Introduction
Colorectal carcinoma (CRC) is one of the most common
types of malignant tumors, ranking third in newly diagnosed cancer
cases and cancer-related deaths, preceded by lung cancer and breast
cancer in humans, thus seriously endangering the safety and lives
of humans (1). In recent years, due
to the rapid development of science and technology, and changes in
people's lifestyles, there is a substantial increasing trend of CRC
incidence in younger population. Although progress has been made in
revealing the molecular mechanism and treatment of CRC, its
prognosis remains very poor, possibly due to the rapid progression
and development of CRC, chemoresistance and lack of effective
screening and treatment approaches (2–4).
Therefore, identifying specific diagnostic biomarkers and effective
therapeutic targets for CRC is of great importance.
Agmatinase (AGMAT), located in mitochondria, is a
key enzyme involved in polyamine metabolism pathway and is mainly
distributed in the kidneys and liver (5). The polyamine metabolic disorder is
commonly caused by the abnormal expression of enzymes involved in
the metabolic pathway. Therefore, the abnormal expression of AGMAT
could promote the occurrence and development of cancer via
affecting polyamine metabolism. A previous study showed that AGMAT
was highly expressed in lung adenocarcinoma tissues and it was
closely associated with tumor stage and prognosis in patient with
lung adenocarcinoma; furthermore, the study also demonstrated that
AGMAT promoted inducible nitric oxide (NO) synthase expression via
activating the MAPK and PI3K/Akt signaling pathways and inducing NO
release to enhance lung cancer cell invasion and metastasis in
vitro (6). Additionally, AGMAT
could promote pancreatic cancer cell proliferation, invasion and
metastasis via the TGF-β/Smad pathway (7). Although AGMAT plays an essential role
in the metabolism of amino acids, its effects on CRC have not been
previously investigated.
With the discovery of microRNAs (miRNAs or miRs),
their role in the onset and progression of CRC has become a
significant area of interest (8).
Increasing evidence has suggested that the dysregulation of miRNAs
is involved in the development of several types of cancer,
including CRC (9). miRNAs also play
a significant role in different biological and cellular processes,
such as in cell proliferation, differentiation, apoptosis, death
and metastasis (10–13). miR-151a-5p is a well-characterized
oncogenic miRNA, which is dysregulated in CRC and is therefore
recognized as a promising biomarker for the early detection and
treatment of CRC (14). As a member
of the miR-151 family, miR-151a-5p is a small non-coding RNA
molecule that acts as a proto-oncogene and is expressed in several
types of cancer, such as lung cancer (15), prostate cancer (16) and lymphoblastic leukemia (17). It has been reported that miR-151a-5p
is involved in numerous biological processes. For instance, a
previous study suggested that miR-151a-5p could be involved in the
regulation of cell respiration and ATP production in mitochondria
via targeting cytochrome B (18).
As a member of housekeeping miRNAs, miR-151a-5p was identified to
be stably expressed in endothelial cells and macrophages in an
inflammatory setting (19).
Furthermore, Guo et al (20)
showed that miR-151a-5p was upregulated in lung cancer, while
miR-151a-5p silencing could inhibit the proliferation, invasion and
migration, and induce the apoptosis of A549 lung cancer cells.
These findings suggested that miR-151a-5p could be a potential
therapeutic target for several types of cancer. Although
miR-151a-5p could be used as a potential non-invasive serological
diagnostic marker for the detection of CRC, the particular effects
and mechanisms of miR-151a-5p in CRC remain unclear (14).
Therefore, the present study aimed to explore the
effect and molecular mechanisms of miR-151a-5p in CRC in
vitro.
Materials and methods
Datasets analysis
Co-expression analyses of the interaction between
miR-151a-5p and AGMAT were downloaded from the starbase database
(http://starbase.sysu.edu.cn/), including
COAD samples (n=450). The potential binding sites between
miR-151a-5p and AGMAT were downloaded from the miRmap database
(http://www.mirmap.ezlab.org). The
expression levels of miR-151a-5p in tumor tissues (n=254) were
directly compared with those in adjacent normal tissues (n=8) using
the expression data from ~32 common types of cancer downloaded from
the Gene Expression Display Server (http://bioinfo.life.hust.edu.cn/web/GEDS/).
Additionally, the expression levels of miR-151a-5p in tumor tissues
(n=445) were also directly compared with those in adjacent normal
tissues (n=8) downloaded from the UCSC Xena database (http://xena.ucsc.edu/).
Cell lines and culture
The human CRC cell lines HCT8, HCT116, HT29, SW480,
SW620, LS174T and DLD1 were purchased from the American Type
Culture Collection. In addition, the normal human colorectal cells
NCM460 and 293T cells were conserved in the central laboratory of
Shanghai Fengxian District Central Hospital. All cell lines were
cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (both from Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin solution in a
humidified incubator at 37°C with 5% CO2.
Plasmids and retroviral infection
The full-length sequence of the human AGMAT gene was
subcloned into the pCD513B vector (Geneppl Technology Co., Ltd.)
and transfected to generate cells stably overexpressing AGMAT. To
stably knock down AGMAT in CRC cells, small hairpin RNAs (shRNAs)
targeting AGMAT were synthesized and subcloned into the lentiviral
pPLK-GFP + Puro vector (Geneppl Technology Co., Ltd.). The
sequences of the shRNA clones used were listed in Table I. The concentrations of shRNA clones
used were as follows: NC, 964 ng/µl; shAGMAT#1, 1,142 ng/µl; and
shAGMAT#3, 795 ng/µl. The mixture of 1 ml DMEM, 10 µg DNA and 10 µl
(µg) GM easy™ Lentivirus mix (Genomeditech) was co-transfected into
293T cells using 60 µl Hg transgene™ reagent (Genomeditech),
according to the manufacturer's instructions (cat. no. GMeasy 40;
Genomeditech). The lentiviral particles were collected at 48 h
following cell transduction.
 | Table I.The sequences of specific primers
used for shRNA and miR-151a-5p. |
Table I.
The sequences of specific primers
used for shRNA and miR-151a-5p.
| Primer | Sequence
(5′→3′) |
|---|
| shAGMAT#1 |
CGATGTGAATGTCAATCTTTA |
| shAGMAT#3 |
CGGGAAGAATCAGTGATGCTT |
| NC |
GTTCTCCGAACGTGTCACGTT |
| miR-151a-5p |
UUGUACUACACAAAAGUACUG |
| mimic NC |
|
| miR-151a-5p
mimics |
UCGAGGAGCUCACAGUCUAGU |
| miR-151a-5p |
CAGUACUUUUGUGUAGUACAA |
| inhibitor NC |
|
| miR-151a-5p
inhibitor |
ACUAGACUGUGAGCUCCUCGA |
Generation of stable cell lines and
miR-151a-5p transfection
When the cell density of ~80–90%, CRC cells seeded
into 6-cm culture-plates were infected with filtered lentiviral
particles (the 2nd generation system) plus 8 µg/ml Polybrene
(Sigma-Aldrich; Merck KGaA) for 24 h. Following transduction, cells
were treated with 0.5 µg/ml puromycin (Invitrogen; Thermo Fisher
Scientific, Inc.) for >7 days to obtain stably transfected
cells; afterwards, the stably transfected cells were maintained
with 0.3 µg/ml puromycin. Subsequently, when reached ~60-70%
confluency in the cell culture plate, HCT116 cells, SW480 cells and
stably AGMAT expressing CRC cells were treated with a mixture
containing 5 µl Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), 100 pmol miR-151a-5p and control
oligonucleotides, according to the manufacturer's instructions.
Following transfection for 48 h, the relative expression levels of
miR-151a-5p were determined by reverse transcription-quantitative
PCR (RT-qPCR). The sequences of miR-151a-5p mimics or inhibitor
used were listed in Table I
(Genomeditech).
Dual luciferase reporter assay
The luciferase reporter constructs were generated by
inserting the wild-type (WT) or mutant (MUT) 3′ untranslated region
(3′ UTR) of AGMAT into the pGL3 luciferase vector (Geneppl
Technology Co., Ltd.). Subsequently, the stable AGMAT
overexpressing CRC cells were co-transfected with 2 µg WT or MUT
AGMAT-3′ UTR, Renilla Luc, miR-151a-5p mimics or miR-NC
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions
(Shanghai Yeasen Biotechnology Co., Ltd.). Following transfection
for 48 h, the firefly and Renilla luciferase activities were
determined by spectrophotometry using the Dual Luciferase Reporter
Assay System (Glomax 96; Promega Corporation). The luciferase
activity was then calculated. The results are expressed as the
ratio of firefly luciferase activity to Renilla luciferase
activity.
Cell counting kit-8 (CCK-8) assay
The cell proliferation ability was assessed using
CCK-8 assay (Dojindo Molecular Technologies, Inc.), according to
the manufacturer's protocol. Briefly, treated CRC cell lines were
seeded into a 96-well plate at a density of 103
cells/well in 100 µl DMEM supplemented with 10% FBS and were then
cultured for 10, 24, 48 and 72 h at 37°C with 5% CO2.
Subsequently, cells were supplemented with 10 µl CCK-8 solution,
followed by incubation for an additional 1 h in the dark. Finally,
the optical density (OD) of each well was measured at a wavelength
of 450 nm using a microplate reader (BioTek Instruments, Inc.).
Colony formation assay
For colony formation assays, treated CRC cells were
seeded into a 12-well plate at a density of 3×103
cells/well and cultured in DMEM supplemented with 10% FBS for 7 or
10 days. The formed colonies were fixed with 4% paraformaldehyde at
room temperature for 30 min and were then stained with 0.1% crystal
violet at room temperature for 1 h followed by washing with
ddH2O. Images of the formed colonies were captured under
an inverted light microscope (Olympus Corporation). Finally, the
number of colony cells (>50 cells per cell colony) were counted
using ImageJ software (version 1.52a; National Institutes of
Health).
Cell migration and invasion assay
The migration and invasion abilities of HCT116 and
SW480 cells transfected with the aforementioned plasmids/miR
mimics/inhibitors for 48 h were evaluated using Transwell chambers
(0.8 µm; cat. no. 353097; Corning, Inc.). Briefly, treated HCT116
and SW480 cells at a density of 105 cells/well were
added to the upper chamber of the Transwell chamber coated or not
with Matrigel (0.8 µm; cat. no. 354480; Corning, Inc.). The upper
chambers of the Transwell chamber with Matrigel were precoated with
500 µl medium without serum for 1 h at 37°C with 5% CO2.
For migration assays, the lower chamber was supplemented with 600
µl medium with 10% FBS, cells were cultured for 48 h, while for the
invasion assays, the lower chamber was supplemented with 600 µl
medium with 20% FBS, cells were cultured for 24 h, the upper
chamber was supplemented without serum. The migratory or invasive
cells were fixed with 4% paraformaldehyde at room temperature for
30 min and were then stained with 0.1% crystal violet at room
temperature for 1 h followed by ddH2O washing. Finally,
migratory or invasive cells were observed and images were captured
under a fluorescence microscope (magnification, ×100).
RT-qPCR analysis
Total RNA was extracted from treated cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The nuclear and cytoplasmic extracts were isolated using the
cytoplasmic and nuclear RNA purification kit (cat. no. 21000;
Norgen Biotek Corp.). Subsequently, 1 µg RNA was reverse
transcribed into cDNA using the Evo M-MLV RT Mix kit with gDNA
Clean for qPCR (cat. no. AG11728; Accurate Bio-Medical Technology
Co., Ltd.) and the miRNA 1st strand cDNA synthesis kit (by
stem-loop) (cat. no. MR101-02; Vazyme Biotech Co., Ltd.). qPCR was
performed on the quantum Studio 6 real-time PCR system (Thermo
Fisher Scientific, Inc.) using the SYBR Green premix Pro Taq HS
qPCR kit (Rox plus) (cat. no. AG1718; Accurate Bio-Medical
Technology Co., Ltd.). The thermocycling conditions used for
reverse transcription PCR were as follows: For gDNA clean, 42°C 2
min, 4°C; for synthesis of first strand cDNA of miR-151a-5p, 25°C
for 5 min, 50°C for 15 min, 85°C for 5 min; for synthesis of common
primer cDNA, 37°C for 15 min, 85°C for 5 min, 4°C. The
thermocycling conditions used for qPCR were as follows: For
miR-151a-5p, 95°C for 5 min, 95°C 10 sec, 60°C for 5 min; for
common primers, 95°C for 30 sec, 95°C for 5 sec, 60°C for 30 sec.
The relative mRNA expression levels of the target genes were
calculated using the 2−ΔΔCq method (21). GAPDH served as an internal reference
gene for AGMAT. The primers for U1 were purchased from Guangzhou
RiboBio Co., Ltd. The primer sequences used are listed in Table II (Sangon Biotech Co., Ltd.).
 | Table II.The sequences of specific primers
used for reverse transcription-quantitative PCR. |
Table II.
The sequences of specific primers
used for reverse transcription-quantitative PCR.
| Gene name | Sequence
(5′→3′) |
|---|
| GAPDH | F:
TTGGTATCGTGGAAGGACTCA |
|
| R:
TGTCATCATATTTGGCAGGTT |
|
microRNA-151a-5p | F:
CGCGTCGAGGAGCTCACAG |
|
| R:
AGTGCAGGGTCCGAGGTATT |
| AGMAT | F:
CTTGTCGAAGTTTCACCACCGTA |
|
| R:
CTTTGGGGAGAGCACATAGCATC |
| AZIN2 | F:
CAACTCAGCCTTGGACCTGTACTTC |
|
| R:
CTGCTCCGTGGATGGTTTCTTCTG |
| AMD1 | F:
CCCGACGCAAACCAAGGATCTG |
|
| R:
TTCAACAGGGGAACCAGTGCTTTC |
| ODC1 | F:
CACTGTTGCTGCTGCCTCTACG |
|
| R:
GGTTCTGGAATTGCTGCATGAGTTG |
| OAZ1 | F:
TCTCCCTCCACTGCTGTAGTAACC |
|
| R:
TGACTATTCCCTCGCCCACCTG |
| SRM | F:
GATGATCGCCAACCTGCCTCTC |
|
| R:
ATCTCACACTGGACCACGGACTC |
| SMS | F:
TTGGCAGAGAGTGATTTGGCATATACC |
|
| R:
CCACCTCCCAGAATGAGTACATCTTTG |
| SAT1 | F:
TGGTTGCAGAAGTGCCGAAAGAG |
|
| R:
ATAACTTGCCAATCCACGGGTCATAG |
| SMOX | F:
CACACCCTCACCTACCCACCTG |
|
| R:
CACTGCCTCGTCATCACACTTCTC |
| PMF1 | F:
GCGATGACACAGCAAATCTATGACAAG |
|
| R:
AGGCATTCAAGACAGCTTCTAGGTTC |
| TP53 | F:
GCCCATCCTCACCATCATCACAC |
|
| R:
GCACAAACACGCACCTCAAAGC |
| PAOX | F:
TTCCAGTGTCGGTAGAGTGTGAGG |
|
| R:
ATCTTCCTGATTGCTTCTGCCTTCTC |
Immunofluorescence assay
The transfected cells were cultured in 48-well
plates at a density of 6×104 cells/well for 24 h.
Following fixation with 4% paraformaldehyde at room temperature for
30 min, the cells were permeabilized with 1% Triton X-100 at room
temperature for 15 min followed by blocking with 3% BSA (cat. no.
4240GR100; Biofroxx; neoFroxx) at 37°C for 1 h. After cell
incubation with primary antibodies at 4°C overnight, the cells were
treated with the corresponding secondary Goat anti-Rabbit IgG (H +
L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 568
(Thermo Fisher Scientific, Inc.) at room temperature for 30 min in
the dark. Nuclei were counterstained with DAPI (5 mg/ml; cat. no.
C1006; Beyotime Institute of Biotechnology) at room temperature for
20 min in the dark. The stained cells were observed under a
fluorescence microscope (magnification, ×200). The antibodies used
for immunofluorescence are listed in Table III.
 | Table III.Antibodies used for immunofluorescent
staining. |
Table III.
Antibodies used for immunofluorescent
staining.
| Target | Supplier | Cat. no. | Dilution | Isotype | Host species |
|---|
| E-cadherin | Proteintech Group,
Inc. | 20874-1-AP | 1:200 | IgG | Rabbit |
| Vimentin | Proteintech Group,
Inc. | 10366-1-AP | 1:200 | IgG | Rabbit |
Western blot analysis
Total proteins were extracted from treated CRC cells
using RIPA lysis buffer (New cell & Molecular Biotech Co.,
Ltd.) supplemented with phosphatase inhibitors (Beyotime Institute
of Biotechnology) and protein concentration was measured with the
BCA Protein Assay Kit (cat. no. P0012; Beyotime Institute of
Biotechnology). Subsequently, the proteins (10–20 µl/lane, 25
mg/ml) were separated by 10% SDS-PAGE (New cell & Molecular
Biotech Co., Ltd.) and were then transferred onto a nitrocellulose
membrane (Pall Corporation). Following blocking with 5% skimmed
milk (Beyotime Institute of Biotechnology) at room temperature for
1 h, the membrane was incubated with primary antibodies at 4°C
overnight. After incubation with the corresponding secondary
antibodies at room temperature for 1 h, the immunoreactive bands
were visualized using an ECL Kit (Vazyme Biotech Co., Ltd.) and
images were captured under the Tanon 4600 system (Tanon Science and
Technology Co., Ltd.). The density of each band was measured using
a computer-assisted imaging analysis system (Adobe Systems Inc.).
The expression levels of the target proteins were normalized to
those of β-tubulin. The following primary and secondary antibodies
were used: Anti-AGMAT, anti-MMP2, anti-MMP9, anti-N-cadherin,
anti-E-cadherin, anti-Vimentin, anti-β-catenin, HRP-conjugated
Affinipure Goat anti-Mouse IgG (H+L) and HRP-conjugated Affinipure
Goat anti-Ribbit IgG (H+L) (Table
IV). The protein expression levels were assessed using ImageJ
software (National Institutes of Health).
 | Table IV.Antibodies used for western blot
analyses. |
Table IV.
Antibodies used for western blot
analyses.
| Primary
antibodies |
|---|
|
|---|
| Target | Supplier | Cat. no. | Dilution | Isotype | Host species | MW (KDa) |
|---|
| AGMAT | Abcam | Ab231894 | 1:2,000 | IgG | Rabbit | 38 |
| MMP2 | Proteintech Group,
Inc. | 10373-2-AP | 1:1,000 | IgG | Rabbit | 60 |
| MMP9 | Proteintech Group,
Inc. | 10375-2-AP | 1:1,000 | IgG | Rabbit | 92 |
| N-cadherin | Proteintech Group,
Inc. | 22018-1-AP | 1:2,000 | IgG | Rabbit | 130 |
| E-cadherin | Proteintech Group,
Inc. | 20874-1-AP | 1:5,000 | IgG | Rabbit | 135 |
| Vimentin | Proteintech Group,
Inc. | 10366-1-AP | 1:2,000 | IgG | Rabbit | 54 |
| β-Catenin | Proteintech Group,
Inc. | 51067-2-AP | 1:5,000 | IgG | Rabbit | 92 |
| β-Tubulin | Proteintech Group,
Inc. | 66240-1-Ig | 1:10,000 | IgG2a | Mouse | 55 |
|
| Secondary
antibodies |
|
| Target |
Supplier | cat.
no. |
Dilution |
|
| HRP-conjugated
Affinipure Goat anti-mouse IgG (H+L) | Proteintech Group,
Inc. | SA00001-1 | 1:5,000 |
| HRP-conjugated
Affinipure Goat anti-rabbit IgG (H+L) | Proteintech Group,
Inc. | SA00001-2 | 1:5,000 |
Statistical analysis
All experiments were repeated at least in
triplicate. Data are expressed as the mean ± SD. All statistical
analyses were carried out using GraphPad Prism 8 (GraphPad
Software, Inc.). The differences between two groups were compared
using unpaired Student's t-test. Comparisons among multiple groups
were conducted using with one- or two-way ANOVA followed by
Dunnett's multiple comparisons test used as post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-151a-5p is upregulated in CRC cell
lines and tissues
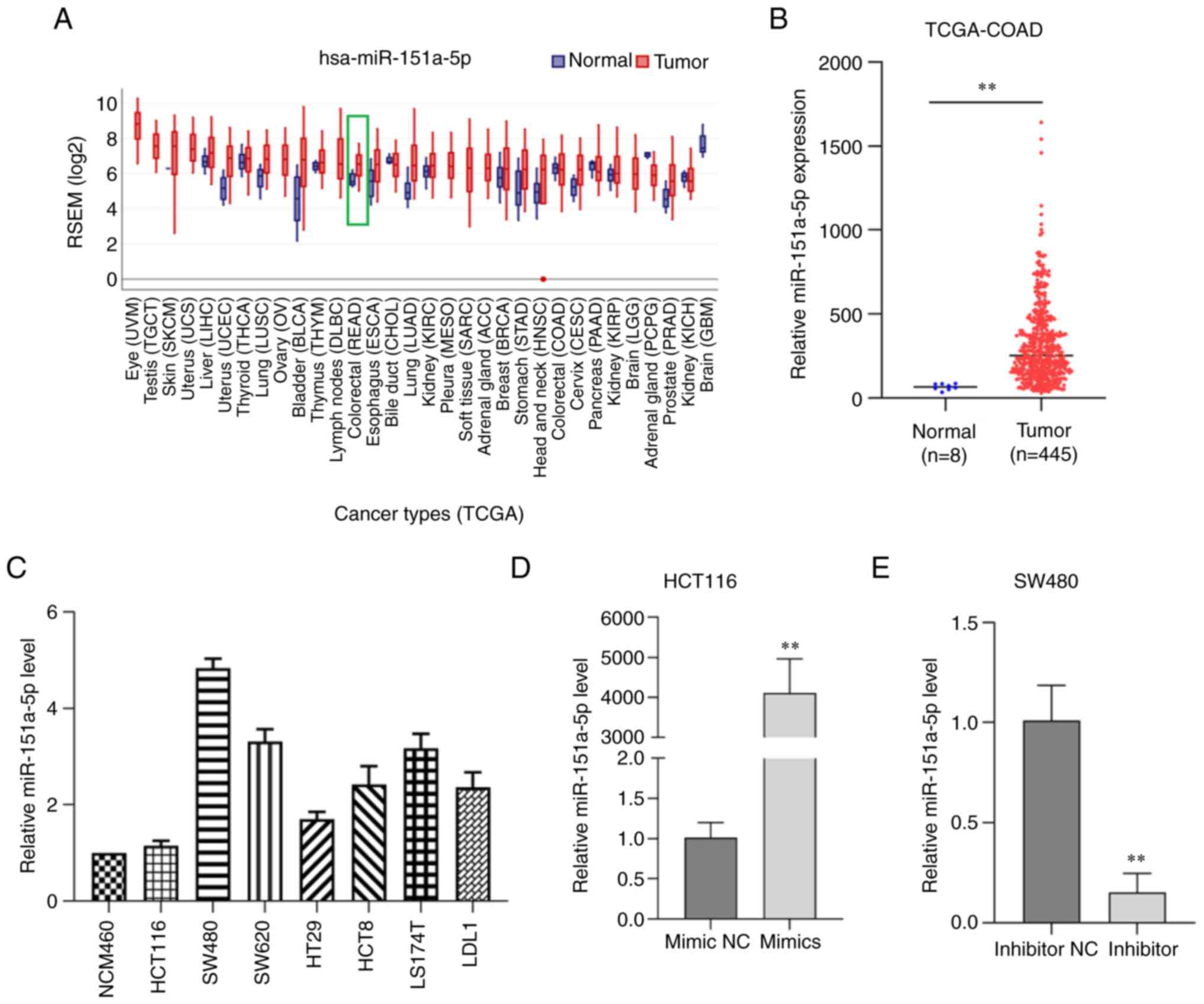
To explore the pathophysiological relevance of
miR-151a-5p in human CRC, the expression levels of miR-151a-5p were
determined in CRC and adjacent normal tissues obtained from UCSC
Xena database and the Gene Expression Display Server. Therefore,
compared with normal colorectal tissues, miR-151a-5p was
significantly upregulated in CRC tissues (Fig. 1A and B). These data suggested that
miR-151a-5p was upregulated in CRC tissues and could therefore
regulate the occurrence and development of CRC via acting as a
tumor-promoting gene. In addition, the expression levels of
miR-151a-5p were further detected in several CRC cell lines by
RT-qPCR. The analysis showed that the expression levels of
miR-151a-5p were higher in CRC cell lines compared with normal
colorectal cells. Among all CRC cell lines examined, SW480 cells
exhibited the highest miR-151a-5p expression levels, while HCT116
cells the lowest one (Fig. 1C).
Therefore, HCT116 and SW480 cells were selected for the subsequent
experiments. Furthermore, the transfection efficiency of
miR-151a-5p in CRC cells was also verified (Fig. 1D and E).
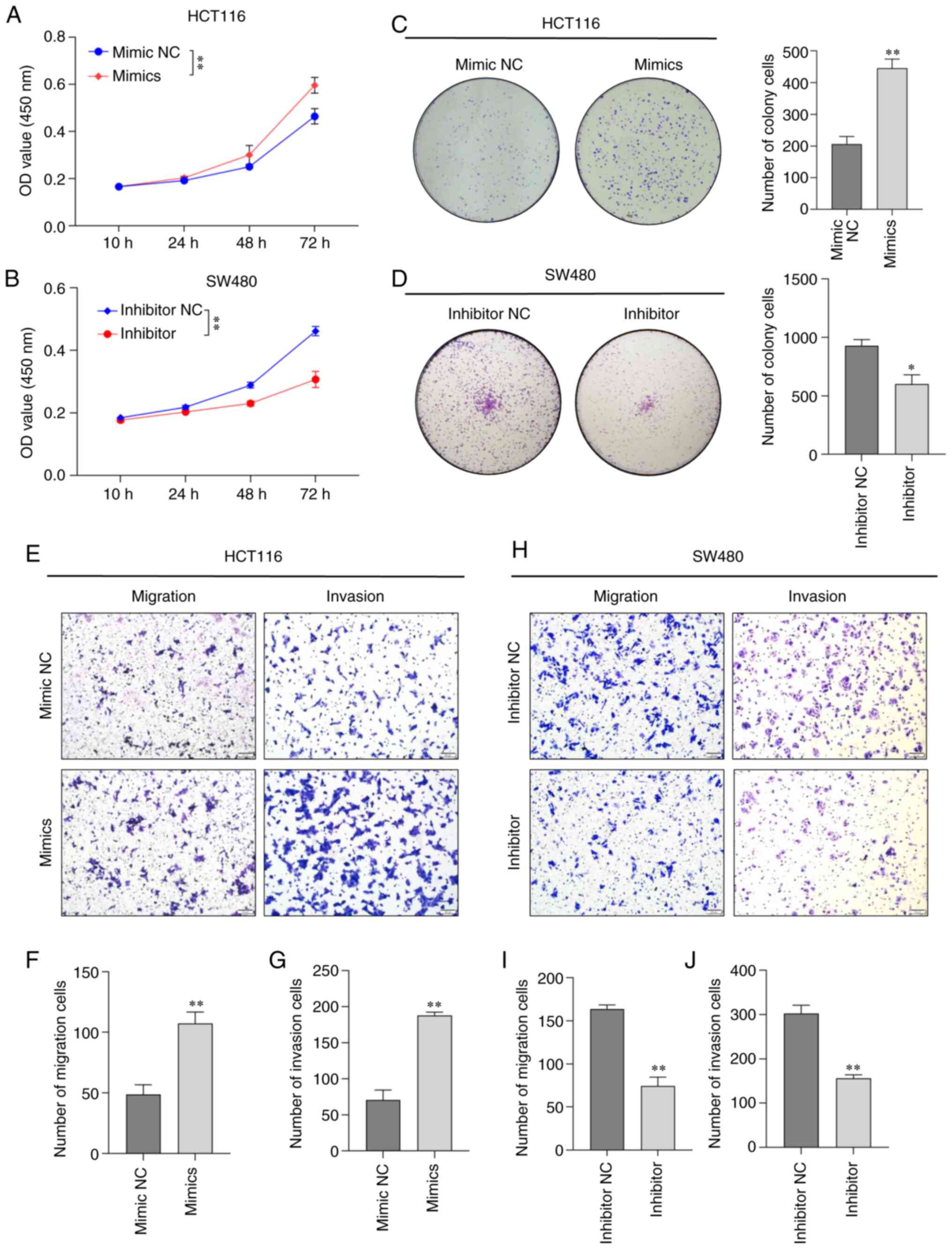
miR-151a-5p promotes CRC cell
proliferation, migration and invasion
To further uncover the potential role of miR-151a-5p
in CRC, HCT116 or SW480 cells were transfected with miR-151a-5p
mimics or inhibitor, respectively. Following cell transfection for
48 h, the cell proliferation ability was assessed by CCK-8 and
colony formation assays. The results showed that compared with the
NC group, the change in the overexpression levels of miR-151a-5p
significantly promoted the proliferation of HCT116 cells in
time-dependent manner (Fig. 2A),
while as predicted, the proliferation capacity of SW480 cells
transfected with miR-151a-5p inhibitor was significantly inhibited
(Fig. 2B). In addition, the effect
of miR-151a-5p on the proliferation of HCT116 and SW480 cells was
evaluated by colony formation assay. The results demonstrated that
the overexpression levels of miR-151a-5p promoted the proliferation
of HCT116 cells (Fig. 2C). By
contrast, the proliferation of SW480 cells was significantly
inhibited after transfection with miR-151a-5p inhibitor (Fig. 2D). The enhanced migration and
invasion abilities of CRC cells could also serve a key role in the
growth of CRC. Therefore, to further evaluate the effect of
miR-151a-5p on CRC cell invasion and migration, Transwell assays
coated with Matrigel or not, respectively, were used. The data
revealed that compared with the corresponding NC group, the
migration and invasion abilities of HCT116 cells were significantly
increased following cell transfection with miR-151a-5p mimics
(Fig. 2E-G). However, the opposite
effect was observed in miR-151a-5p-depleted SW480 cells (Fig. 2H-J). The aforementioned findings
indicated that miR-151a-5p could significantly enhance the
proliferation, migration and invasion abilities of CRC cells.
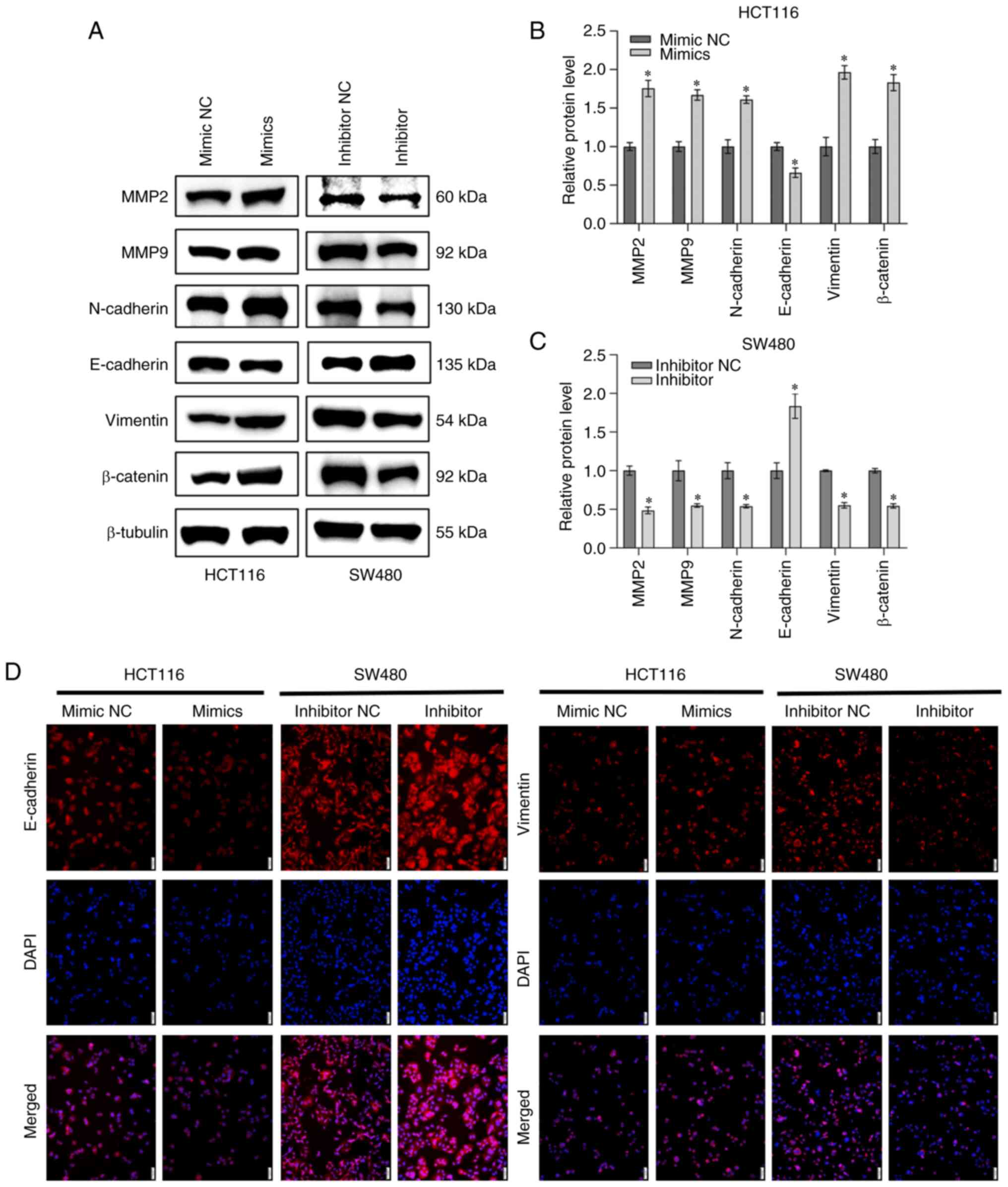
miR-151a-5p enhances the
epithelial-mesenchymal transition (EMT) of CRC cells
Epithelial cells are characterized by mesenchymal
phenotype and changes in the expression of three significant
biomarkers, including E-cadherin, vimentin and N-cadherin,
eventually leading to reduced cell adhesion, loss of polarity and
tight junction (22). Matrix
metalloproteinases (MMPs) are proteins involved in the degradation
of base membranes, thus affecting cell metastasis during the
development of malignant tumors (23). Therefore, western blot analysis was
performed and the result revealed that miR-151a-5p overexpression
in HCT116 cells downregulated E-cadherin and upregulated
N-cadherin, vimentin and β-catenin. Correspondingly, western blot
analysis was also performed to evaluate the effect of miR-151a-5p
on the expression levels of metastasis-related MMPs, such as MMP2
and MMP9. The results demonstrated that compared with the NC group,
miR-151a-5p enhanced the protein expression levels of both MMP2 and
MMP9 in HCT116 cells. By contrast, miR-151a-5p silencing in SW480
cells exhibited the opposite effect (Fig. 3A-C). To further determine the
expression of EMT-related proteins in CRC cell lines, the
expression of E-cadherin and vimentin was assessed by
immunofluorescence analysis. The immunofluorescence analysis
results demonstrated that miR-151a-5p overexpression in HCT116
cells downregulated E-cadherin and upregulated vimentin. However,
miR-151a-5p silencing upregulated E-cadherin and downregulated
vimentin (Fig. 3D). The
aforementioned results verified that miR-151a-5p could promote EMT
in CRC cells.
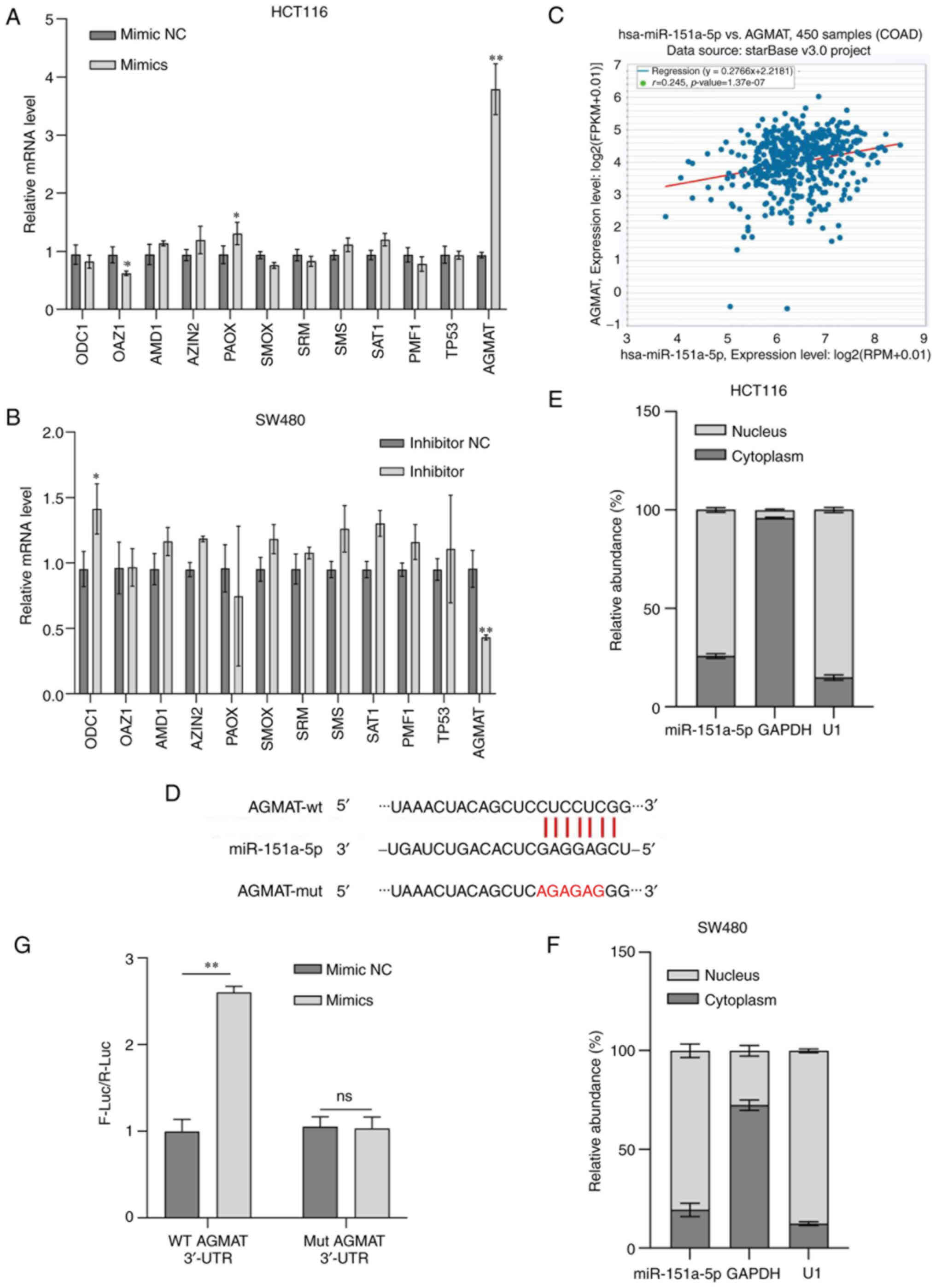
AGMAT is a target gene of miR-151a-5p
and is positively associated with miR-151a-5p expression
Polyamines, such as putrescine, spermidine and
spermine, produced during the metabolic process, are low molecular
weight, aliphatic and polycationic nitrogen compounds containing
two or more amino groups. Currently, the application of new
technologies has increased our understanding of the genetics and
potential molecular biology of polyamine function in normal and
tumor cells. Recognition of the effect of the increased polyamine
levels on tumor transformation and progression has deepen our
understanding of the significant role of polyamines in cancer, thus
providing reasonable targets for intervention (24). It has been reported that miRNAs can
regulate tumor-associated proteins in CRC (25,26).
However, the association between genes and miRNAs is not a
one-to-one correspondence. The results of the current study showed
that following miR-151a-5p overexpression or silencing in CRC cell
lines, the levels of the polyamine metabolism-related metabolites
and key enzymes significantly altered, while the most differences
were observed in the expression levels of AGMAT at the molecular
level (Fig. 4A and B). These
findings indicated that miR-151a-5p could promote polyamine
metabolism via regulating AGMAT. Therefore, subsequently, the
present study aimed to explore whether miR-151a-5p could regulate
CRC progression via targeting AGMAT. Bioinformatic analysis using
the starbase database predicted that AGMAT encompassed a binding
site for miR-151a-5p and revealed that AGMAT was positively
associated with miR-151a-5p expression in CRC (Fig. 4C). In addition, it was further
predicted using the miRmap database that the potential miR-151a-5p
binding site was encompassed in AGMAT-3′ UTR (Fig. 4D). Previous studies suggested that
the majority of miRNAs in the cytoplasm could act as tumor
suppressors, thus resulting in gene silencing via inhibiting gene
translation (27). By contrast,
other studies showed that several miRNAs, located at the nucleus,
could positively regulate gene transcription via binding to and
activating their target enhancers (28,29).
In the present study, RNA isolation from nuclear and cytoplasmic
extracts revealed that miR-151a-5p was mainly localized in the
nucleus, as evidenced by RT-qPCR analysis (Fig. 4E and F). To further verify whether
miR-151a-5p could regulate the expression of AGMAT via binding to
AGMAT-3′ UTR, a dual luciferase reporter assay was performed. As
revealed in Fig. 4G, the luciferase
activity of AGMAT-3′ UTR was significantly increased in cells
transfected with miR-151a-5p mimics. However, miR-151a-5p
overexpression showed a modest effect on luciferase activity in
cells transfected with mutated putative binding site. For the
convenience of follow-up tests, the stable AGMAT expression cell
lines were designed, namely, the overexpression of AGMAT in HCT116
cells and the knockdown of AGMAT in SW480 cells. The transfection
efficiency of AGMAT in stable transmutation cells was determined by
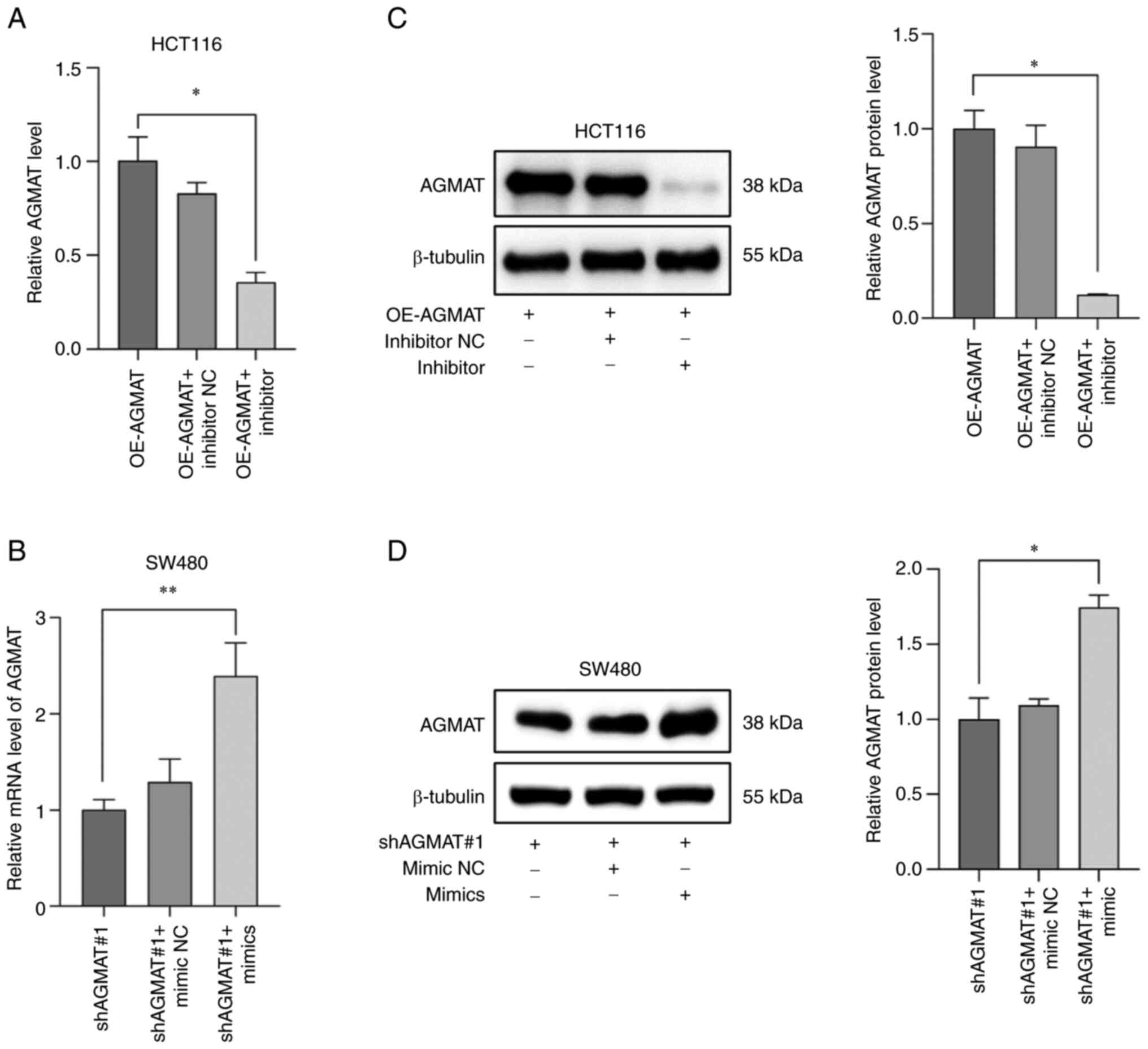
RT-qPCR and western blot analysis. ShAGMAT#1 was selected from the
AGMAT-stably knocked down SW480 cells due to the best knockdown
effect (Fig. S1). Furthermore,
RT-qPCR and western blot analysis were carried out to reveal the
relationship between miR-151a-5p and AGMAT expression. The results
identified that the mRNA expression levels of AGMAT were
significantly reduced in AGMAT-stably overexpressing HCT116 cells
transfected with miR-151a-5p inhibitor (Fig. 5A). However, the opposite effect was
observed in the AGMAT-stably knocked down SW480 cells transfected
with miR-151a-5p mimics (Fig. 5B).
Western blot analysis further verified the positive regulatory
association between miR-151a-5p and AGMAT (Fig. 5C and D). This indicated that
miR-151a-5p could promote the expression of AGMAT in CRC cells. The
aforementioned data suggested that AGMAT could be a potential
target of miR-151a-5p and could be positively associated with
miR-151a-5p expression in CRC.
miR-151a-5p promotes the effects of
AGMAT on the proliferation, migration and invasion of CRC
cells
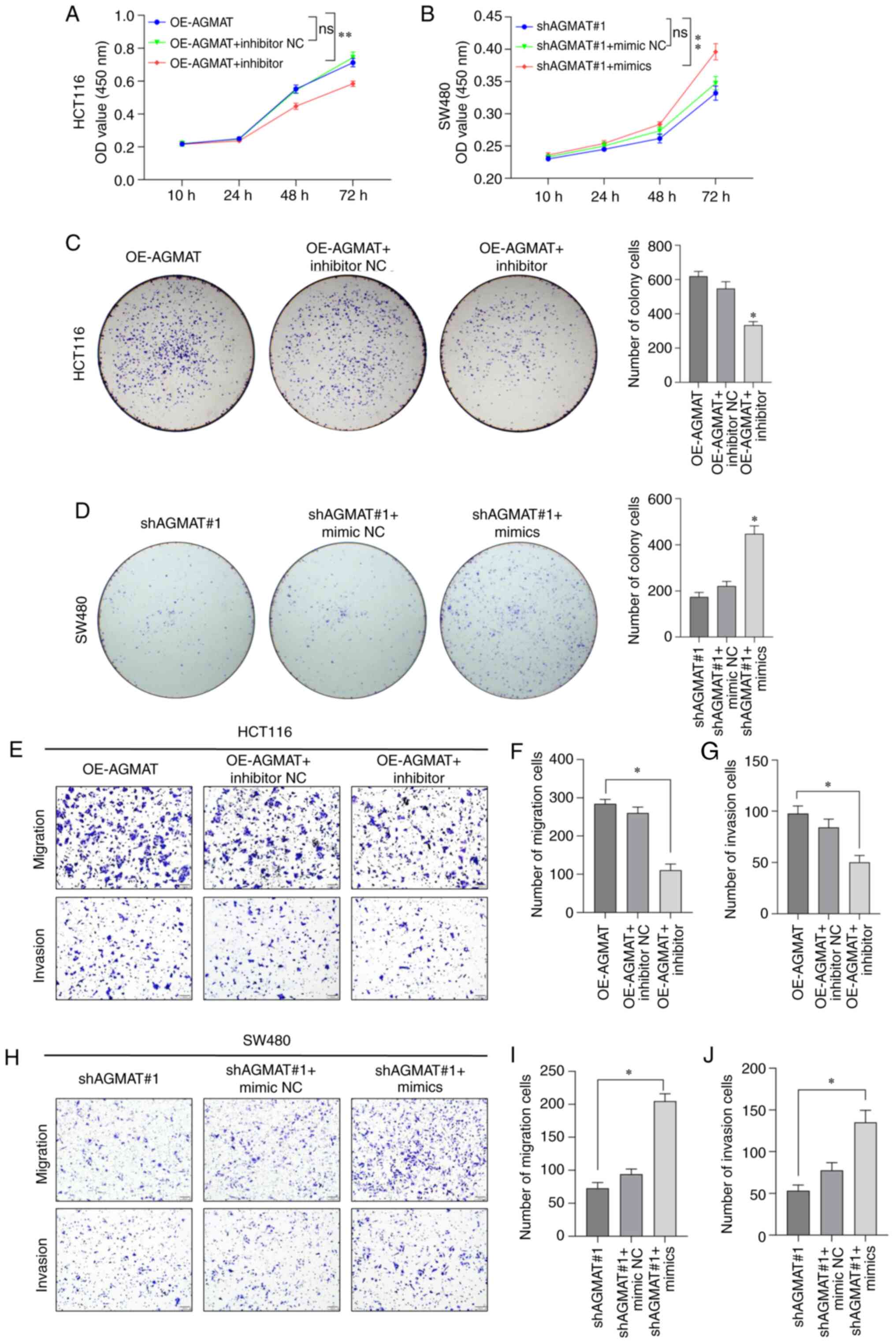
To further verify whether miR-151a-5p could regulate
the potential functions of AGMAT in CRC, AGMAT-stably expressing
HCT116 or SW480 cells were transfected with miR-151a-5p inhibitor
or mimics. Following transfection for 48 h, the proliferation
ability of AGMAT-stably expressing CRC cells was evaluated by CCK-8
and colony formation assays. Therefore, compared with the NC group,
the proliferation of the AGMAT-stably overexpressing HCT116 cells
was significantly decreased after transfection with miR-151a-5p
inhibitor in a time-dependent manner (Fig. 6A). However, the significant opposite
result of the proliferation was observed in a time-dependent manner
in the AGMAT-stably knocked down SW480 cells transfected with
miR-151a-5p mimics (Fig. 6B).
Additionally, the effect of miR-151a-5p targeting AGMAT on the
proliferation of the AGMAT-stably overexpressing HCT116 or SW480
cells was also evaluated by colony formation assays. As expected,
the proliferation ability of the AGMAT-stably overexpressing HCT116
cells transfected with miR-151a-5p inhibitor was significantly
decreased (Fig. 6C), but the
significant opposite result of the proliferation was observed in a
time-dependent manner in the AGMAT-stably knocked down SW480 cells
transfected with miR-151a-5p mimics (Fig. 6D). To further explore whether
miR-151a-5p could regulate CRC cell invasion and migration via
targeting AGMAT, Transwell invasion and migration assays were
performed. The results showed that compared with the NC group, the
migratory and invasive abilities of the AGMAT-stably overexpressing
HCT116 cells transfected with miR-151a-5p inhibitor were
significantly decreased (Fig.
6E-G). The completely opposite effect was obtained in the
AGMAT-stably knocked down SW480 cells transfected with miR-151a-5p
mimics (Fig. 6H-J). The
aforementioned results indicated that miR-151a-5p could enhance the
biofunction of CRC cells via targeting AGMAT.
Discussion
Emerging evidence has suggested that miRNAs can
mediate the development of several diseases, such as CRC, through
multiple genes and biological pathways (30–32).
It has been reported that miRNAs are abnormally expressed in CRC
tissues, thus serving as powerful and promising biomarkers for the
early screening and treatment of CRC (33,34).
For example, Lopez-Camarillo et al (35) investigated the expression of several
miRNAs, known as metaMIRs, which are involved in the initiation of
cell invasion and metastasis via targeting numerous proteins
associated with several cellular processes. Therefore, the
aforementioned study highlighted the potential use of metaMIRs as
novel specific markers for evaluating cancer progression. MicroRNAs
play an important role in regulating gene expression under normal
and pathological conditions, such as cancer (36). The majority of miRNAs act as tumor
suppressors, thus promoting gene silencing through translational
suppression, which mainly occurs in the cytoplasm (27). However, the aberrant overexpression
of other miRNAs, known as super-enhancers, commonly localized in
the nucleus, can promote tumor occurrence, growth and/or metastasis
(28,29). It has been reported that several
miRNAs can act as biomarkers in CRC via negatively regulating the
expression of different genes. However, the application of miRNAs
as super-enhancers in clinical practice remains limited. Previous
studies demonstrated that miR-151a-5p could be used as a potential
non-invasive serological diagnostic marker for the diagnosis of CRC
(14) and endometrial cancer
(37). However, the effect and
underlying mechanism of miR-151a-5p on CRC remains unclear.
Consistent with previous findings, the results of the current study
revealed that miR-151a-5p was significantly upregulated in CRC.
Additionally, miR-151a-5p overexpression enhanced the proliferation
ability of CRC cells. Furthermore, the results demonstrated that
the tumor-promoting effects of miR-151a-5p were mediated via
improving the migration and invasion abilities of CRC cells. MMPs
are significant calcium- and zinc-dependent proteolytic enzymes,
which can degrade the components of the extracellular matrix and
basement membrane. These proteins are involved in cell invasion and
metastasis during cancer progression (38). The present study verified that
miR-151a-5p overexpression notably reinforced the migration and
invasion abilities of CRC cells and increased the protein
expression levels of MMP2 and MMP9. In addition, miR-151a-5p
promoted the EMT of CRC cells.
miRNAs are a class of endogenous small
single-stranded non-encoding RNAs with a length of ~18–24
nucleotides, completely or incompletely complementary with the 3′
UTR of their target mRNAs. miRNAs can negatively regulate the
expression of their target genes at the post-transcriptional level
(39,40). In the present study, bioinformatic
analysis revealed that the AGMAT 3′ UTR encompassed a complementary
binding site for miR-151a-5p and AGMAT positively regulated
miR-151a-5p expression. To further explore the combined effect of
miR-151a-5p and AGMAT on the occurrence and development of CRC, a
dual luciferase reporter assay was performed to verify that
miR-151a-5p could directly target AGMAT. Similar to arginase,
AGMAT, as a metal hydrolase, belongs to the urea hydrolase
superfamily. The catalytic activity of AGMAT depends on
Mn2+ ions (41).
Agmatine, an endogenous polyamine located in mitochondria, is
catalyzed by L-arginine decarboxylase and hydrolyzed by agmatine
enzyme to form urea and putrescine. Previous studies suggested that
AGMAT could not only regulate the biological effects of agmatine in
mammalian cells (42), but it could
also be considered as an alternative mechanism for polyamine
biosynthesis (5). Several studies
have reported the regulatory mechanisms of AGMAT and its analogs in
several diseases (43–45). However, the role of AGMAT in cancer
remains elusive. Recent studies showed that AGMAT could play a
significant role in lung adenocarcinoma (6) and pancreatic adenocarcinoma (7). In the present study, AGMAT was
identified to be positively associated with miR-151a-5p expression.
In addition, transfection of AGMAT-stably knocked down CRC cells
with miR-151a-5p mimics promoted the proliferation, migration and
invasion of CRC cells. The current study was the first to
demonstrate that miR-151a-5p could play a significant role in
regulating the proliferation and metastasis of CRC cells via
targeting AGMAT.
In conclusion, the current study demonstrated that
miR-151a-5p was significantly upregulated in CRC tissues and cell
lines, while its ectopic expression promoted the proliferation,
migration and invasion of CRC cells via targeting AGMAT.
Furthermore, miR-151a-5p could enhance the EMT processes of CRC
cells. Therefore, the miR-151a-5p/AGMAT axis could serve as a
potential therapeutic target for the diagnosis and treatment of
CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Shanghai municipal health
commission Foundation (grant no. 20204Y0181), the Health Commission
research Program of Hunan (grant no. 202203034469), and the
Fengxian Science and Technology Commission Project (grant no.
20211805).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YX, YZ, LC and XL contributed to the experiments. CW
and MH were responsible for the bioinformatic analysis and the
experiment result analysis. YX and XZ confirm the authenticity of
all the raw data. XL and YX contributed to writing the manuscript.
JC and XZ designed project ideas, provided technical guidance and
revised the manuscript. All authors have read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal carcinoma
|
|
AGMAT
|
agmatinase
|
|
TGF
|
transforming growth factor
|
|
GEDS
|
Gene Expression Display Server
|
|
TCGA
|
The Cancer Genome Atlas
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
GFP
|
green fluorescent protein
|
|
DAPI
|
4′,6-diamidino-2-phenylindole
|
|
MMP
|
matrix metalloproteinase
|
|
EMT
|
epithelial-mesenchymal transition
|
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kaplan MA, Isikdogan A, Gumus M, Arslan
UY, Geredeli C, Ozdemir N, Koca D, Dane F, Suner A, Elkiran ET, et
al: Childhood, adolescents, and young adults (≤25 y) colorectal
cancer: Study of Anatolian society of medical oncology. J Pediatr
Hematol Oncol. 35:83–89. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Heervä E, Lavonius M, Jaakkola P, Minn H
and Ristamäki R: Overall survival and metastasis resections in
patients with metastatic colorectal cancer using electronic medical
records. J Gastrointest Cancer. 49:245–251. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang X, Ying W, Dunlap KA, Lin G,
Satterfield MC, Burghardt RC, Wu G and Bazer FW: Arginine
decarboxylase and agmatinase: An alternative pathway for de novo
biosynthesis of polyamines for development of mammalian
conceptuses. Biol Reprod. 90:842014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu HE, Yin JY, Chen DX, He S and Chen H:
Agmatinase promotes the lung adenocarcinoma tumorigenesis by
activating the NO-MAPKs-PI3K/Akt pathway. Cell Death Dis.
10:8542019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Cao L, Xie Y, Wang C, Liu X,
Zhang X and Chen J: Agmatinase facilitates the tumorigenesis of
pancreatic adenocarcinoma through the TGFβ/Smad pathway. Exp Ther
Med. 24:4902022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hosseini M, Khatamianfar S, Hassanian SM,
Nedaeinia R, Shafiee M, Maftouh M, Ghayour-Mobarhan M, ShahidSales
S and Avan A: Exosome-encapsulated microRNAs as potential
circulating biomarkers in colon cancer. Curr Pharm Des.
23:1705–1709. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pidíkova P, Reis R and Herichova I: miRNA
clusters with down-regulated expression in human colorectal cancer
and their regulation. Int J Mol Sci. 21:46332020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Slaby O, Svoboda M, Michalek J and Vyzula
R: MicroRNAs in colorectal cancer: Translation of molecular biology
into clinical application. Mol Cancer. 8:1022009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang H: MicroRNAs and apoptosis in
colorectal cancer. Int J Mol Sci. 21:53532020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu G and Li B: Role of miRNA in
transformation from normal tissue to colorectal adenoma and cancer.
J Cancer Res Ther. 15:278–285. 2019.PubMed/NCBI
|
|
13
|
Huang S, Tan X, Huang Z, Chen Z, Lin P and
Fu SW: microRNA biomarkers in colorectal cancer liver metastasis. J
Cancer. 9:3867–3873. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang H, Zhu M, Shan X, Zhou X, Wang T,
Zhang J, Tao J, Cheng W, Chen G, Li J, et al: A panel of
seven-miRNA signature in plasma as potential biomarker for
colorectal cancer diagnosis. Gene. 687:246–254. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Daugaard I, Sanders KJ, Idica A,
Vittayarukskul K, Hamdorf M, Krog JD, Chow R, Jury D, Hansen LL,
Hager H, et al: miR-151a induces partial EMT by regulating
E-cadherin in NSCLC cells. Oncogenesis. 6:e3662017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fredsøe J, Rasmussen AKI, Mouritzen P,
Borre M, Ørntoft T and Sørensen KD: A five-microRNA model (pCaP)
for predicting prostate cancer aggressiveness using cell-free
urine. Int J Cancer. 145:2558–2567. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Almeida RS, Costa E, Silva M, Coutinho LL,
Garcia Gomes R, Pedrosa F, Massaro JD, Donadi EA and Lucena-Silva
N: MicroRNA expression profiles discriminate childhood T-from
B-acute lymphoblastic leukemia. Hematol Oncol. 37:103–112. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou R, Wang R, Qin Y, Ji J, Xu M, Wu W,
Chen M, Wu D, Song L, Shen H, et al: Mitochondria-related
miR-151a-5p reduces cellular ATP production by targeting CYTB in
asthenozoospermia. Sci Rep. 5:177432015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Link F, Krohn K and Schumann J: Author
correction: Identification of stably expressed housekeeping miRNAs
in endothelial cells and macrophages in an inflammatory setting.
Sci Rep. 9:144662019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guo S, Zhang J, Zhao YY, Zhou LY, Xie Y,
Wu XY, Bian X and Yu XY: The expressions of miR-151a-5p and miR-23b
in lung cancer tissues and their effects on the biological
functions of lung cancer A549 cells. Eur Rev Med Pharmacol Sci.
24:6779–6785. 2020.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baj J, Korona-Glowniak I, Forma A, Maani
A, Sitarz E, Rahnama-Hezavah M, Radzikowska E and Portincasa P:
Mechanisms of the epithelial-mesenchymal transition and tumor
microenvironment in Helicobacter pylori-induced gastric cancer.
Cells. 9:10552020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adnan M, Siddiqui AJ, Hamadou WS, Snoussi
M, Badraoui R, Ashraf SA, Jamal A, Awadelkareem AM, Sachidanandan
M, Hadi S, et al: Deciphering the molecular mechanism responsible
for efficiently inhibiting metastasis of human non-small cell lung
and colorectal cancer cells targeting the matrix metalloproteinases
by selaginella repanda. Plants (Basel). 10:9792021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arruabarrena-Aristorena A, Zabala-Letona A
and Carracedo A: Oil for the cancer engine: The cross-talk between
oncogenic signaling and polyamine metabolism. Sci Adv.
4:eaar26062018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yan L, Yao J and Qiu J: miRNA-495
suppresses proliferation and migration of colorectal cancer cells
by targeting FAM83D. Biomed Pharmacother. 96:974–981. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kang X, Kong B, Chen Q and Zhao S: Low
expression of miR-138 inhibit the proliferation, migration and
invasion of colorectal cancer and affect patient survival by
targeting SIRT1. Transl Cancer Res. 10:3548–3559. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang D, Qiu C, Zhang H, Wang J, Cui Q and
Yin Y: Human microRNA oncogenes and tumor suppressors show
significantly different biological patterns: From functions to
targets. PLoS One. 5:e130672010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xiao M, Li J, Li W, Wang Y, Wu F, Xi Y,
Zhang L, Ding C, Luo H, Li Y, et al: MicroRNAs activate gene
transcription epigenetically as an enhancer trigger. RNA Biol.
14:1326–1334. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Suzuki HI, Young RA and Sharp PA:
Super-enhancer-mediated RNA processing revealed by integrative
MicroRNA network analysis. Cell. 168:1000–1014.e15. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen E, Li Q, Wang H, Yang F, Min L and
Yang J: MiR-92a promotes tumorigenesis of colorectal cancer, a
transcriptomic and functional based study. Biomed Pharmacother.
106:1370–1377. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guo S, Zhu KX, Yu WH, Wang T, Li S, Wang
YX, Zhang CC and Guo JQ: SH3PXD2A-AS1/miR-330-5p/UBA2 ceRNA network
mediates the progression of colorectal cancer through regulating
the activity of the Wnt/β-catenin signaling pathway. Environ
Toxicol. 36:1969–1980. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jafarzadeh M and Soltani BM: MiRNA-Wnt
signaling regulatory network in colorectal cancer. J Biochem Mol
Toxicol. 35:e228832021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang N, Hu X, Du Y and Du J: The role of
miRNAs in colorectal cancer progression and chemoradiotherapy.
Biomed Pharmacother. 134:1110992021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zuo Z, Jiang Y, Zeng S, Li Y, Fan J, Guo Y
and Tao H: The value of microRNAs as the novel biomarkers for
colorectal cancer diagnosis: A meta-analysis. Pathol Res Pract.
216:1531302020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lopez-Camarillo C, Marchat LA,
Arechaga-Ocampo E, Perez-Plasencia C, Del Moral-Hernandez O,
Castaneda-Ortiz EJ and Rodriguez-Cuevas S: MetastamiRs: Non-coding
MicroRNAs driving cancer invasion and metastasis. Int J Mol Sci.
13:1347–1379. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kunej T, Godnic I, Horvat S, Zorc M and
Calin GA: Cross talk between microRNA and coding cancer genes.
Cancer J. 18:223–231. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fan X, Cao M, Liu C, Zhang C, Li C, Cheng
W, Zhang S, Zhang H and Zhu W: Three plasma-based microRNAs as
potent diagnostic biomarkers for endometrial cancer. Cancer
Biomark. 31:127–138. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pellikainen JM, Ropponen KM, Kataja VV,
Kellokoski JK, Eskelinen MJ and Kosma VM: Expression of matrix
metalloproteinase (MMP)-2 and MMP-9 in breast cancer with a special
reference to activator protein-2, HER2, and prognosis. Clin Cancer
Res. 10:7621–7628. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bracken CP, Scott HS and Goodall GJ: A
network-biology perspective of microRNA function and dysfunction in
cancer. Nat Rev Genet. 17:719–732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Uribe E, Reyes MB, Martinez I, Mella K,
Salas M, Tarifeño-Saldivia E, López V, Garcia-Robles M,
Martinez-Oyanedel J, Figueroa M, et al: Functional analysis of the
Mn2+ requirement in the catalysis of ureohydrolases
arginase and agmatinase-a historical perspective. J Inorg Biochem.
202:1108122020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chai J, Luo L, Hou F, Fan X, Yu J, Ma W,
Tang W, Yang X, Zhu J, Kang W, et al: Agmatine reduces
lipopolysaccharide-mediated oxidant response via activating
PI3K/Akt pathway and up-regulating Nrf2 and HO-1 expression in
macrophages. PLoS One. 11:e01636342016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yılmaz E, Şekeroğlu MR, Yılmaz E and
Çokluk E: Evaluation of plasma agmatine level and its metabolic
pathway in patients with bipolar disorder during manic episode and
remission period. Int J Psychiatry Clin Pract. 23:128–133. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dallmann K, Junker H, Balabanov S,
Zimmermann U, Giebel J and Walther R: Human agmatinase is
diminished in the clear cell type of renal cell carcinoma. Int J
Cancer. 108:342–347. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bernstein HG, Stich C, Jäger K, Dobrowolny
H, Wick M, Steiner J, Veh R, Bogerts B and Laube G: Agmatinase, an
inactivator of the putative endogenous antidepressant agmatine, is
strongly upregulated in hippocampal interneurons of subjects with
mood disorders. Neuropharmacology. 62:237–246. 2012. View Article : Google Scholar : PubMed/NCBI
|