Introduction
Lung cancer (LC) is the most common cancer worldwide
and the second largest cause of cancer morbidity and mortality,
with 2.2 million new cases and 1.8 million deaths in 2020 (1). It is predicted that the number of
incident cases of LC will reach 3.8 million by 2050 (2). LC is classified into two types based
on pathological characteristics: small cell lung cancer (SCLC) and
non-small cell lung cancer (NSCLC), with the latter including lung
adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC) and
large cell carcinoma (LCC) (3).
NSCLC is the most common pathological type of LC, which accounts
for ~85% of all cases. The mechanism of leading to LC is the
apoptosis of alveolar epithelial cells mediated by asbestos via
mitochondrial and p53-regulated death pathways in the human body
(4). Additional disorders
associated with LC include chronic obstructive pulmonary disease,
tuberculosis, emphysema and interstitial lung disease (5,6).
Due to the insidious nature of NSCLC, the illness is
typically diagnosed at advanced stages and the 5-year survival rate
is less than 15% (7). Patients
with NSCLC who receive radical surgery at an early stage can have
the 5-year survival rate of 40–70% (8). Therefore, early diagnosis of NSCLC
could significantly reduce patient mortality. Currently, the main
clinical strategy for diagnosing NSCLC is a low-dose computed
tomography (LDCT) scan. However, LDCT has certain drawbacks, such
as overdiagnosis, harmful radiation exposure from repeated
detections, and elevated anxiety in patients. Researchers have
studied certain new biomarkers with high specificity and
sensitivity for NSCLC, such as microRNAs (miRNAs), long non-coding
RNAs (lncRNAs), circular RNAs (circRNAs), circulating tumor cells
and circulating tumor DNA (9–11).
Particularly, miRNAs have attracted more attention in the field of
high-quality biomarkers.
miRNAs are small and evolutionarily conserved class
of non-coding RNAs (ncRNAs) with a size of ~19–25 nucleotides.
miRNAs are key transcriptional regulators and can affect a variety
of biological functions by targeting the 3′ untranslated region
(UTR) of messenger RNA (mRNA) to induce mRNA degradation and
inhibit translation (12). The
first miRNA, lin4, was discovered in Caenorhabditis elegans
in 1993 (13). At present, more
than 2,000 miRNAs have been identified in the human genome, which
are involved in the regulation of a variety of physiological and
pathological processes. Therefore, miRNAs have been widely
researched as potential biomarkers and therapeutic targets
(14).
Exosomes are membrane-enclosed extracellular
vesicles with a diameter of 30–150 nm. In addition to proteins and
lipids, exosomes also contain a rich trove of nucleic acid
metabolites such as miRNAs and lncRNAs (15). Tumor cells generate exosomes that
contain abundant miRNAs, and tumor-specific exosomal miRNAs vanish
when tumor tissue is removed. The expression profiles of exosomal
miRNAs derived from plasma or serum are significantly different
between NSCLC patients and healthy controls (16).
In the present review, a variety of serum and plasma
miRNAs with high specificity and sensitivity that play an important
role in the early diagnosis of NSCLC were summarized. Finally, the
value of exosomal miRNAs as novel biomarkers for NSCLC diagnosis
was emphasized.
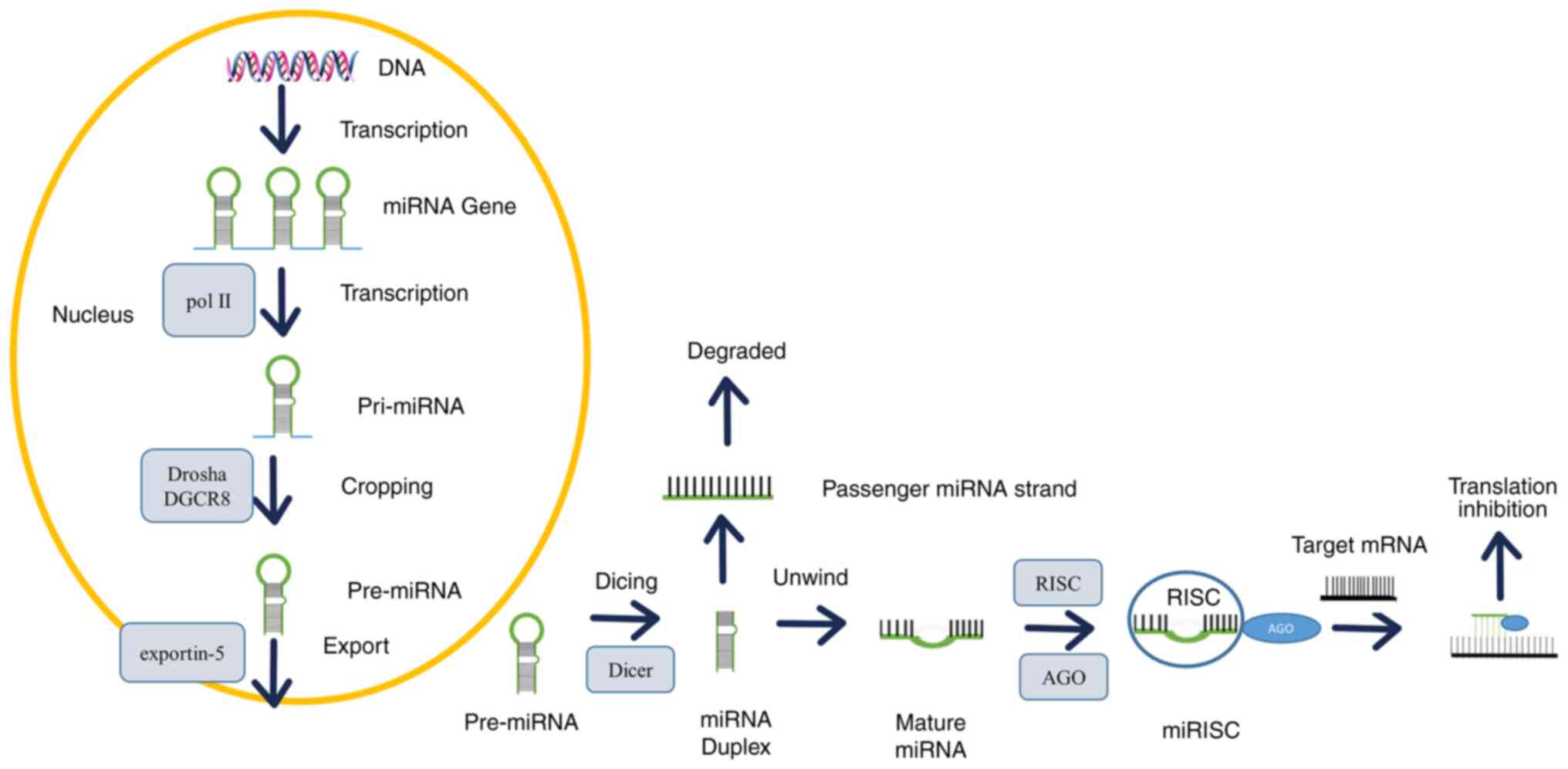
miRNA biogenesis and function
The majority of miRNA genes are located in the
intron and intergenic regions of protein-coding genes (17), which are transcribed by RNA
polymerase II (Pol II) and RNA polymerase III (Pol III) (18). The typical biogenesis of miRNAs
involves the beginning in the nucleus and the ending in the
cytoplasm (Fig. 1), and comprises
three main events: cropping, export and dicing (19). miRNA is typically generated from a
primary miRNA (pri-miRNA) transcript through two consecutive
cutting events. The pri-miRNA is usually added with a 5′ cap
structure and a 3′ poly (A) tail, containing one or more long
hairpin structures. Because the structural characteristics of these
hairpins are unique, they can be distinguished from various RNA
stem ring-like structures in the nucleus. Pri-miRNA hairpins
typically have an imperfect 30 bp stem with flanking
single-stranded RNA fragments at the base (20). Initially, pri-miRNAs are cleaved in
the nucleus by a microprocessor complex, which is composed of the
RNase III enzyme Drosha, the double-stranded RNA-binding protein
(RBP) DiGeorge syndrome critical region gene 8 (DGCR8) and
associated proteins (21). DGCR8
recognizes the connection between the stem and the flanking
single-stranded RNA of the pri-miRNA hairpin, recruits Drosha, cuts
the RNA double strand, and produces a 70-bp stem-loop structure
known as precursor miRNA (pre-miRNA) (22,23).
Methyltransferase-like 3 (METTL3) methylates pri-miRNA, which can
be recognized and processed by DGCR8. METTL3 depletion reduces
DGCR8 and pri-miRNA binding, resulting in a decrease in mature
miRNA and an increase in unprocessed pri-miRNA accumulation
(24). Pre-miRNA is transported to
the cytoplasm by exportin-5, and cleaved by the cytoplasmic RNase
III enzyme Dicer to produce mature double-stranded miRNA. Then the
mature double-stranded miRNA binds to Argonaute (AGO) protein and
forms a miRNA-induced silencing complex (miRISC). The mature chain
is kept in miRISC, while the over-guest chain is released and
degraded (25). By complementary
pairing with the binding site of the 3′-UTR mRNAs, miRNAs lead to
target mRNA degradation and/or translation inhibition of target
genes (26). Under normal
physiological conditions, miRNAs regulate cell biological processes
such as proliferation, differentiation, apoptosis and protein
synthesis. Therefore, its disturbance is involved in the regulation
of tumor development and progression. In addition, a single miRNA
can regulate multiple interaction networks and translation
processes by targeting multiple mRNAs, whereas an mRNA can be
regulated by multiple miRNAs (27).
miRNAs and the pathogenesis of NSCLC
miRNAs regulate cellular processes in both
physiological and pathological conditions. A miRNA can bind to one
or more mRNAs, affecting the expression of oncogenes and tumor
suppressor genes, which is related to the pathogenesis of NSCLC. In
NSCLC, various miRNAs are upregulated or downregulated and serve as
either oncogenic miRNAs or tumor suppressor miRNAs. Several
significant miRNAs implicated in the development of NSCLC are
listed in Table I.
 | Table I.Roles of miRNAs in the occurrence and
progression of NSCLC. |
Table I.
Roles of miRNAs in the occurrence and
progression of NSCLC.
| miRNA | Target gene | Type of
dysregulation | Function | (Refs.) |
|---|
| miR-224 | Sirtuins3 | Up | Promotes NSCLC
cells proliferation, EMT and invasion | (28) |
| miR-629 | FOXO1 | Up | Promotes
proliferation, migration and invasion in NSCLC cells | (29) |
| miR-654-3p | RASAL2 | Down | Suppresses the
viability and induce the apoptosis of NSCLC cells | (30) |
| miR-139-5p | HOXB2 | Down | Inhibits
proliferation and promotes apoptosis, enhances cisplatin
sensitivity in NSCLC cells | (31) |
| miR-144 | TLR2 | Down | Inhibits NSCLC
cells migration, invasion and release of inflammatory cytokines
related to tumor chemical resistance | (32) |
| miR-195-5p | HOXA10 | Down | Inhibits the
growth, invasion and migration of LUAD, increases the proportion of
cells in G1 phase in the cell cycle, promotes apoptosis and
enhances radio sensitivity | (33) |
| miR-196b | AQP4 | Up | Promotes the
invasion and migration of LUAD cells, and the poor prognosis and
low survival in patients with LUAD | (34) |
| miR-20a-5p | KLF9 | Up | Promotes the
proliferation and invasion of NSCLC cells | (35) |
| miR-1 | Mpl | Down | Inhibits NSCLC
growth and angiogenesis | (36) |
| miR-3157-3p | TIMP2/KLF2 | Up | Promotes
angiogenesis and increased vascular permeability | (37) |
| miR-543 | MTA1 | Up | Promotes
proliferation and angiogenesis in NSCLC | (38) |
| miR-153 | Jagged1/Notch1 | Down | Inhibits stem cell
properties and tumor growth of lung cancer cells | (39) |
| miR-582-3p | AXIN2, DKK3 and
SFRP1 | Up | Maintain stem
cell-like characteristics and promote tumorigenesis,
chemoresistance and relapse of NSCLC cells | (40) |
miR-224
miR-224 plays a dual function in various cancer
cells. It plays an oncogenic role in the formation and progression
of numerous kinds of malignant cancers, including NSCLC (28), breast cancer (41) and colorectal carcinogenesis
(42). Otherwise, it functions as
a tumor suppressor and is downregulated in certain patients with
uveal melanoma (43).
Sirtuin3 (SIRT3) is a member of the sirtuin family
of NAD+-dependent deacetylases. By controlling the
acetylation of several mitochondrial proteins, it contributes to
biological processes like energy metabolism and cell aging. It is
also intimately associated to the formation and development of
cancers (44). In comparison to
paracancerous tissues and healthy controls, the expression levels
of SIRT3 have significantly increased in NSCLC tissue and serum.
SIRT3 may function as a tumor suppressor in NSCLC because its level
was inversely related to tumor size, lymph node metastasis and TNM
stage in individuals (45).
miR-224 inhibits expression of SIRT3 and targets its 3′-UTR,
contributing to the development of cancer. The overexpression of
miR-224 may drastically reduce the degree of AMP-activated protein
kinase (AMPK) phosphorylation in the co-culture model of
cancer-associated fibroblasts (CAF) and NSCLC cells.
Simultaneously, forced overexpression of SIRT3 may improve AMPK
activation and counteract miR-224 mediated AMPK suppression.
Additionally, miR-224 can stimulate the mammalian target of
rapamycin/hypoxia inducible factor-1α (mTOR/HIF-1α) signal pathway
to control the growth of NSCLC (28). mTOR is a serine-threonine kinase
that acts as a crucial regulatory protein in typical cell
physiology. The mTOR signaling pathway is crucial for regulating
signals that promote cancer cell growth and survival and is a
significant contributor to the development of NSCLC and other types
of cancer (46). In addition, AMPK
can inhibit mTOR (47). With the
growth of NSCLC tumor, hypoxia aggravates the expression of HIF-1α.
HIF-1α overexpression can activate downstream signaling molecules
like vascular endothelial growth factor A (VEGFA) and accelerate
the growth of tumors in NSCLC (48). Higher levels of HIF-1α stimulate
the production of miR-224, producing the
miR-224-SIRT3-AMPK-mTOR-HIF-1α positive feedback loop, which
promotes tumor development, angiogenesis, and metastasis in NSCLC
cells by targeting SIRT3 and inhibiting AMPK (28).
Phosphatase and tensin homolog (PTEN), as a
prominent negative regulator of cell growth and
phosphatidylinositol-3-kinase/v-akt murine thymoma viral oncogene
homolog (PI3K/AKT) signaling pathway, plays a role as a tumor
suppressor gene. Abnormal pathological features are caused by the
loss of PTEN expression in numerous cancers (49). In serum-starved A549 cells, miR-224
negatively regulates PTEN through PI3K signal pathway to inhibit
cell proliferation and induces apoptosis and autophagy due to the
change of tumor microenvironment (50).
Angiopoietin-likeprotein1 (ANGPTL1) is another
target gene of miR-224. ANGPTL1 prevents angiogenesis and the
spread of cancer by acting as a tumor suppressor and an
anti-angiogenic factor (51).
Overexpression of snail family zinc finger 2 (SLUG) promotes tumor
cell migration. By decreasing the expression of the SLUG, ANGPTL1
inhibits epithelial-mesenchymal transition (EMT) (52). The ectopic expression of miR-224
enhances NSCLC cell proliferation, migration, and lymph node
metastasis by directly targeting ANGPTL1 mRNA (53).
miR-139-5p
miR-139-5p can function as tumor suppressor, which
mainly regulates the translation of mRNA at the
post-transcriptional level and plays an inhibitory role by
mediating a variety of target genes and downstream signal pathways.
The expression of miR-139-5p is decreased in multiple cancer
tissues, such as pancreatic cancer, colorectal cancer and
hepatocellular carcinoma. Therefore, miR-139-5p can be forcedly
overexpressed in tumors to prevent the proliferation, invasion, and
migration of tumor cells (54–56).
As the target gene of miR-139-5p, homeobox B2
(HOXB2) is a member of the homeobox (HOX) transcription factor
family. The majority of the HOX proteins encoded by HOX gene
function as transcription factors, regulating embryonic
development, cell differentiation and carcinogenesis. HOXB2 is a
crucial gene in the regulation of cell differentiation (57). miR-139-5p inhibits HOXB2 expression
when it selectively binds to the 3′-UTR of HOXB2 and decreases
tumor growth by promoting apoptosis in cells and inhibiting cell
proliferation. In NSCLC cells treated with cisplatin (DDP),
overexpression of miR-139-5p overcomes DDP resistance by regulating
the PI3K/AKT/caspase-3 signaling pathway (31). The PI3K/AKT signaling pathway
modulates diverse cellular processes, including cell proliferation.
Caspase-3 is the key execution enzyme in cell survival and
apoptosis (58). Therefore,
overexpression of miR-139-5p inhibits cell proliferation and
promotes cell apoptosis by downregulating the PI3K/AKT signaling
pathway and increasing caspase-3 expression. Overexpression of
miR-139-5p could attenuate paclitaxel (PTX) resistance of NSCLC
cells. Integrin beta-8 (ITGB8) is an integrin family number.
Integrins are located on the surface of cancer cells and promote
tumor metastasis by mediating cell-to-cell adhesion and invasion.
ITGB8 is typically upregulated in cancer and is associated with
cancer metastasis (59).
Delta/Notch-like epidermal growth factor-related receptor (DNER) is
a transmembrane protein involved in the tumor development. CircDNER
has been proved to function as a miRNA sponge to sequester
miR-139-5p away from its target mRNA, thus reducing miRNA-mediated
gene inhibition. ITGB8 is the target gene of miR-139-5p, and
circDNER/miR-139-5p/ITGB8 forms a new regulatory axis. Enforced
overexpression of miR-139-5p in PTX-resistant NSCLC cells reverses
the tumor promoting functions of circDNER on NSCLC cell
proliferation and invasion by targeting and inhibiting ITGB8, while
also slows the growth of tumors and encourages the apoptosis of
PTX-resistant cells (60).
Fine particulate matter (PM2.5) accelerates the
development of NSCLC by suppressing the expression of miR-139-5p.
High dose PM2.5 stimulation causes precancerous lesions such as
bronchial epithelial dysplasia in mice, which promotes the EMT
process and raises the risk of LC by lowering the level of
E-cadherin protein and raising the level of vimentin protein.
Meanwhile, PM2.5 regulates Notch1, the target of miR-139-5p
(61). Notch1 is overexpressed in
NSCLC, and that is associated with the disease's progression and
poor prognosis (62).
Overexpression of miR-139-5p can prevent NSCLC from developing by
suppressing Notch1 expression and reversing PM2.5-induced EMT,
indicating that miR-139-5p has the potential to be a therapeutic
target in NSCLC.
miR-152
miR-152 is considered as a tumor suppressor and its
expression is usually downregulated in different solid tumors.
Tensin1 (TNS1), a member of the focal adhesion-associated proteins
family, is essential for preserving normal tissue and structural
stability. TNS1 is involved in cell proliferation, adhesion,
migration, and regulation of signal transduction pathways (63). miR-152 could directly target and
suppress the expression of TNS1. Therefore, inhibiting the
expression of TNS1 could reduce metastasis and invasion of NSCLC
cells (64). In addition, the core
region of miR-152 is hypermethylated, and the hypermethylation
level is regulated by the DNA methyltransferase 3B (DNMT3B), which
leads to a reduction in miR-152 expression. Simultaneously, miR-152
directly targets and inhibits the expression of neural cell
adhesion molecule 1 (NCAM1). As a highly expressed transmembrane
protein in NSCLC, the NCAM1 gene may promote the proliferation and
metastasis of NSCLC cells (65).
This suggested that inhibiting DNMT3B can reduce the methylation
level of miR-152. The overexpression of miR-152 inhibits NCAM1 and
reduces NSCLC cell proliferation (66).
Fibroblast growth factor 2 (FGF2) is another target
of miR-152, which is a multifunctional cytokine that expresses and
influences multiple biological processes in a variety of cancers.
The FGF/FGFR signaling pathway controls a number of biological
functions, including cell proliferation, differentiation and
migration (67). miR-152
specifically binds and inhibits the expression of FGF2 mRNA and
protein, which participates in preventing NSCLC cell proliferation
and migration (68).
LncRNA, a non-coding RNA, can bind to miRNA but
cannot be transcribed into a protein. LncRNAs play crucial roles in
a variety of biological processes, including cell proliferation,
differentiation, apoptosis, and its dysregulation can lead to
cancer. It is reported that lncRNA colon cancer-associated
transcript 1 (CCAT1) sponge stimulates NSCLC cell growth and
migration by suppressing the expression of miR-152. CCAT1 promotes
EMT with the downregulation of E-cadherin (69). LncRNA prostate cancer gene
expression marker-1 (PCGEM1) is correlated with lymph node
metastasis and TNM stage in NSCLC. PCGEM1 targets and inhibits
miR-152-3p to promote NSCLC proliferation and migration (70).
miRNAs as biomarkers for NSCLC
miRNAs have the potential to become high-quality
biomarkers with the advantages of high stability, non-invasive,
convenient, and efficient screening methods. In several studies,
researchers used miRNA microarray or reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) to
analyze serum miRNA levels for patients with NSCLC, benign lung
disease (BLD) and healthy subjects, to select specific miRNAs for
routine examination to improve NSCLC sensitivity and specificity
for early diagnosis (Table
II).
 | Table II.miRNA as a biomarker for early
diagnosis and prognostic of NSCLC. |
Table II.
miRNA as a biomarker for early
diagnosis and prognostic of NSCLC.
| Up miRNA | Down miRNA | Sample type | Selection
cohort | Validation
cohort | Analytical
technique | Normalization | Effect | (Refs.) |
|---|
| miR-339-3p | - | Serum | 25 NSCLC | 117 NSCLC | TaqMan
low-density | RNU6 | Diagnostic
biomarker | (71) |
|
|
|
| 30 OLs | 113 OLs | arrays |
| for NSCLC |
|
|
|
|
| 19 HC |
| RT-qPCR |
|
|
|
| miR-762 | - | Serum | 148 NSCLC | - | RT-qPCR | cel-miR-39 | Diagnostic and
prognostic | (72) |
|
|
|
| 60 HC |
|
|
| biomarker for
NSCLC |
|
| miR-146b | - | Serum | 63 NSCLC | 65 NSCLC | RT-qPCR | mean of CT of
all | Early diagnosis
and | (74) |
| miR-205 |
|
| 15 HC | 17 COPD |
| healthy
controls | prognosis biomarker
for |
|
| miR-29c |
|
|
| 15 HC |
|
| NSCLC |
|
| miR-30b |
|
|
|
|
|
|
|
|
| miR-337 |
|
|
|
|
|
|
|
|
| miR-411 |
|
|
|
|
|
|
|
|
| miR-1247-5p | - | Plasma | 154 NSCLC | - | miRNA
microarray | U6 snRNA | Early diagnosis
for | (11) |
| miR-301b-3p |
|
| 146 HC |
| RT-qPCR |
| NSCLC |
|
| miR-105-5p |
|
|
|
|
|
|
|
|
| miR-16-5p | - | Plasma | 38 NSCLC | 40 NSCLC | Nanostring | The exogenous | Early detection for
LC | (75) |
| miR-92a-3p |
|
| 21 HC | 40 HC |
nCounter® assay | ath-miR-159 |
|
|
| miR-451a |
|
|
|
| RT-qPCR |
|
|
|
| miR-210 | - | Plasma | 40 NSCLC | 88 NSCLC | RT-qPCR | U6 snRNA | Early diagnosis
and | (76) |
| miR-1290 |
|
| 20 HC | 50 BLD |
|
| prognosis for
NSCLC |
|
| miR-150 |
|
|
| 40 HC |
|
|
|
|
| miR-21-5p |
|
|
|
|
|
|
|
|
| miR-760 | miR-139-3p | Plasma | 220 high-risk | 203 high-risk | miRNA
microarray | miR-1228 | Diagnosis of LC
and | (77) |
|
| miR-17 |
| individuals | individuals | RT-qPCR |
| further
discrimination of |
|
|
| miR-19a |
| 156 AC | 133 AC |
|
| SCLC and NSCLC |
|
|
| miR-26b |
| 67 SCLC | 49 SCLC |
|
|
|
|
|
| miR-451 |
| 122 SQ | 76 SQ |
|
|
|
|
| miR-31-5p | - | Sputum and | 76 NSCLC | 56 NSCLC | RT-qPCR | U6 snRNA | Early detection
for | (78) |
| miR-210-3p |
| Plasma | 72 HC | 55 HC |
|
| NSCLC |
|
| miR-21-5p |
|
|
|
|
|
|
|
|
| miR-486-5p |
|
|
|
|
|
|
|
|
| - | miR-186 | Serum and
exhaled | 62 NSCLC | - | RT-qPCR | cel-miR-39 | The diagnosis
and | (79) |
|
|
| breath
condensate | 60 HC |
|
|
| severity
assessment |
|
|
|
|
|
|
|
|
| of NSCLC |
|
| miR-21 | miR-126 | Serum | 60 NSCLC | - | RT-qPCR | U6 snRNA | Early diagnosis for
LC | (80) |
| miR-143 | miR-155 |
| 60 HC |
|
|
|
|
|
| miR-145 | let-7a |
|
|
|
|
|
|
|
|
| miR-146 |
|
|
|
|
|
|
|
|
| miR-31 |
|
|
|
|
|
|
|
|
| miR-182 |
|
|
|
|
|
|
|
|
| let-7g |
|
|
|
|
|
|
|
|
| miR-19b |
|
|
|
|
|
|
|
|
|
| Serum | 166 NSCLC | - | RT-qPCR | - | Prognostic
biomarker | (81) |
| miR-629 |
|
| 70 NMLDs |
|
|
| for NSCLC |
|
|
|
|
| 100 HC |
|
|
|
|
|
In a previous study, Trakunram et al
(71) used a TaqMan low-density
array to compare the expression levels of 745 miRNAs in NSCLC, BLD
and healthy subjects, and selected miR-339-3p through verification
set and diagnostic evaluation. The area under the curve (AUC) of
the miRNA is 0.616, indicating that it has guiding significance in
the diagnosis of NSCLC. Chen et al (72) used RT-qPCR to profile miRNAs in 148
NSCLC patients and healthy controls. The high level of miR-762 was
related to an advanced stage, poor tumor grade and positive lymph
node metastasis. Combined detection of miR-762, carcinoembryonic
antigen (CEA), and cytokeratin fragment antigen 21-1 (CYFRA21-1)
could improve the diagnostic accuracy for NSCLC. Furthermore,
miR-762 expression can be used as a predictive biomarker for
gefitinib resistance, and high expression predicts poor therapeutic
effect (73).
In addition, Yang et al (74) selected serum miRNAs for NSCLC early
diagnosis, and 8 miRNAs were selected and validated by training set
and validation set, ultimately obtaining the best predictive model
composed of miR-146b, miR-205, miR-29c and miR-30b. The combination
could be used not only in the diagnosis of NSCLC patients but also
in NSCLC subtypes analysis and TNM staging. AUC of the combined
training and verification sets was estimated to be 0.96 with 95.31%
sensitivity and 82.98% specificity.
The researchers measured the expression of specific
miRNAs in plasma, not just the serum sample. Dong et al
(11) used a miRNA chip to examine
the miRNAs in plasma from NSCLC patients and healthy volunteers.
RT-qPCR was used to evaluate the expression of 11 upregulated
miRNAs. Three plasma miRNAs (miR-1247-5p, miR-301b-3p and
miR-105-5p) were selected and finally determined to distinguish
between early NSCLC patients and healthy individuals, and their AUC
are 0.769, 0.761 and 0.777, respectively. Reis et al
(75) performed a study based on
the NSCLC subtypes. They used Nanostring nCounter
®technology to evaluate the expression of miRNA in LUAD,
LUSC and healthy controls, and identified a correlation between the
expression of the majority of miRNAs in the two histological
subtypes. A total of 12 differentially expressed miRNAs were
selected for verification and the expression level of 11 miRNAs
were consistent with the found set. Furthermore, 3 miRNAs
(miR-16-5p, miR-92a-3p and miR-451a) with the best statistical
performance were selected for pathway enrichment analysis, and it
was found that the 3 miRNAs were related to the LC pathways such as
epidermal growth factor receptor (EGFR) and PI3K/AKT. Concurrently,
these 3 miRNAs can predict NSCLC with high specificity and
sensitivity. Moreover, the researchers selected 12 previously
reported aberrantly expressed miRNAs in NSCLC. A total of 4 miRNAs
(miR-210, miR-1290, miR-150 and miR-21-5p) obtained from test set
and verification set could distinguish NSCLC, BLD and healthy
individuals. In the study of postoperative NSCLC patients, it was
found that the significantly decreased expression of the 4 miRNAs
were predictors of prolonged disease-free survival. A total of 2
miRNAs (miR-210 and miR-150) could predict patient's prognosis even
if their expression levels do not significantly alter as NSCLC
progresses (76). Based on miRNA
chip, Lu et al (77)
identified 6 miRNAs (miR-17, miR-190b, miR-19a, miR-19b, miR-26b
and miR-375). These 6 miRNAs could distinguish between LC and
asymptomatic high-risk patients through screening in three stages
of discovery, training and verification. Further research showed
that 3 miRNAs (miR-17, miR-190b and miR-375) could accurately
differentiate SCLC from NSCLC.
Previous studies have indicated that combining
different fluid biopsies could improve the accuracy of NSCLC
detection. For example, Liao et al (78) used the Taqman miRNA assay to detect
the expression of 2 miRNAs (miR-31-5p and miR-210p-3p) in sputum
and 3 miRNAs (miR-21-5p, miR-210-3p and miR-486-5p) in plasma. The
logical regression model with limited parameters in least absolute
shrinkage and selection operator was used to optimize the miRNA
detection panel. The detection of 2 sputum miRNAs (miR-31-5p and
miR-210-3p) and 1 plasma miRNA (miR-21-5p) in the combined model
had a synergistic effect on the diagnosis of NSCLC. The combination
study proved that the analysis of 2 sputum miRNA biomarkers and 1
plasma miRNA biomarker had improved performance than a single class
of miRNA biomarkers. Similarly, the study by Xie et al
(79) revealed a positive
correlation between the expression of miR-186 in serum and exhaled
breath condensate, and the combination of decreased miR-186 and
increased IL-1β were used for the diagnosis and severity evaluation
of NSCLC.
Researchers typically assessed the effectiveness of
the miRNA diagnostic model for the study of early diagnosis of
miRNA by using logical regression analysis and receiver operating
characteristic (ROC) curve (78,80).
The majority of miRNAs have been studied to distinguish early NSCLC
patients from BLD patients or healthy individuals, while certain
miRNAs have been studied to identify NSCLC subtypes. Previous
studies (11,74) revealed that miRNAs have high
specificity and sensitivity, indicating that there is considerable
potential for using miRNA in the early detection of NSCLC. However,
it has poor repeatability for miRNA detection. The reasons may be
the heterogeneity of NSCLC patients and the regulation of tumor
formation by multiple genes. The selected miRNA should be validated
in large-scale NSCLC patients utilizing standard operating
procedures in the future. Based on understanding the pathway of
miRNA mechanism, the best miRNAs combination for NSCLC diagnosis
would be found.
In addition, miRNAs can serve as prognostic
biomarkers. Higher levels of the serum miR-629 have been associated
with poor differentiation, lymph node metastases and advanced
clinical stage in patients with NSCLC compared with those with
non-malignant lung disease and healthy controls (81). miRNAs differentially expressed in
serum samples provide a novel basis for predicting the prognosis of
NSCLC patients.
Exosomal miRNAs as potential biomarkers for
NSCLC
Focus has been addressed on exosomal miRNAs as
potential biomarkers since they are one of the major components of
exosomes and play functional roles in cell-to-cell communication.
Exosomal miRNAs may be used as prognostic and diagnostic biomarkers
for NSCLC (Table III).
 | Table III.Exosomal miRNAs as diagnostic markers
for NSCLC. |
Table III.
Exosomal miRNAs as diagnostic markers
for NSCLC.
| Down exosomal
miRNA | Up exosomal
miRNA | Sample type | Selection
cohort | Validation
cohort | Technique | Normalization | Clinical value | (Refs.) |
|---|
| miR-21 | - | Serum | 7 NSCLC | - | Conductive
polymer- | - | Ultrasensitive
and | (82) |
| miR-155 |
|
| 1 HC |
| based
exo-miRNA |
| reproducible |
|
| miR-205 |
|
|
|
| array sensor |
|
electrochemical |
|
| let-7b |
|
|
|
|
|
| detection of
LC |
|
| - | miR-5684 | Serum | 2 NSCLC | 330 NSCLC | miRNA
microarray | U6 snRNA | Diagnostic and | (83) |
|
| miR-125b-5p |
| 1 HC | 312 HC | RT-qPCR |
| prognostic
biomarkers |
|
|
|
|
|
|
|
|
| for NSCLC |
|
| miR-17-5p | - | Serum | 100 NSCLC | 72 NSCLC | RT-qPCR | miR-16-5p | Early diagnostic
marker | (84) |
|
|
|
| 90 HC | 47 HC |
|
| for NSCLC |
|
| miR-1246 | - | Serum | 150 NSCLC | - | RT-qPCR | cel-miR-39 | Useful diagnosis
and | (16) |
|
|
|
| 50 NMRD |
|
|
| prognosis for
NSCLC |
|
|
|
|
| 50 HC |
|
|
|
|
|
| - | miR-4448 | Plasma | 2 NSCLC
metastasis | 20 NSCLC | miRNA
microarray | U6 snRNA | Diagnostic marker
for | (86) |
|
|
|
| 2 NSCLC | metastasis | RT-qPCR |
| metastatic
adenocarcinoma |
|
|
|
|
| N-metastasis | 20 NSCLC |
|
|
|
|
|
|
|
| 2 HC | N-metastasis |
|
|
|
|
|
|
|
|
| 20 HC |
|
|
|
|
| miR-23b-3p | - | Plasma | 10 AC | 196 NSCLC | RT-qPCR | let-7a-5p | Prognostic
biomarkers | (87) |
| miR-10b-5p |
|
| 10 HC | 11 non-tumor |
|
| for NSCLC |
|
| miR-21-5p |
|
|
| respiratory |
|
|
|
|
|
|
|
|
| diseases |
|
|
|
|
|
|
|
|
| 10 HC |
|
|
|
|
| miR-451a | - | Plasma | 6 NSCLC | 285 NSCLC | miRNA
microarray | RNU6B | Biomarker for
recurrence | (88) |
|
|
|
| 3 HC | 24 HC | RT-qPCR |
| and prognosis of
NSCLC |
|
| miR-21 | - | Plasma | 6 NSCLC | 195 NSCLC | miRNA
microarray | miR-16a | Biomarker for
the | (89) |
| miR-4257 |
|
| 3 HC | 30 HC | RT-qPCR |
| recurrence of
NSCLC |
|
Exosomal miRNAs in blood have been extensively
studied as biomarkers for the diagnosis of NSCLC. A novel
immunomagnetic separation technique was used to selectively extract
exosomes from serum of patients, which is more specific than
traditional ultracentrifugation, and a multiplexed array sensor is
used to simultaneously detect 4 exo-miRNAs (miR-21, miR-155,
miR-205 and miR-let-7b) (82). A
microarray-based study found that the combination of exosomal
miR-5684 and miR-125b-5p had effective diagnostic value (AUC=0.744)
for patients with NSCLC. Notably, in the tumor staging studies, it
was found that exosomal miR-125b-5p is highly diagnostic in
distinguishing between early and late stage, lymph node metastasis
and distant metastasis (83). In
comparison with traditional tumor markers, the level of miR-17-5p
was significantly increased in NSCLC patients compared with healthy
controls, and the detection performance of miR-17-5p was superior
to CEA, CYFRA21-1 and squamous cell carcinoma antigen. The
combination of these 4 tumor markers outperforms a single exosomal
miR-17-5p in terms of diagnostic performance, indicating that the
combination of exosomal miRNA and conventional tumor markers have
significant clinical utility for the diagnosis of NSCLC (84).
Exosomes are circulating membrane-enclosed vesicles
that contain miRNA, RNA, lipids, and proteins. By adhering to the
target cell membrane, exosomes can transport miRNA and other
contents from donor cells to recipient cells, which is relevant to
the diagnosis and prognosis of NSCLC patients (85). For example, an increased level of
miR-1246 isolated from serum exosomes was associated with a lower
survival rate in patients with NSCLC (16). Plasma exosomal miR-4448 was shown
to be decreased in patients with metastatic LUAD. Exosomal miR-4448
could be used as a diagnostic marker for patients with metastatic
LUAD (86). In addition, elevated
levels of exosomal miR-23b-3p, miR-10b-5p and miR-21-5p were
independently associated with poor overall survival in patients
with NSCLC (87). Plasma exosomal
miR-451a was evaluated to be a reliable biomarker for predicting
recurrence and prognosis in NSCLC patients with stage I, II or III
cancer (88). Similarly, plasma
exosomal miR-4257 and miR-21 have been identified as biomarkers of
recurrence and TNM stage in NSCLC patients (89).
Comparison of circulating miRNAs with
exosomal miRNAs
With the further development of miRNA research,
circulating miRNAs and exosomal miRNAs may become the primary
research forms in the early diagnosis and prognosis of NSCLC and
other cancers. Few studies have simultaneously compared their
detection performance. Exosomal miRNA may be more stable than
circulating miRNA due to the protection of lipid bilayer. The
distribution of miR-126 in the circulation of NSCLC patients at the
early and advanced stages of the disease was evaluated, and it was
found that miR-126 is primarily found in exosomes in the early and
late stages of NSCLC. The levels of miR-126 increased in exosomes
while they decreased in the exosome-free serum (90). Similarly, compared with whole
plasma, the content of miR-21 in the exosomes of patients with
hepatoblastoma was higher (91).
The findings revealed a difference in the distribution of specific
miRNAs between circulating miRNAs and exosomal miRNAs. The levels
of miRNAs were higher in exosomes than serum or other bodily
fluids.
Circulating miRNAs and exosomal miRNAs were
distributed differentially in NSCLC patients. Several studies
simultaneously investigated the changes in circulating miRNA and
exosomal miRNA in cancer. In ovarian cancer, 5 miRNAs (miR-200c-3p,
miR-346, miR-127-3p, miR-143-3p and miR-205-5p) were significantly
upregulated in serum and exosomes (92). Wu et al (93) measured the expression levels of 8
serum miRNAs and their corresponding exosomal miRNAs in NSCLC,
benign pulmonary lesions and healthy subjects. The AUC values of
exosomal miR-146a-5p and miR-486-5p were found to be over 0.8, but
the AUC values of 4 serum miRNAs (miR-21-5p, miR-141-3p, miR-222-3p
and miR-486-5p) were all less than 0.8. The present study
demonstrated that exosomal miRNA had an improved detection
performance than circulating serum miRNA in identifying cancer
samples from healthy control samples. The detection performance of
the same miRNA in different studies was compared, and it was
identified that the diagnostic value of exosomal miR-1246
(AUC=0.827) was greater than that of circulating plasma (AUC=0.641)
(16,94). Exosomal miR-205-5p diagnostic value
(AUC=0.806) was similar to that in circulating serum (AUC=0.8250)
(95,96). To obtain more accurate detection
results, it is necessary to detect circulating free and exosomal
miRNA for one patient at the same time, and then identify which
sample type is reasonable for miRNA detection.
Future prospects and conclusion
Numerous studies have shown that the dysregulation
of miRNA is an important driver of NSCLC progression and plays
crucial roles in the early diagnosis, treatment and prognosis of
NSCLC (97,98). As there is a wide variety of
miRNAs, there is also diversity in the roles of these miRNAs in
NSCLC. The miRNAs in tissue or blood of patients with NSCLC, BLD,
and healthy controls were examined using microarray, RT-qPCR, and
next-generation sequencing (11,99,100). It has been found that a multitude
of miRNAs have notable changes in NSCLC, suggesting that particular
miRNAs can be used to diagnose NSCLC. Despite the existence of
numerous studies on miRNAs, the mechanism of miRNAs in various
tissue subtypes of NSCLC remains unknown due to the diversity of
miRNA action mechanisms and the heterogeneity of NSCLC patients.
The mechanisms of miRNAs should be studied more extensively and
systematically in order to improve the use of miRNAs in clinical
treatment. Additionally, the uniform operational procedure should
be established for the repeatability of miRNAs detection in order
to apply the miRNA detection with favorable performance for the
clinical diagnosis of NSCLC.
In the present review, focus was addressed on the
biological functions of miRNAs and their molecular mechanisms in
the occurrence and progression of NSCLC, as well as the importance
of various miRNAs in the diagnosis and prognosis of NSCLC. Although
over 2,000 human miRNAs have been identified, most studies have
focused on a single signaling pathway mechanism between a specific
miRNA and its target gene. Future research should concentrate on
the network of interactions between different miRNAs. In one word,
miRNAs are well-known to exist in plasma and other bodily fluids
and are one promising biomarker for NSCLC diagnosis.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
XL wrote the manuscript and revised it. QW designed
and supervised the study. YW designed the tables and figure. SL
edited and critically revised the article for intellectual content.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pilleron S, Soto-Perez-de-Celis E, Vignat
J, Ferlay J, Soerjomataram I, Bray F and Sarfati D: Estimated
global cancer incidence in the oldest adults in 2018 and
projections to 2050. Int J Cancer. 148:601–608. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Osmani L, Askin F, Gabrielson E and Li QK:
Current WHO guidelines and the critical role of immunohistochemical
markers in the subclassification of non-small cell lung carcinoma
(NSCLC): Moving from targeted therapy to immunotherapy. Semin
Cancer Biol. 52:103–109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu G, Cheresh P and Kamp DW: Molecular
basis of asbestos-induced lung disease. Annu Rev Pathol. 8:161–187.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Choi WI, Park SH, Park BJ and Lee CW:
Interstitial lung disease and lung cancer development: A 5-year
nationwide population-based study. Cancer Res Treat. 50:374–381.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Seijo LM and Zulueta JJ: Understanding the
links between lung cancer, COPD, and emphysema: A Key to more
effective treatment and screening. Oncology (Williston Park).
31:93–102. 2017.PubMed/NCBI
|
|
7
|
Ben Amar J, Ben Safta B, Zaibi H, Dhahri
B, Baccar MA and Azzabi S: Prognostic factors of advanced stage
non-small-cell lung cancer. Tunis Med. 94:360–367. 2016.PubMed/NCBI
|
|
8
|
Naylor EC: Adjuvant therapy for stage I
and II non-small cell lung cancer. Surg Oncol Clin N Am.
25:585–599. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Liu L and Song X, Wang K, Niu L, Xie
L and Song X: TEP linc-GTF2H2-1, RP3-466P17.2, and lnc-ST8SIA4-12
as novel biomarkers for lung cancer diagnosis and progression
prediction. J Cancer Res Clin Oncol. 147:1609–1622. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maly V, Maly O, Kolostova K and Bobek V:
Circulating tumor cells in diagnosis and treatment of lung cancer.
In Vivo. 33:1027–1037. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dong X, Chang M and Song X, Ding S, Xie L
and Song X: Plasma miR-1247-5p, miR-301b-3p and miR-105-5p as
potential biomarkers for early diagnosis of non-small cell lung
cancer. Thorac Cancer. 12:539–548. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iqbal MA, Arora S, Prakasam G, Calin GA
and Syed MA: MicroRNA in lung cancer: Role, mechanisms, pathways
and therapeutic relevance. Mol Aspects Med. 70:3–20. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:(Database Issue).
D140–D144. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu J, Ren L, Li S, Li W, Zheng X, Yang Y,
Fu W, Yi J, Wang J and Du G: The biology, function, and
applications of exosomes in cancer. Acta Pharm Sin B. 11:2783–2797.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang D and Qu D: Early diagnostic and
prognostic value of serum exosomal miR-1246 in non-small cell lung
cancer. Int J Clin Exp Pathol. 13:1601–1607. 2020.PubMed/NCBI
|
|
17
|
de Rie D, Abugessaisa I, Alam T, Arner E,
Arner P, Ashoor H, Åström G, Babina M, Bertin N, Burroughs AM, et
al: An integrated expression atlas of miRNAs and their promoters in
human and mouse. Nat Biotechnol. 35:872–878. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Solé C and Lawrie CH: MicroRNAs in
metastasis and the tumour microenvironment. Int J Mol Sci.
22:48592021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Starega-Roslan J, Koscianska E, Kozlowski
P and Krzyzosiak WJ: The role of the precursor structure in the
biogenesis of microRNA. Cell Mol Life Sci. 68:2859–2871. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shang R, Baek SC, Kim K, Kim B, Kim VN and
Lai EC: Genomic clustering facilitates nuclear processing of
suboptimal Pri-miRNA loci. Mol Cell. 78:303–316.e4. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yeom KH, Lee Y, Han J, Suh MR and Kim VN:
Characterization of DGCR8/Pasha, the essential cofactor for Drosha
in primary miRNA processing. Nucleic Acids Res. 34:4622–4629. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi
S, Xie H, Peng X, Yin W, Tao Y and Wang X: miRNA-based biomarkers,
therapies, and resistance in cancer. Int J Biol Sci. 16:2628–2647.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Alarcón CR, Lee H, Goodarzi H, Halberg N
and Tavazoie SF: N6-methyladenosine marks primary microRNAs for
processing. Nature. 519:482–485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Wang Z and Gemeinhart RA:
Progress in microRNA delivery. J Control Release. 172:962–974.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Matsuyama H and Suzuki HI: Systems and
synthetic microRNA biology: From biogenesis to disease
pathogenesis. Int J Mol Sci. 21:1322019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Haneklaus M, Gerlic M, O'Neill LA and
Masters SL: miR-223: Infection, inflammation and cancer. J Intern
Med. 274:215–226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang J, Han L, Yu J, Li H and Li Q:
miR-224 aggravates cancer-associated fibroblast-induced progression
of non-small cell lung cancer by modulating a positive loop of the
SIRT3/AMPK/mTOR/HIF-1α axis. Aging (Albany NY). 13:10431–10449.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu L, Chen Y, Liu J, Nie K, Xiao Y and Yu
H: MicroRNA-629 promotes the tumorigenesis of non-small-cell lung
cancer by targeting FOXO1 and activating PI3K/AKT pathway. Cancer
Biomark. 29:347–357. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiong J, Xing S, Dong Z, Niu L, Xu Q, Li
Y, Liu P and Yang P: miR-654-3p suppresses cell viability and
promotes apoptosis by targeting RASAL2 in non-small-cell lung
cancer. Mol Med Rep. 23:1242021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Du H, Bao Y, Liu C, Zhong A, Niu Y and
Tang X: miR-139-5p enhances cisplatin sensitivity in non-small cell
lung cancer cells by inhibiting cell proliferation and promoting
apoptosis via the targeting of homeobox protein Hox-B2. Mol Med
Rep. 23:1042021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pu R, Pu M, Huang H and Cui Y: MicroRNA
144 inhibits cell migration and invasion and regulates inflammatory
cytokine secretion through targeting toll like receptor 2 in
non-small cell lung cancer. Arch Med Sci. 17:1028–1037. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yuan C, Bai R, Gao Y, Jiang X, Li S, Sun
W, Li Y, Huang Z, Gong Y and Xie C: Effects of MicroRNA-195-5p on
biological behaviors and radiosensitivity of lung adenocarcinoma
cells via targeting HOXA10. Oxid Med Cell Longev. 2021:45222102021.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu X, Wu G, Zhang H, Peng X, Huang B,
Huang M, Ding J, Mao C and Peng C: MiR-196b promotes the invasion
and migration of lung adenocarcinoma cells by targeting AQP4.
Technol Cancer Res Treat. 20:15330338209858682021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fang QY, Deng QF, Luo J and Zhou CC:
MiRNA-20a-5p accelerates the proliferation and invasion of
non-small cell lung cancer by targeting and downregulating KLF9.
Eur Rev Med Pharmacol Sci. 24:2548–2556. 2020.PubMed/NCBI
|
|
36
|
Korde A, Jin L, Zhang JG, Ramaswamy A, Hu
B, Kolahian S, Guardela BJ, Herazo-Maya J, Siegfried JM, Stabile L,
et al: Lung endothelial MicroRNA-1 regulates tumor growth and
angiogenesis. Am J Respir Crit Care Med. 196:1443–1455. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ma Z, Wei K, Yang F, Guo Z, Pan C, He Y,
Wang J, Li Z, Chen L, Chen Y and Xia Y: Tumor-derived exosomal
miR-3157-3p promotes angiogenesis, vascular permeability and
metastasis by targeting TIMP/KLF2 in non-small cell lung cancer.
Cell Death Dis. 12:8402021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang D, Cai L, Tian X and Li W: MiR-543
promotes tumorigenesis and angiogenesis in non-small cell lung
cancer via modulating metastasis associated protein 1. Mol Med.
26:442020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao G, Zhang Y, Zhao Z, Cai H, Zhao X,
Yang T, Chen W, Yao C, Wang Z, Wang Z, et al: MiR-153 reduces stem
cell-like phenotype and tumor growth of lung adenocarcinoma by
targeting Jagged1. Stem Cell Res Ther. 11:1702020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fang L, Cai J, Chen B, Wu S, Li R, Xu X,
Yang Y, Guan H, Zhu X, Zhang L, et al: Aberrantly expressed
miR-582-3p maintains lung cancer stem cell-like traits by
activating Wnt/β-catenin signalling. Nat Commun. 6:86402015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang L, Zhang X, Wang X, He M and Qiao S:
MicroRNA-224 promotes tumorigenesis through downregulation of
caspase-9 in triple-negative breast cancer. Dis Markers.
2019:73789672019.PubMed/NCBI
|
|
42
|
Fassan M, Cui R, Gasparini P, Mescoli C,
Guzzardo V, Vicentini C, Munari G, Loupakis F, Lonardi S, Braconi
C, et al: miR-224 Is significantly upregulated and targets
caspase-3 and caspase-7 during colorectal carcinogenesis. Transl
Oncol. 12:282–291. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li J, Liu X, Li C and Wang W: miR-224-5p
inhibits proliferation, migration, and invasion by targeting
PIK3R3/AKT3 in uveal melanoma. J Cell Biochem. 120:12412–12421.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Alhazzazi TY, Kamarajan P, Verdin E and
Kapila YL: SIRT3 and cancer: Tumor promoter or suppressor? Biochim
Biophys Acta. 1816:80–88. 2011.PubMed/NCBI
|
|
45
|
Tao F, Gu C, Li N, Ying Y, Feng Y, Ni D,
Zhang Q and Xiao Q: SIRT3 acts as a novel biomarker for the
diagnosis of lung cancer: A retrospective study. Medicine
(Baltimore). 100:e265802021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Marinov M, Fischer B and Arcaro A:
Targeting mTOR signaling in lung cancer. Crit Rev Oncol Hematol.
63:172–182. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhan P, Zhao S, Yan H, Yin C, Xiao Y, Wang
Y, Ni R, Chen W, Wei G and Zhang P: α-enolase promotes
tumorigenesis and metastasis via regulating AMPK/mTOR pathway in
colorectal cancer. Mol Carcinog. 56:1427–1437. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lu X and Kang Y: Hypoxia and
hypoxia-inducible factors: Master regulators of metastasis. Clin
Cancer Res. 16:5928–5935. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen CY, Chen J, He L and Stiles BL: PTEN:
Tumor suppressor and metabolic regulator. Front Endocrinol
(Lausanne). 9:3382018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang G, Han J, Zhuang L, Li S, Gong Q and
Chen Y: Serum starvation induces cell death in NSCLC via miR-224.
Onco Targets Ther. 12:3953–3962. 2019.PubMed/NCBI
|
|
51
|
Endo M: The roles of ANGPTL families in
cancer progression. J UOEH. 41:317–325. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kuo TC, Tan CT, Chang YW, Hong CC, Lee WJ,
Chen MW, Jeng YM, Chiou J, Yu P, Chen PS, et al: Angiopoietin-like
protein 1 suppresses SLUG to inhibit cancer cell motility. J Clin
Invest. 123:1082–1095. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Han H, Pan B, Liang F, Wu L, Liu X, Yang Y
and Chen J: MiR-224 promotes lymphatic metastasis by targeting
ANGPTL1 in non-small-cell lung carcinoma. Cancer Biomark.
34:431–441. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhu JH, De Mello RA, Yan QL, Wang JW, Chen
Y, Ye QH, Wang ZJ, Tang HJ and Huang T: MiR-139-5p/SLC7A11 inhibits
the proliferation, invasion and metastasis of pancreatic carcinoma
via PI3K/Akt signaling pathway. Biochim Biophys Acta Mol Basis Dis.
1866:1657472020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Song M, Yin Y, Zhang J, Zhang B, Bian Z,
Quan C, Zhou L, Hu Y, Wang Q, Ni S, et al: MiR-139-5p inhibits
migration and invasion of colorectal cancer by downregulating AMFR
and NOTCH1. Protein Cell. 5:851–861. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Qiu G, Lin Y, Zhang H and Wu D: miR-139-5p
inhibits epithelial-mesenchymal transition, migration and invasion
of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2.
Biochem Biophys Res Commun. 463:315–321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Pan X, Liu W, Chai Y, Wang J and Zhang Y:
Genetic and clinical characterization of HOXB2 in glioma. Onco
Targets Ther. 13:10465–10473. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Faes S and Dormond O: PI3K and AKT:
Unfaithful partners in cancer. Int J Mol Sci. 16:21138–21152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Huang L, Cai JL, Huang PZ, Kang L, Huang
MJ, Wang L and Wang JP: miR19b-3p promotes the growth and
metastasis of colorectal cancer via directly targeting ITGB8. Am J
Cancer Res. 7:1996–2008. 2017.PubMed/NCBI
|
|
60
|
Li J, Zhu T, Weng Y, Cheng F, Sun Q, Yang
K, Su Z and Ma H: Exosomal circDNER enhances paclitaxel resistance
and tumorigenicity of lung cancer via targeting miR-139-5p/ITGB8.
Thorac Cancer. 13:1381–1390. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang Y, Zhong Y, Zhang C, Liao J and Wang
G: PM2.5 downregulates MicroRNA-139-5p and induces EMT in
bronchiolar epithelium cells by targeting Notch1. J Cancer.
11:5758–5767. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Donnem T, Andersen S, Al-Shibli K, Al-Saad
S, Busund LT and Bremnes RM: Prognostic impact of Notch ligands and
receptors in nonsmall cell lung cancer: Coexpression of Notch-1 and
vascular endothelial growth factor-A predicts poor survival.
Cancer. 116:5676–5685. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liao YC and Lo SH: Tensins-emerging
insights into their domain functions, biological roles and disease
relevance. J Cell Sci. 134:jcs2540292021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Duan J, Wang L, Shang L, Yang S, Wu H,
Huang Y and Miao Y: miR-152/TNS1 axis inhibits non-small cell lung
cancer progression through Akt/mTOR/RhoA pathway. Biosci Rep.
41:BSR202015392021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hinsby AM, Berezin V and Bock E: Molecular
mechanisms of NCAM function. Front Biosci. 9:2227–2244. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yang B, Huang S, Chen H, Li R, Hou S, Zhao
J and Li Y: DNMT3B regulates proliferation of A549 cells through
the microRNA-152-3p/NCAM1 pathway. Oncol Lett. 23:112022.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yang L, Zhou F, Zheng D, Wang D, Li X,
Zhao C and Huang X: FGF/FGFR signaling: From lung development to
respiratory diseases. Cytokine Growth Factor Rev. 62:94–104. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cheng Z, Ma R, Tan W and Zhang L: MiR-152
suppresses the proliferation and invasion of NSCLC cells by
inhibiting FGF2. Exp Mol Med. 46:e1122014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zong D, Liu X, Li J, Long Y, Ouyang R and
Chen Y: LncRNA-CCAT1/miR-152-3p is involved in CSE-induced
inflammation in HBE cells via regulating ERK signaling pathway. Int
Immunopharmacol. 109:1088182022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Huang J, Lou J, Liu X and Xie Y: LncRNA
PCGsEM1 contributes to the proliferation, migration and invasion of
non-small cell lung cancer cells via acting as a sponge for
miR-152-3p. Curr Pharm Des. 27:4663–4670. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Trakunram K, Chaniad P, Geater SL,
Keeratichananont W, Chittithavorn V, Uttayamakul S, Buya S,
Raungrut P and Thongsuksai P: Serum miR-339-3p as a potential
diagnostic marker for non-small cell lung cancer. Cancer Biol Med.
17:652–663. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chen L, Li Y and Lu J: Identification of
circulating miR-762 as a novel diagnostic and prognostic biomarker
for non-small cell lung cancer. Technol Cancer Res Treat.
19:15330338209642222020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ge P, Cao L, Chen X, Jing R and Yue W:
miR-762 activation confers acquired resistance to gefitinib in
non-small cell lung cancer. BMC Cancer. 19:12032019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yang X, Zhang Q, Zhang M, Su W, Wang Z, Li
Y, Zhang J, Beer DG, Yang S and Chen G: Serum microRNA signature is
capable of early diagnosis for non-small cell lung cancer. Int J
Biol Sci. 15:1712–1722. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Reis PP, Drigo SA, Carvalho RF, Lopez Lapa
RM, Felix TF, Patel D, Cheng D, Pintilie M, Liu G and Tsao MS:
Circulating miR-16-5p, miR-92a-3p, and miR-451a in plasma from lung
cancer patients: Potential application in early detection and a
regulatory role in tumorigenesis pathways. Cancers (Basel).
12:20712020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Jiang HG, Dai CH, Xu YP, Jiang Q, Xia XB,
Shu Y and Li J: Four plasma miRNAs act as biomarkers for diagnosis
and prognosis of non-small cell lung cancer. Oncol Lett.
22:7922021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Lu S, Kong H, Hou Y, Ge D, Huang W, Ou J,
Yang D, Zhang L, Wu G, Song Y, et al: Two plasma microRNA panels
for diagnosis and subtype discrimination of lung cancer. Lung
Cancer. 123:44–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Liao J, Shen J, Leng Q, Qin M, Zhan M and
Jiang F: MicroRNA-based biomarkers for diagnosis of non-small cell
lung cancer (NSCLC). Thorac Cancer. 11:762–768. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Xie H and Chen J, Lv X, Zhang L, Wu J, Ge
X, Yang Q, Zhang D and Chen J: Clinical value of serum and exhaled
breath condensate miR-186 and IL-1β levels in non-small cell lung
cancer. Technol Cancer Res Treat. 19:15330338209474902020.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Tulinsky L, Dzian A, Matakova T and Ihnat
P: Overexpression of the miR-143/145 and reduced expression of the
let-7 and miR-126 for early lung cancer diagnosis. J Appl Biomed.
20:1–6. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Liu F, Li T, Hu P and Dai L: Upregulation
of serum miR-629 predicts poor prognosis for non-small-cell lung
cancer. Dis Markers. 2021:88199342021.PubMed/NCBI
|
|
82
|
Mahmudunnabi RG, Umer M, Seo KD, Park DS,
Chung JH, Shiddiky MJA and Shim YB: Exosomal microRNAs array sensor
with a bioconjugate composed of p53 protein and hydrazine for the
specific lung cancer detection. Biosens Bioelectron.
207:1141492022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhang Z, Tang Y and Song X, Xie L, Zhao S
and Song X: Tumor-derived exosomal miRNAs as diagnostic biomarkers
in non-small cell lung cancer. Front Oncol. 10:5600252020.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang Y, Zhang Y, Yin Y and Li S:
Detection of circulating exosomal miR-17-5p serves as a novel
non-invasive diagnostic marker for non-small cell lung cancer
patients. Pathol Res Pract. 215:1524662019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang
G, Song J, Li Z, Zhang Z and Yuan W: Effect of exosomal miRNA on
cancer biology and clinical applications. Mol Cancer. 17:1472018.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Xu Z, Wang Z, Sun H and Xin H: Evaluation
of exosomal miRNA in Blood as a potential diagnostic biomarker for
human non-small cell lung cancer. Med Sci Monit. 26:e9247212020.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z,
Xiang Y, Wu N, Wu L, Bai L and Li Y: Circulating exosomal microRNAs
as prognostic biomarkers for non-small-cell lung cancer.
Oncotarget. 8:13048–13058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Kanaoka R, Iinuma H, Dejima H, Sakai T,
Uehara H, Matsutani N and Kawamura M: Usefulness of plasma exosomal
MicroRNA-451a as a noninvasive biomarker for early prediction of
recurrence and prognosis of non-small cell lung cancer. Oncology.
94:311–323. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Dejima H, Iinuma H, Kanaoka R, Matsutani N
and Kawamura M: Exosomal microRNA in plasma as a non-invasive
biomarker for the recurrence of non-small cell lung cancer. Oncol
Lett. 13:1256–1263. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Grimolizzi F, Monaco F, Leoni F, Bracci M,
Staffolani S, Bersaglieri C, Gaetani S, Valentino M, Amati M,
Rubini C, et al: Exosomal miR-126 as a circulating biomarker in
non-small-cell lung cancer regulating cancer progression. Sci Rep.
7:152772017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Liu W, Chen S and Liu B: Diagnostic and
prognostic values of serum exosomal microRNA-21 in children with
hepatoblastoma: A Chinese population-based study. Pediatr Surg Int.
32:1059–1065. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wang W, Wu LR, Li C, Zhou X, Liu P, Jia X,
Chen Y and Zhu W: Five serum microRNAs for detection and predicting
of ovarian cancer. Eur J Obstet Gynecol Reprod Biol X.
3:1000172019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Wu Q, Yu L, Lin X, Zheng Q, Zhang S, Chen
D, Pan X and Huang Y: Combination of serum miRNAs with serum
exosomal miRNAs in early diagnosis for non-small-cell lung cancer.
Cancer Manag Res. 12:485–495. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Li M, Shan W, Hong B, Zou J, Li H, Han D,
Zhang Y, Li L, Li D and Lin W: Circulating miR-92b and miR-375 for
monitoring the chemoresistance and prognosis of small cell lung
cancer. Sci Rep. 10:127052020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhao YL, Zhang JX, Yang JJ, Wei YB, Peng
JF, Fu CJ, Huang MH, Wang R, Wang PY, Sun GB and Xie SY: MiR-205-5p
promotes lung cancer progression and is valuable for the diagnosis
of lung cancer. Thorac Cancer. 13:832–843. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wang X, Jiang X, Li J, Wang J, Binang H,
Shi S, Duan W, Zhao Y and Zhang Y: Serum exosomal miR-1269a serves
as a diagnostic marker and plays an oncogenic role in non-small
cell lung cancer. Thorac Cancer. 11:3436–3447. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhou C, Chen Z, Zhao L, Zhao W, Zhu Y, Liu
J and Zhao X: A novel circulating miRNA-based signature for the
early diagnosis and prognosis prediction of non-small-cell lung
cancer. J Clin Lab Anal. 34:e235052020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yang Y, Jia Y, Xiao Y, Hao Y, Zhang L,
Chen X, He J, Zhao Y and Qian Z: Tumor-targeting anti-MicroRNA-155
delivery based on biodegradable poly(ester amine) and hyaluronic
acid shielding for lung cancer therapy. Chemphyschem. 19:2058–2069.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Jin X, Chen Y, Chen H, Fei S, Chen D, Cai
X, Liu L, Lin B, Su H, Zhao L, et al: Evaluation of tumor-derived
exosomal miRNA as potential diagnostic biomarkers for early-stage
non-small cell lung cancer using next-generation sequencing. Clin
Cancer Res. 23:5311–5319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Migdalska-Sęk M, Modrzewska B, Kordiak J,
Pastuszak-Lewandoska D, Kiszałkiewicz JM, Bielec F, Antczak A and
Brzeziańska-Lasota E: Diagnostic value of PPARδ and miRNA-17
expression levels in patients with non-small cell lung cancer. Sci
Rep. 11:241362021. View Article : Google Scholar : PubMed/NCBI
|