Introduction
Colorectal cancer (CRC), the third most common
malignant cancer, exhibits high mortality worldwide (1). It was revealed that a total of 53,200
individuals succumbed to CRC in the United States, in 2020
(2). Although the diagnosis and
treatment of CRC have improved, patients particularly those with
advanced CRC still have a poor prognosis (3). Therefore, it is worth clarifying the
molecular mechanism of CRC for effective treatment.
Recently, long non-coding RNAs (lncRNAs) have been
demonstrated as the pivotal regulators of various tumorigenic
processes such as cell proliferation, migration, and apoptosis
(4–6). Research has revealed that several
lncRNAs, such as lncRNA GASS, ADIPOQ, FEZF1-AS1 and GLCC1, are
aberrantly expressed in CRC and play vital roles in the development
of CRC (7–10). Among lncRNAs, NONHSAG028908.3 also
known as LINC01123, has been demonstrated to promote cell
proliferation, migration and invasion in different types of cancer,
including non-small cell lung cancer, osteosarcoma and
hepatocellular carcinoma (11–13),
however the role of NONHSAG028908.3 in CRC progression is poorly
understood. Data obtained from The Cancer Genome Atlas (TCGA)
(http://cancergenome.nih.gov/) database
reveals that the expression of NONHSAG028908.3 is significantly
upregulated in CRC tissues compared with normal tissues
(P<0.001). In addition, patients with high expression of
NONHSAG028908.3 may have low survival probability. Thus, it is
hypothesized that NONHSAG028908.3 contributes to the progression of
CRC.
In addition, lncRNAs act as a sponge to bind with
microRNAs (miRNAs or miRs), mRNA or proteins to perform biological
functions (14). Studies indicate
that the aberrant expression of miRNAs participates in regulating
tumorigenic processes in CRC (15).
For instance, Huang et al reported that reduced expression
of miR-4319 was correlated with poor prognosis in patients with
CRC, and miR-4319 suppressed CRC progression by targeting ABTB1
(16). Although some studies have
revealed that miR-34a-5p is usually downregulated in CRC (17–20),
relevant molecular mechanisms of miR-34a-5p in CRC have not been
fully elucidated. The regulatory association between
NONHSAG028908.3 and miR-34a-5p in CRC remains unclear.
Aberrant metabolism, particularly abnormal
activation of the glycolytic pathway, is a general feature of
cancer. Cancer cells usually exhibit increased glycolysis to
produce more energy (21).
Moreover, a high rate of glycolysis has been demonstrated to be
associated with a poor prognosis in patients with cancer (22). As a glycolytic enzyme, aldolase,
fructose-bisphosphate A (ALDOA), has been demonstrated to be
upregulated in various types of cancer (23–25).
Nevertheless, the role of ALDOA in CRC is controversial. Li et
al reported that downregulation of ALDOA promoted the migration
of CRC cells (SW480 and SW620) (26). By contrast, Dai et al
revealed that ALDOA was highly expressed in CRC tissues and was
associated with a poor prognosis of CRC (27). Therefore, it is necessary to reveal
the function of ALDOA in CRC. Moreover, whether NONHSAG028908.3
targets miR-34a-5p/ALDOA to regulate tumorigenic processes in CRC
warrants investigation.
The present study explored the function of
NONHSAG028908.3 in CRC, and further clarified its regulatory effect
on the miR-34a-5p/ALDOA axis.
Materials and methods
Cell culture
The normal human colon mucosal epithelial cell line
(NCM460; cat. no. CP-H040) and human CRC cell lines (LoVo, cat. no.
CL-0144; HCT116, cat. no. CL-0096; and Caco-2, cat. no. CL-0050)
were purchased from Procell Life Science & Technology Co., Ltd.
HT-29 (cat. no. ZQ0057), a CRC cell line, was obtained from
Shanghai Zhong Qiao Xin Zhou Biotechnology Co., Ltd. The cell lines
used have been authenticated using STR profiling. NCM460 cells were
incubated in human colonic mucosal epithelial cell complete medium
(cat. no. CM-H040; Procell Life Science & Technology Co.,
Ltd.). HT-29 and Caco-2 cells were incubated in DMEM (cat. no.
SH30022; Hyclone; Cytiva) and MEM (cat. no. 41500-067; Gibco;
Thermo Fisher Scientific, Inc.) medium supplemented with 10% fetal
bovine serum (FBS; cat. no. 04-011-1A, Biological Industries;
Sartorius AG), respectively. The LoVo cells were incubated in Ham's
F-12K (cat. no. PM150910; Procell Life Science & Technology
Co., Ltd.) medium which contained 10% FBS. McCoy's 5A (cat. no.
PM150710; Procell Life Science & Technology Co., Ltd.) medium
including 10% FBS was used to culture HCT116 cells. Penicillin (100
U/ml) and streptomycin (0.1 mg/ml) were added into all the
aforementioned media. The cells were maintained in a humidified
incubator at a temperature of 37°C and a CO2 ratio of
5%. Cells growing in the logarithmic phase, were used in the
subsequent experiments.
Cell transfection
NONHSAG028908.3 siRNAs [si-NONHSAG028908.3-1 forward
(F), 5′-GCUAGAUUGCUAUAGUCUATT-3′ and reverse (R),
5′-UAGACUAUAGCAAUCUAGCTT-3′; and si-NONHSAG028908.3-2 F,
5′-GAGAAGUUCUGCAGAUGUATT-3′ and R, 5′-UACAUCUGCAGAACUUCUCTT-3′],
ALDOA siRNA (si-ALDOA F, 5′-GGAGGAGUAUGUCAAGCGAGCTT-3′ and R,
5′-UCGCUUGACAUACUCCUCCUGTT-3′); negative control (si-NC F,
5′-UUCUCCGAACGUGUCACGUTT-3′ and R, 5′-ACGUGACACGUUCGGAGAATT-3′);
miR-34a-5p mimics (F, 5′-UGGCAGUGUCUUAGCUGGUUGU-3′ and R,
5′-AACCAGCUAAGACACUGCCAUU-3′); NC mimics (F,
5′-UUCUCCGAACGUGUCACGUTT-3′ and R, 5′-ACGUGACACGUUCGGAGAATT-3′);
miR-34a-5p inhibitor (F, 5′-ACAACCAGCUAAGACACUGCCA-3′); and NC
inhibitor (F, 5′-UUGUACUACACAAAAGUACUG-3′) were obtained from JTS
scientific; http://www.jtsbio.com/. The
NONHSAG028908.3-overexpressing plasmid and corresponding empty
vector were prepared by General Bio Co., Ltd. Lipofectamine 2000
reagent (cat. no. 11668-019; Invitrogen; Thermo Fisher Scientific,
Inc.) was used for transfection following the manufacturer's
protocols.
When the degree of cell confluence reached 70%,
cells in 6-well plates were cultured with serum-free medium for 1
h. Subsequently, 100 µl Opti-MEM was used to dilute 8 µl
Lipofectamine 2000 reagent, which was incubated at room temperature
for 5 min. A total of 100 pmol of each construct was diluted with
100 µl Opti-MEM following incubation for 5 min, and then was
transfected into cells. All dilutions were performed at room
temperature for 10 min. Following the addition of the dilutions to
the cells for 4 h, the medium was replaced with complete medium and
further incubated for 48 h.
Reverse transcription-quantitative PCR
(RT-qPCR)
The total RNA from cultured cells was extracted via
RNzol-Total RNA isolation Kit (cat. no. RP1001; BioTeke
Corporation), and the concentration of RNA was detected using an
ultraviolet spectrophotometer NANO 2000 (Thermo Fisher Scientific,
Inc.). MiR-34a-5p was extended with a specific loop primer,
5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACAACC-3′. The total
RNA was reverse transcribed into complementary DNA (cDNA) using
Super M-MLV reverse transcriptase (cat. no. PR6502; BioTeke
Corporation). Real-time PCR was performed using SYBR Green (cat.
no. S9430; MilliporeSigma). The signals were measured via
Exicycler™ 96 PCR instrument (Bioneer Corporation) and the
thermocycling program consisted of holding at 94°C for 5 min,
followed by 40 cycles of 15 sec at 94°C, 25 sec at 60°C, and 30 sec
at 72°C. Gene expression was normalized to β-actin or U6. The
relative mRNA expression was calculated via the 2−ΔΔCq
method (28). All primer sequences
are presented in Table I.
 | Table I.Primer sequences of reverse
transcription-quantitative PCR. |
Table I.
Primer sequences of reverse
transcription-quantitative PCR.
| Gene | Primers 5′-3′ |
|---|
|
NONHSAG028908.3 | F:
TACTTTGCCTTGCTTACACG |
|
| R:
AGGAGCCAGTTCCAGACC |
| miR-34a-5p | F:
TGGCAGTGTCTTAGCTGGTTGT |
|
| R:
GCAGGGTCCGAGGTATTC |
| ALDOA | F:
CCCTGACCTTCTCCTACGG |
|
| R:
GGCTCGCTTGACATACTCCT |
| β-actin | F:
CACTGTGCCCATCTACGAGG |
|
| R:
TAATGTCACGCACGATTTCC |
| U6 | F:
GCTTCGGCAGCACATATACT |
|
| R:
GCAGGGTCCGAGGTATTC |
MTT assay
The LoVo, HCT116, and NCM460 cells (3×103
cells/well) were seeded into 96-well plates. Cells were transfected
with corresponding si-NC, si-NONHSAG028908.3-1,
si-NONHSAG028908.3-2, vector, NONHSAG028908.3, NC inhibitor,
miR-34a-5p inhibitor, NC mimics, miR-34a-5p mimics or si-ALODA
after cell adherence. The MTT reagent (0.5 mg/ml; cat. no. M-2128;
MilliporeSigma) was added to the plates according to the protocol
at 0, 24, 48 and 72 h, respectively. After incubation at 37°C for 4
h, the formazan crystals were dissolved using DMSO. Subsequently,
the OD values were determined using a microplate reader (ELX-800;
BioTek Instruments, Inc.) at 570 nm.
BrdU analysis
The LoVo and HCT116 cells were treated with 10 µM
BrdU solution (cat. no. B110731; Aladdin Biochemical Technology
Co., Ltd) for 1 h at post transfection. Cells were fixed using 4%
paraformaldehyde for 15 min at room temperature, and then 0.1%
Triton X-100 (cat. no. ST795; Beyotime Insitute of Biotechnology)
was added for 30 min. Subsequently, 10% goat serum (cat. no. SL038;
Beijing Solarbio Science & Technology Co., Ltd.) was used for
blocking at room temperature for 15 min. Accordingly, the cells
were then incubated at 4°C overnight, with anti-BrdU antibody
(1:200; cat. no. 66241-1; ProteinTech Group, Inc.). Following
incubation with a fluorescence secondary antibody (1:200; cat. no.
A0521; Beyotime Institute of Biotechnology), the cells were stained
using DAPI (cat. no. C1002; Beyotime Institute of Biotechnology).
The images were captured via fluorescence microscope at a
magnification of ×400.
Flow cytometry
Following transfection for 48 h, 2×104
cells (LoVo and HCT116) were collected and washed with PBS.
Pre-cooled ethanol (concentration, 70%) was used for fixing cells
at 4°C for 12 h. Following centrifugation at 1,000 × g at 4°C for 5
min, cells were resuspended in 500 µl cell staining buffer (cat.
no. C1052-1; Beyotime Institute of Biotechnology). Subsequently,
the cells were stained with propidium iodide (25 µl; cat. no.
C1052-2; Beyotime Institute of Biotechnology) and RNase A (10 µl;
cat. no. C1052-3; Beyotime Institute of Biotechnology) at 37°C for
30 min in the dark according to the manufacturer's instructions.
The cell cycle was then assessed via flow cytometry (NovoCyte;
Agilent Technologies, Inc.) using NovoExpress software (version
1.4.1; Agilent Technologies, Inc.).
Western blotting
Proteins from cells were extracted using RIPA lysis
buffer with 1% PMSF (cat. nos. P0013B and ST506, respectively;
Beyotime Institute of Biotechnology). A BCA kit (cat. no. P0009;
Beyotime Institute of Biotechnology) was used to assess the protein
concentration. Total protein (30 µg per lane) was separated using
10–12% SDS-PAGE (cat. no. P0015; Beyotime Institute of
Biotechnology) and accordingly transferred to a PVDF membrane (cat.
no. LC2005; Thermo Fisher Scientific, Inc.). Subsequently, the
membrane was placed in Tris-buffered saline with 1.5% Tween-20
(TBST) for 5 min, and then incubated with primary antibody
anti-cyclin D1 (1:1,000; cat. no. A19038; ABclonal Biotech Co.,
Ltd.), anti-CDK4 (1:1,000; cat. no. A0366; ABclonal Biotech Co.,
Ltd.), anti-mature matrix metalloproteinase (MMP)-2 (1:1,000;
10373–2-AP; ProteinTech Group, Inc.), anti-mature MMP-9 (1:500;
cat. no. 10375-2-AP; Proteintech Group, Inc.) and anti-ALDOA
(1:1,000; cat. no. A1142; ABclonal Biotech Co., Ltd.) overnight at
4°C. The membrane was washed using TBST following treatment with
HRP-labeled secondary antibody (1:10,000; cat. no. SA00001-2;
Proteintech Group, Inc.) at 37°C for 40 min. ECL (cat. no. E003;
7sea Technology Co., Ltd.) was then added into membranes to
visualize the protein bands. The relative density was evaluated via
Gel-Pro-Analyzer software (version 4.0; Beijing Liuyi Biotechnology
Co., Ltd.).
Wound healing assay
When LoVo and HCT116 cells reached 100% confluency,
the medium was replaced with serum-free medium including 1 µg/ml
mitomycin C (cat. no. M0503; MilliporeSigma) for 1 h. A 200-µl
pipette tip was used to a scratch the layer of the cells at 0 h,
and the images were captured with microscope at a magnification of
×100. The cells were then incubated at 37°C for 24 h. Subsequently,
the images were captured.
Transwell assay
The invasion capabilities of LoVo and HCT116 cells
were determined via 24-well Transwell chamber assays (cat. no.
3422, Corning, Inc.). LoVo and HCT116 cells were collected at 48 h
post transfection. A cell suspension (2×104 cells; 200
µl without FBS) was added to the upper chamber, which was precoated
with 40 µl Matrigel for 2 h at 37°C, while the lower chamber
contained 30% FBS. After the cells were incubated for 24 h, they
were fixed with 4% paraformaldehyde at room temperature for 15 min.
Crystal violet (0.4%; cat. no. 0528; Amresco, LLC) was used to
stain the cells at room temperature for 5 min, and then the number
of cells passing through the Matrigel was determined. Five fields
were randomly selected for cell counting, and the experiment was
repeated three times.
Dual-luciferase reporter assay
The sequences of wild-type (wt) NONHSAG028908.3
(wt-NONHSAG028908.3), mutant (mut) NONHSAG028908.3
(mut-NONHSAG028908.3), wt-ALDOA or mut-ALDOA were subcloned into
pmirGLO vectors (GenScript Biotechnology Co., Ltd.; http://www.genscript.com.cn/). The molecular sequences
were as follows: wt-NONHSAG028908.3, 5′-CCCAUCAGCAGCCACUGCCC-3′;
mut-NONHSAG028908.3, 5′-CCCAUGUCGUCCGUGACGGC-3′; wt-ALDOA,
5′-CACCCUUUCCGGCACACUGCCA-3′; mut-ALDOA,
5′-CACCCUUUCCGGCUGUGACGGA-3′. Accordingly, these reporter vectors
with miR-34a-5p mimics or NC mimics were co-transfected into 293T
cells (cat. no. ZQ0033; Shanghai Zhong Qiao Xin Zhou Biotechnology
Co., Ltd.) using Lipofectamine 2000 (cat. no. 11668-019;
Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, the
transfected cells were harvested, and the luciferase activities
were measured using the dual-luciferase reporter kit (cat. no.
KGAF040; Nanjing KeyGen Biotech Co., Ltd.) normalized to
Renilla luciferase activity.
Bioinformatics analysis
The expression pattern of NONHSAG028908.3 in CRC
tissues and normal tissues was predicted using the online database,
TCGA. The data of the correlation between NONHSAG028908.3 and
miR-34a-5p and/or ALDOA expression levels in CRC cells or tissues
were obtained from the Cancer Cell Line Encyclopedia (CCLE;
http://sites.broadinstitute.org/ccle)
and TCGA datasets.
Statistical analysis
Results were conducted using GraphPad 8.0 software
(GraphPad software Inc.). The unpaired Student's t-test was
performed to analyze the differences between two groups. One-way
ANOVA followed by Bonferroni's post hoc test was used to analyze
multiple comparisons. Pearson's correlation coefficient analysis
was used to analyze correlations. Data is presented as the mean ±
standard deviation (SD). P<0.05 was considered to indicate a
statistically significant difference.
Results
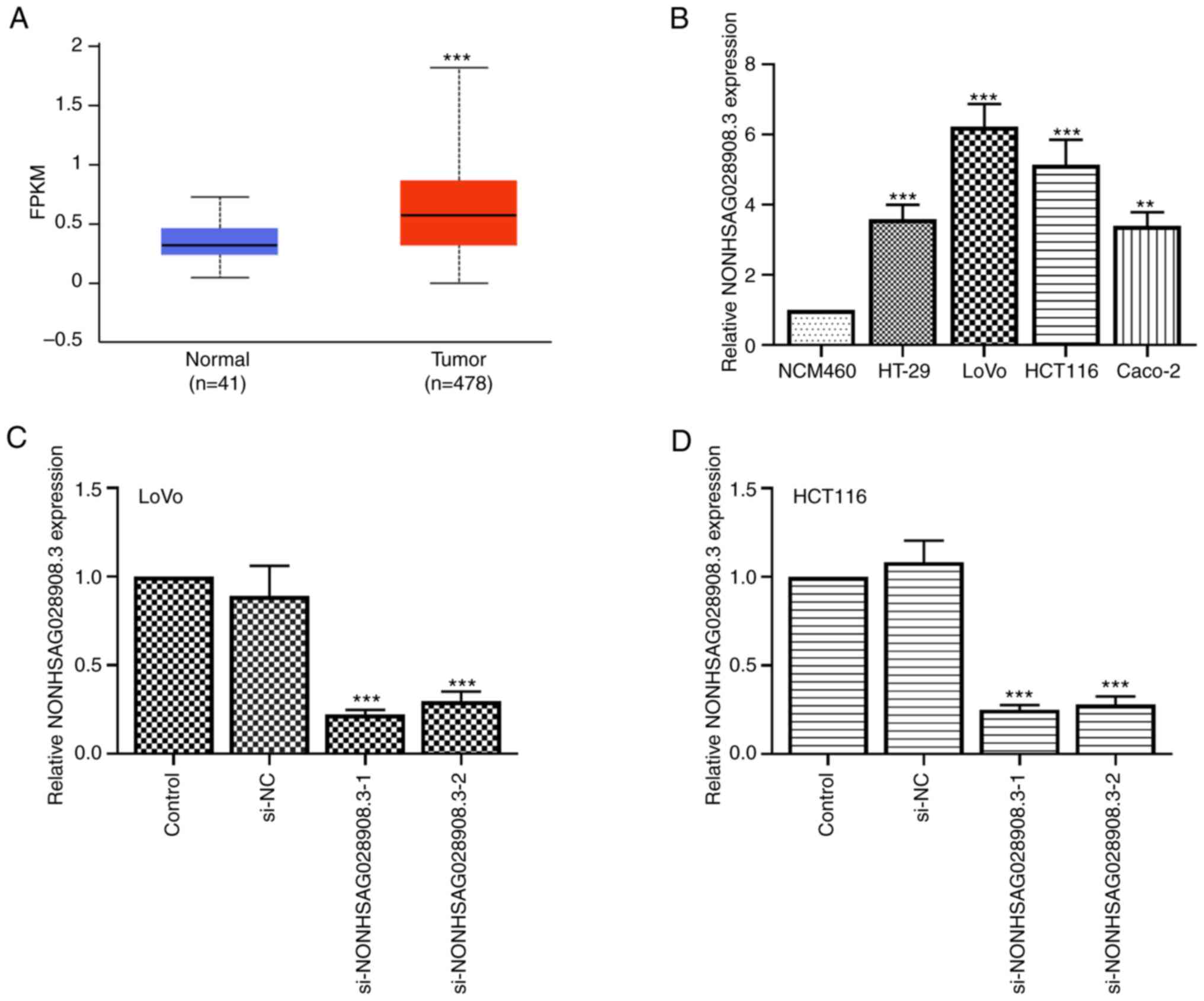
NONHSAG028908.3 is upregulated in CRC
tissues and cells
To determine the expression level of NONHSAG028908.3
(www.noncode.org) in CRC tissues and cells, TCGA
database was employed and it was revealed that NONHSAG028908.3 was
markedly upregulated in CRC tissues compared with that in normal
tissues (Fig. 1A; P<0.001). In
addition, normal colorectal cell line (NCM460) and four types of
CRC cell lines were selected to determine the expression of
NONHSAG028908.3 using RT-qPCR. The results revealed that the
expression of NONHSAG028908.3 was also increased in HT-29, LoVo,
HCT116 and Caco-2 cells compared with NCM460 cells (Fig. 1B; P<0.01). Among these cell
lines, two NONHSAG028908.3 high-expressing cell lines, LoVo and
HCT116, were selected to generate NONHSAG028908.3-silencing cell
lines. RT-qPCR assay indicated that the expression of
NONHSAG028908.3 was downregulated both in LoVo and HCT116 cells
transfected with si-NONHSAG028908.3-1/2 (Fig. 1C and D; P<0.001).
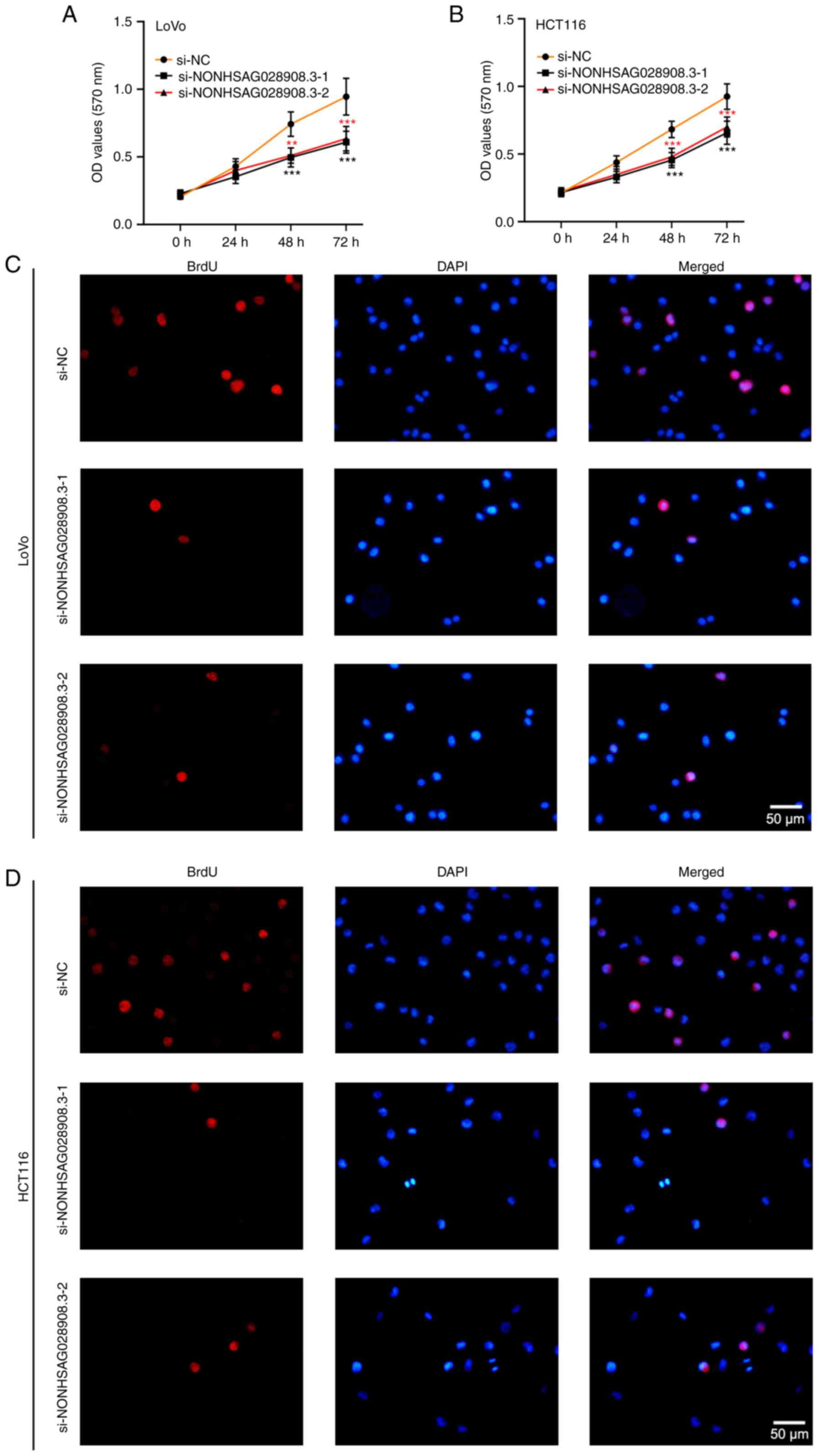
Silencing of NONHSAG028908.3
suppresses the proliferation of CRC cells
To reveal the function of NONHSAG028908.3 on cell
proliferation, an MTT assay was carried out on LoVo, HCT116 and
NCM460 cells after transfection. The results of the MTT assay
demonstrated that proliferation abilities of CRC cells (LoVo and
HCT116) were decreased once transfected with the
si-NONHSAG028908.3-1/2 (Fig. 2A and
B; P<0.01). Overexpression of NONHSAG028908.3 promoted cell
growth in NCM460 cells, a normal colorectal cell line (Fig. S1). In addition, it was found that
the number of stained BrdU-positive cells was observably reduced
under NONHSAG028908.3 silencing (Fig.
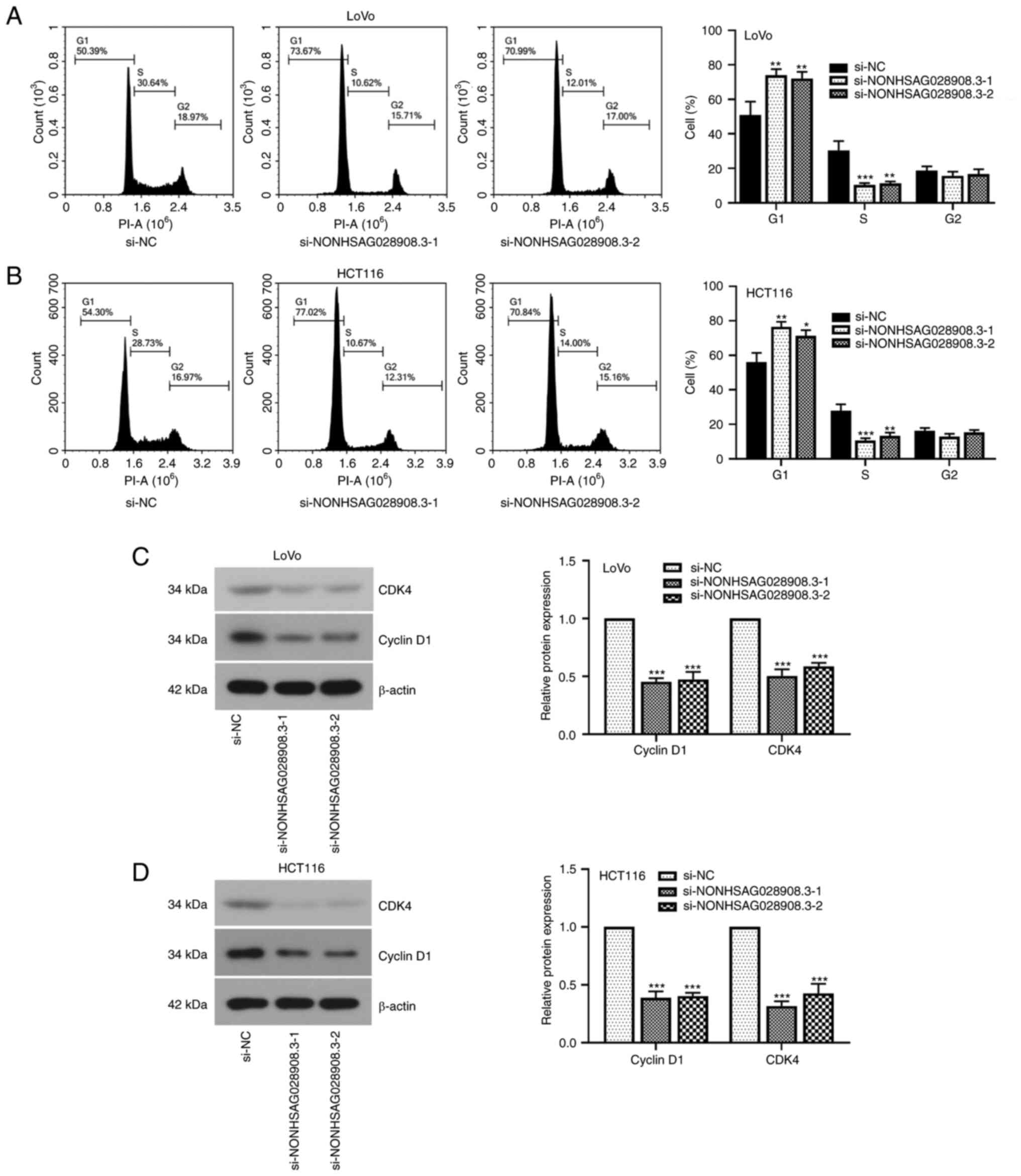
2C and D). In addition, the results of the flow cytometric
assays demonstrated that knockdown of NONHSAG028908.3 blocked the
cell cycle in the G1 phase (Fig. 3A and
B; P<0.05). When the expression of NONHSAG028908.3 was
inhibited, the levels of cell cycle-related proteins, cyclin D1 and
CDK4, were downregulated in LoVo and HCT116 cells (Fig. 3C and D; P<0.001).
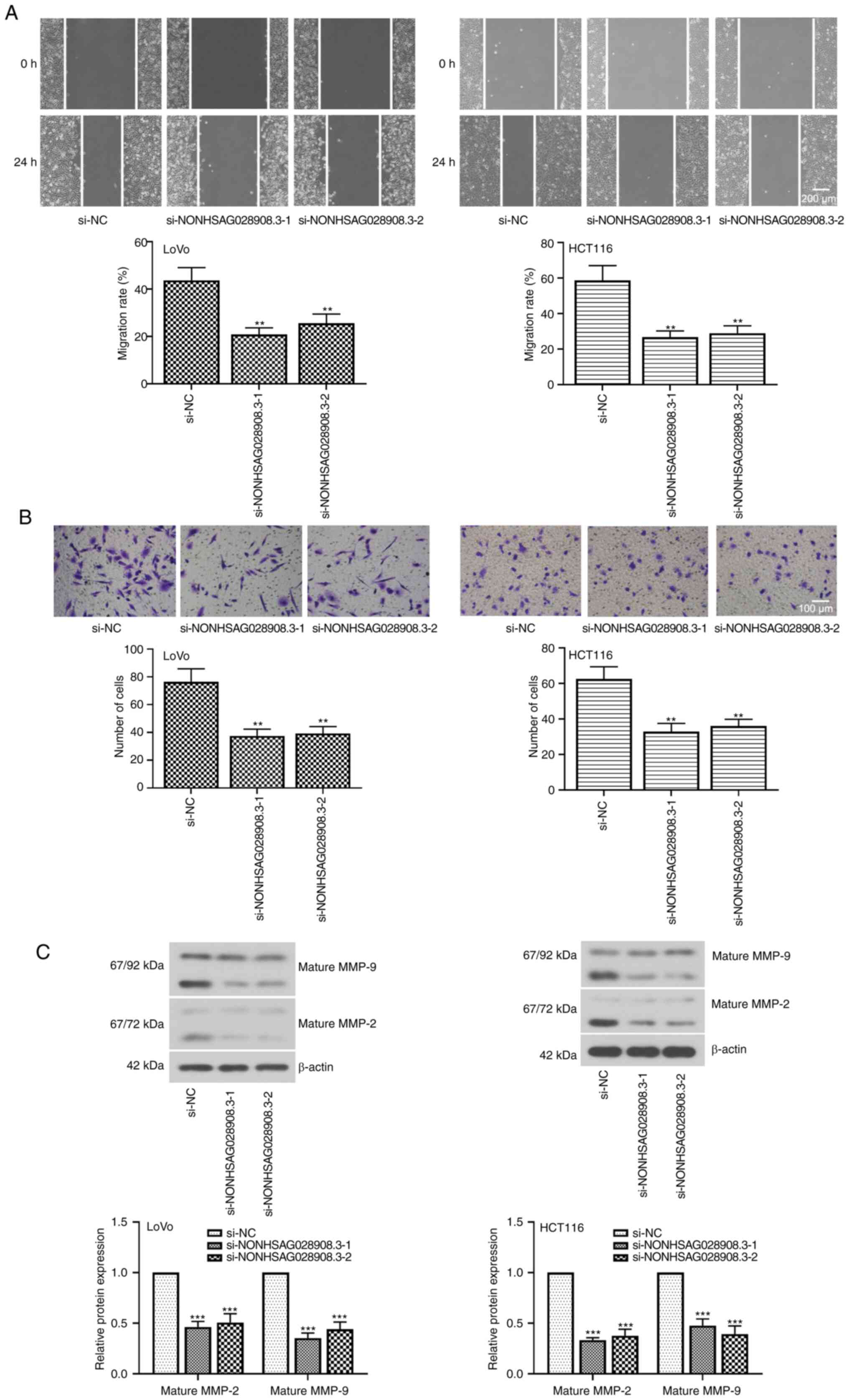
Knockdown of NONHSAG028908.3 restrains
CRC cell migration and invasion
To further characterize the role of NONHSAG028908.3
in CRC, wound healing and Transwell assays were performed for
monitoring cell migration and cell invasion, respectively. The
findings of the wound healing assays revealed that the migration
rate of CRC cells was significantly suppressed after inhibition of
NONHSAG028908.3 (Fig. 4A;
P<0.01). Similarly, the results of the Transwell invasion assays
revealed that the number of cells passing through the membranes was
markedly diminished when CRC cells were transfected with
si-NONHSAG028908.3 (Fig. 4B;
P<0.01). Furthermore, the results of the western blot assays
indicated that knockdown of NONHSAG028908.3 downregulated the
levels of mature MMP-2 and mature MMP-9 (Fig. 4C; P<0.001).
NONHSAG028908.3 serves as a sponge to
bind with miR-34a-5p
Previous research has revealed that lncRNAs can
sponge miRNAs to regulate metastasis (29). In the present study, it was
hypothesized that NONHSAG028908.3 served as a sponge to combine
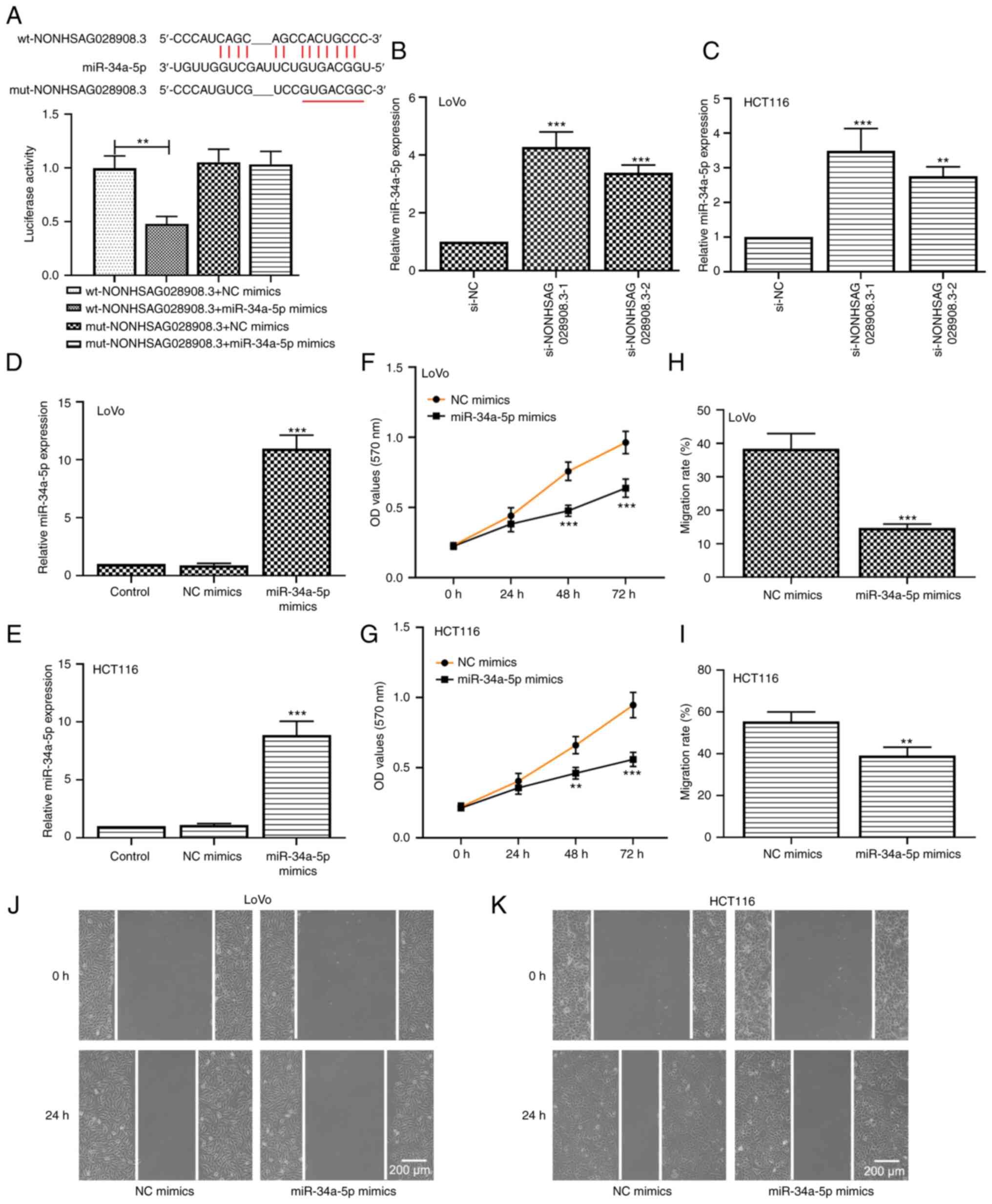
with miR-34a-5p, and their binding sites are presented in Fig. 5A. Accordingly, a luciferase reporter
assay was carried out to verify the aforementioned hypothesis. The
data demonstrated that miR-34a-5p mimics significantly diminished
the luciferase activity of the wt-NONHSAG028908.3 reporter but did
not suppress that of the mut-NONHSAG028908.3 reporter (Fig. 5A; P<0.01). Moreover, it was found
that the knockdown of NONHSAG028908.3 significantly enhanced
miR-34a-5p expression (Fig. 5B and
C; P<0.01). These data indicated that NONHSAG028908.3
functioned as a sponge for miR-34a-5p. Additionally, the results of
RT-qPCR confirmed the transfection efficiency of miR-34a-5p mimics
both in LoVo and HCT116 cells (Fig. 5D
and E; P<0.001). Overexpression of miR-34a-5p inhibited the
CRC cell growth of LoVo and HCT116 cells according to the data of
the MTT assays (Fig. 5F and G). The
results of the wound healing assays demonstrated that the ability
of cell migration was decreased in miR-34a-5p-overexpressing CRC
cells compared with that in cells transfected with NC mimics
(Fig. 5H-K).
MiR-34a-5p silencing abolishes the
inhibitory effect of NONHSAG028908.3 knockdown on the malignant
behavior of cells
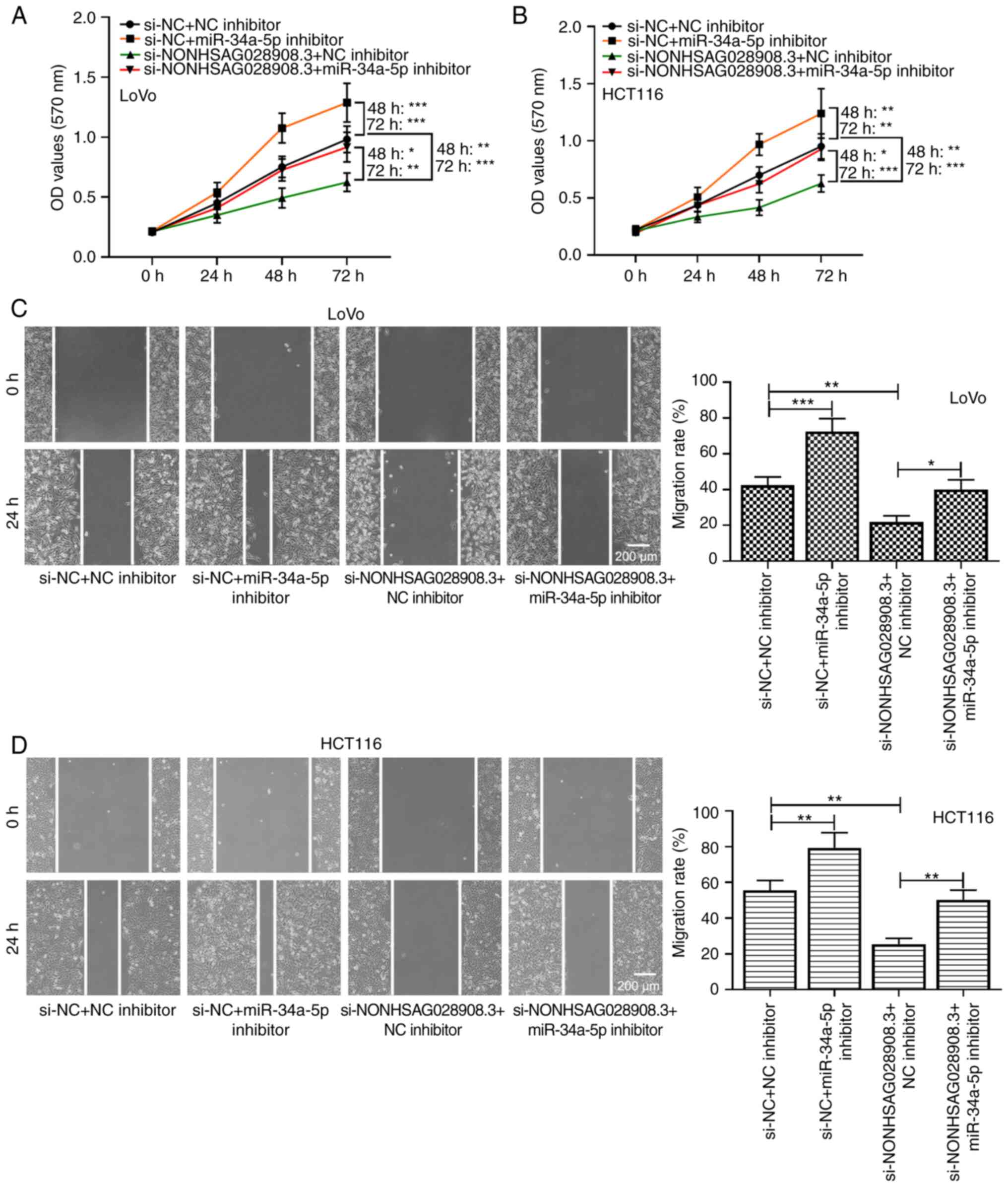
To further explore the association between
NONHSAG028908.3 and miR-34a-5p in CRC cells, rescue experiments
were performed. Data from MTT assays demonstrated that the
inhibitory effect of si-NONHSAG028908.3 on cell proliferation was
reversed by silencing of miR-34a-5p in LoVo and HCT116 cells
(Fig. 6A and B; P<0.05). Wound
healing assay also confirmed this finding. In brief,
NONHSAG028908.3 knockdown significantly suppressed cell
proliferation and migration, whereas silencing of miR-34a-5p
abolished the suppressive effect of NONHSAG028908.3 knockdown
(Fig. 6C and D; P<0.05).
ALDOA directly targets miR-34a-5p and
is positively correlated with NONHSAG028908.3
For the next experiment, pmirGLO luciferase reporter
vectors including wt-ALDOA and mut-ALDOA were constructed. When the
293T cells were co-transfected with wt-ALDOA and miR-34a-5p mimics,
the luciferase activity was significantly decreased (Fig. 7A; P<0.001). However, the
luciferase activity of mut-ALDOA underwent no obvious change.
Furthermore, the expression of ALDOA was assessed in
miR-34a-5p-overexpressing cells. The results of RT-qPCR and western
blot assays demonstrated that overexpression of miR-34a-5p
negatively regulated the expression level of ALDOA in LoVo and
HCT116 cells (Fig. 7B and C;
P<0.001). Furthermore, knockdown of NONHSAG028908.3
significantly decreased the mRNA and protein levels of ALDOA, which
were rescued via silencing of miR-34a-5p (Fig. 7D and E; P<0.05). The results
obtained from TCGA and CCLE datasets revealed that the expression
level of NONHSAG028908.3 was positively correlated with the
expression level of ALDOA in CRC (Fig.
S2; P=0.001). Collectively, these findings indicated that
NONHSAG028908.3 may positively regulate ALDOA expression through
sponging miR-34a-5p.
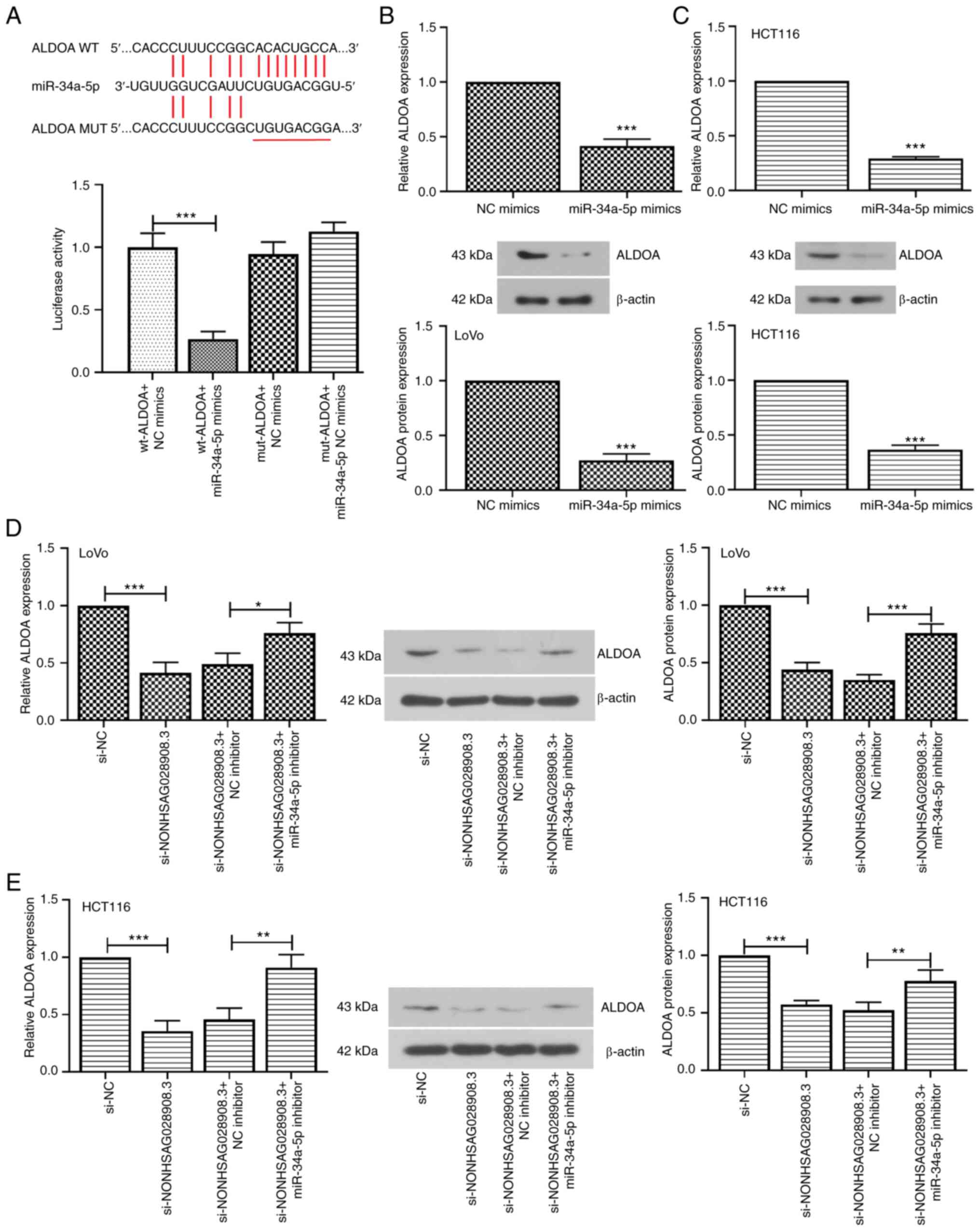 | Figure 7.ALDOA is a direct target of
miR-34a-5p. (A) The binding sites of miR-34a-5p to ALDOA were
predicted. A luciferase assay confirmed that miR-34a-5p mimics
decreased the luciferase activity of wt-ALDOA, whereas the
luciferase activity of mut-ALDOA underwent no obvious change. (B
and C) The LoVo and HCT116 cells were transfected with NC mimics or
miR-34a-5p mimics. RT-qPCR and western blotting were used to
determine the mRNA and protein expression of ALDOA, respectively.
(D and E) The expression of ALDOA was detected in LoVo and HCT116
cells transfected with si-NC or si-NONHSAG028908.3 and NC inhibitor
or miR-34a-5p inhibitor. The results are presented as the mean ±
SD. *P<0.05, **P<0.01 and ***P<0.001. ALDOA, aldolase,
fructose-bisphosphate A; miR-34a-5p, microRNA-34a-5p; wt,
wild-type; mut, mutant; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; si-, siRNA. |
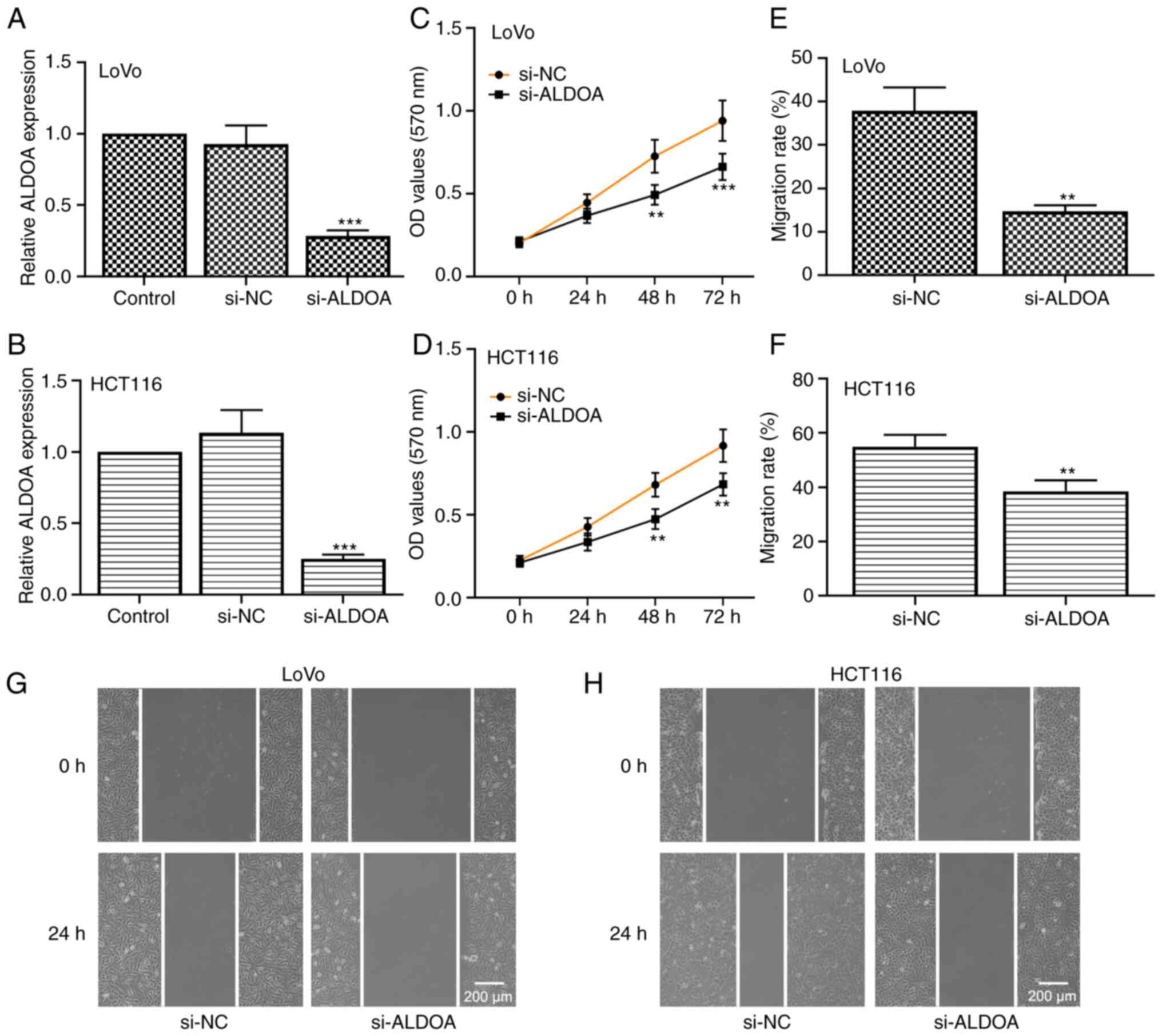
ALDOA silencing suppresses CRC cell
growth and migration
Furthermore, ALDOA was successfully knocked down in
LoVo and HCT116 CRC cells (Fig. 8A and
B; P<0.001). The results of MTT assays demonstrated that
silencing of ALDOA inhibited CRC cell proliferation (Fig. 8C and D). In addition, the migration
rate of CRC cells transfected with si-ALDOA was markedly reduced
compared with that in cells transfected with si-NC (Fig. 8E and H; P<0.01). Thus, the data
indicated that knockdown of ALDOA suppressed CRC cell proliferation
and migration.
Discussion
Although treatment of CRC has greatly improved, CRC
often results in an unfavorable outcome due to the malignant
metastasis of CRC cells. In consequence, researchers are urgently
seeking new biomarkers for the diagnosis and prognosis of CRC.
Numerous studies have demonstrated that tumor-associated genes are
frequently aberrantly expressed. A number of genomes are
transcribed as noncoding RNAs, of which lncRNAs and miRNAs make up
the majority of noncoding RNAs (30,31).
Research has revealed that lncRNAs participate in a metastatic
cascade and a high abundance of cancer-related lncRNAs regulate CRC
pathogenesis (32,33).
To the best of our knowledge, the function of
NONHSAG028908.3 has not been fully clarified in human types of
cancer. In the present study, data from TCGA database was
retrieved, and the results revealed that NONHSAG028908.3 was
upregulated in CRC tissues. In addition, a high level of
NONHSAG028908.3 was also detected in CRC cells. Furthermore, the
promoting effect of NONHSAG028908.3 on cell proliferation and
migration was revealed. The aggregation of cyclin D1 and CDK4 has
also been demonstrated to be of great importance in the cell cycle
(34). Upregulation of cyclin D1
has been revealed to cause dysregulated CDK activity, rapid cell
growth under conditions of restricted mitogenic signaling, and
finally, neoplastic growth (34).
In the present study it was revealed that NONHSAG028908.3 silencing
led to G1 cell cycle arrest involving the alleviation of cyclin D1
and CDK4. Moreover, previous studies indicated that MMP-2 and MMP-9
accelerated cell invasion via participating in extracellular matrix
degradation (35). Similarly, the
data of the present revealed that inhibition of NONHSAG028908.3
suppressed cell invasion, partly by a mechanism associated to the
decrease of mature MMP-2 and MMP-9. All aforementioned findings
demonstrated the oncogenic property of NONHSAG028908.3 in CRC cell
growth.
Mechanistic insights using a luciferase reporter
assay confirmed that NONHSAG028908.3 exhibited a high affinity to
interact with miR-34a-5p. It has been established that miR-34a-5p
is often downregulated and plays a tumor suppressive role in
multiple cancers, including CRC. Moreover, a previous study
demonstrated that miR-34a-5p was downregulated in CRC tissues, and
miR-34a-5p expression was positively related with disease-free
survival (17). The data of the
present study also confirmed that overexpression of miR-34a-5p
suppressed CRC cell proliferation and migration. In addition, the
findings of other studies are consistent with the findings of the
present study. For instance, Li et al demonstrated that
miR-34a-5p was negatively associated with lncARSR expression, and
inhibited CRC cell invasion and metastasis (18). Gao et al revealed that
miR-34a-5p restrained recurrence of CRC by facilitating cell
apoptosis in a p53-dependent manner (17). Bao et al reported that
inhibiting miR-34a expression aggravated CRC cell invasion via
promoting epithelial-mesenchymal transition (36). Additionally, the results of the
present study also revealed that depletion of NONHSAG028908.3
restrained cell proliferation and migration, which was abrogated
via miR-34a-5p inhibition. This indicated that NONHSAG028908.3
served as a sponge to bind to miR-34a-5p, thus facilitating cell
proliferation and migration.
Previous research demonstrated that increased
glycolysis promotes cancer cell growth and survival (37). Thus, targeting glycolysis to inhibit
cancer appears to be an effective method. In the present study, it
was demonstrated that miR-34a-5p directly targeted ALDOA, which was
regulated by NONHSAG028908.3. ALDOA, a pivotal glycolytic enzyme,
acts as a catalyst in a conversion reaction of fructose-1,
6-bisphosphate (38). As
aforementioned, the function of ALDOA in CRC has not been fully
elucidated. Dai et al revealed that the expression of ALDOA
was increased in CRC tissues and liver metastatic CRC tissues, and
high expression of ALDOA was correlated with poor prognosis of CRC
(27). Kawai et al
demonstrated that ALDOA was positively associated with CRC cell
proliferation and invasion (39).
Similar to these previous studies, it was observed in the present
study that ALDOA was upregulated in CRC cells (LoVo and HCT116).
Knockdown of ALDOA inhibited CRC cell growth and migration. In
addition, silencing of NONHSAG028908.3 attenuated the expression
level of ALDOA, which was reversed by suppressing miR-34a-5p.
However, Li et al reported that downregulation of ALDOA
promoted the migration of CRC cells, SW480 and SW620 (26). Since the aforementioned study used
different cells from the ones used in the present study, it is
hypothesized that the different results may be caused by cell
differences. In addition to ALDOA, other targets of miR-34a-5p,
including TNFAIP8, flotillin-2, and hexokinase-1 have been revealed
(18,40,41).
Among them, Li et al reported that hexokinase-1 is also the
key enzyme of glycolysis and participates in CRC progression
(18). However, it is not yet clear
whether increased ALDOA expression is related to changes in other
miR-34a-5p targets. Therefore, in future experiments, this will be
investigated. In addition, lack of an animal study is a limitation
of the present study. In a future study, an animal model will be
established to completely confirm the function of NONHSAG028908.3
on tumor growth and metastasis in vivo. In addition, a
sufficient number of clinical samples will be collected to further
detect the expression levels of NONHSAG028908.3, miR-34a-5p, and
ALDOA in CRC tissues and adjacent normal tissues, and analyze their
correlation.
Collectively, the data of the present study revealed
the importance of the NONHSAG028908.3/miR-34a-5p/ALDOA axis in
modulating CRC progression. Upregulation of NONHSAG028908.3 was
associated with the aggressive phenotypes of CRC. NONHSAG028908.3
served as a sponge to combine with miR-34a-5p and formed a positive
feedback loop to regulate ALDOA. The present study provides
insights into and a new mechanism for CRC treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 81470086 and 81871465).
Availability of data and materials
The datasets generated during and/or analyzed during
the current study are available from the corresponding author on
reasonable request.
Authors' contributions
CW performed experiments, analyzed the data and
wrote the manuscript. HX and GY performed experiments and analyzed
the data. ZL designed the experiments and revised the manuscript.
CW and ZL confirm the authenticity of all the raw data. All authors
read and approved the final manuscript and agree to be accountable
for all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim C, Kim WR, Kim KY, Chon HJ, Beom SH,
Kim H, Jung M, Shin SJ, Kim NK and Ahn JB: Predictive nomogram for
recurrence of stage I colorectal cancer after curative resection.
Clin Colorectal Cancer. 17:e513–e518. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Forrest ME and Khalil AM: Review:
Regulation of the cancer epigenome by long non-coding RNAs. Cancer
Lett. 407:106–112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou
A, Liu J, Che L and Li J: Long noncoding RNA GAS5 inhibits
progression of colorectal cancer by interacting with and triggering
YAP phosphorylation and degradation and is negatively regulated by
the m6A reader YTHDF3. Mol Cancer. 18:1432019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang HQ, Meng YL, Lu QL, Dou YY, Liang LL
and Luo Y: Decreased long noncoding RNA ADIPOQ promoted cell
proliferation and metastasis via miR-219c-3p/TP53 pathway in
colorectal carcinoma. Eur Rev Med Pharmacol Sci. 24:7645–7654.
2020.PubMed/NCBI
|
|
9
|
Bian Z, Zhang J, Li M, Feng Y, Wang X,
Zhang J, Yao S, Jin G, Du J, Han W, et al: LncRNA-FEZF1-AS1
promotes tumor proliferation and metastasis in colorectal cancer by
regulating PKM2 signaling. Clin Cancer Res. 24:4808–4819. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tang J, Yan T, Bao Y, Shen C, Yu C, Zhu X,
Tian X, Guo F, Liang Q, Liu Q, et al: LncRNA GLCC1 promotes
colorectal carcinogenesis and glucose metabolism by stabilizing
c-Myc. Nat Commun. 10:34992019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hua Q, Jin M, Mi B, Xu F, Li T, Zhao L,
Liu J and Huang G: LINC01123, a c-Myc-activated long non-coding
RNA, promotes proliferation and aerobic glycolysis of non-small
cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol.
12:912019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pan X, Tan J, Tao T, Zhang X, Weng Y, Weng
X, Xu J, Li H, Jiang Y, Zhou D and Shen Y: LINC01123 enhances
osteosarcoma cell growth by activating the Hedgehog pathway via the
miR-516b-5p/Gli1 axis. Cancer Sci. 112:2260–2271. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang M, Han Y, Zheng Y, Zhang Y, Zhao X,
Gao Z and Liu X: ZEB1-activated LINC01123 accelerates the
malignancy in lung adenocarcinoma through NOTCH signaling pathway.
Cell Death Dis. 11:9812020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li JH, Liu S, Zheng LL, Wu J, Sun WJ, Wang
ZL, Zhou H, Qu LH and Yang JH: Discovery of protein-lncRNA
interactions by integrating large-scale CLIP-Seq and RNA-Seq
datasets. Front Bioeng Biotechnol. 2:882015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu X, Yan F, Wang L, Sun G, Liu J, Qu M,
Wang Y and Li T: MicroRNA: Another pharmacological avenue for
colorectal cancer? Front Cell Dev Biol. 8:8122020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang L, Zhang Y, Li Z, Zhao X, Xi Z, Chen
H, Shi H, Xin T, Shen R and Wang T: MiR-4319 suppresses colorectal
cancer progression by targeting ABTB1. United European
Gastroenterol J. 7:517–528. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao J, Li N, Dong Y, Li S, Xu L, Li X, Li
Y, Li Z, Ng SS, Sung JJ, et al: miR-34a-5p suppresses colorectal
cancer metastasis and predicts recurrence in patients with stage
II/III colorectal cancer. Oncogene. 34:4142–4152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li S, Zhu K, Liu L, Gu J, Niu H and Guo J:
lncARSR sponges miR-34a-5p to promote colorectal cancer invasion
and metastasis via hexokinase-1-mediated glycolysis. Cancer Sci.
111:3938–3952. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, Zeng C, Hu J, Pan Y, Shan Y, Liu B
and Jia L: Long non-coding RNA-SNHG7 acts as a target of miR-34a to
increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in
colorectal cancer progression. J Hematol Oncol. 11:892018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rokavec M, Öner MG, Li H, Jackstadt R,
Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et
al: IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated
colorectal cancer invasion and metastasis. J Clin Invest.
124:1853–1867. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gatenby RA and Gillies RJ: Why do cancers
have high aerobic glycolysis? Nat Rev Cancer. 4:891–899. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ji S, Zhang B, Liu J, Qin Y, Liang C, Shi
S, Jin K, Liang D, Xu W, Xu H, et al: ALDOA functions as an
oncogene in the highly metastatic pancreatic cancer. Cancer Lett.
374:127–135. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fu H, Gao H, Qi X, Zhao L, Wu D, Bai Y, Li
H, Liu X, Hu J and Shao S: Aldolase A promotes proliferation and
G1/S transition via the EGFR/MAPK pathway in non-small
cell lung cancer. Cancer Commun (Lond). 38:182018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kukita A, Yoshida MC, Fukushige S,
Sakakibara M, Joh K, Mukai T and Hori K: Molecular gene mapping of
human aldolase A (ALDOA) gene to chromosome 16. Hum Genet.
76:20–26. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li H, Zhang X, Jin Z, Yin T, Duan C, Sun
J, Xiong R and Li Z: MiR-122 promotes the development of colon
cancer by targeting ALDOA in vitro. Technol Cancer Res Treat.
18:15330338198713002019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dai L, Pan G, Liu X, Huang J, Jiang Z, Zhu
X, Gan X, Xu Q and Tan N: High expression of ALDOA and DDX5 are
associated with poor prognosis in human colorectal cancer. Cancer
Manag Res. 10:1799–1806. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang XJ, Wang W and Hann SS: Interactions
among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie.
163:58–72. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Heneghan HM, Miller N and Kerin MJ: MiRNAs
as biomarkers and therapeutic targets in cancer. Curr Opin
Pharmacol. 10:543–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun DE and Ye SY: Emerging roles of long
noncoding RNA regulator of reprogramming in cancer treatment.
Cancer Manag Res. 12:6103–6112. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin X, Zhuang S, Chen X, Du J, Zhong L,
Ding J, Wang L, Yi J, Hu G, Tang G, et al: lncRNA ITGB8-AS1
functions as a ceRNA to promote colorectal cancer growth and
migration through integrin-mediated focal adhesion signaling. Mol
Ther. 30:688–702. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan
C, Xu M, Sun H, Liu C, Wei P and Du X: The lncRNA NEAT1 activates
Wnt/β-catenin signaling and promotes colorectal cancer progression
via interacting with DDX5. J Hematol Oncol. 11:1132018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qie S and Diehl JA: Cyclin D1, cancer
progression, and opportunities in cancer treatment. J Mol Med
(Berl). 94:1313–1326. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dragutinović VV, Radonjić NV, Petronijević
ND, Tatić SB, Dimitrijević IB, Radovanović NS and Krivokapić ZV:
Matrix metalloproteinase-2 (MMP-2) and −9 (MMP-9) in preoperative
serum as independent prognostic markers in patients with colorectal
cancer. Mol Cell Biochem. 355:173–178. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bao Y, Lu Y, Feng W, Yu H, Guo H, Tao Y,
Shi Q, Chen W and Wang X: COUP-TFII promotes epithelial-mesenchymal
transition by inhibiting miR-34a expression in colorectal cancer.
Int J Oncol. 54:1337–1344. 2019.PubMed/NCBI
|
|
37
|
Lunt SY and Vander Heiden MG: Aerobic
glycolysis: Meeting the metabolic requirements of cell
proliferation. Annu Rev Cell Dev Biol. 27:441–464. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tochio T, Tanaka H, Nakata S and Hosoya H:
Fructose-1,6-bisphosphate aldolase A is involved in HaCaT cell
migration by inducing lamellipodia formation. J Dermatol Sci.
58:123–129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kawai K, Uemura M, Munakata K, Takahashi
H, Haraguchi N, Nishimura J, Hata T, Matsuda C, Ikenaga M, Murata
K, et al: Fructose-bisphosphate aldolase A is a key regulator of
hypoxic adaptation in colorectal cancer cells and involved in
treatment resistance and poor prognosis. Int J Oncol. 50:525–534.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li X, Zhao S, Fu Y, Zhang P, Zhang Z,
Cheng J, Liu L and Jiang H: miR-34a-5p functions as a tumor
suppressor in head and neck squamous cell cancer progression by
targeting Flotillin-2. Int J Biol Sci. 17:4327–4339. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang X, Sun Q, Song Y and Li W: circHUWE1
exerts an oncogenic role in inducing DDP-resistant NSCLC
progression depending on the regulation of miR-34a-5p/TNFAIP8. Int
J Genomics. 2021:39970452021. View Article : Google Scholar : PubMed/NCBI
|