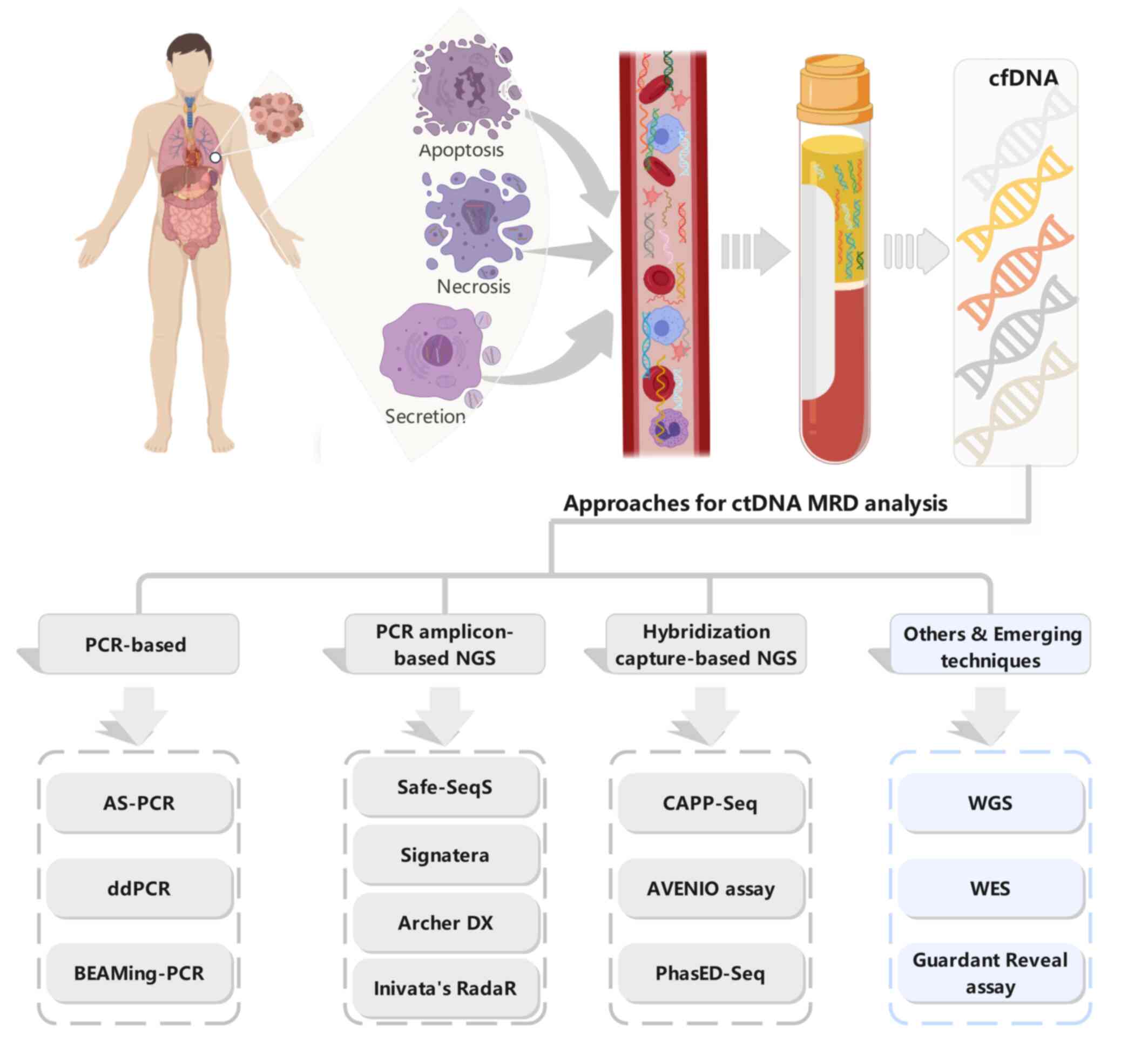
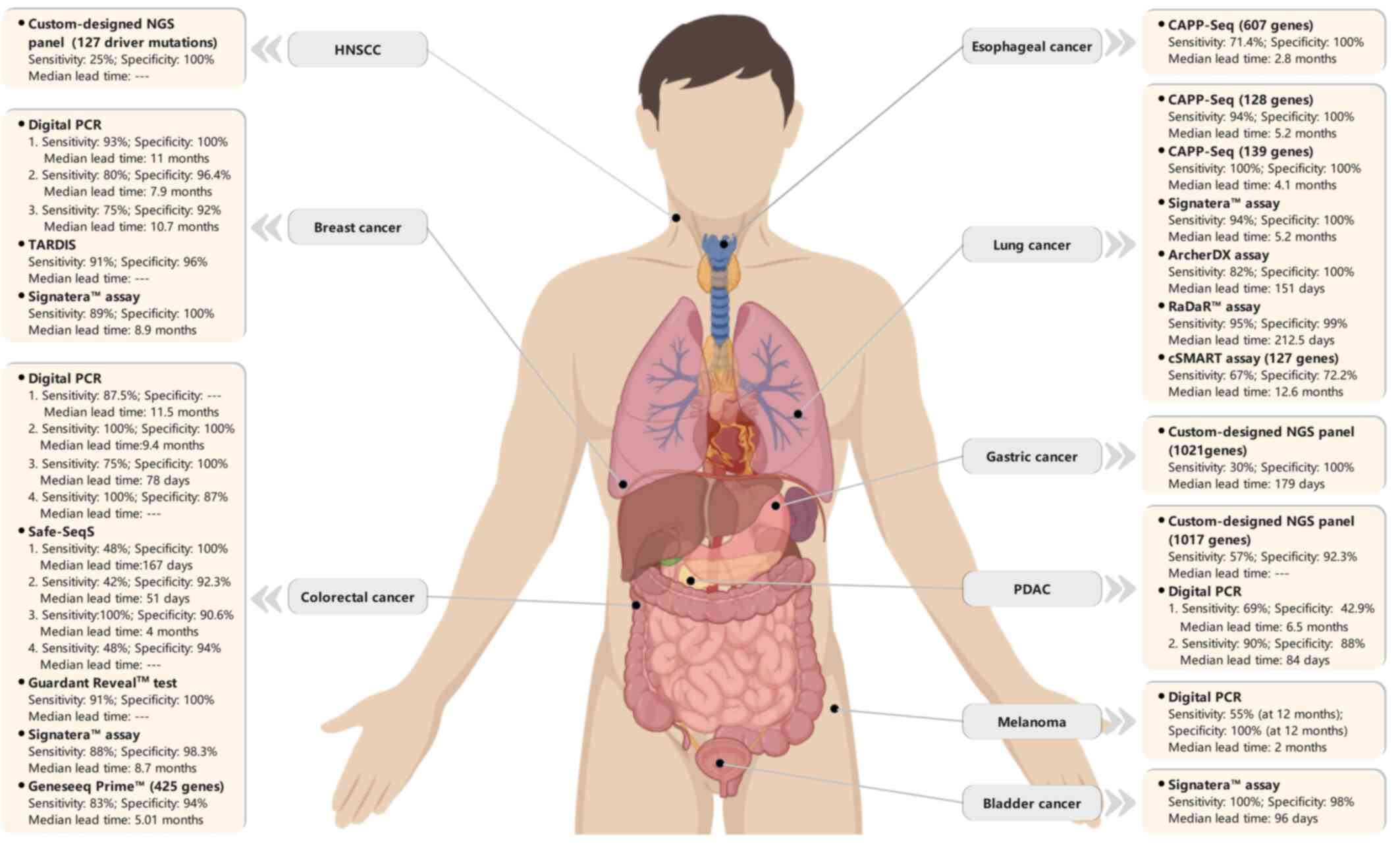
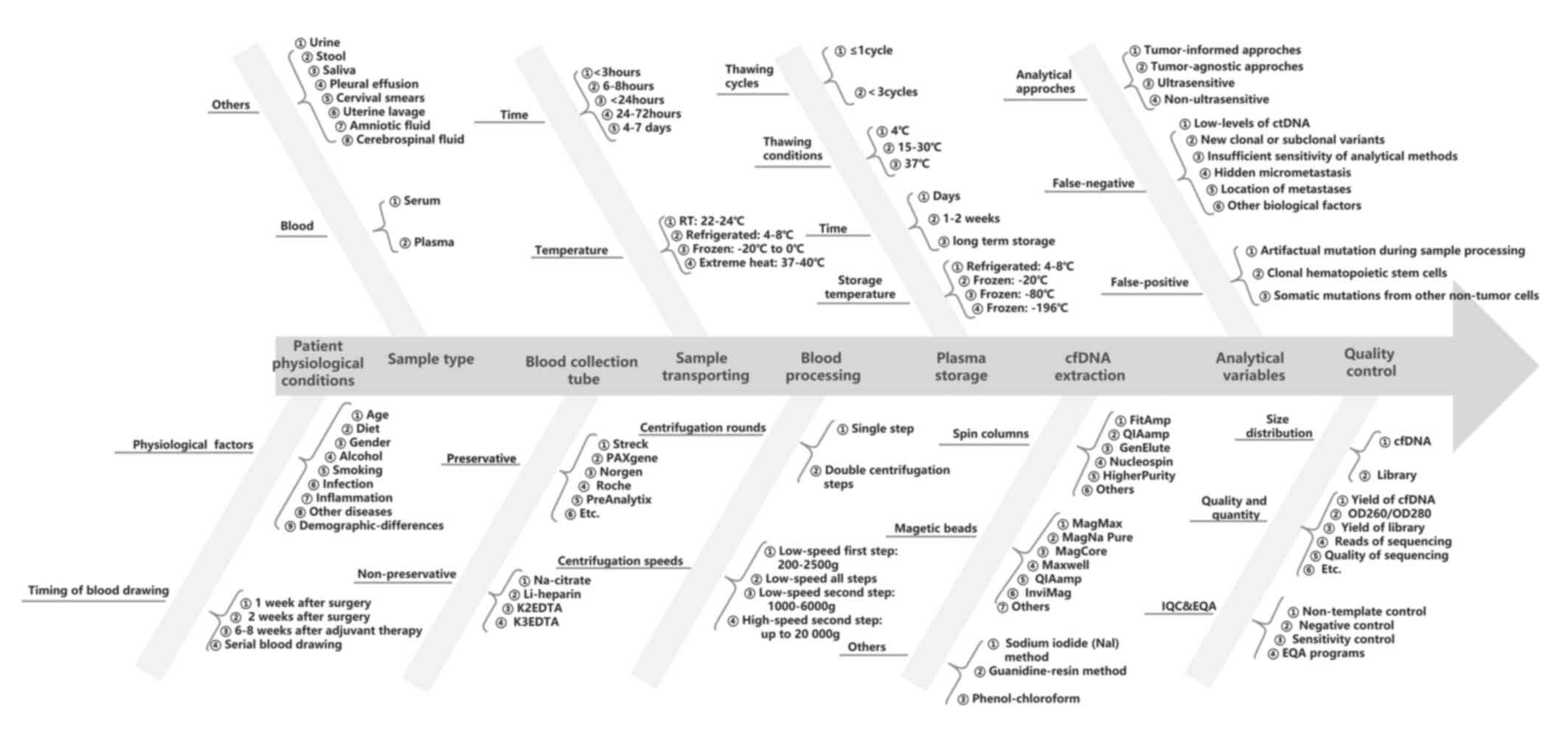
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qiu B, Guo W, Zhang F, Lv F, Ji Y, Peng Y,
Chen X, Bao H, Xu Y, Shao Y, et al: Dynamic recurrence risk and
adjuvant chemotherapy benefit prediction by ctDNA in resected
NSCLC. Nat Commun. 12:6770–6780. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li H, Ma ZL, Li B, Pan YJ, Xiang JQ, Zhang
YW, Sun YH, Hou T, Lizaso A, Chen Y, et al: Potential utility of
longitudinal somatic mutation and methylation profiling for
predicting molecular residual disease in postoperative non-small
cell lung cancer patients. Cancer Med. 10:8377–8386. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chaudhuri AA, Chabon JJ, Lovejoy AF,
Newman AM, Stehr H, Azad TD, Khodadoust MS, Esfahani MS, Liu CL,
Zhou L, et al: Early detection of molecular residual disease in
localized lung cancer by circulating tumor DNA profiling. Cancer
Discov. 7:1394–1403. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gale D, Heider K, Ruiz-Valdepenas A,
Hackinger S, Perry M, Marsico G, Rundell V, Wulff J, Sharma G,
Knock H, et al: Residual ctDNA after treatment predicts early
relapse in patients with early-stage non-small cell lung cancer.
Ann Oncol. 33:500–510. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tarazona N, Gimeno-Valiente F, Gambardella
V, Zuñiga S, Rentero-Garrido P, Huerta M, Roselló S,
Martinez-Ciarpaglini C, Carbonell-Asins JA, Carrasco F, et al:
Targeted next-generation sequencing of circulating-tumor DNA for
tracking minimal residual disease in localized colon cancer. Ann
Oncol. 30:1804–1812. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gögenur M, Hadi NA, Qvortrup C, Andersen
CL and Gögenur I: ctDNA for risk of recurrence assessment in
patients treated with neoadjuvant treatment: A systematic review
and meta-analysis. Ann Surg Oncol. 29:8666–8674. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cullinane C, Fleming C, O'Leary DP, Hassan
F, Kelly L, O'Sullivan MJ, Corrigan MA and Redmond HP: Association
of circulating tumor DNA with disease-free survival in breast
cancer: A systematic review and meta-analysis. JAMA Netw Open.
3:e20269212020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pantel K and Alix-Panabières C: Liquid
biopsy and minimal residual disease-latest advances and
implications for cure. Nat Rev Clin Oncol. 16:409–424. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Heitzer E, Haque IS, Roberts CES and
Speicher MR: Current and future perspectives of liquid biopsies in
genomics-driven oncology. Nat Rev Genet. 20:71–88. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reinert T, Henriksen TV, Christensen E,
Sharma S, Salari R, Sethi H, Knudsen M, Nordentoft I, Wu HT, Tin
AS, et al: Analysis of plasma cell-free DNA by ultradeep sequencing
in patients with stages I to III colorectal cancer. JAMA Oncol.
5:1124–1131. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang J, Gong Y, Lam VK, Shi Y, Guan Y,
Zhang Y, Ji L, Chen Y, Zhao Y, Qian F, et al: Deep sequencing of
circulating tumor DNA detects molecular residual disease and
predicts recurrence in gastric cancer. Cell Death Dis. 11:346–354.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
MandeL P and Metais P: Les acides
nucléiques du plasma sanguin chez l'homme (Nuclear Acids in Human
Blood Plasma). C R Seances Soc Biol Fil. 142:241–243. 1948.(In
French). PubMed/NCBI
|
|
14
|
Jahr S, Hentze H, Englisch S, Hardt D,
Fackelmayer FO, Hesch RD and Knippers R: DNA fragments in the blood
plasma of cancer patients: Quantitations and evidence for their
origin from apoptotic and necrotic cells. Cancer Res. 61:1659–1665.
2001.PubMed/NCBI
|
|
15
|
Thakur BK, Zhang H, Becker A, Matei I,
Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, et
al: Double-stranded DNA in exosomes: A novel biomarker in cancer
detection. Cell Res. 24:766–779. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chin RI, Chen K, Usmani A, Chua C, Harris
PK, Binkley MS, Azad TD, Dudley JC and Chaudhuri AA: Detection of
solid tumor molecular residual disease (MRD) using circulating
tumor DNA (ctDNA). Mol Diagn Ther. 23:311–331. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei F, Lin CC, Joon A, Feng Z, Troche G,
Lira ME, Chia D, Mao M, Ho CL, Su WC, et al: Noninvasive
saliva-based EGFR gene mutation detection in patients with lung
cancer. Am J Respir Crit Care Med. 190:1117–1126. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dudley JC, Schroers-Martin J, Lazzareschi
DV, Shi WY, Chen SB, Esfahani MS, Trivedi D, Chabon JJ, Chaudhuri
AA, Stehr H, et al: Detection and surveillance of bladder cancer
using urine tumor DNA. Cancer Discov. 9:500–509. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
De Mattos-Arruda L, Mayor R, Ng CKY,
Weigelt B, Martínez-Ricarte F, Torrejon D, Oliveira M, Arias A,
Raventos C, Tang J, et al: Cerebrospinal fluid-derived circulating
tumour DNA better represents the genomic alterations of brain
tumours than plasma. Nat Commun. 6:88392015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Husain H, Nykin D, Bui N, Quan D, Gomez G,
Woodward B, Venkatapathy S, Duttagupta R, Fung E, Lippman SM, et
al: Cell-free DNA from ascites and pleural effusions: Molecular
insights into genomic aberrations and disease biology. Mol Cancer
Ther. 16:948–955. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang P and Lo Y MD: The long and short of
circulating cell-free DNA and the ins and outs of molecular
diagnostics. Trends Genet. 32:360–371. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yao W, Mei C, Nan X and Hui L: Evaluation
and comparison of in vitro degradation kinetics of DNA in serum,
urine and saliva: A qualitative study. Gene. 590:142–148. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lo YM, Zhang J, Leung TN, Lau TK, Chang AM
and Hjelm NM: Rapid clearance of fetal DNA from maternal plasma. Am
J Hum Genet. 64:218–224. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wong FC, Sun K, Jiang P, Cheng YK, Chan
KC, Leung TY, Chiu RW and Lo YM: Cell-free DNA in maternal plasma
and serum: A comparison of quantity, quality and tissue origin
using genomic and epigenomic approaches. Clin Biochem.
49:1379–1386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lam WKJ, Gai W, Sun K, Wong RSM, Chan RWY,
Jiang P, Chan NPH, Hui WWI, Chan AWH, Szeto CC, et al: DNA of
erythroid origin is present in human plasma and informs the types
of anemia. Clin Chem. 63:1614–1623. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Razavi P, Li BT, Brown DN, Jung B, Hubbell
E, Shen R, Abida W, Juluru K, De Bruijn I, Hou C, et al:
High-intensity sequencing reveals the sources of plasma circulating
cell-free DNA variants. Nat Med. 25:1928–1937. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Breitbach S, Tug S, Helmig S, Zahn D,
Kubiak T, Michal M, Gori T, Ehlert T, Beiter T and Simon P: Direct
quantification of cell-free, circulating DNA from unpurified
plasma. PLoS One. 9:e878382014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tug S, Helmig S, Deichmann ER,
Schmeier-Jürchott A, Wagner E, Zimmermann T, Radsak M, Giacca M and
Simon P: Exercise-induced increases in cell free DNA in human
plasma originate predominantly from cells of the haematopoietic
lineage. Exerc Immunol Rev. 21:164–173. 2015.PubMed/NCBI
|
|
29
|
Lo YM, Corbetta N, Chamberlain PF, Rai V,
Sargent IL, Redman CW, Redman CW and Wainscoat JS: Presence of
fetal DNA in maternal plasma and serum. Lancet. 350:485–487. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leon SA, Shapiro B, Sklaroff DM and Yaros
MJ: Free DNA in the serum of cancer patients and the effect of
therapy. Cancer Res. 37:646–450. 1977.PubMed/NCBI
|
|
31
|
Stroun M, Anker P, Maurice P, Lyautey J,
Lederrey C and Beljanski M: Neoplastic characteristics of the DNA
found in the plasma of cancer patients. Oncology. 46:318–322. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Diehl F, Schmidt K, Choti MA, Romans K,
Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al:
Circulating mutant DNA to assess tumor dynamics. Nat Med.
14:985–990. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo H, Wei W, Ye Z, Zheng J and Xu RH:
Liquid biopsy of methylation biomarkers in cell-free DNA. Trends
Mol Med. 27:482–500. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Underhill HR, Kitzman JO, Hellwig S,
Welker NC, Daza R, Baker DN, Gligorich KM, Rostomily RC, Bronner MP
and Shendure J: Fragment length of circulating tumor DNA. PLoS
Genet. 12:e10061622016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dressman D, Yan H, Traverso G, Kinzler KW
and Vogelstein B: Transforming single DNA molecules into
fluorescent magnetic particles for detection and enumeration of
genetic variations. Proc Natl Acad Sci USA. 100:8817–8822. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tie J, Cohen JD, Wang Y, Christie M,
Simons K, Lee M, Wong R, Kosmider S, Ananda S, McKendrick J, et al:
Circulating tumor DNA analyses as markers of recurrence risk and
benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol.
5:1710–1717. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kurtz DM, Soo J, Co Ting Keh L, Alig S,
Chabon JJ, Sworder BJ, Schultz A, Jin MC, Scherer F, Garofalo A, et
al: Enhanced detection of minimal residual disease by targeted
sequencing of phased variants in circulating tumor DNA. Nat
Biotechnol. 39:1537–1547. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
McDuff SGR, Hardiman KM, Ulintz PJ, Parikh
AR, Zheng H, Kim DW, Lennerz JK, Hazar-Rethinam M, Van Seventer EE,
Fetter IJ, et al: Circulating tumor DNA predicts pathologic and
clinical outcomes following neoadjuvant chemoradiation and surgery
for patients with locally advanced rectal cancer. JCO Precis Oncol.
5:PO.20.00220. 2021.PubMed/NCBI
|
|
39
|
Guerrini F, Paolicchi M, Ghio F, Ciabatti
E, Grassi S, Salehzadeh S, Ercolano G, Metelli MR, Del Re M, Iovino
L, et al: The droplet digital PCR: A new valid molecular approach
for the assessment of B-RAF V600E mutation in hairy cell
leukemia. Front Pharmacol. 7:3632016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huerta M, Roselló S, Sabater L, Ferrer A,
Tarazona N, Roda D, Gambardella V, Alfaro-Cervelló C, Garcés-Albir
M, Cervantes A and Ibarrola-Villava M: Circulating tumor DNA
detection by digital-droplet PCR in pancreatic ductal
adenocarcinoma: A systematic review. Cancers (Basel). 13:9942021.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Elazezy M and Joosse SA: Techniques of
using circulating tumor DNA as a liquid biopsy component in cancer
management. Comput Struct Biotechnol J. 16:370–378. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hu Y, Ulrich BC, Supplee J, Kuang Y,
Lizotte PH, Feeney NB, Guibert NM, Awad MM, Wong KK, Jänne PA, et
al: False-positive plasma genotyping due to clonal hematopoiesis.
Clin Cancer Res. 24:4437–4443. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Abbosh C, Swanton C and Birkbak NJ: Clonal
haematopoiesis: A source of biological noise in cell-free DNA
analyses. Ann Oncol. 30:358–359. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Flach S, Howarth K, Hackinger S, Pipinikas
C, Ellis P, McLay K, Marsico G, Forshew T, Walz C, Reichel CA, et
al: Liquid BIOpsy for MiNimal RESidual DiSease Detection in head
and neck squamous cell carcinoma (LIONESS)-a personalised
circulating tumour DNA analysis in head and neck squamous cell
carcinoma. Br J Cancer. 126:1186–1195. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
McDonald BR, Contente-Cuomo T, Sammut SJ,
Odenheimer-Bergman A, Ernst B, Perdigones N, Chin SF, Farooq M,
Mejia R, Cronin PA, et al: Personalized circulating tumor DNA
analysis to detect residual disease after neoadjuvant therapy in
breast cancer. Sci Transl Med. 11:eaax73922019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Magbanua MJM, Swigart LB, Wu HT, Hirst GL,
Yau C, Wolf DM, Tin A, Salari R, Shchegrova S, Pawar H, et al:
Circulating tumor DNA in neoadjuvant-treated breast cancer reflects
response and survival. Ann Oncol. 32:229–239. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Loomis B, Ishii J, Wolchok JD, Boland G,
Robine N, Altorki NK and Landau DA: Genome-wide cell-free DNA
mutational integration enables ultra-sensitive cancer monitoring.
Nat Med. 26:1114–1124. 2020. View Article : Google Scholar
|
|
48
|
Wan JCM, Heider K, Gale D, Murphy S,
Fisher E, Mouliere F, Ruiz-Valdepenas A, Santonja A, Morris J,
Chandrananda D, et al: ctDNA monitoring using patient-specific
sequencing and integration of variant reads. Sci Transl Med.
12:eaaz80842020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kasi PM, Fehringer G, Taniguchi H,
Starling N, Nakamura Y, Kotani D, Powles T, Li BT, Pusztai L,
Aushev VN, et al: Impact of circulating tumor DNA-Based detection
of molecular residual disease on the conduct and design of clinical
trials for solid tumors. JCO Precis Oncol. 6:e21001812022.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Abbosh C, Birkbak NJ, Wilson GA,
Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA,
Veeriah S, Rosenthal R, et al: Phylo genetic ctDNA analysis depicts
early-stage lung cancer evolution. Nature. 545:446–451. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Olsson E, Winter C, George A, Chen Y,
Howlin J, Tang MH, Dahlgren M, Schulz R, Grabau D, van Westen D, et
al: Serial monitoring of circulating tumor DNA in patients with
primary breast cancer for detection of occult metastatic disease.
EMBO Mol Med. 7:1034–1047. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zehir A, Benayed R, Shah RH, Syed A,
Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et
al: Mutational landscape of metastatic cancer revealed from
prospective clinical sequencing of 10,000 patients. Nat Med.
23:703–713. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Honoré N, Galot R, van Marcke C, Limaye N
and Machiels JP: Liquid biopsy to detect minimal residual disease:
Methodology and impact. Cancers (Basel). 13:53642021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Deveson IW, Gong B, Lai K, LoCoco JS,
Richmond TA, Schageman J, Zhang Z, Novoradovskaya N, Willey JC,
Jones W, et al: Evaluating the analytical validity of circulating
tumor DNA sequencing assays for precision oncology. Nat Biotechnol.
39:1115–1128. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Stahlberg A, Krzyzanowski PM, Egyud M,
Filges S, Stein L and Godfrey TE: Simple multiplexed PCR-based
barcoding of DNA for ultrasensitive mutation detection by
next-generation sequencing. Nat Protoc. 12:664–682. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Newman AM, Bratman SV, To J, Wynne JF,
Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, et
al: An ultrasensitive method for quantitating circulating tumor DNA
with broad patient coverage. Nat Med. 20:548–554. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tewhey R, Warner JB, Nakano M, Libby B,
Medkova M, David PH, Kotsopoulos SK, Samuels ML, Hutchison JB,
Larson JW, et al: Microdroplet-based PCR enrichment for large-scale
targeted sequencing. Nat Biotechnol. 27:1025–1031. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dudley JC and Diehn M: Detection and
diagnostic utilization of cellular and cell-free tumor DNA. Annu
Rev Pathol. 16:199–222. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Parikh AR, Van Seventer EE, Siravegna G,
Hartwig AV, Jaimovich A, He Y, Kanter K, Fish MG, Fosbenner KD,
Miao B, et al: Minimal residual disease detection using a
plasma-only circulating tumor DNA assay in patients with colorectal
cancer. Clin Cancer Res. 27:5586–5594. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sethi H, Salari R, Navarro S, Natarajan P,
Srinivasan R, Dashner S, Tin T, Balcioglu M, Swenerton R and
Zimmermann B: Abstract 4542: Analytical validation of the
SignateraTM RUO assay, a highly sensitive patient-specific
multiplex PCR NGS-based noninvasive cancer recurrence detection and
therapy monitoring assay. Cancer Res. 78:45422018. View Article : Google Scholar
|
|
61
|
Christensen E, Birkenkamp-Demtröder K,
Sethi H, Shchegrova S, Salari R, Nordentoft I, Wu HT, Knudsen M,
Lamy P, Lindskrog SV, et al: Early detection of metastatic relapse
and monitoring of therapeutic efficacy by Ultra-Deep sequencing of
plasma cell-Free DNA in patients with urothelial bladder carcinoma.
J Clin Oncol. 37:1547–1557. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Jamal-Hanjani M, Wilson GA, McGranahan N,
Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al: Tracking the evolution of non-small-cell lung
cancer. N Engl J Med. 376:2109–2121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Coombes RC, Page K, Salari R, Hastings RK,
Armstrong A, Ahmed S, Ali S, Cleator S, Kenny L, Stebbing J, et al:
Personalized detection of circulating tumor DNA antedates breast
cancer metastatic recurrence. Clin Cancer Res. 25:4255–4263. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lipsyc-Sharf M, de Bruin EC, Santos K,
McEwen R, Stetson D, Patel A, Kirkner GJ, Hughes ME, Tolaney SM,
Partridge AH, et al: Circulating Tumor DNA and Late Recurrence in
High-Risk Hormone Receptor-Positive, Human Epidermal Growth Factor
Receptor 2-Negative Breast Cancer. J Clin Oncol. 40:2408–2419.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li R, Bonora G, Dai C, Xiang B, Zheng T,
Mo W, Wang X, Zhou K, Jia S, Luo S, et al: 911P the development and
application of a baseline-agnostic minimal residual disease assay.
Ann Oncol. 33:S9642022. View Article : Google Scholar
|
|
66
|
Pellini B, Pejovic N, Feng W, Earland N,
Harris PK, Usmani A, Szymanski JJ, Qaium F, Mudd J, Petty M, et al:
ctDNA MRD detection and personalized oncogenomic analysis in
oligometastatic colorectal cancer from plasma and urine. JCO Precis
Oncol. 5:PO.20.00276. 2021.PubMed/NCBI
|
|
67
|
Abbosh C, Frankell A, Garnett A, Harrison
T, Weichert M, Licon A, Veeriah S, Daber B, Moreau M, Chesh A, et
al: Abstract CT023: Phylogenetic tracking and minimal residual
disease detection using ctDNA in early-stage NSCLC: A lung TRACERx
study. Cancer Res. 80:CT0232020. View Article : Google Scholar
|
|
68
|
Peng M, Huang Q, Yin W, Tan S, Chen C, Liu
W, Tang J, Wang X, Zhang B, Zou M, et al: Circulating tumor DNA as
a prognostic biomarker in localized Non-small Cell lung cancer.
Front Oncol. 10:5615982020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Groot VP, Mosier S, Javed AA, Teinor JA,
Gemenetzis G, Ding D, Haley LM, Yu J, Burkhart RA, Hasanain A, et
al: Circulating tumor DNA as a clinical test in resected pancreatic
cancer. Clin Cancer Res. 25:4973–4984. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Duineveld LA, van Asselt KM, Bemelman WA,
Smits AB, Tanis PJ, van Weert HC and Wind J: Symptomatic and
asymptomatic colon cancer recurrence: A multicenter cohort study.
Ann Fam Med. 14:215–220. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tie J, Wang Y, Tomasetti C, Li L, Springer
S, Kinde I, Silliman N, Tacey M, Wong HL, Christie M, et al:
Circulating tumor DNA analysis detects minimal residual disease and
predicts recurrence in patients with stage II colon cancer. Sci
Transl Med. 8:346ra922016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Tie J, Wang Y, Cohen J, Li L, Hong W,
Christie M, Wong HL, Kosmider S, Wong R, Thomson B, et al:
Circulating tumor DNA dynamics and recurrence risk in patients
undergoing curative intent resection of colorectal cancer liver
metastases: A prospective cohort study. PLoS Med. 18:e10036202021.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Garcia-Murillas I, Schiavon G, Weigelt B,
Ng C, Hrebien S, Cutts RJ, Cheang M, Osin P, Nerurkar A, Kozarewa
I, et al: Mutation tracking in circulating tumor DNA predicts
relapse in early breast cancer. Sci Transl Med. 7:302ra13322015.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chen YH, Hancock BA, Solzak JP, Brinza D,
Scafe C, Miller KD and Radovich M: Next-generation sequencing of
circulating tumor DNA to predict recurrence in triple-negative
breast cancer patients with residual disease after neoadjuvant
chemotherapy. NPJ Breast Cancer. 3:242017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Garcia-Murillas I, Chopra N, Comino-Méndez
I, Beaney M, Tovey H, Cutts RJ, Swift C, Kriplani D, Afentakis M,
Hrebien S, et al: Assessment of molecular relapse detection in
early-stage breast cancer. JAMA Oncol. 5:1473–1478. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Parsons HA, Rhoades J, Reed SC, Gydush G,
Ram P, Exman P, Xiong K, Lo CC, Li T, Fleharty M, et al: Sensitive
detection of minimal residual disease in patients treated for
early-stage breast cancer. Clin Cancer Res. 26:2556–2564. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Azad TD, Chaudhuri AA, Fang P, Qiao Y,
Esfahani MS, Chabon JJ, Hamilton EG, Yang YD, Lovejoy A, Newman AM,
et al: Circulating tumor DNA analysis for detection of minimal
residual disease after chemoradiotherapy for localized esophageal
cancer. Gastroenterology. 158:494–505. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Jiang J, Ye S, Xu Y, Chang L, Hu X, Ru G,
Guo Y, Yi X, Yang L and Huang D: Circulating tumor DNA as a
potential marker to detect minimal residual disease and predict
recurrence in pancreatic cancer. Front Oncol. 10:12202020.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Leal A, van Grieken NCT, Palsgrove DN,
Phallen J, Medina JE, Hruban C, Broeckaert MAM, Anagnostou V,
Adleff V, Bruhm DC, et al: White blood cell and cell-free DNA
analyses for detection of residual disease in gastric cancer. Nat
Commun. 11:5252020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Hilke FJ, Muyas F, Admard J, Kootz B, Nann
D, Welz S, Rieß O, Zips D, Ossowski S, Schroeder C and Clasen K:
Dynamics of cell-free tumour DNA correlate with treatment response
of head and neck cancer patients receiving radiochemo therapy.
Radiother Oncol. 151:182–189. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Tan L, Sandhu S, Lee RJ, Li J, Callahan J,
Ftouni S, Dhomen N, Middlehurst P, Wallace A, Raleigh J, et al:
Prediction and monitoring of relapse in stage III melanoma using
circulating tumor DNA. Ann Oncol. 30:804–814. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Moding EJ, Nabet BY, Alizadeh AA and Diehn
M: Detecting liquid remnants of solid tumors: circulating tumor DNA
minimal residual disease. Cancer Discov. 11:2968–2986. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Pascual J, Attard G, Bidard FC, Curigliano
G, De Mattos-Arruda L, Diehn M, Italiano A, Lindberg J, Merker JD,
Montagut C, et al: ESMO recommendations on the use of circulating
tumour DNA assays for patients with cancer: A report from the ESMO
Precision Medicine Working Group. Ann Oncol. 33:750–768. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Andersson D, Kristiansson H, Kubista M and
Stahlberg A: Ultrasensitive circulating tumor DNA analysis enables
precision medicine: Experimental workflow considerations. Expert
Rev Mol Diagn. 21:299–310. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Nikolaev S, Lemmens L, Koessler T, Blouin
JL and Nouspikel T: Circulating tumoral DNA: Preanalytical
validation and quality control in a diagnostic laboratory. Anal
Biochem. 542:34–39. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ungerer V, Bronkhorst AJ and Holdenrieder
S: Preanalytical variables that affect the outcome of cell-free DNA
measurements. Crit Rev Clin Lab Sci. 57:484–507. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Risberg B, Tsui DWY, Biggs H,
Ruiz-Valdepenas Martin de Almagro A, Dawson SJ, Hodgkin C, Jones L,
Parkinson C, Piskorz A, Marass F, et al: Effects of collection and
processing procedures on plasma circulating cell-free DNA from
cancer patients. J Mol Diagn. 20:883–892. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Chiu RW, Lui WB, El-Sheikhah A, Chan AT,
Lau TK, Nicolaides KH and Lo YM: Comparison of protocols for
extracting circulating DNA and RNA from maternal plasma. Clin Chem.
51:2209–2210. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Vidal J, Muinelo L, Dalmases A, Jones F,
Edelstein D, Iglesias M, Orrillo M, Abalo A, Rodríguez C, Brozos E,
et al: Plasma ctDNA RAS mutation analysis for the diagnosis and
treatment monitoring of metastatic colorectal cancer patients. Ann
Oncol. 28:1325–1332. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Tie J, Cohen JD, Lo SN, Wang Y, Li L,
Christie M, Lee M, Wong R, Kosmider S, Skinner I, et al: Prognostic
significance of postsurgery circulating tumor DNA in nonmetastatic
colorectal cancer: Individual patient pooled analysis of three
cohort studies. Int J Cancer. 148:1014–1026. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Abbosh C, Birkbak NJ and Swanton C: Early
stage NSCLC-challenges to implementing ctDNA-based screening and
MRD detection. Nat Rev Clin Oncol. 15:577–586. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Mansukhani S, Barber LJ, Kleftogiannis D,
Moorcraft SY, Davidson M, Woolston A, Proszek PZ, Griffiths B,
Fenwick K, Herman B, et al: Ultra-sensitive mutation detection and
genome-wide DNA copy number reconstruction by error-corrected
circulating tumor DNA sequencing. Clin Chem. 64:1626–1635. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Bolton KL, Ptashkin RN, Gao T, Braunstein
L, Devlin SM, Kelly D, Patel M, Berthon A, Syed A, Yabe M, et al:
Cancer therapy shapes the fitness landscape of clonal
hematopoiesis. Nat Genet. 52:1219–1226. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Coombs CC, Zehir A, Devlin SM, Kishtagari
A, Syed A, Jonsson P, Hyman DM, Solit DB, Robson ME, Baselga J, et
al: Therapy-related clonal hematopoiesis in patients with
non-hematologic cancers is common and associated with adverse
clinical outcomes. Cell Stem Cell. 21:374–382. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Chabon JJ, Hamilton EG, Kurtz DM, Esfahani
MS, Moding EJ, Stehr H, Schroers-Martin J, Nabet BY, Chen B,
Chaudhuri AA, et al: Integrating genomic features for non-invasive
early lung cancer detection. Nature. 580:245–251. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chen K, Shields MD, Chauhan PS, Ramirez
RJ, Harris PK, Reimers MA, Zevallos JP, Davis AA, Pellini B and
Chaudhuri AA: Commercial ctDNA assays for minimal residual disease
detection of solid tumors. Mol Diagn Ther. 25:757–774. 2021.
View Article : Google Scholar : PubMed/NCBI
|