is provided to the culture. Other advantages include the
requirement of a smaller number of cells and reagents, reduced risk
of contamination and efficient high-speed experimentation; however,
it is a complicated system that requires skills and a trained user
(54). Sample retrieval is also
difficult for further analysis. Magnetic levitation and gel
embedding methodology also have several limitations, such as poor
mechanical strength of cells and difficulty in performing
immunohistochemistry for studying biomarkers (55). By contrast, AXTEX-4D is a simple,
less expensive and user-friendly platform for performing downstream
experiments, such as fluorescence microscopy, confocal microscopy
and ELISA. However, the platform has several limitations, such as
no continuous supply of media and removal of waste, thus limiting
its automation capability (2). In
addition, the present study did not perform downstream assays on
patient-derived tumors, which is an important aspect of any cell
culture platform (56–59). Therefore, further advancement of
the model described in the present study is needed. We aim to
refine our platform with patient-derived xenografts. Furthermore,
the present study did not explore signaling pathways associated
with drug resistance.
Acknowledgements
We acknowledge Mr. Manish Kumar (CSIR-Institute of
Genomics and Integrative Biology, New Delhi) for helping in imaging
of tissueoids. We also want to acknowledge Mr. Rajan (Regional
Centre for Biotechnology, India) for their assistance in image
analysis and Mr. Avijit Das (Premas Biotech Pvt Ltd.) for their
critical discussion on the experimental study.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AB and SS designed and planned the experiments,
conducted the experiments, analyzed the results and wrote the
manuscript. SM, SK and BPDP were responsible for concept design. PK
studied the concept, designed the study, interpreted the data and
provided scientific inputs. RG analyzed data and drafted the
manuscript. NMA studied the concept, designed the study,
interpreted the data and provided scientific inputs and drafted the
manuscript. All authors read and approved the final manuscript. AB,
NM and PK confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Experiments on the HUVEC line used in the present
study were carried out after approval from the Premas Ethics
Committee.
Patient consent for publication
Not applicable.
Competing interests
All authors are employees of Premas Biotech Pvt Ltd.
A patent for the AXTEX-4D platform has been filed (patent no. US
20200326330 A1; application filed, January 29, 2020; publication
date, October 15, 2020) and is currently in the exploratory
phase.
References
|
1
|
Kapałczyńska M, Kolenda T, Przybyła W,
Zajączkowska M, Teresiak A, Filas V, Ibbs M, Bliźniak R, Łuczewski
Ł and Lamperska K: 2D and 3D cell cultures-a comparison of
different types of cancer cell cultures. Arch Med Sci. 14:910–919.
2018.PubMed/NCBI
|
|
2
|
Boghaert ER, Lu X, Hessler PE, McGonigal
TP, Oleksijew A, Mitten MJ, Foster-Duke K, Hickson JA, Santo VE,
Brito C, et al: The volume of three-dimensional cultures of cancer
cells in vitro influences transcriptional profile differences and
similarities with monolayer cultures and xenografted tumors.
Neoplasia. 19:695–706. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hirschhaeuser F, Menne H, Dittfeld C, West
J, Mueller-Klieser W and Kunz-Schughart LA: Multicellular tumor
spheroids: An underestimated tool is catching up again. J
Biotechnol. 148:3–15. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Valkenburg KC, de Groot AE and Pienta KJ:
Targeting the tumour stroma to improve cancer therapy. Nat Rev Clin
Oncol. 15:366–381. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hay M, Thomas DW, Craighead JL, Economides
C and Rosenthal J: Clinical development success rates for
investigational drugs. Nat Biotechnol. 32:40–51. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martin L, Hutchens M, Hawkins C and Radnov
A: How much do clinical trials cost? Nat Rev Drug Discov.
16:381–382. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baru A, Mazumder S, Kundu PK, Sharma S,
Das Purkayastha BP, Khan S, Gupta R and Mehrotra Arora N:
Recapitulating tumor microenvironment using AXTEX-4DTM for
accelerating cancer research and drug screening. Asian Pac J Cancer
Prev. 23:561–571. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Haisler WL, Timm DM, Gage JA, Tseng H,
Killian TC and Souza GR: Three-dimensional cell culturing by
magnetic levitation. Nat Protoc. 8:1940–1949. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lv D, Hu Z, Lu L, Lu H and Xu X:
Three-dimensional cell culture: A powerful tool in tumor research
and drug discovery. Oncol Lett. 14:6999–7010. 2017.PubMed/NCBI
|
|
10
|
Imamura Y, Mukohara T, Shimono Y,
Funakoshi Y, Chayahara N, Toyoda M, Kiyota N, Takao S, Kono S,
Nakatsura T and Minami H: Comparison of 2D- and 3D-culture models
as drug-testing platforms in breast cancer. Oncol Rep.
33:1837–1843. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Baru A, Sharma S, Purakayastha BP, Khan S,
Mazumdar S, Gupta R, Kundu PK and Arora NM: AXTEX-4D: A
three-dimensional ex vivo platform for preclinical investigations
of immunotherapy agents. Assay Drug Dev Technol. 19:361–372. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Baru A, Mazumdar S, Kundu P, Sharma S,
Purakayastha BP, Khan S, Gupta R and Arora NM: Recapitulating tumor
microenvironment using preclinical 3D tissueoids model for
accelerating cancer research and drug screening. bioRxiv. Dec
22–2020.(Epub ahead of print).
|
|
13
|
Bray LJ, Secker C, Murekatete B, Sievers
J, Binner M, Welzel PB and Werner C: Three-dimensional in vitro
hydro- and cryogel-based cell-culture models for the study of
breast-cancer metastasis to bone. Cancers (Basel). 10:2922018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu Y, Wu H, Zeng J, Pluimer B, Dong S, Xie
X, Guo X, Ge T, Liang X, Feng S, et al: Mild traumatic brain injury
induces microvascular injury and accelerates Alzheimer-like
pathogenesis in mice. Acta Neuropathol Commun. 9:742021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eilenberger C, Kratz SRA, Rothbauer M,
Ehmoser EK, Ertl P and Küpcü S: Optimized alamarBlue assay protocol
for drug dose-response determination of 3D tumor spheroids.
MethodsX. 5:781–787. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lazzari G, Nicolas V, Matsusaki M, Akashi
M, Couvreur P and Mura S: Multicellular spheroid based on a triple
co-culture: A novel 3D model to mimic pancreatic tumor complexity.
Acta Biomater. 78:296–307. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lamichhane SP, Arya N, Kohler E, Xiang S,
Christensen J and Shastri VP: Recapitulating epithelial tumor
microenvironment in vitro using three dimensional tri-culture of
human epithelial, endothelial, and mesenchymal cells. BMC Cancer.
16:5812016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shah PP, Dupre TV, Siskind LJ and Beverly
LJ: Common cytotoxic chemotherapeutics induce
epithelial-mesenchymal transition (EMT) downstream of ER stress.
Oncotarget. 8:22625–22639. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bai C, Yang M, Fan Z, Li S, Gao T and Fang
Z: Associations of chemo- and radio-resistant phenotypes with the
gap junction, adhesion and extracellular matrix in a
three-dimensional culture model of soft sarcoma. J Exp Clin Cancer
Res. 34:582015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Senthebane DA, Jonker T, Rowe A, Thomford
NE, Munro D, Dandara C, Wonkam A, Govender D, Calder B, Soares NC,
et al: The role of tumor microenvironment in chemoresistance: 3D
extracellular matrices as accomplices. Int J Mol Sci. 19:28612018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ellis MJ, Suman VJ, Hoog J, Goncalves R,
Sanati S, Creighton CJ, DeSchryver K, Crouch E, Brink A, Watson M,
et al: Ki67 proliferation index as a tool for chemotherapy
decisions during and after neoadjuvant aromatase inhibitor
treatment of breast cancer: Results from the American college of
surgeons oncology group Z1031 trial (alliance). J Clin Oncol.
35:1061–1069. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Plant AL, Bhadriraju K, Spurlin TA and
Elliott JT: Cell response to matrix mechanics: Focus on collagen.
Biochim Biophys Acta. 1793:893–902. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mak KM and Mei R: Basement membrane type
IV collagen and laminin: An overview of their biology and value as
fibrosis biomarkers of liver disease. Anat Rec (Hoboken).
300:1371–1390. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nugroho RWN, Harjumäki R, Zhang X, Lou YR,
Yliperttula M, Valle-Delgado JJ and Österberg M: Quantifying the
interactions between biomimetic biomaterials-collagen I, collagen
IV, laminin 521 and cellulose nanofibrils-by colloidal probe
microscopy. Colloids Surf B Biointerfaces. 173:571–580. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Koeck S, Zwierzina M, Lorenz E, Gamerith
G, Zwierzina H and Amann A: Infiltration of immune cells into
cancer cell/stroma cell 3D microtissues. J Immunother Cancer. 3
(Suppl 2):P752015. View Article : Google Scholar
|
|
26
|
Bray LJ, Binner M, Körner Y, von Bonin M,
Bornhäuser M and Werner C: A three-dimensional ex vivo tri-culture
model mimics cell-cell interactions between acute myeloid leukemia
and the vascular niche. Haematologica. 102:1215–1226. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bruce A, Evans R, Mezan R, Shi L, Moses
BS, Martin KH, Gibson LF and Yang Y: Three-dimensional microfluidic
tri-culture model of the bone marrow microenvironment for study of
acute lymphoblastic leukemia. PLoS One. 10:e01405062015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liston DR and Davis M: Clinically relevant
concentrations of anticancer drugs: A guide for nonclinical
studies. Clin Cancer Res. 23:3489–3498. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Balaburski GM, Dardes RC, Johnson M,
Haddad B, Zhu F, Ross EA, Sengupta S, Klein-Szanto A, Liu H, Lee
ES, et al: Raloxifene-stimulated experimental breast cancer with
the paradoxical actions of estrogen to promote or prevent tumor
growth: A unifying concept in anti-hormone resistance. Int J Oncol.
37:387–398. 2010.PubMed/NCBI
|
|
30
|
Brady H, Desai S, Gayo-Fung LM,
Khammungkhune S, McKie JA, O'Leary E, Pascasio L, Sutherland MK,
Anderson DW, Bhagwat SS and Stein B: Effects of SP500263, a novel,
potent antiestrogen, on breast cancer cells and in xenograft
models. Cancer Res. 62:1439–1442. 2002.PubMed/NCBI
|
|
31
|
Schafer JM and Jordan VC: Models of
hormone resistance in vitro and in vivo. Methods Mol Med.
120:453–464. 2006.PubMed/NCBI
|
|
32
|
Azab SS, Salama SA, Hassan MH, Khalifa AE,
El-Demerdash E, Fouad H, Al-Hendy A and Abdel-Naim AB:
2-Methoxyestradiol reverses doxorubicin resistance in human breast
tumor xenograft. Cancer Chemother Pharmacol. 62:893–902. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mansoori B, Mohammadi A, Davudian S,
Shirjang S and Baradaran B: The different mechanisms of cancer drug
resistance: A brief review. Adv Pharm Bull. 7:339–348. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Walter M, Liang S, Ghosh S, Hornsby PJ and
Li R: Interleukin 6 secreted from adipose stromal cells promotes
migration and invasion of breast cancer cells. Oncogene.
28:2745–2755. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Neiva KG, Warner KA, Campos MS, Zhang Z,
Moren J, Danciu TE and Nör JE: Endothelial cell-derived
interleukin-6 regulates tumor growth. BMC Cancer. 14:992014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Baghban R, Roshangar L, Jahanban-Esfahlan
R, Seidi K, Ebrahimi-Kalan A, Jaymand M, Kolahian S, Javaheri T and
Zare P: Tumor microenvironment complexity and therapeutic
implications at a glance. Cell Commun Signal. 18:592020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamaguchi H and Sakai R: Direct
interaction between carcinoma cells and cancer associated
fibroblasts for the regulation of cancer invasion. Cancers (Basel).
7:2054–2062. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Akino T, Hida K, Hida Y, Tsuchiya K,
Freedman D, Muraki C, Ohga N, Matsuda K, Akiyama K, Harabayashi T,
et al: Cytogenetic abnormalities of tumor-associated endothelial
cells in human malignant tumors. Am J Pathol. 175:2657–2667. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Winkler J, Abisoye-Ogunniyan A, Metcalf KJ
and Werb Z: Concepts of extracellular matrix remodelling in tumour
progression and metastasis. Nat Commun. 11:51202020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Grigioni WF, Garbisa S, D'Errico A,
Baccarini P, Stetler-Stevenson WG, Liotta LA and Mancini AM:
Evaluation of hepatocellular carcinoma aggressiveness by a panel of
extracellular matrix antigens. Am J Pathol. 138:647–654.
1991.PubMed/NCBI
|
|
41
|
Nürnberg E, Vitacolonna M, Klicks J, von
Molitor E, Cesetti T, Keller F, Bruch R, Ertongur-Fauth T, Riedel
K, Scholz P, et al: Routine optical clearing of 3D-cell cultures:
Simplicity forward. Front Mol Biosci. 7:202020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Imbalzano KM, Tatarkova I, Imbalzano AN
and Nickerson JA: Increasingly transformed MCF-10A cells have a
progressively tumor-like phenotype in three-dimensional basement
membrane culture. Cancer Cell Int. 9:72009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Alessandri K, Sarangi BR, Gurchenkov VV,
Sinha B, Kießling TR, Fetler L, Rico F, Scheuring S, Lamaze C,
Simon A, et al: Cellular capsules as a tool for multicellular
spheroid production and for investigating the mechanics of tumor
progression in vitro. Proc Natl Acad Sci USA. 110:14843–14848.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Raghavan S, Mehta P, Horst EN, Ward MR,
Rowley KR and Mehta G: Comparative analysis of tumor spheroid
generation techniques for differential in vitro drug toxicity.
Oncotarget. 7:16948–16961. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Loessner D, Stok KS, Lutolf MP, Hutmacher
DW, Clements JA and Rizzi SC: Bioengineered 3D platform to explore
cell-ECM interactions and drug resistance of epithelial ovarian
cancer cells. Biomaterials. 31:8494–8506. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Karlsson H, Fryknäs M, Larsson R and
Nygren P: Loss of cancer drug activity in colon cancer HCT-116
cells during spheroid formation in a new 3-D spheroid cell culture
system. Exp Cell Res. 318:1577–1585. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Trédan O, Galmarini CM, Patel K and
Tannock IF: Drug resistance and the solid tumor microenvironment. J
Natl Cancer Inst. 99:1441–1454. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Barbosa MA, Xavier CP, Pereira RF,
Petrikaite V and Vasconcelos MH: 3D cell culture models as
recapitulators of the tumor microenvironment for the screening of
anti-cancer drugs. Cancers (Basel). 14:1902021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Edmondson R, Broglie JJ, Adcock AF and
Yang L: Three-dimensional cell culture systems and their
applications in drug discovery and cell-based biosensors. Assay
Drug Dev Technol. 12:207–218. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yip D and Cho CH: A multicellular 3D
heterospheroid model of liver tumor and stromal cells in collagen
gel for anti-cancer drug testing. Biochem Biophys Res Commun.
433:327–332. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Benton G, DeGray G, Kleinman HK, George J
and Arnaoutova I: In vitro microtumors provide a physiologically
predictive tool for breast cancer therapeutic screening. PLoS One.
10:e01233122015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cornelison RC, Yuan JX, Tate KM, Petrosky
A, Beeghly GF, Bloomfield M, Schwager SC, Berr AL, Cimini D,
Bafakih FF, et al: A patient-designed tissue-engineered model of
the infiltrative glioblastoma microenvironment. bioRxiv. Jul
29–2020.(Epub ahead of print).
|
|
53
|
Wan X, Hou J, Liu S, Zhang Y, Li W, Zhang
Y and Ding Y: Estrogen receptor α mediates doxorubicin sensitivity
in breast cancer cells by regulating E-cadherin. Front Cell Dev
Biol. 9:5835722021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Halldorsson S, Lucumi E, Gómez-Sjöberg R
and Fleming RM: Advantages and challenges of microfluidic cell
culture in polydimethylsiloxane devices. Biosens Bioelectron.
63:218–231. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ventola CL: Medical applications for 3D
printing: Current and projected uses. P T. 39:704–711.
2014.PubMed/NCBI
|
|
56
|
Marangoni E, Vincent-Salomon A, Auger N,
Degeorges A, Assayag F, de Cremoux P, de Plater L, Guyader C, De
Pinieux G, Judde JG, et al: A new model of patient tumor-derived
breast cancer xenografts for preclinical assays. Clin Cancer Res.
13:3989–3998. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cottu P, Marangoni E, Assayag F, de
Cremoux P, Vincent-Salomon A, Guyader Ch, de Plater L, Elbaz C,
Karboul N, Fontaine JJ, et al: Modeling of response to endocrine
therapy in a panel of human luminal breast cancer xenografts.
Breast Cancer Res Treat. 133:595–606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang X, Claerhout S, Prat A, Dobrolecki
LE, Petrovic I, Lai Q, Landis MD, Wiechmann L, Schiff R, Giuliano
M, et al: A renewable tissue resource of phenotypically stable,
biologically and ethnically diverse, patient-derived human breast
cancer xenograft models. Cancer Res. 73:4885–4897. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fong EL, Martinez M, Yang J, Mikos AG,
Navone NM, Harrington DA and Farach-Carson MC: Hydrogel-based 3D
model of patient-derived prostate xenograft tumors suitable for
drug screening. Mol Pharm. 11:2040–2050. 2014. View Article : Google Scholar : PubMed/NCBI
|