Introduction
Oral cancer is a major health problem. According to
the Globocan report (2020), an estimated 377,713 new cases of oral
cancer and 177,757 deaths due to oral cancer occur annually across
the world (1). Oral squamous cell
carcinoma (OSCC) is the most common oral cancer which is
characterized by a predilection for cervical lymph node (LN)
metastases. OSCC is associated with poor clinical outcomes due to
frequent nodal involvement and locoregional recurrence, resulting
in survival rates of <50% (2).
Along with LN metastasis, perineural invasion (PNI), another form
of metastasis, has been widely recognized as a negative prognostic
factor in several neurotropic cancers including OSCC (3). Oral tongue squamous cell carcinoma
(OTSCC) is particularly neurotropic compared with OSCC in other
regions due to its anatomical structure (4). PNI has been shown to correlate with
the nodal status at the time of diagnosis, presence of occult neck
metastases, and neck recurrence in head and neck SCC, particularly
of the oral cavity origin (5).
Therefore, developing optimal biomarkers related to both LN
metastasis and PNI of OSCC may help predict the clinical outcomes,
guide treatment decision-making and develop the therapeutic targets
for the treatment of OSCC.
L1 cell adhesion molecule (L1CAM) is a 200–220 kDa
transmembrane glycoprotein of the immunoglobulin superfamily that
plays a role in neural development by promoting neural cell
adhesion and migration (6). L1CAM
also exerts its functions as a soluble form released from the cell
surface by ADAM (a disintegrin and metalloproteinase) family of
proteinases (7). Besides its
expression in neural tissues, L1CAM is known to be expressed in
normal tissues such as endothelial cells, renal collecting ducts
and skin mast cells (8,9). During the past 20 years, research on
L1CAM has expanded from the field of neurobiology to tumor biology.
A growing body of evidence has revealed that L1CAM overexpression
is associated with progression and metastasis of numerous human
cancers, including melanoma and ovarian, endometrial and pancreatic
cancers (10–13). In addition, L1CAM overexpression has
been identified to correlate with PNI in certain malignancies,
including extrahepatic cholangiocarcinoma and pancreatic ductal
adenocarcinoma (13,14). Thus, a linkage of L1CAM with LN
metastasis and PNI is plausible.
L1CAM is known to activate different signaling
pathways involved in tumor progression, such as PI3K/AKT and ERK
pathways in several malignancies (9,15–17).
Both these pathways have been mainly implicated in the pathogenesis
of human cancers by regulating cell growth, differentiation,
proliferation, migration and apoptosis (18). Nevertheless, whether L1CAM functions
through these pathways in OSCC remains unclear. In the first study
on the role of L1CAM in OSCC, Hung et al (19) demonstrated that L1CAM knockdown
induced retardation of cell cycle at the G1 phase and inhibition of
cell proliferation as well as attenuated the migration and invasion
of OSCC cells; however, they did not investigate the related
signaling pathway. Overexpression or knockdown of L1CAM in
vitro has been demonstrated to induce changes in the expression
levels of epithelial and mesenchymal markers, suggesting the
involvement of L1CAM in the epithelial-mesenchymal transition (EMT)
process of breast, endometrium, lung and oral cavity carcinomas
(19–22). EMT is involved in cancer progression
and metastasis and is regulated by multiple intracellular signaling
networks, including PI3K/AKT and RAS/RAF/ERK axes (23). Thus, it can be hypothesized that
L1CAM is involved in the progression and metastasis of OSCC by
activating the PI3K/AKT/ERK pathways. If so, L1CAM appears to be a
potential therapeutic target for the treatment of OSCC.
In the present study, the clinical significance of
L1CAM was first investigated using OTSCC tissue samples. Next,
in vitro functional assays were performed to elucidate the
biological roles of L1CAM in the progression and metastasis of OSCC
and to evaluate the possibility as a potential therapeutic target.
Finally, the downstream pathways regulated by L1CAM in OSCC were
explored in relation to EMT and PI3K/AKT/ERK pathways.
Materials and methods
Patients and tissue samples
Formalin-fixed paraffin-embedded tissue samples of
80 patients with OTSCC who were surgically treated at the
Department of Oral and Maxillofacial Surgery at the Seoul National
University Dental Hospital between January 2008 and December 2012
were included in the present study. The age of patients ranged from
27–83 years, with a median age of 56 years. Tumors were staged
according to the TNM system recommended by the 8th Edition of the
American Joint Committee on Cancer Staging Manual, Head and Neck
Section. The clinicopathological characteristics of the patients
including age, sex, differentiation status, tumor size (pT), depth
of invasion (DOI), LN metastasis (including late metastasis), PNI,
TNM stage, and recurrence are shown in Table I. All procedures followed in the
present study were in accordance with the guidelines (approval no.
CRI 20003) of the Institutional Review Board of the Seoul National
University Dental Hospital (Seoul, Korea).
 | Table I.Clinicopathological characteristics
of 80 patients with oral tongue squamous cell carcinoma. |
Table I.
Clinicopathological characteristics
of 80 patients with oral tongue squamous cell carcinoma.
| Clinicopathological
characteristics | Number |
|---|
| Median age at
diagnosis, years | 56 |
| Range | 27-83 |
| Sex |
|
|
Male | 48 |
|
Female | 32 |
|
Differentiation |
|
|
Well | 60 |
|
Moderately | 20 |
| Perineural
invasion |
|
| No | 48 |
|
Yes | 32 |
| Tumor size |
|
| T1 | 21 |
| T2 | 38 |
| T3 | 16 |
| T4 | 5 |
| Lymph node
metastasis |
|
| No | 44 |
|
Yes | 36 |
| Stage |
|
| I | 19 |
| II | 24 |
|
III | 11 |
| IV | 26 |
Immunohistochemistry
Tissue specimens were sectioned at 4-µm thickness,
deparaffinized in Neo-clear (Merck KGaA) for 25 min, and rehydrated
by passage through a graded alcohol series for 25 min. Heat-induced
epitope retrieval was performed in Target Retrieval Solution pH 9
(Dako; Agilent Technologies, Inc.) for 10 min using a microwave.
Endogenous peroxidase was inactivated by incubation in 3%
H2O2 solution for 10 min. Immunohistochemical
staining for L1CAM was performed using a mouse monoclonal
anti-human L1CAM antibody (1:100; clone 14.10; cat. no. 826701;
BioLegend Inc.) for 1 h at room temperature (RT). Slides were
rinsed in DAKO wash buffer and then incubated for 30 min with
peroxidase-labeled polymer conjugated to anti-mouse immunoglobulins
(EnVision Detection System; Dako; Agilent Technologies, Inc.). The
chromogenic reaction was carried out with DAB chromogen. All
sections were counterstained with Mayer's hematoxylin for 1 min at
RT.
Evaluation of immunohistochemical
staining
All samples stained with anti-L1CAM antibody were
independently evaluated by two oral pathologists (K-YO and H-JY)
using a light microscope (BX53; Olympus Corporation). Nerve tissues
in each slide were used as an internal positive control. L1CAM
expression was considered positive if 10% or more of the tumor
cells showed moderate to strong membranous staining, as previously
described (12).
Cell lines
The Ca9-22, HSC-2, HSC-3, HSC-4 and SAS cell lines
were kindly provided by professor Masanobu Shindoh of Hokkaido
University (Hokkaido, Japan). The HN22 cell line was generously
provided by the School of Dentistry, DanKook University (Cheonan,
Korea). The cell lines from Japan were authenticated by Japanese
Collection of Research Bioresources (JCRB) Cell Bank using the SRT
profiling. Ca9-22 (cat. no. JCRB0625) cell line was derived from
gingiva SCC; HN22 and HSC-2 (cat. no. JCRB0622) cell lines from
unknown sites of oral cavity; and HSC-3 (cat. no. JCRB0623), HSC-4
(cat. no. JCRB0624) and SAS (cat. no. JCRB0260) cell lines from
tongue SCC. All OSCC cell lines were cultured in Dulbecco's
Modified Eagle's Medium (DMEM)/F-12 (Welgene, Inc.) containing 10%
fetal bovine serum (FBS; Welgene, Inc.) and 100 U/ml penicillin and
streptomycin in a humidified atmosphere with 5% CO2 at
37°C.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted using TRIzol®
Reagent (Thermo Fisher Scientific, Inc.). RNA (1 µg) was
reverse-transcribed using an AMPIGENE cDNA Synthesis Kit (Enzo Life
Sciences, Inc.) according to the manufacturer's instructions.
RT-qPCR for analyzing L1CAM expression level was performed using
Applied Biosystems StepOne Plus Real-time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Two independent
experiments were performed in triplicate. The primer sequences used
for RT-qPCR were as follows: L1CAM forward,
5′-ACGAGGGATGGTGTCCACTTCAAA-3′ and reverse,
5′-TTATTGCTGGCAAAGCAGCGGTAG-3′; and β-actin forward,
5′-CACTCTTCCAGCCTTCCTTC-3′ and reverse, 5′-AGCACTGTGTTGGCGTACAG-3′.
L1CAM mRNA expression was measured using a SYBR Premix Ex Taq™ kit
(Takara Bio, Inc.). β-actin expression was used as a reference. The
thermocycling conditions suitable for L1CAM mRNA were the
following: 95°C for 2 min, followed by 40 cycles of denaturation at
95°C for 10 sec and synthesis at 60°C for 30 sec. The relative
level of L1CAM mRNA was normalized to that of β-actin and
calculated by the 2−∆∆Cq method (24).
Western blotting
Whole cells were lysed in RIPA buffer and protein
concentration was determined using the DC protein assay kit II
(Bio-Rad Laboratories, Inc.). A total of 30 µg protein extract was
separated by 10% SDS-PAGE and transferred to a polyvinylidene
fluoride membrane. After being blocked by 5% non-fat milk at RT for
1 h, the membranes were incubated with the following primary
antibodies diluted at 1:1,000: L1CAM (cat. no. SC-53386; Santa Cruz
Biotechnology, Inc.), E-cadherin (cat. no. BD610181; BD
Biosciences), N-cadherin (cat. no. BD610920; BD Biosciences),
vimentin (cat. no. BD550513; BD Biosciences), Snail (cat. no.
CST3879; Cell Signaling Technology, Inc.), Slug (cat. no. CST9585;
Cell Signaling Technology, Inc.), Twist (cat. no. SC-81417; Santa
Cruz Biotechnology, Inc.), AKT (cat. no. CST9272; Cell Signaling
Technology, Inc.), p-AKT (cat. no. CST9271; Cell Signaling
Technology, Inc.), and β-actin (cat. no. SC-47778; Santa Cruz
Biotechnology, Inc.). After incubation with goat anti-rabbit
(1:3,000; cat. no. GTX213110-01; GeneTex, Inc.) or anti-mouse
secondary antibody (1:3,000; cat. no. GTX213111-01; GeneTex, Inc.)
at RT for 2 h, the proteins were identified by SuperSignal West
Pico Chemiluminescent Substrate (cat. no. SC-2048; Santa Cruz
Biotechnology, Inc.), and immunoreactive bands were visualized
using ImageQuant LAS 500 (Cytiva). Densitometric analysis of
western blot bands was carried out using ImageJ software (version
1.51 k; National Institutes of Health).
L1CAM overexpression and
knockdown
Recombinant human L1CAM (rhL1CAM) was purchased from
R&D Systems, Inc. Cell viability was first assessed at
concentrations of 0, 50, 100, 200, and 400 ng/ml, and a titer of
100 ng/ml of rhL1CAM was used for subsequent experiments.
ON-TARGETplus SMARTpool small interfering RNA
(siRNA) targeting L1CAM (siL1CAM) and Non-targeting Control Pool
(siControl) were purchased from GE Healthcare Dharmacon, Inc. This
SMARTpool siRNA contains four pooled siRNAs, each targeting a
separate region of the L1CAM RNA sequence. ON-TARGETplus
Non-targeting siRNA (D-001810-10-20) was also used as a
non-targeting control. The SMARTpool siRNA and non-targeting siRNA
target sequences are as follows: ON-TARGETplus SMARTpool siRNA
J-011069-05, L1CAM Target Sequence: CACUACACCUUUAGGGUUA;
ON-TARGETplus SMARTpool siRNA J-011069-06, L1CAM Target Sequence:
GCAAGAGACAUAUCCACAA; ON-TARGETplus SMARTpool siRNA J-011069-07,
L1CAM Target Sequence: GAUACAAUGUGACGUACUG; ON-TARGETplus SMARTpool
siRNA J-011069-08, L1CAM Target Sequence: ACACAAUGGUGACCCAAUG; and
ON-TARGETplus Non-targeting pool Target Sequences:
UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA and
UGGUUUACAUGUUUUCCUA.
Transfection was performed using Lipofectamine
2000® (Invitrogen; Thermo Fisher Scientific, Inc.).
Briefly, anti-L1CAM siRNA and Lipofectamine 2000 were mixed
according to the protocol and allowed to form the
siRNA-Lipofectamine 2000 lipoplexes at RT for 20 min. HSC-4 or HN22
cells was seeded on 60-mm plates and transfected transiently with
100 nM transfection complex. After 24 h of transfection, subsequent
wound healing or Transwell migration/invasion experiment was
performed.
Cell proliferation assay
A total of 3×105 cells were seeded in
six-well plates and incubated overnight. After treatment with 100
ng/ml of rhL1CAM or 100 nM of siL1CAM for 24 h, cells were
incubated with 5% CO2 at 37°C for 24, 48 and 72 h.
Viable cells were stained with 0.4% trypan blue solution (Gibco;
Thermo Fisher Scientific, Inc.) at RT for 5 min and counted using a
hemocytometer at each time-point.
Wound healing assay
Wound healing assay was carried out as previously
described (25). Ca9-22, HSC-2,
HSC-4, and HN22 (3×105 cells) were seeded in six-well
plates. After being cultured for 24 h, cells were treated with
rhL1CAM (100 ng/ml) or siL1CAM (100 nM) using Opti-MEM medium
(Gibco; Thermo Fisher Scientific, Inc.). After 24 h, the center of
the cell monolayers was scratched vertically with a sterilized
100-µl pipette tip. After rinsing with PBS three times for removing
cell debris, the cells were incubated in 5% FBS-containing medium
for 12 h to enable wound healing. Images were captured at 0 and 12
h using an inverted microscope (CKX53; Olympus Corporation) and the
wound dimensions were measured using ImageJ software (version 1.51
k; National Institutes of Health).
Transwell migration and Matrigel
invasion assays
For Transwell migration assay, Ca9-22
(0.8×105), HSC-2 (0.8×105), HSC-4
(1.8×105), and HN22 (1.8×105) cells in
serum-free medium were seeded in the upper chambers of 24 well
plate with Collagen type I-coated PET membrane of 8.0-µm pore size
(BD Biosciences). For invasion assay, culture inserts of 24-well
plate were coated with Matrigel (BD Biosciences) in a 37°C
incubator for 2 h. Ca9-22 (1.0×105), HSC-2
(1.0×105), HSC-4 (1.2×105), and HN22
(1.2×105) cells in serum-free medium were seeded in the
upper chambers. Lower chambers were filled with the media
containing 10% FBS as a chemoattractant. After incubation for 24 h,
the non-migratory or non-invasive cells in upper chamber were
removed with a cotton swab. Cells on the lower surface of the
filter were fixed with 100% methanol for 2 min and stained with
H&E solution. Images of migratory or invasive cells were
captured under an inverted light microscope (CKX53; Olympus
Corporation) and the number of migratory or invasive cells were
counted in randomly selected areas in three different microscopic
fields (magnification, ×100).
Statistical analysis
Statistical analyses were performed using SPSS
software (version 26.0; IBM Corp.). Correlation of L1CAM expression
with clinicopathological parameters was assessed with Pearson's
Chi-squared test or Fisher's exact test. Survival curves were
generated using the Kaplan-Meier method and between-group
differences assessed using the log-rank test. For in vitro
experiments, the mean ± standard deviation values from three
independent experiments are reported. Statistical significance was
analyzed using a two-tailed Student's t-test for paired samples.
One-way ANOVA followed by Tukey's post hoc test analyses were used
for multiple-group comparisons. For all analyses, P<0.05 was
considered to indicate a statistically significant difference.
Results
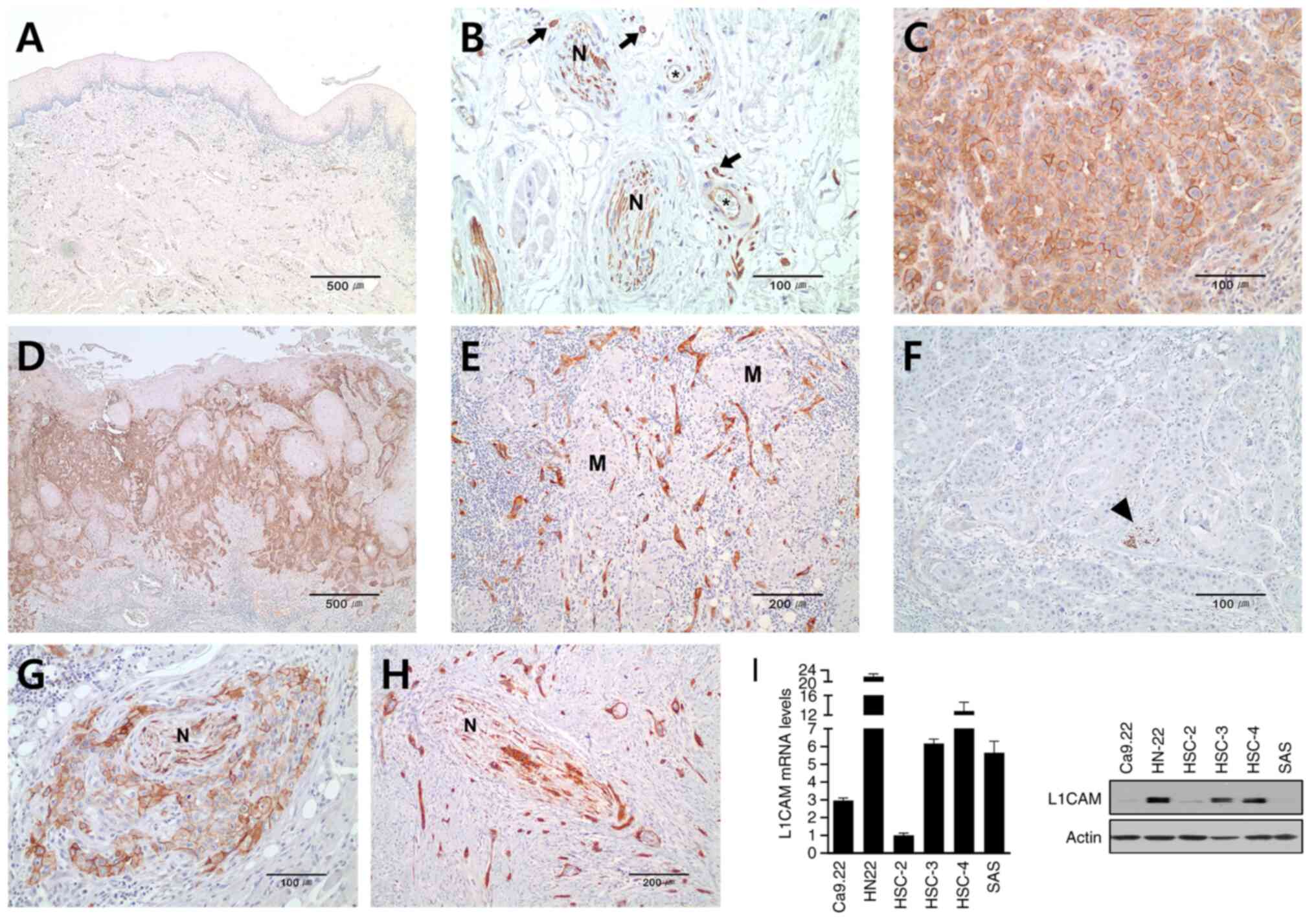
L1CAM expression in OSCC tissues and
cell lines
L1CAM expression was not detected in the epithelium
of the normal oral mucosa (Fig.
1A), but was strongly expressed in the peripheral nerve tissue,
vessels and mast cells (Fig. 1B).
In OTSCC tissue samples, tumor cells showed a membranous pattern of
expression of L1CAM (Fig. 1C). Of
the 80 clinical samples, 26 (32.5%) samples were positive for
L1CAM. Tumor cells exhibiting moderate to strong expression were
frequently found at the invasive front of tumors or in the
less-differentiated and usually spindle-shaped tumor cells
(Fig. 1D and E), while negative
expression was observed in relatively well-differentiated tumors
(Fig. 1F). In the area of PNI,
tumor cells expressing L1CAM were found in the perineural tissues
or even inside the nerves (Fig. 1G and
H).
mRNA and protein levels of L1CAM were examined in
six OSCC cell lines (Fig. 1I).
HSC-3, HSC-4 and HN-22 presented relatively high levels of L1CAM
compared with other cell lines (Ca9.22, HSC-2 and SAS). Thus,
Ca9.22 and HSC-2 were used for confirming the effect of L1CAM
upregulation induced by treatment with rhL1CAM. By contrast, HSC-4
and HN-22 were used for determining the effect of L1CAM
downregulation using siL1CAM.
L1CAM overexpression in OSCC
significantly correlates with poor clinical outcomes
To assess the correlation of L1CAM expression with
clinicopathological parameters, including PNI, PNI was determined
more objectively according to the commonly accepted definition by
Liebig et al (3): i) Tumor
in close proximity to nerve and involving at least 33% of its
perimeter or ii) tumor cells within any of the three layers of the
nerve sheath. PNI was detected in 40.0% of 80 OTSCC samples. PNI,
which is a known adverse prognostic factor in OSCC, revealed a
significant correlation with pT (P=0.001), DOI (P<0.001), LN
metastasis (P<0.001), advanced clinical stage (P=0.001) and
survival status (P=0.002) (Table
II). L1CAM expression showed significant correlation with high
histologic grade (P=0.013), greater DOI (P=0.009), LN metastasis
(P<0.001), presence of PNI (P<0.001), advanced stage
(P=0.004), and survival status (P=0.018) (Table III). These findings suggested high
clinical relevance of L1CAM in relation to cancer invasion,
metastasis, and progression.
 | Table II.Correlation of PNI with
clinicopathologic parameters in oral tongue squamous cell
carcinoma. |
Table II.
Correlation of PNI with
clinicopathologic parameters in oral tongue squamous cell
carcinoma.
|
|
| PNI |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | Total cases | Negative (%) | Positive (%) | P-value |
|---|
|
| n=80 | n=48 | n=32 |
|
| Age |
|
|
| 0.235a |
|
<56 | 39 | 26 (66.7) | 13 (33.3) |
|
|
≥56 | 41 | 22 (53.7) | 19 (46.3) |
|
| Sex |
|
|
| 0.192a |
|
Male | 48 | 26 (54.2) | 22 (45.8) |
|
|
Female | 32 | 22 (68.8) | 10 (31.3) |
|
|
Differentiation |
|
|
| 0.292a |
|
Well | 60 | 38 (63.3) | 22 (36.7) |
|
|
Moderately | 20 | 10 (50.0) | 10 (50.0) |
|
| Tumor size |
|
|
| 0.001a |
| T1 +
T2 | 59 | 42 (71.2) | 17 (28.8) |
|
| T3 +
T4 | 21 | 6 (28.6) | 15 (71.4) |
|
| Depth of
invasion |
|
|
|
<0.001a |
| ≤0.5
cm | 30 | 28 (93.3) | 2 (6.7) |
|
| 0.5~1.0
cm | 25 | 15 (60.0) | 10 (40.0) |
|
| >1.0
cm | 25 | 5 (20.0) | 20 (80.0) |
|
| Lymph node
metastasis |
|
|
|
<0.001a |
| No | 44 | 35 (79.5) | 9 (20.5) |
|
|
Yes | 36 | 13 (36.1) | 23 (63.9) |
|
| Stage |
|
|
| 0.001a |
| I +
II | 43 | 33 (76.7) | 10 (23.3) |
|
| III +
IV | 37 | 15 (40.5) | 22 (59.5) |
|
| Local
recurrence |
|
|
| 0.734b |
| No | 71 | 42 (59.2) | 29 (40.8) |
|
|
Yes | 9 | 6 (66.7) | 3 (33.3) |
|
| Survival
status |
|
|
| 0.002a |
|
Alive | 62 | 43 (69.4) | 19 (30.6) |
|
|
Dead | 18 | 5 (27.8) | 13 (72.2) |
|
 | Table III.Correlation of L1CAM expression with
clinicopathologic parameters in oral tongue squamous cell
carcinoma. |
Table III.
Correlation of L1CAM expression with
clinicopathologic parameters in oral tongue squamous cell
carcinoma.
|
|
| L1CAM |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | Total cases | Negative (%) | Positive (%) | P-value |
|---|
| Age |
|
|
| 0.747a |
|
<56 | 39 | 27 (69.2) | 12 (30.8) |
|
|
≥56 | 41 | 27 (65.9) | 14 (34.1) |
|
| Sex |
|
|
| 0.079a |
|
Male | 48 | 36 (75.0) | 12 (25.0) |
|
|
Female | 32 | 18 (56.3) | 14 (43.8) |
|
|
Differentiation |
|
|
| 0.013a |
|
Well | 60 | 45 (75.0) | 15 (25.0) |
|
|
Moderately | 20 | 9 (45.0) | 11 (55.0) |
|
| Tumor size |
|
|
| 0.238a |
| T1 +
T2 | 59 | 42 (71.2) | 17 (28.8) |
|
| T3 +
T4 | 21 | 12 (57.1) | 9 (42.9) |
|
| Depth of
invasion |
|
|
| 0.009a |
| ≤0.5
cm | 30 | 26 (86.7) | 4 (13.3) |
|
| 0.5~1.0
cm | 25 | 16 (64.0) | 9 (36.0) |
|
| >1.0
cm | 25 | 12 (48.0) | 13 (52.0) |
|
| Lymph node
metastasis |
|
|
|
<0.001a |
| No | 44 | 37 (84.1) | 7 (15.9) |
|
|
Yes | 36 | 17 (47.2) | 19 (52.8) |
|
| Perineural
invasion |
|
|
|
<0.001a |
| No | 48 | 40 (83.3) | 8 (16.7) |
|
|
Yes | 32 | 14 (43.8) | 18 (56.3) |
|
| Stage |
|
|
| 0.004a |
| I +
II | 43 | 35 (81.4) | 8 (18.6) |
|
| III +
IV | 37 | 19 (51.4) | 18 (48.6) |
|
| Local
recurrence |
|
|
| 0.710b |
| No | 71 | 47 (66.2) | 24 (33.8) |
|
|
Yes | 9 | 7 (77.8) | 2 (22.2) |
|
| Survival
status |
|
|
| 0.018a |
|
Alive | 62 | 46 (74.2) | 16 (25.8) |
|
|
Dead | 18 | 8 (44.4) | 10 (55.6) |
|
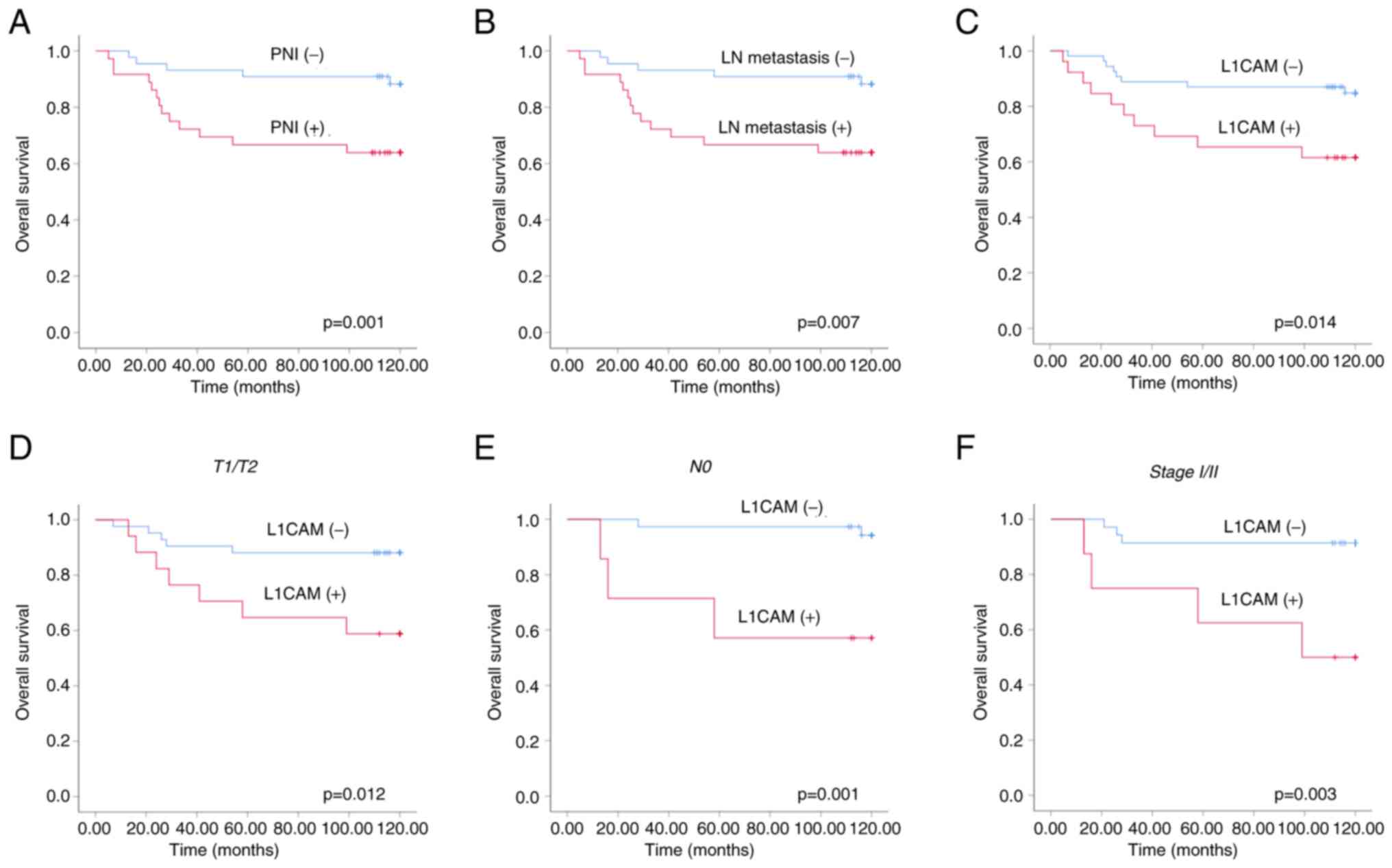
Kaplan-Meier survival analysis was performed to
evaluate the prognostic significance of LN metastasis, PNI, and
L1CAM expression for overall survival (OS) of patients with OSCC.
OTSCC patients with PNI or LN metastasis demonstrated significantly
worse OS rate (P=0.001 and P=0.007, respectively; Fig. 2A and B). As for a novel marker,
L1CAM, patients with positive expression of L1CAM showed
significantly lower OS than those with negative expression
(P=0.014; Fig. 2C). L1CAM
expression was found to have a more significant effect on the
decrease of OS rate in patients with T1/2 tumor, N0 tumor, or stage
I/II tumor (P=0.012, P=0.001 and P=0.003, respectively; Fig. 2D-F). These findings suggested that
expression of L1CAM in early OSCC may be an indicator of poor
prognosis.
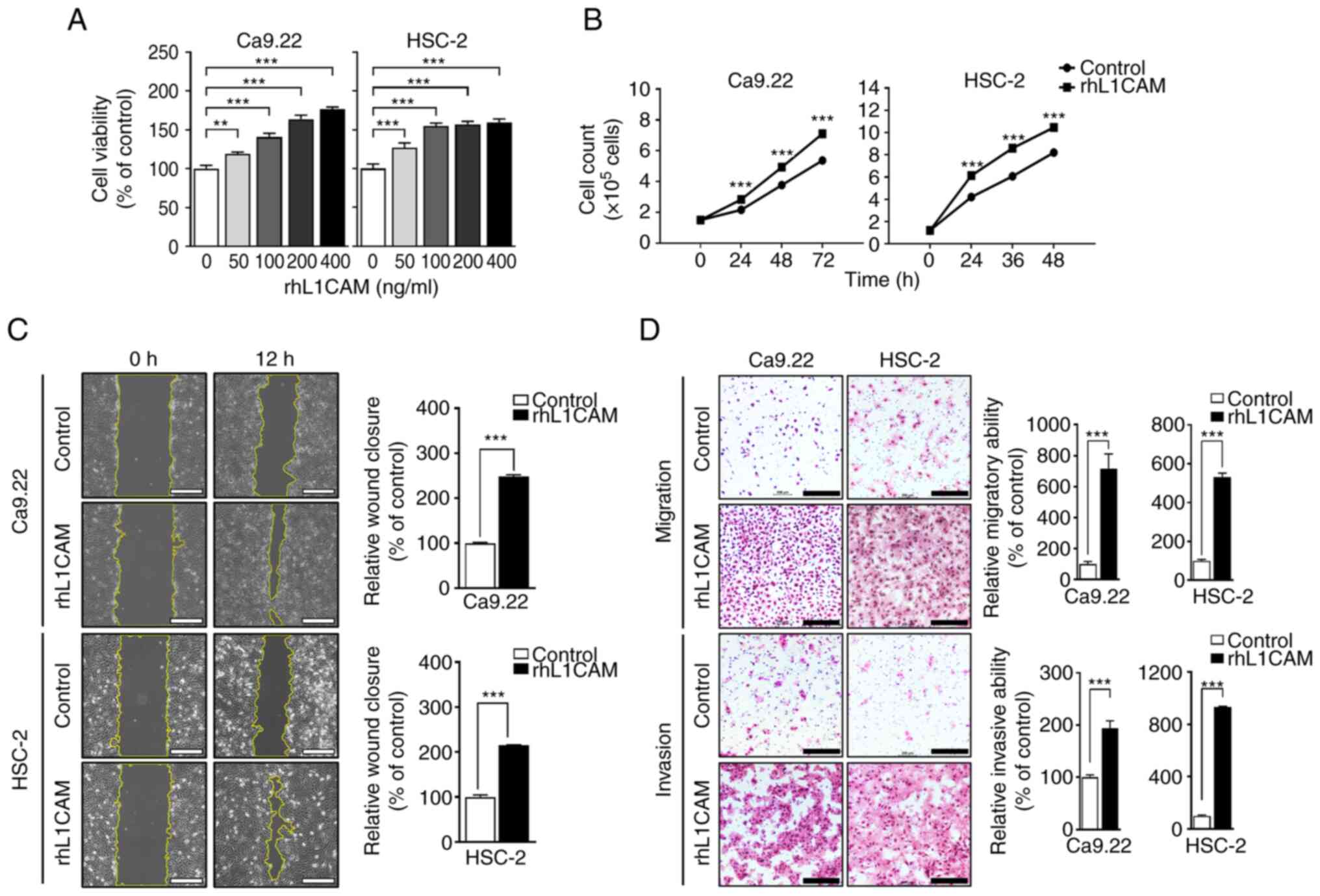
Overexpression of L1CAM increases cell
proliferation, migration and invasion of OSCC cells in vitro
To evaluate the function of L1CAM in cell
proliferation, migration and invasion in OSCC, Ca9-22 and HSC-2
cell lines were treated with rhL1CAM. The viability of Ca9-22 cells
increased in a concentration-dependent manner up to 400 ng/ml of
concentration of rhL1CAM, while viability of HSC-2 cells increased
gradually up to 100 ng/ml of concentration and there was no change
>100 ng/ml (**P<0.01 and ***P<0.001; Fig. 3A). Overexpression of L1CAM
significantly increased proliferation capacity of both cell lines
at each time-point (***P<0.001; Fig.
3B). Next, wound healing assay revealed that upregulation of
L1CAM significantly increased the migration of both cell lines
(***P<0.001; Fig. 3C). Wound
closure in the rhL1CAM-treated groups increased more than two-fold
compared with that of control groups in both cell lines. Lastly,
Transwell migration and Matrigel invasion assays also revealed
significantly increased migration and invasion ability in
rhL1CAM-treated groups of both cell lines compared with the
corresponding control groups (***P<0.001; Fig. 3D). Collectively, these findings
indicated that overexpression of L1CAM can promote cell
proliferation, migration, and invasion, suggesting its important
role in the progression of OSCC.
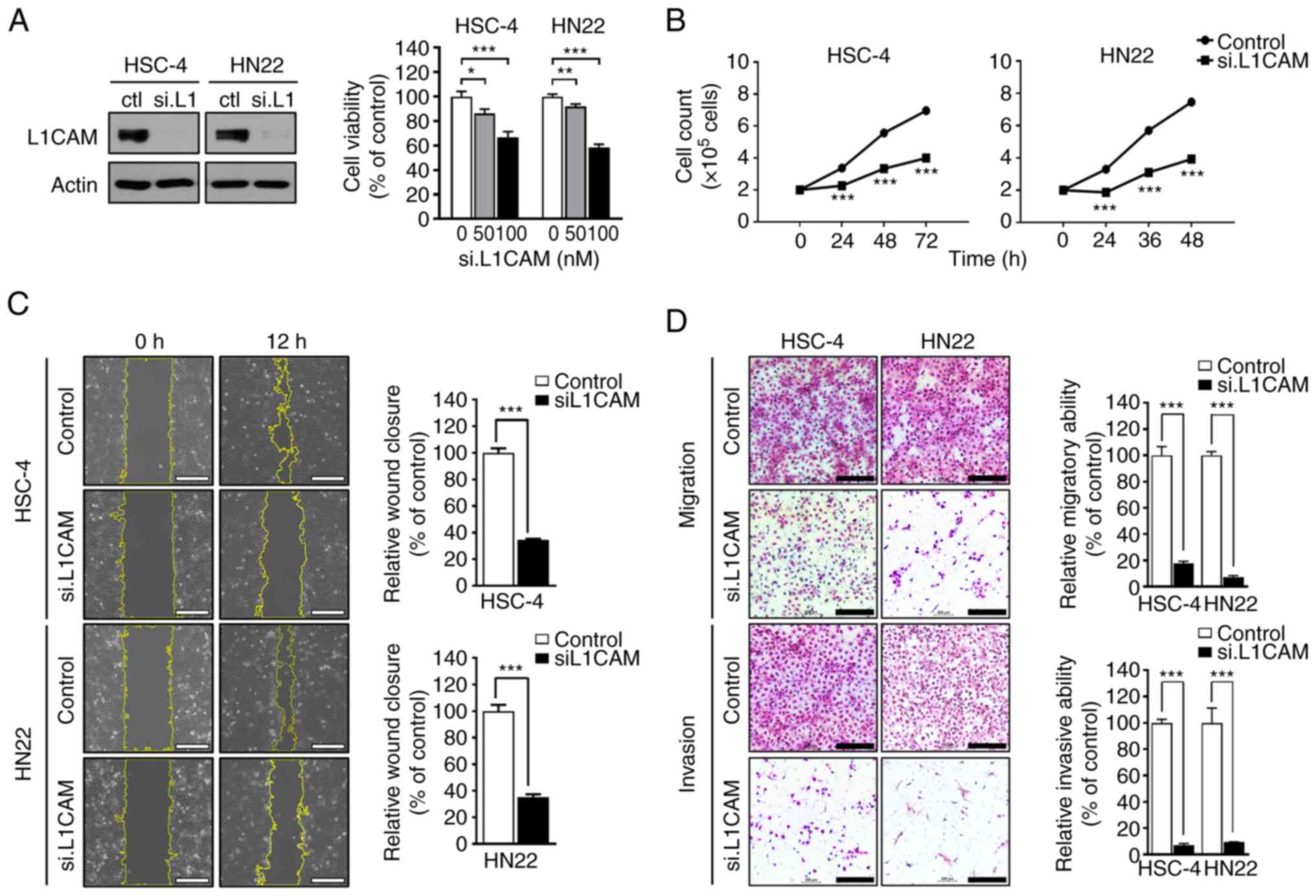
Downregulation of L1CAM inhibits cell
proliferation, migration and invasion and induces apoptosis of OSCC
cells in vitro
To assess the effect of L1CAM knockdown, both HSC-4
and HN22 cells were treated with 100 nM siL1CAM for 24 h. HSC-4 and
HN22 cell lines revealed decreased cell viability in a
concentration-dependent manner (*P<0.05, **P<0.01 and
***P<0.001; Fig. 4A).
Suppression of L1CAM expression significantly decreased the
proliferation capacity of both cell lines at each time-point
(***P<0.001; Fig. 4B). Next, in
wound healing assay, L1CAM knockdown significantly affected the
migration ability of both cell lines. The ability of wound closure
in siL1CAM-treated groups of both cell lines decreased more than
three-fold compared with that in the corresponding control groups
(***P<0.001; Fig. 4C). Lastly,
Transwell migration and Matrigel invasion assays demonstrated
significantly decreased cell migration and invasion ability in
siL1CAM-treated groups of both cell lines compared with the control
groups (***P<0.001; Fig. 4D).
These results indicated that knockdown of L1CAM can inhibit
proliferation, migration and invasion, suggesting its possibility
to serve as a therapeutic target.
L1CAM is involved in EMT and PI3K/AKT
and ERK signaling pathways during tumor progression
Based on the effect of L1CAM in enhancing the
migration and invasion of OSCC cells, the association of L1CAM with
EMT was assessed. After L1CAM knockdown, changes in the expression
of EMT-associated markers were assessed by western blotting. As
revealed in Fig. 5A, expression of
epithelial cell marker E-cadherin was increased, whereas
mesenchymal markers including N-cadherin, vimentin, snail, slug and
twist were significantly decreased after L1CAM knockdown. In other
words, suppression of L1CAM was found to reverse EMT. These results
suggested the involvement of L1CAM in the EMT process of OSCC
cells.
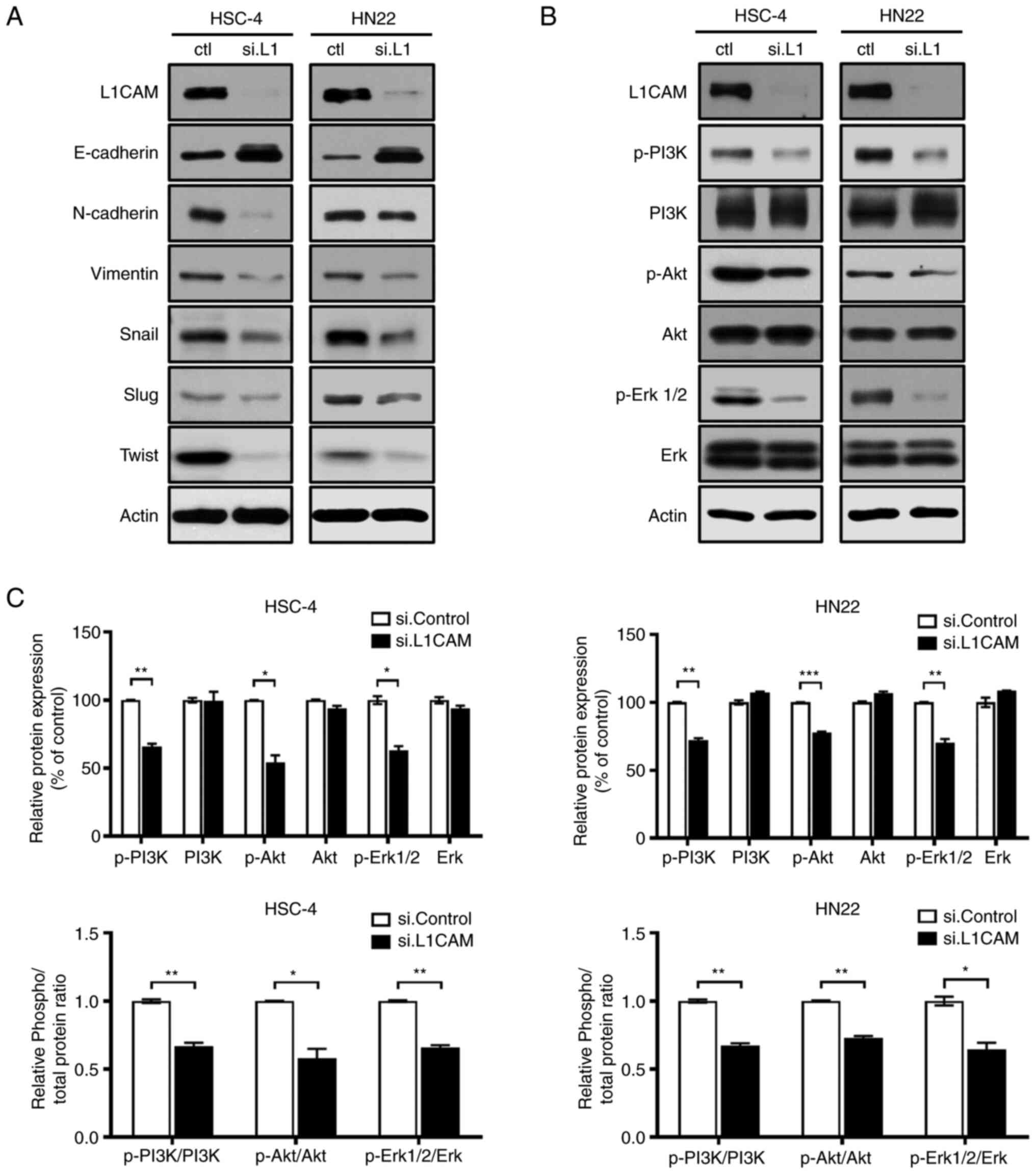 | Figure 5.L1CAM is involved in the
epithelial-mesenchymal transition process and functions through the
PI3K/AKT and ERK signaling pathways. (A) When L1CAM expression is
knocked down, expression of E-cadherin is significantly increased,
whereas expression levels of N-cadherin, vimentin, snail, slug, and
twist are decreased correspondingly. (B) Knockdown of L1CAM
expression induces a remarkable decrease of p-PI3K, p-AKT, and
p-ERK1/2 levels. (C) Relative protein expression levels and the
ratio of phosphorylated/total protein were shown as bar-chart
graphs (*P<0.05, **P<0.01 and ***P<0.001). L1CAM, L1 cell
adhesion molecule; p-, phosphorylated; si-, small interfering. |
The PI3K/AKT and ERK pathways are the main signaling
pathways regulating cell proliferation, differentiation, survival,
motility and apoptosis. Dysregulation of these pathways is known to
contribute to carcinogenesis and cancer progression (18). In order to determine whether the
functions of L1CAM in OSCC are mediated through the PI3K/AKT and
ERK signaling pathways, the effect of L1CAM knockdown on the
activation of these pathways was investigated. As revealed in
Fig. 5B, no change was observed in
the expression of PI3K/AKT and ERK1/2 in the HSC-4 and HN22 cell
lines; however, there was significant decrease of p-PI3K, p-AKT and
p-ERK1/2 levels. According to the densitometric analysis, relative
protein expression levels and the ratio of p-/total protein were
evaluated with bar-chart graphs (*P<0.05, **P<0.01 and
***P<0.001; Fig. 5C). These data
indicated that L1CAM likely plays a role in the progression of OSCC
via both the PI3K/AKT and ERK pathways.
Discussion
Over the past two decades, L1CAM overexpression has
been identified in numerous different types of cancer including
ovarian, endometrial, pancreatic, colorectal and gastric cancers.
Moreover, L1CAM expression has been identified to be a valuable
prognostic marker in these cancers (6,11–13,16,26).
In these tumors, L1CAM expression is significantly related with
aggressive tumor characteristics including high histologic grade,
LN metastasis, advanced stage and worse survival. Moreover, L1CAM
expression has been shown to significantly correlate with the
presence of PNI in extrahepatic cholangiocarcinoma and pancreatic
ductal adenocarcinoma (13,14). In a previous study on OSCC by Hung
et al (19), L1CAM
overexpression was closely associated with high histological grade
of the tumor; however, the association of L1CAM overexpression with
other clinicopathological parameters was not assessed. In addition,
a recent study by Adnan et al (27) demonstrated that L1CAM expression was
not correlated with any clinicopathological parameters and survival
of patients in OSCC. In the present study, however, a significant
correlation between L1CAM expression and high histologic grade,
greater DOI, cervical LN metastasis, presence of PNI and advanced
TNM stage was observed in OTSCC. These results were inconsistent
with those of Adnan et al (27). This may be due to difference in the
scoring method of immunohistochemical staining. Adnan et al
(27) scored by multiplying the
intensity of staining with the percentage of positive cells,
whereas a cutoff (10% of tumor cells) was used in the present study
as it was used in most studies of endometrial carcinoma in which
the prognostic value of L1CAM has been well established (12,28,29).
Although Adnan et al (27)
reported that L1CAM had no effect on either OS or disease-free
survival in OSCC, our survival data from 10 years of follow-up
showed significantly reduced OS in patients with positive L1CAM
(P=0.014). In the present study, the most notable result was that
L1CAM expression was more significantly associated with worse OS
rate in patients with early OTSCC, such as T1/2, N0, or stage I/II
tumors (P=0.012, P=0.001 and P=0.003, respectively). To solve the
discrepancy between the different studies, further studies on the
reliable evaluation method for L1CAM immunostaining in OSCC are
needed in the future. According to the present results, L1CAM
appears to be a potential biomarker that is closely related to the
progression of OSCC and predict poor prognosis in patients with
OSCC.
Although OSCC cell lines originating from various
parts of the oral cavity were used for in vitro experiments,
the cohort was intentionally constituted of patients only with
OTSCC due to the anatomical preference of tongue for PNI and the
easiness to observe the resected whole specimen by serial sections.
Gingival SCC tissues were excluded to avoid errors in PNI detection
resulting from demineralization process which could interfere the
observation of H&E-stained soft tissue and induce inaccurate
results of immunohistochemical staining. In the present study, a
significant association was observed between L1CAM and PNI in OTSCC
(P<0.001). This is very notable due to several reasons. First,
PNI is a key pathologic feature of head and neck SCC; however, the
reported rates of PNI have ranged from 5.2–90% due to
methodological inconsistencies (e.g., detection in a limited number
of H&E-stained slides and subjective interpretation of PNI)
(5). Therefore, auxiliary
diagnostic markers are required for a more objective detection of
PNI. Although the association between L1CAM and PNI has been
reported only in certain cancers (13,14),
L1CAM is considered as one of the molecules involved in the
development of PNI through the paracrine interaction between
Schwann cells and cancer cells (30). This suggests a potential role of
L1CAM as a marker of PNI. Second, PNI has been considered as an
independent predictor of cervical metastasis, neck recurrence and
worse survival in head and neck SCC, including OTSCC (4,5,31).
Particularly, cN0 T1/2 OTSCC with PNI was suggested as an
indication for adjuvant radiation or elective neck dissection due
to the high risk of occult metastasis leading to neck recurrence
(32,33). Therefore, use of potential molecular
marker of PNI, possibly L1CAM, may help inform more rigorous
evaluation of PNI by microscopy and facilitate optimal neck
management.
Regarding the expression pattern in OTSCC tissue
samples, L1CAM expression was more frequently found at the invasive
front or in less-differentiated, spindled tumor cells. These
findings are consistent with those of previous studies on
endometrial, pancreatic and colorectal cancers (21,34,35).
Moreover, L1CAM expression was significantly associated with DOI
(P=0.009) and presence of LN metastasis (P<0.001) in the present
study of OTSCC, suggesting that L1CAM may play a role in the
invasion and metastasis of OSCC cells during tumor progression. The
present in vitro functional assays revealed the direct
effects of L1CAM on the migration and invasion of OSCC cells.
Overexpression of L1CAM induced significant increase in
proliferation, migration and invasion of both Ca9.22 and HSC-2
cells, whereas suppression of L1CAM significantly reduced these
attributes in both HSC-4 and HN22 cells. In a previous study by
Hung et al (19), shRNA
knockdown of L1CAM in SCC4 cells (tongue SCC cell line)
overexpressing L1CAM strongly attenuated cell proliferation,
migration and invasion. Besides, it was found that these phenomena
were parallel to changes in the expression of EMT-related molecules
in SCC4 cells. These findings are consistent with the current
results that L1CAM knockdown could reverse the EMT phenotype,
possibly resulting in significant inhibition of migration and
invasion of tongue SCC cell line (HSC-4 cell line). The ability of
L1CAM to promote migration and invasion has also been confirmed in
breast, gastric, pancreatic and esophageal cancer cell lines
(15,20–22,36,37).
Several studies have also demonstrated the relationship between
L1CAM and EMT process of carcinoma (20–22).
Shtutman et al (20)
suggested for the first time the functions of L1CAM during EMT
process in MCF7 breast carcinoma cells. In case of endometrial
carcinomas, L1CAM was upregulated at the invasive front of tumor,
whereas expression of E-cadherin, one of key epithelial markers,
was downregulated in the same area (21). In lung SCC tissues, L1CAM expression
was increased at the tumor-stroma interface rather than at the
tumor center, and at the same time, E-cadherin expression was
decreased, and slug expression was increased in the same area
(22). Collectively, L1CAM can play
a role in the progression of OSCC by participating in the EMT
process similar to that observed in other malignancies.
Given the involvement of L1CAM in the PI3K/AKT or
ERK signaling pathways in several cancers (9,13,15–17,26),
it was investigated whether inhibition of L1CAM expression affects
the activity of those downstream molecules in OSCC cell lines.
Downregulation of L1CAM reduced the phosphorylation of PI3K, AKT
and ERK, indicating that L1CAM is linked to both signaling pathways
in OSCC and regulates cell proliferation, migration and invasion
possibly through these pathways. In the study by Silletti et
al (38), L1CAM was revealed to
induce sustained activation of the ERK pathway and the concomitant
expression of ERK-related gene products such as integrin
αvß3, resulting in increasing cell mobility
and invasion. Additional studies have demonstrated a close
association between L1CAM and ERK in pancreatic, gastric and
colorectal cancers (13,15,16).
Moreover, L1CAM-dependent activation of the PI3K/AKT signaling
pathway has been reported in bile duct and gastric cancers
(17,26). L1CAM overexpression or knockdown was
found to increase or decrease phosphorylation of AKT in
cholangiocarcinoma (17). In
gastric cancer, inhibition of PI3K or AKT suppressed L1CAM-induced
cancer cell migration and invasion (26). Given that inhibition of ERK/PI3K/AKT
pathway-associated molecules is an attractive anticancer
therapeutic strategy, further in-depth studies are needed on the
exact mechanism by which L1CAM promotes OSCC progression through
these pathways.
Considering that L1CAM contributes to various
cellular events during tumor progression, in vitro and in
vivo studies have been conducted to develop the therapeutic
approach targeting L1CAM (17,39,40).
In the present study, inhibition of L1CAM by siRNA significantly
reduced proliferation and migration/invasion of OSCC cells in
vitro, which suggests that L1CAM may be a potential molecular
target to hinder OSCC progression. Hung et al (19) also demonstrated that downregulation
of L1CAM by shRNA could suppress the tumor growth and metastasis of
OSCC cells in an animal model. Arlt et al (39) investigated the effects of anti-L1CAM
monoclonal antibodies on ovarian carcinoma. It was found that
L1CAM-directed antibody significantly inhibited the proliferation
of ovarian carcinoma cells in vitro; in addition, it reduced
pT and inhibited peritoneal growth and dissemination of cancer
cells in ovarian carcinoma-bearing mice. Several subsequent studies
using tumor xenograft models have demonstrated the inhibitory
effect of L1CAM-blocking antibodies on tumor growth and metastasis
in intrahepatic cholangiocarcinoma, pancreatic and ovarian
carcinoma (17,40). Schäfer et al (40) reported the therapeutic potential of
combined treatment with L1CAM antibodies and cytostatic drugs in
pancreatic and ovarian carcinomas. These cancers are representative
of tumors that are associated with poor prognosis and show poor
response to conventional chemotherapy; therefore, a novel antibody
treatment targeting L1CAM may overcome the limitation of
traditional treatment. In addition to antibody-based approach,
in vivo targeting by intratumoral administration of
liposome-encapsulated L1CAM siRNAs effectively inhibited prostate
cancer growth in mouse bone (41).
These findings strongly suggested the plausibility of L1CAM being a
major driver of tumor growth and metastasis, and it is likely to
serve as a promising therapeutic target in OSCC. However, further
in vivo studies are required to confirm the effect of L1CAM
inhibition on OSCC. Subsequent study using mouse model is in
progress in the authors' lab.
In conclusion, expression of L1CAM in OSCC tissues
showed significant correlation with aggressive tumor
characteristics including DOI, LN metastasis, presence of PNI and
poor survival rate. Based on the high association of L1CAM with LN
metastasis and PNI, L1CAM could be considered a potential
prognostic biomarker in patients with OSCC, particularly in those
with early tongue tumor. Furthermore, L1CAM was found to play a
role in the progression of OSCC by promoting cell proliferation,
migration and invasion, likely via both the PI3K/AKT and ERK
signaling pathways. These findings suggested that L1CAM could serve
as a promising therapeutic target in OSCC.
Acknowledgements
The present study has been previously published on
the Seoul National University website as the doctoral thesis of
Kwang-Won Lee at https://s-space.snu.ac.kr/bitstream/10371/178702/1/000000168528.pdf.
Funding
The present study was supported by the National Research
Foundation of Korea (NRF) grant funded by the Korea government
(MSIT) (grant no. 2020R1A2C1102907).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JHK contributed to the conduction of in vitro
experiments, data acquisition and interpretation, and preparation
of the manuscript. KWL contributed to the conduction of
immunohistochemical study, data acquisition and analysis, and
preparation of the manuscript. DGA contributed the collection of
clinical data and statistical analysis. KYO contributed to the
editing of figures and statistical analysis. HJY contributed to the
conception, design of experiment, revision of the manuscript, and
supervision of the overall aspects of the project. KYO and HJY
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
All procedures followed in the present study were in
accordance with the guidelines (approval no. CRI 20003) of the
Institutional Review Board of the Seoul National University Dental
Hospital (Seoul, Korea). A waiver of informed consent was approved
by the IRB due to the retrospective nature of the study and the
analysis of anonymized data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Ervik M, Lam F, Colombet M, Mery
L, Piñeros M, Znaor A, Soerjomataram I and Bray F: Global cancer
observatory: Cancer today. International Agency for Research on
Cancer; Lyon: 2020, Available from:. https://gco.iarc.fr/today
|
|
2
|
Noguti J, De Moura CF, De Jesus GP, Da
Silva VH, Hossaka TA, Oshima CT and Ribeiro DA: Metastasis from
oral cancer: an overview. Cancer Genomics Proteomics. 9:329–335.
2012.PubMed/NCBI
|
|
3
|
Liebig C, Ayala G, Wilks JA, Berger DH and
Albo D: Perineural invasion in cancer: A review of the literature.
Cancer. 115:3379–3391. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tarsitano A, Tardio ML and Marchetti C:
Impact of perineural invasion as independent prognostic factor for
local and regional failure in oral squamous cell carcinoma. Oral
Surg Oral Med Oral Pathol Oral Radiol. 119:221–228. 2015.
View Article : Google Scholar
|
|
5
|
Schmitd LB, Scanlon CS and D'Silva NJ:
Perineural invasion in head and neck cancer. J Dent Res.
97:742–750. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Altevogt P, Doberstein K and Fogel M:
L1CAM in human cancer. Int J Cancer. 138:1565–1576. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kiefel H, Bondong S, Hazin J, Ridinger J,
Schirmer U, Riedle S and Altevogt P: L1CAM: A major driver for
tumor cell invasion and motility. Cell Adh Migr. 6:374–384. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Inaguma S, Wang Z, Lasota JP and Miettinen
MM: Expression of neural cell adhesion molecule L1 (CD171) in
neuroectodermal and other tumors: An immunohistochemical study of
5155 tumors and critical evaluation of CD171 prognostic value in
gastrointestinal stromal tumors. Oncotarget. 7:55276–55289. 2016.
View Article : Google Scholar
|
|
9
|
Gschwandtner M, Paulitschke V, Mildner M,
Brunner PM, Hacker S, Eisenwort G, Sperr WR, Valent P, Gerner C and
Tschachler E: Proteome analysis identifies L1CAM/CD171 and
DPP4/CD26 as novel markers of human skin mast cells. Allergy.
72:85–97. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thies A, Schachner M, Moll I, Berger J,
Schulze HJ, Brunner G and Schumacher U: Overexpression of the cell
adhesion molecule L1 is associated with metastasis in cutaneous
malignant melanoma. Eur J Cancer. 38:1708–1716. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fogel M, Gutwein P, Mechtersheimer S,
Riedle S, Stoeck A, Smirnov A, Edler L, Ben-Arie A, Huszar M and
Altevogt P: L1 expression as a predictor of progression and
survival in patients with uterine and ovarian carcinomas. Lancet.
362:869–875. 2003. View Article : Google Scholar
|
|
12
|
Zeimet AG, Reimer D, Huszar M, Winterhoff
B, Puistola U, Azim SA, Müller-Holzner E, Ben-Arie A, van Kempen
LC, Petru E, et al: L1CAM in early-stage type I endometrial cancer:
results of a large multicenter evaluation. J Natl Cancer Inst.
105:1142–1150. 2013. View Article : Google Scholar
|
|
13
|
Ben QW, Wang JC, Liu J, Zhu Y, Yuan F, Yao
WY and Yuan YZ: Positive expression of L1-CAM is associated with
perineural invasion and poor outcome in pancreatic ductal
adenocarcinoma. Ann Surg Oncol. 17:2213–2221. 2010. View Article : Google Scholar
|
|
14
|
Li S, Jo YS, Lee JH, Min JK, Lee ES, Park
T, Kim JM and Hong HJ: L1 cell adhesion molecule is a novel
independent poor prognostic factor of extrahepatic
cholangiocarcinoma. Clin Cancer Res. 15:7345–7351. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ito T, Yamada S, Tanaka C, Ito S, Murai T,
Kobayashi D, Fujii T, Nakayama G, Sugimoto H, Koike M, et al:
Overexpression of L1CAM is associated with tumor progression and
prognosis via ERK signaling in gastric cancer. Ann Surg Oncol.
21:560–568. 2014. View Article : Google Scholar
|
|
16
|
Fang QX, Zheng XC and Zhao HJ: L1CAM is
involved in lymph node metastasis via ERK1/2 signaling in
colorectal cancer. Am J Transl Res. 12:837–846. 2020.PubMed/NCBI
|
|
17
|
Min JK, Kim JM, Li S, Lee JW, Yoon H, Ryu
CJ, Jeon SH, Lee JH, Kim JY, Yoon HK, et al: L1 cell adhesion
molecule is a novel therapeutic target in intrahepatic
cholangiocarcinoma. Clin Cancer Res. 16:3571–3580. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
De Luca A, Maiello MR, D'Alessio A,
Pergameno M and Normanno N: The RAS/RAF/MEK/ERK and the PI3K/AKT
signalling pathways: Role in cancer pathogenesis and implications
for therapeutic approaches. Expert Opin Ther Targets. 16 (Suppl
2):S17–S27. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hung SC, Wu IH, Hsue SS, Liao CH, Wang HC,
Chuang PH, Sung SY and Hsieh CL: Targeting l1 cell adhesion
molecule using lentivirus-mediated short hairpin RNA interference
reverses aggressiveness of oral squamous cell carcinoma. Mol Pharm.
7:2312–2323. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shtutman M, Levina E, Ohouo P, Baig M and
Roninson IB: Cell adhesion molecule L1 disrupts
E-cadherin-containing adherens junctions and increases scattering
and motility of MCF7 breast carcinoma cells. Cancer Res.
66:11370–11380. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huszar M, Pfeifer M, Schirmer U, Kiefel H,
Konecny GE, Ben-Arie A, Edler L, Münch M, Müller-Holzner E,
Jerabek-Klestil S, et al: Up-regulation of L1CAM is linked to loss
of hormone receptors and E-cadherin in aggressive subtypes of
endometrial carcinomas. J Pathol. 220:551–561. 2010. View Article : Google Scholar
|
|
22
|
Tischler V, Pfeifer M, Hausladen S,
Schirmer U, Bonde AK, Kristiansen G, Sos ML, Weder W, Moch H,
Altevogt P and Soltermann A: L1CAM protein expression is associated
with poor prognosis in non-small cell lung cancer. Mol Cancer.
10:1272011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iwatsuki M, Mimori K, Yokobori T, Ishi H,
Beppu T, Nakamori S, Baba H and Mori M: Epithelial-mesenchymal
transition in cancer development and its clinical significance.
Cancer Sci. 101:293–299. 2010. View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Martinotti S and Ranzato E: Scratch wound
healing assay. Methods Mol Biol. 2109:225–229. 2020. View Article : Google Scholar
|
|
26
|
Chen DL, Zeng ZL, Yang J, Ren C, Wang DS,
Wu WJ and Xu RH: L1cam promotes tumor progression and metastasis
and is an independent unfavorable prognostic factor in gastric
cancer. J Hematol Oncol. 6:432013. View Article : Google Scholar
|
|
27
|
Adnan Y, Ali SMA, Farooqui HA, Kayani HA,
Idrees R and Awan MS: High CD44 immunoexpression correlates with
poor overall survival: Assessing the role of cancer stem cell
markers in oral squamous cell carcinoma patients from the high-risk
population of Pakistan. Int J Surg Oncol. 2022:99904892022.
|
|
28
|
Kommoss FK, Karnezis AN, Kommoss F,
Talhouk A, Taran FA, Staebler A, Gilks CB, Huntsman DG, Krämer B,
Brucker SY, et al: L1CAM further stratifies endometrial carcinoma
patients with no specific molecular risk profile. Br J Cancer.
119:480–486. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Abdelrahman AE, Salem A, Al Attar AZ,
Elsebai E, Samy W, Ibrahim MA and Ibrahim HM: p53, Pirh2, and L1CAM
as promising prognostic biomarkers of endometrial carcinoma: An
immunohistochemical and genetic study. Appl Immunohistochem Mol
Morphol. 30:713–725. 2022. View Article : Google Scholar
|
|
30
|
Amit M, Na'ara S and Gil Z: Mechanisms of
cancer dissemination along nerves. Nat Rev Cancer. 16:399–408.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shen WR, Wang YP, Chang JY, Yu SY, Chen HM
and Chiang CP: Perineural invasion and expression of nerve growth
factor can predict the progression and prognosis of oral tongue
squamous cell carcinoma. J Oral Pathol Med. 43:258–264. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tai SK, Li WY, Chu PY, Chang SY, Tsai TL,
Wang YF and Huang JL: Risks and clinical implications of perineural
invasion in T1-2 oral tongue squamous cell carcinoma. Head Neck.
34:994–1001. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chinn SB, Spector ME, Bellile EL, McHugh
JB, Gernon TJ, Bradford CR, Wolf GT, Eisbruch A and Chepeha DB:
Impact of perineural invasion in the pathologically N0 neck in oral
cavity squamous cell carcinoma. Otolaryngol Head Neck Surg.
149:893–899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tsutsumi S, Morohashi S, Kudo Y, Akasaka
H, Ogasawara H, Ono M, Takasugi K, Ishido K, Hakamada K and Kijima
H: L1 cell adhesion molecule (L1CAM) expression at the cancer
invasive front is a novel prognostic marker of pancreatic ductal
adenocarcinoma. J Surg Oncol. 103:669–673. 2011. View Article : Google Scholar
|
|
35
|
Kajiwara Y, Ueno H, Hashiguchi Y, Shinto
E, Shimazaki H, Mochizuki H and Hase K: Expression of l1 cell
adhesion molecule and morphologic features at the invasive front of
colorectal cancer. Am J Clin Pathol. 136:138–144. 2011. View Article : Google Scholar
|
|
36
|
Na'ara S, Amit M and Gil Z: L1CAM induces
perineural invasion of pancreas cancer cells by upregulation of
metalloproteinase expression. Oncogene. 38:596–608. 2019.
View Article : Google Scholar
|
|
37
|
Guo JC, Xie YM, Ran LQ, Cao HH, Sun C, Wu
JY, Wu ZY, Liao LD, Zhao WJ, Fang WK, et al: L1CAM drives
oncogenicity in esophageal squamous cell carcinoma by stimulation
of ezrin transcription. J Mol Med (Berl). 95:1355–1368. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Silletti S, Yebra M, Perez B, Cirulli V,
McMahon M and Montgomery AMP: Extracellular signal-regulated kinase
(ERK)-dependent gene expression contributes to L1 cell adhesion
molecule-dependent motility and invasion. J Biol Chem.
279:28880–28888. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Arlt MJE, Novak-Hofer I, Gast D, Gschwend
V, Moldenhauer G, Grünberg J, Honer M, Schubiger PA, Altevogt P and
Krüger A: Efficient inhibition of intra-peritoneal tumor growth and
dissemination of human ovarian carcinoma cells in nude mice by
anti-L1-cell adhesion molecule monoclonal antibody treatment.
Cancer Res. 66:936–943. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Schäfer H, Dieckmann C, Korniienko O,
Moldenhauer G, Kiefel H, Salnikov A, Krüger A, Altevogt P and
Sebens S: Combined treatment of L1CAM antibodies and cytostatic
drugs improve the therapeutic response of pancreatic and ovarian
carcinoma. Cancer Lett. 319:66–82. 2012. View Article : Google Scholar
|
|
41
|
Sung SY, Wu IH, Chuang PH, Petros JA, Wu
HC, Zeng HJ, Huang WC, Chung LW and Hsieh CL: Targeting L1 cell
adhesion molecule expression using liposome-encapsulated siRNA
suppresses prostate cancer bone metastasis and growth. Oncotarget.
5:9911–9929. 2014. View Article : Google Scholar
|