Introduction
Esophageal carcinoma is one of the leading causes of
cancer-related death worldwide. According to the Global Cancer
Statistics 2020, esophageal carcinoma is the seventh most common
and sixth most lethal type of cancer (1). Two most common subtypes of esophageal
cancer are esophageal squamous cell carcinoma (ESCC) and esophageal
adenocarcinoma, and the prevalence varies by country (2). ~90% of esophageal cancer cases are
ESCC, which has a poor prognosis and a high mortality rate
(3). It is worth noting that China
accounts for ~50% of all esophageal cancer cases worldwide, and in
certain areas, ESCC is one of the leading causes of cancer-related
death (4,5). The prognosis of patients with ESCC is
poor, with <20% of patients surviving >5 years after
diagnosis (6). Therefore, there is
a need to identify novel therapeutic targets in ESCC.
Aurora kinase A (AURKA) is a serine/threonine kinase
that is involved in mitosis progression, centrosome
maturation/separation, and mitotic spindle function. In addition to
its important role in the cell cycle, an increasing number of
studies have shown that AURKA participates in cancer development
and progression in several types of cancer and upregulated AURKA
expression has consistently been shown to be associated with a poor
prognosis, including in stomach (7), bladder (8), breast (9), colorectal (10) and ovarian cancer (11). AURKA can promote cancer cell
proliferation and migration, inhibit apoptosis, and is associated
with resistance to radiotherapy and chemotherapy. Given its
significance in cancer development, AURKA may serve as a candidate
for kinase inhibition in the management of cancer. Over the past
decades, a range of AURKA inhibitors have been developed and
several of these have been assessed in clinical studies (12).
Although several studies have indicated that AURKA
can act as a valuable biomarker and therapeutic target, there is a
lack of comprehensive research on AURKA expression, function, and
the molecular mechanism involved in ESCC. In the present study, it
was revealed that AURKA expression was upregulated in ESCC, and its
clinical value in cancer diagnosis was determined based on data
obtained from The Cancer Genome Atlas (TCGA) and tissue array
analysis. In addition, AURKA was identified to enhance ESCC cell
proliferation and colony formation in vitro and promoted
ESCC tumor growth in vivo. Both bioinformatics analysis and
pull-down assays identified TPX2 as an interacting and correlated
protein of AURKA. AURKA cooperated with TPX2 to promote ESCC
proliferation through the PI3K/Akt pathway. These findings
highlighted the AURKA/TPX2 axis as a useful candidate for ESCC
diagnosis and therapy.
Materials and methods
Cell culture and reagents
All ESCC cell lines (KYSE30, KYSE70, KYSE140,
KYSE150, KYSE410, KYSE450, KYSE510 and EC109) and 293T cells were
purchased from the Type Culture Collection of the Chinese Academy
of Sciences (Shanghai, China) and were cultured at 37°C in a 5%
CO2 humidified incubator and were not maintained for
>2 months. All ESCC cells were maintained in RPMI-1640, while
293T cells were cultured in DMEM supplemented with penicillin (100
units/ml), streptomycin (100 µg/ml) and 10% FBS (Biological
Industries). All cell lines were mycoplasma-free and were
authenticated using short tandem repeat DNA typing. The human
immortalized normal esophageal epithelial cell line SHEE was kindly
gifted by Dr Enmin Li from the Laboratory of Tumor Pathology
(Shantou University Medical College, Shantou, Guangdong, China).
Transfection was performed using Lipofectamine® 2000
(cat. no. 11668-019; Invitrogen; Thermo Fisher Scientific, Inc.)
for ESCC cell lines and Simple-Fect Reagent (Signaling Dawn
Biotech) for 293T cells according to the manufacturer's protocol.
AURKA inhibitor alisertib was purchased from Selleck Chemicals
(cat. no. S1133).
The antibodies used in the present study were as
follows: Anti-AURKA (cat. no. ab13824; Abcam), anti-Flag (cat. no.
F1804; MilliporeSigma), anti-HA (cat. no. 3724; Cell Signaling
Technology, Inc.), anti-His (cat. no. ab137839; Abcam),
anti-phosphorylated (p)-PI3K (Tyr467) (cat. no. sc-293115; Santa
Cruz Biotechnology, Inc.), anti-p-AKT (Ser473) (cat. no. 4060; Cell
Signaling Technology, Inc.), anti-p-Akt (Thr308) (cat. no. 13038;
Cell Signaling Technology, Inc.), anti-PI3Kp85 (cat. no. 4257; Cell
Signaling Technology, Inc.), anti-Akt (pan) (cat. no. 4691; Cell
Signaling Technology, Inc.), anti-p21 (cat. no. sc-397; Santa Cruz
Biotechnology, Inc.), anti-p27 (cat. no. 3686; Cell Signaling
Technology, Inc.), anti-p53 (cat. no. sc-126; Santa Cruz
Biotechnology, Inc.), anti-TPX2 (cat. no.122455; Cell Signaling
Technology, Inc.), anti-RPS6 (cat. no. 2217; Cell Signaling
Technology, Inc.), anti-HSP27 (cat. no. sc-13132; Santa Cruz
Biotechnology, Inc.), anti-VDAC (cat. no. 4866; Cell Signaling
Technology, Inc.), anti-RPL7 (cat. no. 2415; Cell Signaling
Technology, Inc.) and anti-GAPDH (cat. no. HRP-60004; ProteinTech
Group, Inc.).
AURKA knockdown using short hairpin
(sh)RNA
Gene knockdown was performed using specific shRNAs
kindly provided by Professor Xiang Li from Zhengzhou University.
The target sequence for shAURKA-1 is ‘TGGCTCTTAAAGTGTTATTTA’ and
for shAURKA-3 is ‘GCAGAGAACTGCTACTTATAT’. Briefly, 4 µg pMD2.G, 4
µg psPAX2 packaging plasmids, and 4 µg pLKO.1 plasmid containing
the specific shRNAs were transfected into 293T cells. A total of 2
days after transfection, the viral particles were collected and
filtered using a 0.45-mm filter. To obtain stable AURKA knockdown
ESCC cell lines, the lentivirus was transfected with a MOI of 50
using 8 mg/ml polybrene (cat. no. 107689; MilliporeSigma) and
selected using 2 µg/ml puromycin.
Western blotting
Cells were lysed using RIPA lysis buffer (50 mM
Tris-HCl pH 7.4, 1 mM EDTA, 0.25% deoxycholic acid disodium salt,
1% NP40, 150 mM NaCl and 0.1% SDS). After sonication, cells were
centrifuged at 12,000 × g for 15 min at 4°C. Protein concentration
was determined using a BCA assay. Then, 30 µg of cell extracts were
subjected to 10–12% SDS-PAGE electrophoresis and transferred to
PVDF membranes. Following protein transfer, the membranes were
blocked with 5% milk and then incubated with specific primary
antibodies (1:1,000) at 4°C overnight followed by a horseradish
peroxidase-conjugated secondary antibody (1:5,000; cat. nos. ab6721
and ab6728; Abcam) at room temperature for 2 h. Protein bands were
visualized using an enhanced chemiluminescence reagent (cat. no.
32106; Thermo Fisher Scientific, Inc.).
Immunoprecipitation (IP)
Cell lysates were prepared using lysis buffer
containing 50 mM Tris-HCl pH 7.4, 1 mM EDTA, 1% Triton X-100, 150
mM NaCl, protease inhibitors and PMSF for 30 min at 4°C and
centrifuged at 12,000 g for 15 min at 4°C. Co-IP was performed
using 2 µg antibodies and 500 mg lysate. Cell lysates were
incubated with specific antibodies at 4°C overnight and then
combined with 20 µl protein A/G agarose beads (cat. no. sc-2003;
Santa Cruz Biotechnology, Inc.) for 3 h at 4°C. The beads were
washed twice with lysis buffer and twice with PBS, and the immune
complexes were eluted with sample loading buffer for 5 min at 95°C.
The immunoprecipitated proteins were assessed using western
blotting.
MTT and colony formation assays
Viable cells were seeded into 96-well transparent
walled plates at a density of 1–2×103 cells per well,
depending on the growth kinetics of the specific cell line. Cell
proliferation was determined using MTT assays according to the
manufacturer's, protocol (cat. no. M1020; Beijing Solarbio Science
& Technology Co., Ltd.). Namely, 10% of 5 mg/ml MTT solution
was added to cells for a duration of 4 h at the time point of 0,
24, 48, 72, or 96 h. The medium was then discarded, and 110-µl
formazan solution was added to dissolve the MTT crystals. Cell
proliferation was next measured at an absorbance of 490 nm. For the
colony formation assay, 200 viable cancer cells were seeded into
six-well plates in 2-ml cell culture medium in triplicate. After
1–2 weeks depending on the growth kinetics size of the cell line,
colonies were stained with 0.2% crystal violet at room temperature
for 15 min and images were captured under the light microscope. The
number of colonies which contained at least 20 cells was counted
manually and analyzed.
Soft agar colony formation assay
A total of 8×103 viable cancer cells were
seeded in triplicate in 1-ml cell culture medium with 2 mM
glutamine, 5 µg/ml gentamycin and 0.3% soft agar. The mixture was
layered onto 0.5% solidified agar in cell culture medium in
six-well plates. After 1–3 weeks depending on the growth kinetics
of the cells, images of the colonies were captured under a light
microscope with ×100 magnification and counted using the Image-Pro
Plus software (version 6.0; Media Cybernetics, Inc.).
Immunohistochemical (IHC)
analysis
A human ESCC tissue array was purchased from
Shanghai Outdo Biotech Company. The slides were deparaffinized in
xylene, rehydrated using a series of decreasing alcohol solutions,
immersed in 3% hydrogen peroxide (cat. no. PV-6002 Kit; OriGene
Technologies, Inc.) for 10 min to block endogenous peroxidase
activity, and antigen-retrieval was performed using a pressure
cooker for 3 min in citrate buffer (pH=6). Subsequently,
non-specific binding was blocked using 10% normal goat serum (cat.
no. ZLI-9022; OriGene Technologies, Inc.) at room temperature for
30 min. Then, the slides were incubated overnight at 4°C in a
humidified chamber with an anti-AURKA antibody (1:200). After
washing, the slides were incubated with the horseradish
peroxidase-labeled secondary antibody (cat. no. PV-6002 Kit;
OriGene Technologies, Inc.) for 30 min at room temperature. Slides
were stained using DAB Kit (cat. no. ZLI-9017; OriGene
Technologies, Inc.) and finally counterstained with hematoxylin.
After staining, images of the sections were captured under a light
microscope and analyzed using Image-Pro Plus.
Purification of recombinant AURKA
protein
AURKA was subcloned into the pet28a vector and
transformed into E. coli strain BL-21. Then, protein
expression in BL21 was induced at 16°C overnight using 0.5 mM
isopropyl-D-thiogalactopyranoside. Cells were collected and
resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM
imidazole, 1 mM phenylmethanesulfonylfluoride, 1% Triton X-100).
After sonication, the supernatants of the cell lysates were
incubated with NI-NTA agarose (Qiagen GmbH) at 4°C for 4 h. The
beads with bound AURKA were washed four times with washing buffer
(50 mM NaH2PO4, 300 mM NaCl, 60 mM imidazole). Protein purity was
determined using SDS-PAGE followed by Coomassie brilliant blue
(CBB) staining.
Pull down assay and mass
spectrometry
The purified AURKA-NI-NTA agarose complex was
incubated with KYSE410 cell lysates for 4 h at 4°C. The complex was
subsequently washed four times in washing buffer (50 mM NaH2PO4,
300 mM NaCl, 60 mM imidazole) and subjected to SDS/PAGE. After CBB
staining, discrepant gel lanes were cut down and prepared for mass
spectrometry analysis.
Xenograft mouse model
The present study was approved (approval no.
NYISTIRB-2021-005) by the Ethics Committee of Nanyang Institute of
Technology (Nanyang, China). A total of 12 6–8-week-old NU/NU
female mice (weight, 22±2 g; Beijing Vital River Laboratory Animal
Technology Co., Ltd.) were used for animal experiments. All mice
were housed in a specific pathogen-free sterile environment with a
constant temperature of 25°C, a relative humidity of 50–70%, with
free access to sterilized food and autoclaved water supply and mice
health and behaviour were monitored daily. Mice were randomly
divided into three groups according to body weight (scramble group:
n=4; shAURKA-1 group: n=4; shAURKA-3 group: n=4). KYSE450 cells
infected with the indicated lentivirus (1×107 cells)
were injected subcutaneously into the right flank of each mouse.
The tumor volumes were determined using calipers to measure the
longest (length) and shortest (diameter) every 4 days and were
calculated as follows: length × diameter2 ×0.5. The
humane endpoint was determined as a tumor volume of 1
cm3 and all the mice were alive until this time point. A
total of 4 weeks after tumor injection, all mice were humanely
sacrificed by intraperitoneal injection of 2% sodium pentobarbital
(100 mg/kg), and the death was confirmed by the cessation of
breathing and heartbeat. Next, the tumor samples were harvested
from sacrificed mice, and tumor weight was measured.
Data acquisition
All bioinformatics data on esophageal carcinoma were
obtained from TCGA and data analysis was performed using R version
3.6.3. AURKA and TPX2 expression in paired tissues were visualized
using the ggplot2 package and ROC curves for AURKA and TPX2 were
generated using the pROC package. For the correlation heat map,
data was obtained from TIMER2.0 (timer.cistrome.org/) and
visualized using the ggplot2 and ggdendro packages. The expression
of AURKA and TPX2 and its clinic correlation was from UALCAN
(http://ualcan.path.uab.edu).
AURKA-binding proteins were derived from STRING (https://string-db.org/) and GEPIA2.0 (http://gepia2.cancer-pku.cn/#index) was used to
get AURKA-correlated genes. The GSE161533 dataset, downloaded from
Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/), contained the
expression profiles from paired normal tissues, para-tumor tissues,
and tumor tissues of 28 ESCC patients. For Gene Ontology (GO)
(http://geneontology.org/) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg/) functional enrichment,
the clusterProfiler package was used for analysis, and the
org.Hs.eg.db package for ID transition. For GSEA, data was obtained
from MSigDB Collections (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) and
the clusterProfiler package was used for analysis. The ggplot2 and
ggdendro packages were used for the visualization of the enriched
data. Immune cell infiltration analysis was performed using the R
package GSVA and ggplot2.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism version 6 (Dotmatics). All statistical tests were two-sided,
and P<0.05 was considered to indicate a statistically
significant difference. All data are presented as the mean ± SD.
Gene expression in paired tissues was analyzed using a Student's
paired t-test. Gene expression in unpaired tissues was analyzed
using an unpaired Student's t-test. Kaplan-Meier analysis was used
for the overall survival analysis followed by the log-rank test. An
unpaired Student's t-test was also used to analyze the data of the
functional assays between control and AURKA knockdown or
overexpression cells.
Results
AURKA expression is increased in
esophageal carcinoma
First, AURKA expression was explored in cancer based
on data obtained from TCGA. Upregulation of AURKA was observed in
several cancer types and AURKA exhibited different mutation rates
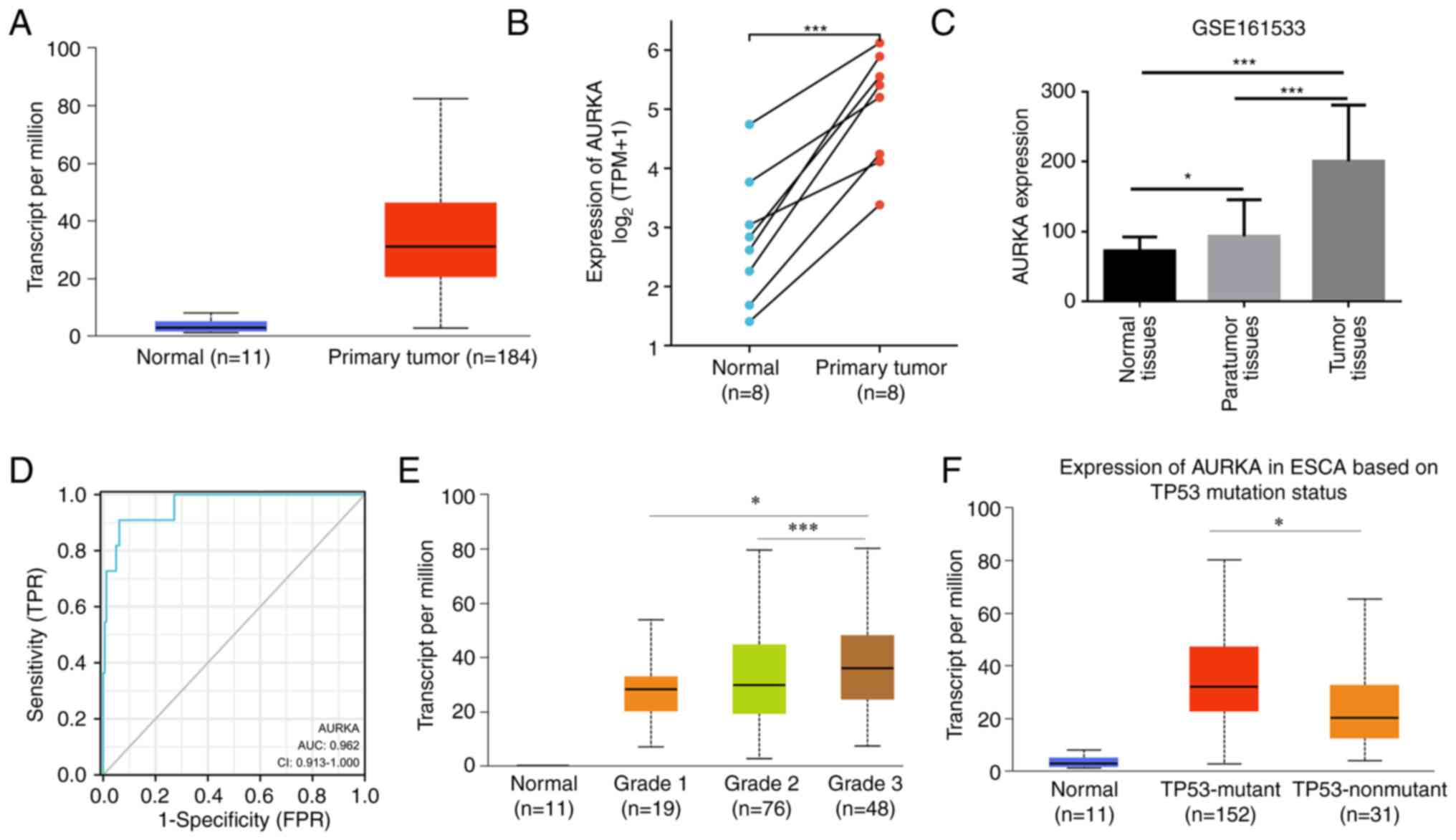
in the different types of various cancer (Fig. S1A and B). As shown in Fig. 1A, AURKA mRNA expression was
significantly higher in primary esophageal carcinoma tumor tissues
than in the respective normal tissues. In addition, the expression
levels of AURKA were upregulated in esophageal carcinoma tumor
tissues compared with the paired normal tissues based on the data
obtained from TCGA (Fig. 1B). Based
on the data obtained from GEO, AURKA expression was highest in ESCC
tumor tissues compared with the paired para-tumor tissues and
normal tissues, and the para-tumor tissues exhibited higher AURKA
expression levels than the normal tissues (Fig. 1C). Furthermore, ROC diagrams for
AURKA demonstrated that AURKA expression levels served as an
excellent biomarker for stratifying esophageal carcinoma patients
from healthy individuals (AUC=0.962, Fig. 1D). Next, the relationship between
AURKA expression and certain clinicopathological characteristics in
TCGA was assessed. It was revealed that patients with tumor grade
3, which is poorly differentiated, exhibited higher AURKA
expression than grade 1 and grade 2 tumors (Fig. 1E). Notably, it was found that AURKA
expression was associated with p53 mutation status. In p53 mutant
esophageal carcinoma tissues, AURKA revealed significantly higher
expression than p53 non-mutant tissues (Fig. 1F).
AURKA expression is correlated with a
poorer prognosis
To further explore AURKA expression and its
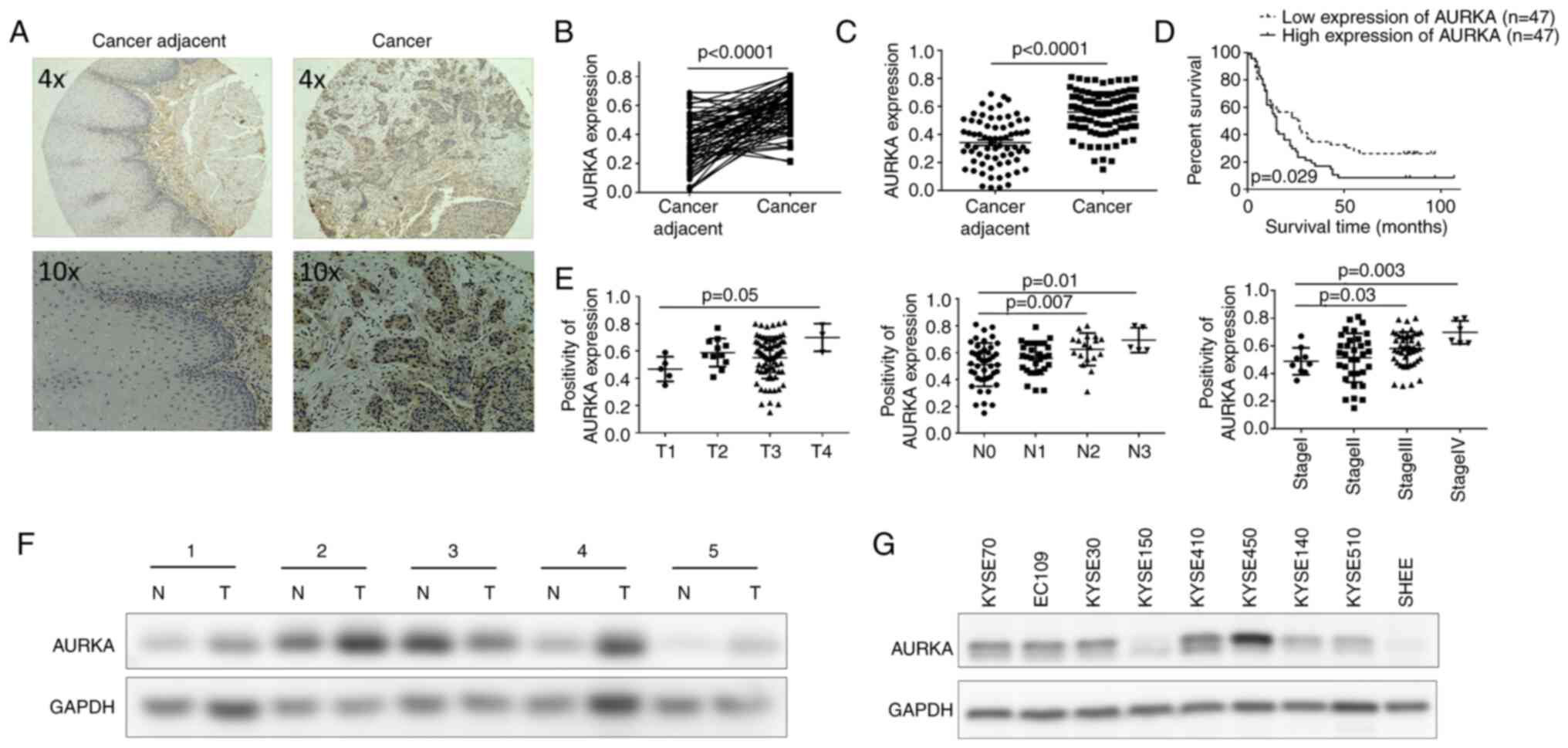
significance in ESCC, AURKA expression was measured in a human ESCC
tissue array using IHC staining. The results demonstrated that the
expression levels of AURKA were significantly upregulated in ESCC
tissues compared with the paired (Fig.
2A and B) and unpaired (Fig.
2C) cancer adjacent tissues. Next, the association between
AURKA expression and the survival of patients with ESCC was
examined. ESCC patients with upregulated AURKA expression levels
had a significantly shorter overall survival than those with lower
AURKA expression levels based on the Kaplan-Meier analysis
(Fig. 2D). Furthermore, the
clinicopathologic correlation analysis revealed that AURKA
expression was significantly higher in patients with a larger tumor
size, lymph node metastasis, or more advanced clinical stage cancer
(Fig. 2E; Table I). These data showed a critical role
for AURKA upregulation in the progression of ESCC. To further
validate the expression of AURKA in ESCC, western blotting was used
to measure AURKA protein levels. It was identified that in 80% of
ESCC tumor tissues, AURKA expression was significantly higher than
that in the paired adjacent normal tissues (Fig. 2F). Finally, AURKA expression in ESCC
cell lines and in the SHEE normal esophageal epithelial cells was
also determined. A general trend of increased AURKA expression in
ESCC cell lines compared with the control SHEE cells was observed
(Fig. 2G). Collectively, these
results suggested that AURKA expression in ESCC was upregulated and
was correlated with patient survival and clinical stage, serving as
an informative prognostic factor in ESCC.
 | Table I.Correlation between AURKA expression
and clinicopathologic characteristics of esophageal squamous cell
carcinoma. |
Table I.
Correlation between AURKA expression
and clinicopathologic characteristics of esophageal squamous cell
carcinoma.
|
| Expression levels
of AURKA |
|
|---|
|
|
|
|
|---|
|
Characteristics | Low (n=47) (%) | High (n=47)
(%) | P-value |
|---|
| Sex |
|
| 0.194 |
|
Male | 33 (70.21) | 37 (78.72) |
|
|
Female | 14 (29.79) | 10 (21.28) |
|
| Age, years |
|
| 0.169 |
|
≤60 | 12 (25.53) | 17 (36.17) |
|
|
>60 | 35 (74.47) | 30 (63.83) |
|
| Histological
grade |
|
| 0.876 |
|
Well/moderately | 34 (72.34) | 33 (70.21) |
|
|
Poorly | 13 (27.66) | 14 (29.79) |
|
| T
classification |
|
| 0.02 |
| T1 | 4 (8.51) | 1 (2.13) |
|
| T2 | 6 (12.77) | 5 (10.64) |
|
| T3 | 37 (78.72) | 38 (80.85) |
|
| T4 | 0 (0.00) | 3 (6.38) |
|
| N
classification |
|
| <0.0001 |
| N0 | 27 (57.45) | 16 (34.04) |
|
| N1 | 15 (31.91) | 13 (27.66) |
|
| N2 | 5 (10.64) | 13 (27.66) |
|
| N3 | 0 (0.00) | 5 (10.64) |
|
| Clinical stage |
|
| <0.0001 |
| I | 7 (14.90) | 2 (4.26) |
|
| II | 20 (42.55) | 13 (27.66) |
|
|
III | 20 (42.55) | 26 (55.32) |
|
| IV | 0 (0.00) | 6 (12.76) |
|
AURKA promotes ESCC proliferation via
the PI3K/Akt pathway
To investigate the contribution of AURKA in ESCC
progression, the cell proliferation and colony formation potential
of ESCC cells after AURKA overexpression was first assessed. The
results indicated that AURKA was effectively transfected into
KYSE450, KYSE30, and KYSE150 cells based on the western blotting
(Fig. S2A). Additionally, ectopic
overexpression of AURKA significantly increased cell growth based
on the results of the MTT assay (Fig.
S2B). Moreover, an increased number and larger colonies were
formed in ESCC cells overexpressing AURKA (Fig. S2C).
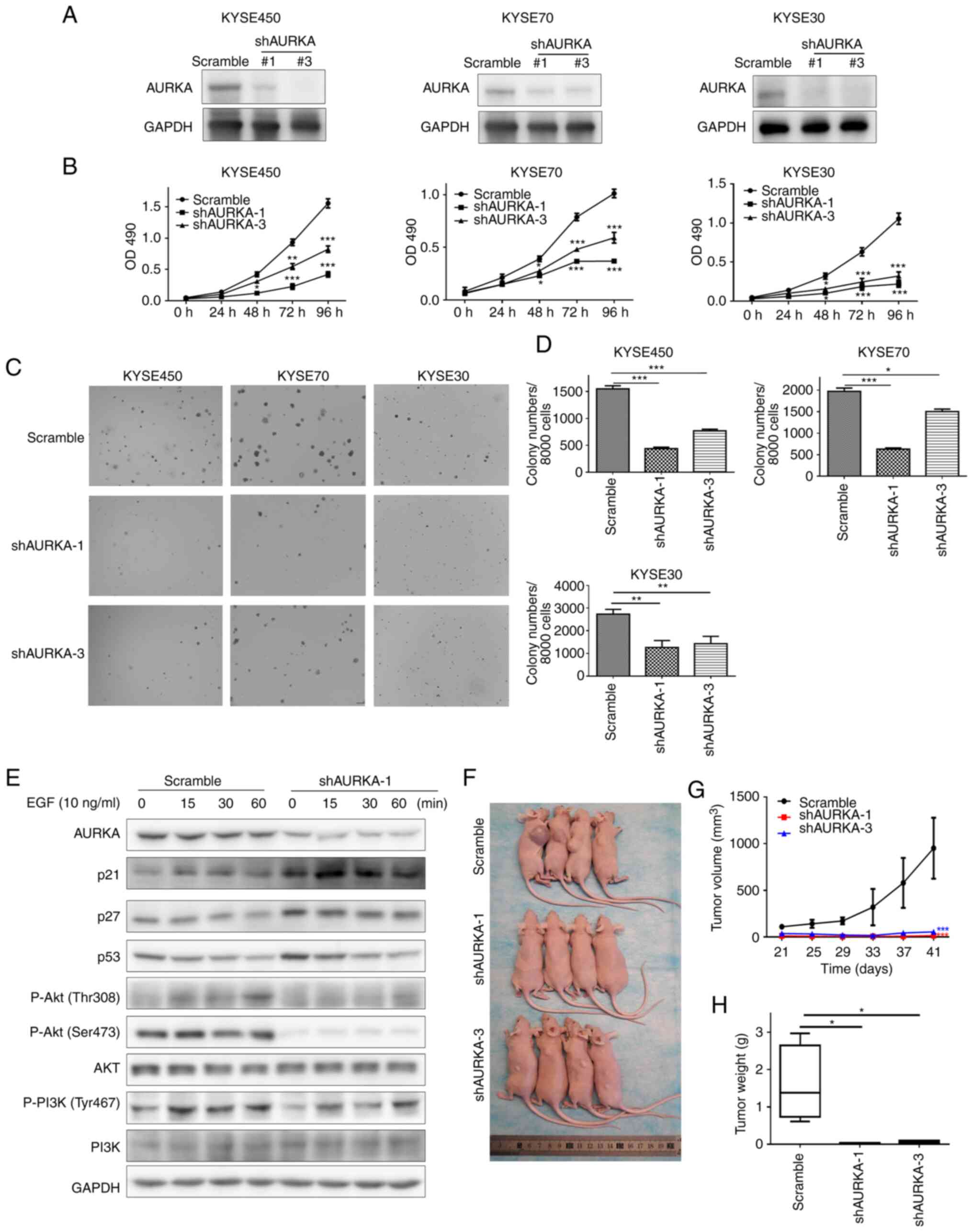
To further confirm the oncogenic role of AURKA in
ESCC, specific shRNAs targeting AURKA were used to generate stable
AURKA-knockdown cells. Both shAURKA-1 and shAURKA-3 effectively
reduced AURKA expression in ESCC cells (Fig. 3A). Next, MTT and soft agar colony
formation assays were used to assess whether AURKA affected cell
proliferation and colony formation of the ESCC cells. The cell
growth and colony formation capacity were significantly reduced
following AURKA knockdown (Fig.
3B-D).
Next, the AURKA downstream signaling pathway in ESCC
was assessed based on EGF stimulation. Western blotting revealed
that AURKA knockdown reduced phosphorylation of Akt and PI3K
(Fig. 3E). Furthermore, increased
expression of p21, p53, and p27 was also observed after knockdown
of AURKA expression (Fig. 3E).
Finally, to determine whether AURKA could contribute to ESCC tumor
growth in vivo, control cells or AURKA knockdown cells were
subcutaneously injected into nude mice. The results showed that
AURKA knockdown significantly inhibited tumor growth based on the
reduced tumor volume and tumor weight (Fig. 3F-H). Taken together, AURKA was found
to be associated with the acquisition of an oncogenic phenotype in
ESCC.
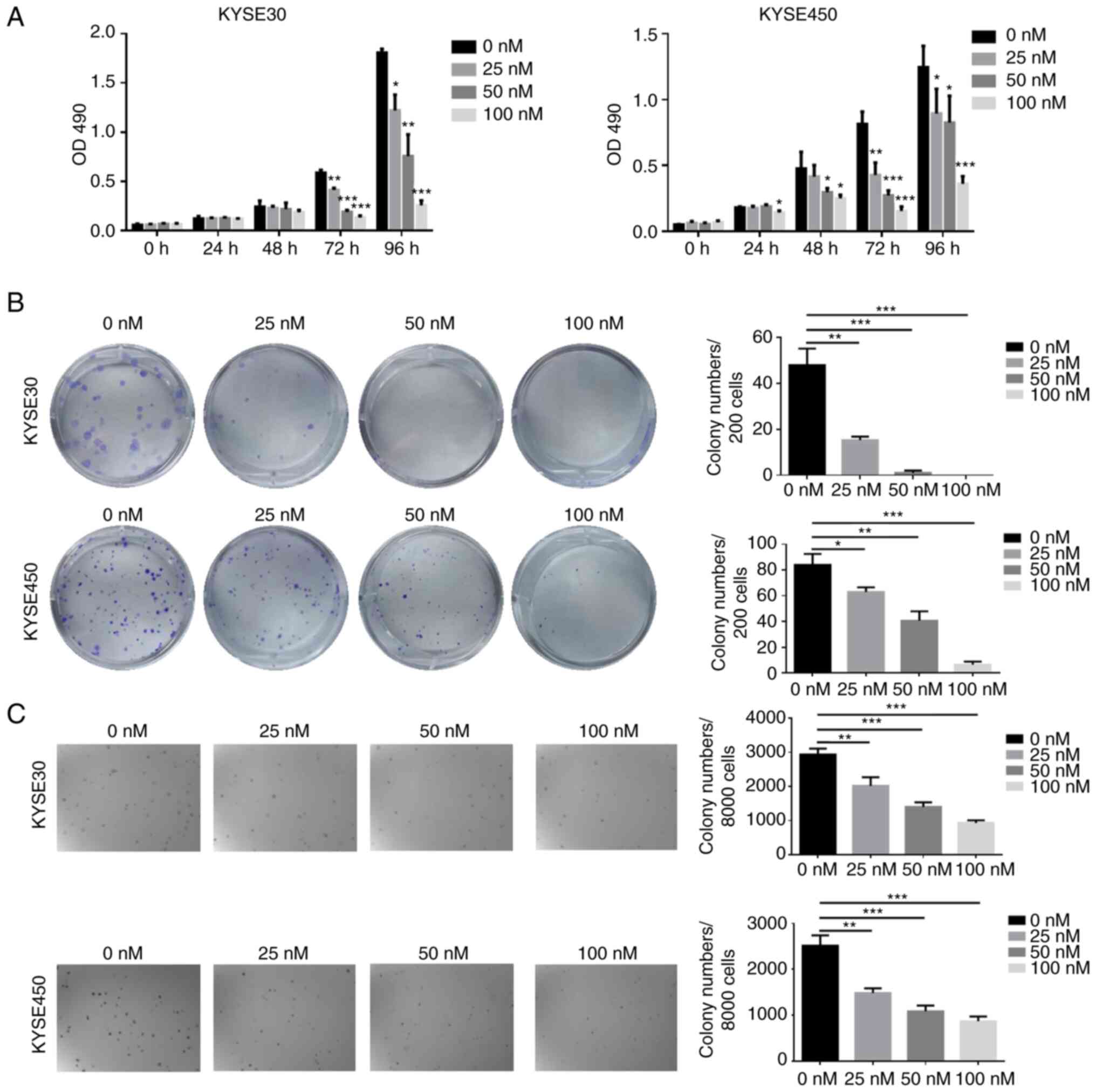
Targeting AURKA with alisertib
inhibits ESCC cell growth
To further verify the significance of targeting
AURKA in ESCC, the antitumor effects of alisertib, an AURKA
inhibitor, were determined. The MTT assay demonstrated that
alisertib treatment significantly inhibited cell proliferation in a
concentration-dependent manner and the inhibitory effects at 100 nM
alisertib were ~85% in KYSE30 cells and 70% in KYSE450 cells
(Fig. 4A). Furthermore, alisertib
significantly inhibited KYSE30 and KYSE450 cell colony formation
(Fig. 4B). In the
anchorage-independent cell growth assay, alisertib exhibited
inhibitory effects consistent with its suppressive effects on cell
proliferation in KYSE30 and KYSE450 cells (Fig. 4C).
AURKA is involved in multiple
biological processes and pathways in ESCC
To study the molecular mechanism of AURKA in ESCC
tumorigenesis, the AURKA-binding proteins and the genes correlated
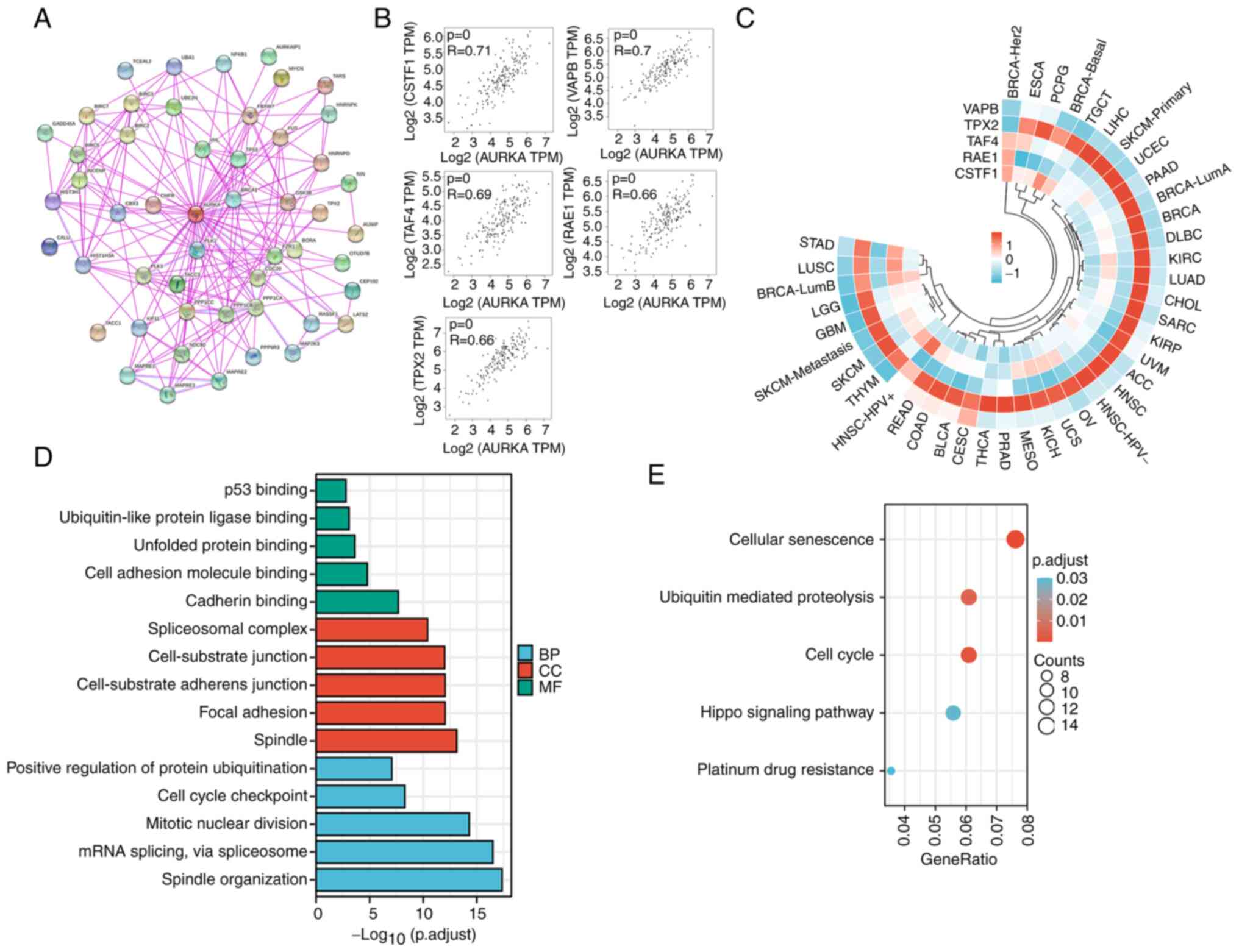
with AURKA expression were screened. First, a total of 50
AURKA-binding proteins were obtained, which were supported by
experimental evidence using STRING. The interaction network of
these proteins is shown in Fig. 5A.
Next, to identify the AURKA-binding proteins, a His-pull down assay
was used. AURKA was purified using a prokaryotic expression system
as demonstrated in Fig. S3A, and
the purified AURKA protein was verified by western blotting
(Fig. S3B). A total of 239
proteins were identified via mass spectrometry following the His
pull-down assay (Fig. S3C and
Table SI). Immunoprecipitation
assays were performed to verify several AURKA-interacting proteins
based on the mass spectrometry analysis. The results revealed that
AURKA could bind with RPS6, HSP27, VDAC and RPL7 (Fig. S3D).
Next, the GEPIA2.0 tool was used to obtain the top
100 genes that were correlated with AURKA expression in esophageal
carcinoma. The five most positively related genes were CSTF1
(R=0.71, P<0.001), VAPB (R=0.7, P<0.001), TAF4 (R=0.69,
P<0.001), RAE1 (R=0.66, P<0.001) and TPX2 (R=0.66,
P<0.001) (Fig. 5B). Furthermore,
the heat map of the correlation of these genes in various types of
cancer showed that TPX2 was correlated with AURKA in different
types of cancer (Fig. 5C). Data
obtained from GEPIA2 and STRING was combined with the results of
the His-pull-down assay to perform KEGG and GO enrichment analyses.
As demonstrated in Fig. 5D, the GO
enrichment analysis data indicated that the majority of these genes
were associated with the biological process or pathways of
cell-substrate junction (including cell adhesion molecule binding,
cadherin binding, focal adhesion) and cell cycle regulation (such
as cell cycle checkpoint, mitotic nuclear division and spindle
organization). The KEGG data suggested that ‘cellular senescence’,
‘ubiquitin mediated proteolysis’, ‘cell cycle’, ‘hippo signaling
pathway’ and ‘platinum drug resistance’ may be involved in the
effect of AURKA on tumor pathogenesis (Fig. 5E).
AURKA-mediated effects on ESCC
progression involve TPX2
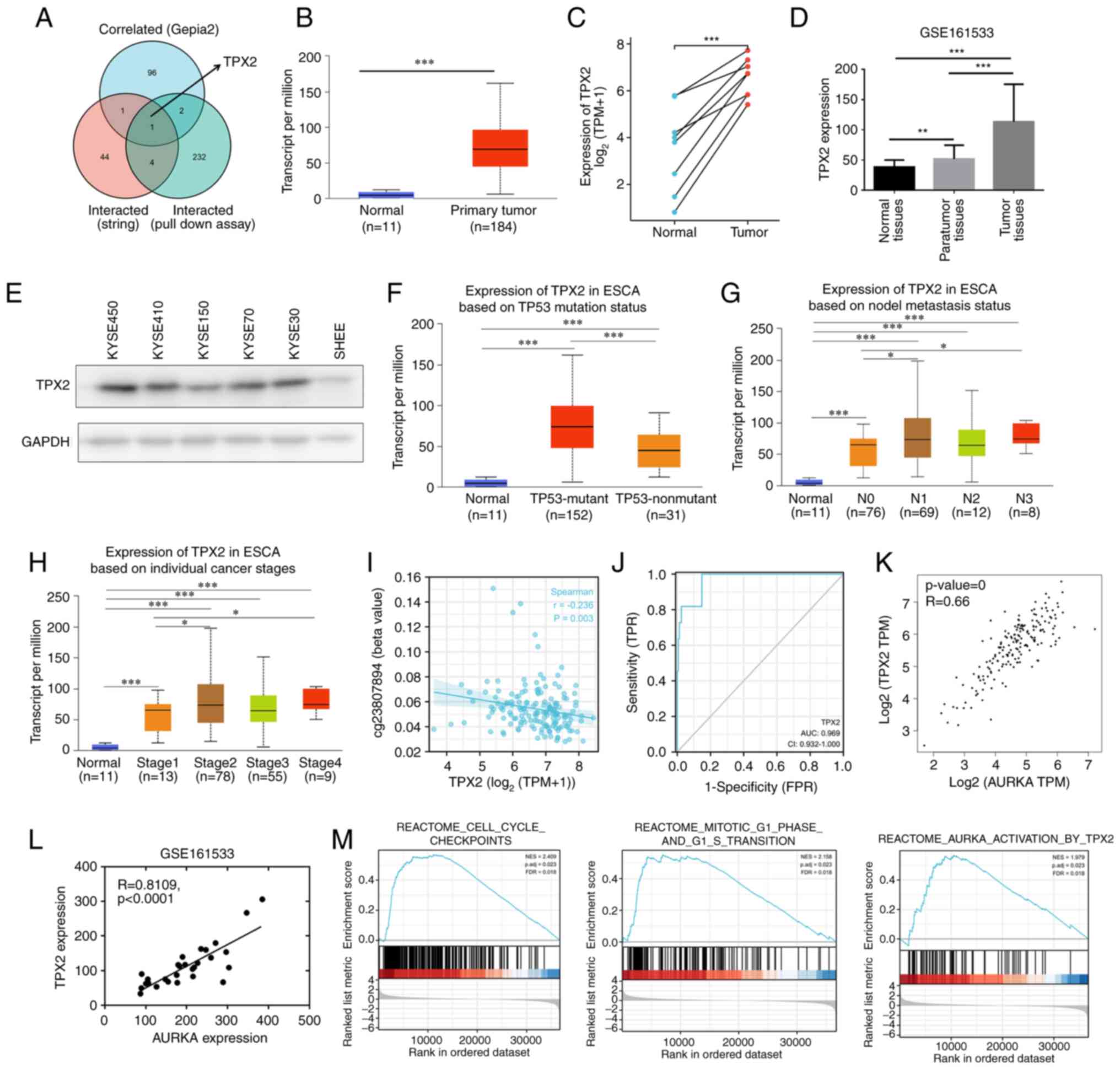
Among AURKA-binding and AURKA expression-correlated
proteins, an intersection analysis of STRING, GEPIA, and
His-pull-down proteins showed one common member, namely, TPX2
(Fig. 6A). Additionally, aberrantly
high TPX2 expression was observed in several different types of
cancer (Fig. S4), and TPX2
expression was higher in esophageal carcinoma compared with the
normal controls (Fig. 6B and C).
According to a GEO dataset, TPX2 expression was upregulated in ESCC
tumor tissues compared with the para-tumor tissues and normal
tissues (Fig. 6D). Additionally,
western blotting revealed increased TPX2 expression in ESCC cell
lines compared with the normal esophageal epithelium SHEE cells
(Fig. 6E). The association between
TPX2 expression and clinicopathological features was further
evaluated based on the UALCAN database. Similar to AURKA, TPX2
demonstrated significantly higher expression in p53 mutant tissues
than in the p53 wild-type tissues (Fig.
6F). Furthermore, TPX2 expression was correlated with lymph
node metastasis and advanced-clinical stage cancer (Fig. 6G and H). Next, TPX2 promoter
methylation status was examined in esophageal carcinoma based on
data obtained from TCGA, and the results showed that TPX2
expression was negatively associated with promoter methylation at
the −513 region (Fig. 6I). More
importantly, TPX2 was found to serve as an excellent diagnostic
indicator for esophageal carcinoma (AUC=0.969, Fig. 6J). Finally, the correlation between
AURKA and TPX2 expression was assessed. Based on GEPIA, AURKA
expression was positively associated with TPX2 in esophageal
carcinoma (R=0.66, P<0.001, Fig.
6K). Based on the GSE161533 dataset, TPX2 expression was
associated with AURKA expression in ESCC tumor tissues (R=0.81,
P<0.001, Fig. 6L).
Next, GSEA enrichment was performed to analyze the
potential function of TPX2 in ESCC. The results showed that TPX2
was involved in cell cycle checkpoints, in mitotic G1 phase and in
G1 to S transition (Fig. 6M).
Additionally, TPX2 was related to AURKA activation in ESCC
(Fig. 6M). To further confirm
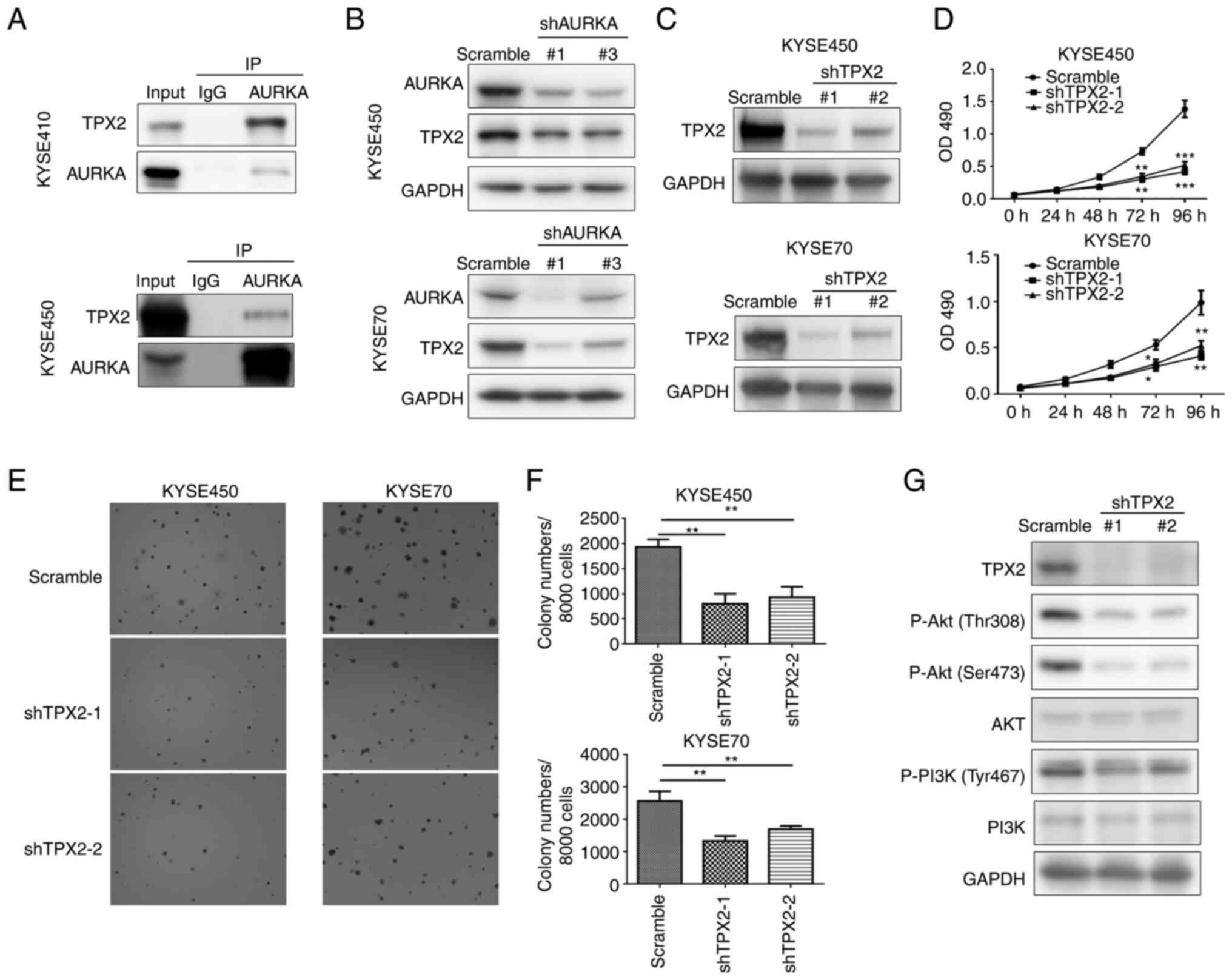
whether TPX2 was both an AURKA-interacting and AURKA-correlated
protein, an immunoprecipitation assay was performed, and the
results revealed that AURKA could bind with TPX2 in KYSE410 and
KYSE450 cells (Fig. 7A).
Furthermore, decreased expression of TPX2 was observed following
AURKA knockdown (Fig. 7B). To
further confirm TPX2 was involved in the oncogenic role of AURKA,
the function of TPX2 in ESCC was also measured. It was shown that
TPX2 knockdown significantly reduced ESCC cell proliferation and
colony formation (Fig. 7C-F).
Additionally, the expression of PI3K/Akt pathway-related proteins
were evaluated after TPX2 knockdown, and the results demonstrated
that the activity of the PI3K/Akt pathway was reduced when TPX2
expression was knocked down (Fig.
7G). Accordingly, AURKA may co-operate with TPX2 to regulate
ESCC progression via the PI3K/Akt pathway.
AURKA or TPX2 expression levels are
associated with tumor immune cell infiltration
Immune cells in the tumor microenvironment affect
cancer patient survival. Among these immune cells, T cell-mediated
immune responses have been used as therapeutic targets clinically
(13,14). Thus, the relationship between AURKA
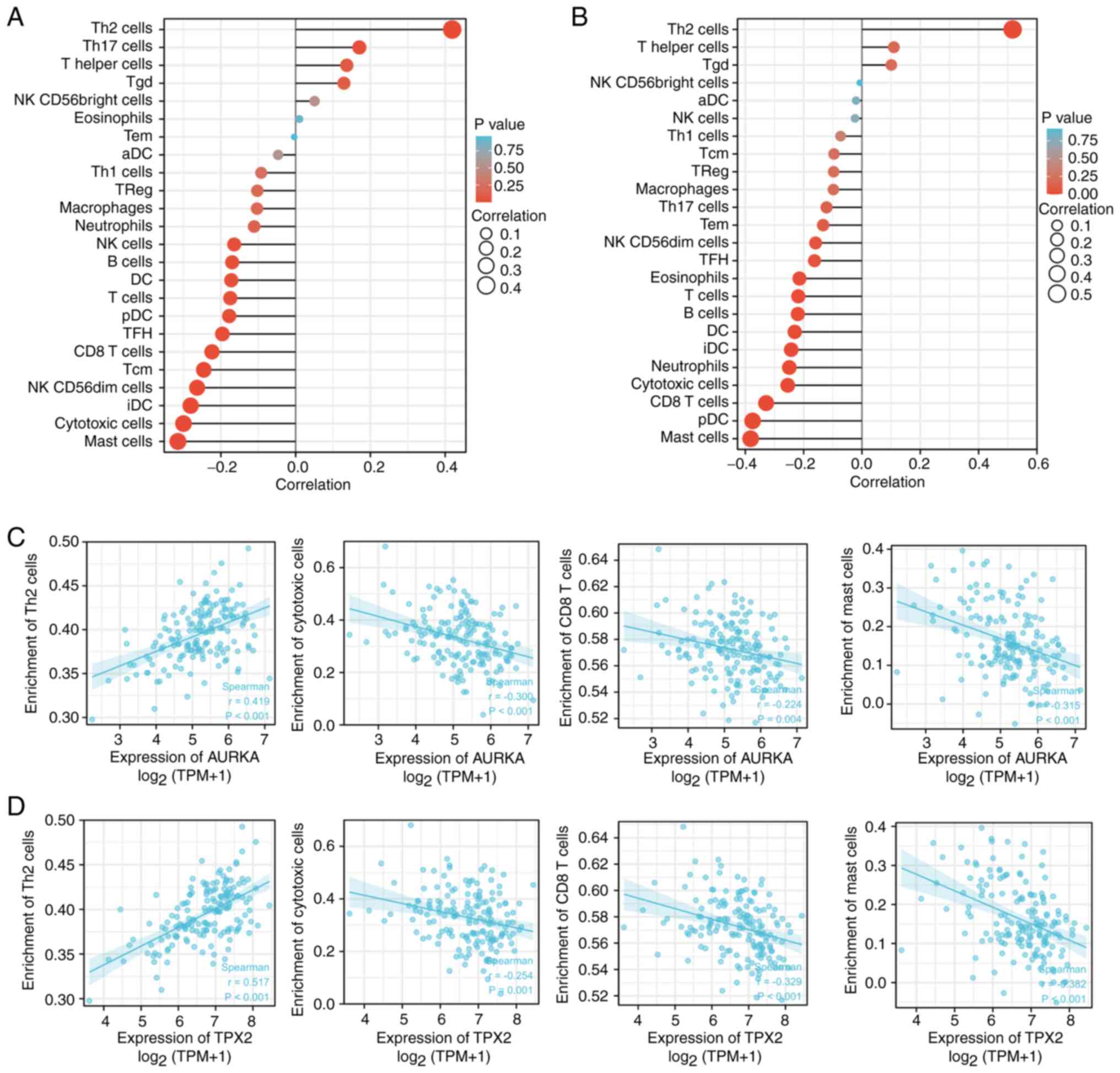
and TPX2 expression levels with the levels of CD4+ T
cells was examined. There was a significantly positive relationship
between AURKA and TPX2 expression with the infiltration of Th2
cells in humans pan-cancer (Fig.
S5). Next, the relationship between AURKA and TPX2 expression
levels with the different subsets of immune cells was examined.
AURKA and TPX2 expression levels were positively associated with
Th2 cells, T helper cells, and Tgd (Fig. 8A and B). In addition, AURKA was
positively associated with Th17 cells (Fig. 8A). AURKA and TPX2 expression were
negatively associated with the majority of the remaining types of
immune cells including mast cells, cytotoxic cells, and
CD8+ T cells, amongst others (Fig. 8A and B). The correlation analysis
between AURKA and TPX2 expression with the Th2 cells, cytotoxic
cells, CD8+ T cells, and mast cells is shown in Fig. 8C and D. Collectively, these results
strongly suggested that AURKA and TPX2 exerted vital roles in tumor
immunity.
Discussion
ESCC is an aggressive upper gastrointestinal tumor
whose 5-year survival is <20%, given that several cases are
diagnosed in the first instance at an advanced stage at the time of
diagnosis and the lack of specific targeted drugs. Although the
molecular signatures of ESCC have been well defined including the
most frequently mutated and amplified genes, there are still no
effective therapeutic targets (5,15,16).
Therefore, studies elucidating the actionable targets in ESCC are
urgently required. AURKA upregulation has been indicated in several
types of cancer and was shown to be inversely related to disease
prognosis. It has been reported that upregulation of AURKA in ESCC
was related to distant lymph node metastasis and a poorer prognosis
(4). The results of the present
study highlighted the oncogenic role of AURKA in ESCC both through
bioinformatics analysis and experimental verification. Upregulated
AURKA expression in ESCC was associated with advanced-stage disease
and a poorer prognosis, indicating that AURKA may serve as an
excellent therapeutic target. AURKA was initially shown to serve as
a regulator of mitosis and was subsequently considered to regulate
a malignant phenotype of cancer cells. The results also showed that
AURKA promoted ESCC cell proliferation, and AURKA knockdown
significantly suppressed ESCC tumor growth. These results strongly
indicated that AURKA can be used as a prognostic and diagnostic
biomarker for ESCC.
TPX2 has been studied as a factor critical for
mitosis and spindle assembly. Additionally, TPX2 was verified as a
potential therapeutic target in pancreatic (17), breast (18), and cervical cancer (19), amongst others. Based on the data
obtained from TCGA, TPX2 was identified to be significantly
upregulated in esophageal cancer and can serve as a diagnostic
marker. Previous studies indicated that TPX2 can stimulate
autophosphorylation and autoactivation of AURKA, resulting in AURKA
activation (20,21). Bioinformatics analysis from TCGA
data and pull-down assays suggested that TPX2 may both interact
with AURKA and was correlated with AURKA in esophageal cancer. It
was further verified that AURKA interacted with TPX2 through
immunoprecipitation assays. In addition, AURKA expression was
positively correlated with TPX2 based on data obtained from TCGA,
and AURKA knockdown decreased TPX2 protein expression. AURKA and
TPX2 may form a positive feedback loop to regulate ESCC
progression, although this requires further confirmation. In
colorectal cancer, AURKA was found to coordinate with TPX2 to drive
tumor progression (22,23). Additionally, TPX2 was reported to
regulate the PI3K/Akt pathway to promote hepatocellular carcinoma
progression and act as a STAT3 regulator (24,25).
Based on the results of the present study, AURKA knockdown
inhibited the EGF-stimulated PI3K/Akt pathway activation, and TPX2
knockdown also impeded PI3K/Akt signaling activation. According to
the AURKA interactome from previous studies, AURKA-interacting
proteins were enriched in the PI3K/Akt pathway (12). Thus, AURKA may cooperate with TPX2
to promote ESCC development through the PI3K/Akt pathway.
AURKA is a serine/threonine kinase that is crucial
in regulating the cell cycle. p53, a primary tumor suppressor,
induces cell cycle arrest through transcriptional downregulation of
several cell cycle-associated genes and therefore impedes tumor
cell cycle progression (26–28).
In the present study, it was revealed that AURKA knockdown
increased p53 expression, suggesting that AURKA may regulate the
cell cycle through p53 in ESCC. In several types of cancer, TP53 is
the most commonly mutated gene, and the majority of tumors have
lost p53 function due to mutations (29). In ESCC, p53 mutations have been
observed in as high as 93% of patients (5). According to the UALCAN database, AURKA
expression was significantly higher in p53 mutant tissues compared
with p53 non-mutant tissues. Thus, AURKA may result in the loss of
the tumor-inhibiting role of p53. Furthermore, it was demonstrated
that AURKA suppression also induced p21 and p27 expression, which
are both tumor suppressors and CDK inhibitors. Taken together, it
was shown that AURKA regulated cell cycle related genes and thus
promoted ESCC proliferation.
A series of inhibitors targeting AURKA have been
explored as previously described in a review by the authors,
amongst which alisertib is the most popular (12). Alisertib has finished phase III
clinical assessment in patients with relapsed/refractory peripheral
T-cell lymphoma and showed no significant improvement over
chemotherapy (30). However,
alisertib showed relatively good effects in solid tumors, including
breast and small-cell lung cancer in a multicenter phase II study
(31). In the present study, it was
demonstrated that 25 nM alisertib significantly inhibited ESCC cell
proliferation and colony formation. These data support the
necessity of further investigation of alisertib in ESCC therapy.
Another strategy targeting AURKA is to disrupt the interaction
between AURKA and its activators including TPX2. Withanone, a
herbal ligand isolated from ashwagandha, exerted an anticancer
effect by binding to the TPX2/AURKA complex and dissociate TPX2
from AURKA (32). Therefore,
targeting the AURKA/TPX2 axis in ESCC is a promising strategy for
ESCC therapy.
The tumor microenvironment has been gaining
increasing interest in research circles in recent years.
Tumor-infiltrating lymphocytes have been suggested to be an
independent predictor of a patient's, prognosis in several types of
cancer (33–35). AURKA was reported to participate in
TCR activation (36) and determine
CD8+ T cell cytotoxic activity and antiviral responses
(37). Furthermore, AURKA
inhibition promoted CD8+ T-cell infiltration and
activation by inducing IL-10 production (38). In breast cancer, AURKA inhibitor
alisertib eliminated myeloid cell-mediated immunosuppression and
enhanced anti-PD-L1 therapy (39).
It was also found that AURKA inhibitors enhanced T-cell
cytotoxicity in vitro and could potentiate antitumor
immunity in vivo (40).
CD4+ T cells and CD8+ T cells are crucial
members of the tumor microenvironment that participate in specific
antitumor immune responses (41).
Among the different subsets of CD4+ T cells, Th1 and Th2
cells are the most important classes of CD4+ T cells.
Based on the results of the present study, both expression levels
of AURKA and TPX2 were positively associated with Th2 cell
infiltration and negatively associated with cytotoxic cells and
CD8+ T cell infiltration. Th2 cell aggregation resulted
in the dysfunction of CD8+ T cells and cytotoxic T cells
and ultimately contributed to immune escape. Therefore, the role of
AURKA or TPX2 in tumorigenesis may be associated with increased Th2
cell and decreased CD8+ T and cytotoxic T cell
infiltration, which require further study. Mast cells are immune
cells present in all classes of vertebrates that have also been
identified in tumor tissues. The pro- or antitumorigenic roles of
mast cells in different tumors are cancer type-specific (42,43).
It was reported that mast cells expressing IL-17 was predictive of
a favorable prognosis in ESCC (44). It was observed that there was a
negative correlation between AURKA or TPX2 expression and
infiltration of mast cells, which may indicate the possible
pro-tumorigenic role of mast cells in ESCC. Collectively, these
observations may help to reveal the role of the AURKA/TPX2 axis in
the tumor microenvironment and provides a reference for a deeper
study of AURKA or TPX2 in tumor immunity.
In conclusion, an AURKA/TPX2 axis was elucidated
that may be used as a potential target in ESCC using both
bioinformatics analysis and experimental validation. AURKA
expression was upregulated in ESCC and was predictive of poorer
patient survival. Upregulated AURKA expression promoted ESCC cell
proliferation and tumor growth via the PI3K/Akt pathway. Moreover,
targeting AURKA with alisertib inhibited ESCC cell proliferation
and colony formation. Mechanistic analysis suggested AURKA
interacted and correlated with TPX2 which was upregulated in ESCC.
Furthermore, both TPX2 and AURKA were positively associated with
Th2 cell infiltration and negatively associated with cytotoxic,
CD8+ T and mast cell infiltration. These findings may
assist in elucidating the role of the AURKA/TPX2 axis in
tumorigenesis and development, whilst providing a reference for the
realization of more precise and personalized therapy in the
future.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 82273058 and 82002592), the Science
and Technology Project of Henan Province (grant no. 222102310156),
the Doctoral Research Start-up Fund Project of Nanyang Institute of
Technology (grant no. NGBJ-2022-05) and the Talent Program of
Central China: Science and Technology Innovation Leading Talent
(grant no. 234200510006).
Availability of data and materials
The datasets used and analyzed in the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
HB and LH designed the project. RJD performed the
cell functional experiments and data analysis and wrote the
article. KL, ZJZ and YLH performed the molecular biology
experiment. KLG and HZ focused on the animal experiment. ZGC and
XLZ finished the bioinformatic analysis and revised the article.
All authors read and edited the manuscript. All authors read and
approved the final manuscript. RJD and LH confirmed the
authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved (approval no.
NYISTIRB-2021-005) by the Ethics Committee of Nanyang Institute of
Technology (Nanyang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AURKA
|
Aurora kinase A
|
|
ESCC
|
esophageal squamous cell carcinoma
|
|
IHC
|
immunohistochemistry
|
|
IP
|
immunoprecipitation
|
|
GEO
|
Gene Expression Omnibus
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arnold M, Ferlay J, van Berge Henegouwen
MI and Soerjomataram I: Global burden of oesophageal and gastric
cancer by histology and subsite in 2018. Gut. 69:1564–1571. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tramontano AC, Chen Y, Watson TR, Eckel A,
Hur C and Kong CY: Esophageal cancer treatment costs by phase of
care and treatment modality, 2000–2013. Cancer Med. 8:5158–5172.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tran GD, Sun XD, Abnet CC, Fan JH, Dawsey
SM, Dong ZW, Mark SD, Qiao YL and Taylor PR: Prospective study of
risk factors for esophageal and gastric cancers in the Linxian
general population trial cohort in China. Int J Cancer.
113:456–463. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun
ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY, et al: Genetic landscape of
esophageal squamous cell carcinoma. Nat Genet. 46:1097–1102. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thrift AP: Global burden and epidemiology
of Barrett oesophagus and oesophageal cancer. Nat Rev Gastroenterol
Hepatol. 18:432–443. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jung J, Jeong H, Choi JW, Kim HS, Oh HE,
Lee ES, Kim YS and Lee JH: Increased expression levels of AURKA and
KIFC1 are promising predictors of progression and poor survival
associated with gastric cancer. Pathol Res Pract. 224:1535242021.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
de Martino M, Shariat SF, Hofbauer SL,
Lucca I, Taus C, Wiener HG, Haitel A, Susani M and Klatte T: Aurora
A kinase as a diagnostic urinary marker for urothelial bladder
cancer. World J Urol. 33:105–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miyoshi Y, Iwao K, Egawa C and Noguchi S:
Association of centrosomal kinase STK15/BTAK mRNA expression with
chromosomal instability in human breast cancers. Int J Cancer.
92:370–373. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Belt EJ, Brosens RP, Delis-van Diemen PM,
Bril H, Tijssen M, van Essen DF, Heymans MW, Beliën JA, Stockmann
HB, Meijer S and Meijer GA: Cell cycle proteins predict recurrence
in stage II and III colon cancer. Ann Surg Oncol. 19 (Suppl
3):S682–S692. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang G, Chang B, Yang F, Guo X, Cai KQ,
Xiao XS, Wang H, Sen S, Hung MC, Mills GB, et al: Aurora kinase A
promotes ovarian tumorigenesis through dysregulation of the cell
cycle and suppression of BRCA2. Clin Cancer Res. 16:3171–3181.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Du R, Huang C, Liu K, Li X and Dong Z:
Targeting AURKA in cancer: Molecular mechanisms and opportunities
for cancer therapy. Mol Cancer. 20:152021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kraehenbuehl L, Weng CH, Eghbali S,
Wolchok JD and Merghoub T: Enhancing immunotherapy in cancer by
targeting emerging immunomodulatory pathways. Nat Rev Clin Oncol.
19:37–50. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Finck AV, Blanchard T, Roselle CP,
Golinelli G and June CH: Engineered cellular immunotherapies in
cancer and beyond. Nat Med. 28:678–689. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Talukdar FR, di Pietro M, Secrier M,
Moehler M, Goepfert K, Lima SSC, Pinto LFR, Hendricks D, Parker MI
and Herceg Z: Molecular landscape of esophageal cancer:
Implications for early detection and personalized therapy. Ann N Y
Acad Sci. 1434:342–359. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng C, Zhou Y, Li H, Xiong T, Li S, Bi
Y, Kong P, Wang F, Cui H, Li Y, et al: Whole-genome sequencing
reveals diverse models of structural variations in esophageal
squamous cell carcinoma. Am J Hum Genet. 98:256–274. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Warner SL, Stephens BJ, Nwokenkwo S,
Hostetter G, Sugeng A, Hidalgo M, Trent JM, Han H and Von Hoff DD:
Validation of TPX2 as a potential therapeutic target in pancreatic
cancer cells. Clin Cancer Res. 15:6519–6528. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang Y, Liu Y, Tan X, Yu S and Luo J:
TPX2 as a novel prognostic indicator and promising therapeutic
target in triple-negative breast cancer. Clin Breast Cancer.
19:450–455. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang H, Wang J, Tian Y, Xu J, Gou X and
Cheng J: The TPX2 gene is a promising diagnostic and therapeutic
target for cervical cancer. Oncol Rep. 27:1353–1359.
2012.PubMed/NCBI
|
|
20
|
Eyers PA, Erikson E, Chen LG and Maller
JL: A novel mechanism for activation of the protein kinase Aurora
A. Curr Biol. 13:691–697. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bayliss R, Sardon T, Vernos I and Conti E:
Structural basis of Aurora-A activation by TPX2 at the mitotic
spindle. Mol Cell. 12:851–862. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sillars-Hardebol AH, Carvalho B, Tijssen
M, Beliën JA, de Wit M, Delis-van Diemen PM, Pontén F, van de Wiel
MA, Fijneman RJ and Meijer GA: TPX2 and AURKA promote 20q
amplicon-driven colorectal adenoma to carcinoma progression. Gut.
61:1568–1575. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takahashi Y, Sheridan P, Niida A, Sawada
G, Uchi R, Mizuno H, Kurashige J, Sugimachi K, Sasaki S, Shimada Y,
et al: The AURKA/TPX2 axis drives colon tumorigenesis cooperatively
with MYC. Ann Oncol. 26:935–942. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang DH, Jian J, Li S, Zhang Y and Liu
LZ: TPX2 silencing exerts anti-tumor effects on hepatocellular
carcinoma by regulating the PI3K/AKT signaling pathway. Int J Mol
Med. 44:2113–2122. 2019.PubMed/NCBI
|
|
25
|
Nagel S, Pommerenke C, MacLeod RAF, Meyer
C, Kaufmann M and Drexler HG: The NKL-code for innate lymphoid
cells reveals deregulated expression of NKL homeobox genes HHEX and
HLX in anaplastic large cell lymphoma (ALCL). Oncotarget.
11:3208–3226. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Engeland K: Cell cycle arrest through
indirect transcriptional repression by p53: I have a DREAM. Cell
Death Differ. 25:114–132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Krause K, Wasner M, Reinhard W, Haugwitz
U, Dohna CL, Mössner J and Engeland K: The tumour suppressor
protein p53 can repress transcription of cyclin B. Nucleic Acids
Res. 28:4410–4418. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Engeland K: Cell cycle regulation:
p53-p21-RB signaling. Cell Death Differ. 29:946–960. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bykov VJN, Eriksson SE, Bianchi J and
Wiman KG: Targeting mutant p53 for efficient cancer therapy. Nat
Rev Cancer. 18:89–102. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
O'Connor OA, Özcan M, Jacobsen ED, Roncero
JM, Trotman J, Demeter J, Masszi T, Pereira J, Ramchandren R,
Beaven A, et al: Randomized phase III study of alisertib or
investigator's choice (selected single agent) in patients with
relapsed or refractory peripheral T-cell lymphoma. J Clin Oncol.
37:613–623. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Melichar B, Adenis A, Lockhart AC,
Bennouna J, Dees EC, Kayaleh O, Obermannova R, DeMichele A,
Zatloukal P, Zhang B, et al: Safety and activity of alisertib, an
investigational aurora kinase A inhibitor, in patients with breast
cancer, small-cell lung cancer, non-small-cell lung cancer, head
and neck squamous-cell carcinoma, and gastro-oesophageal
adenocarcinoma: A five-arm phase 2 study. Lancet Oncol. 16:395–405.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Grover A, Singh R, Shandilya A, Priyandoko
D, Agrawal V, Bisaria VS, Wadhwa R, Kaul SC and Sundar D:
Ashwagandha derived withanone targets TPX2-Aurora A complex:
Computational and experimental evidence to its anticancer activity.
PLoS One. 7:e308902012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Federico L, McGrail DJ, Bentebibel SE,
Haymaker C, Ravelli A, Forget MA, Karpinets T, Jiang P, Reuben A,
Negrao MV, et al: Distinct tumor-infiltrating lymphocyte landscapes
are associated with clinical outcomes in localized non-small-cell
lung cancer. Ann Oncol. 33:42–56. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Loi S, Michiels S, Adams S, Loibl S,
Budczies J, Denkert C and Salgado R: The journey of
tumor-infiltrating lymphocytes as a biomarker in breast cancer:
Clinical utility in an era of checkpoint inhibition. Ann Oncol.
32:1236–1244. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Okadome K, Baba Y, Yagi T, Kiyozumi Y,
Ishimoto T, Iwatsuki M, Miyamoto Y, Yoshida N, Watanabe M and Baba
H: Prognostic nutritional index, tumor-infiltrating lymphocytes,
and prognosis in patients with esophageal cancer. Ann Surg.
271:693–700. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Blas-Rus N, Bustos-Morán E, Pérez de
Castro I, de Cárcer G, Borroto A, Camafeita E, Jorge I, Vázquez J,
Alarcón B, Malumbres M, et al: Aurora A drives early signalling and
vesicle dynamics during T-cell activation. Nat Commun. 7:113892016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bustos-Morán E, Blas-Rus N, Alcaraz-Serna
A, Iborra S, González-Martínez J, Malumbres M and Sánchez-Madrid F:
Aurora A controls CD8+ T cell cytotoxic activity and
antiviral response. Sci Rep. 9:22112019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han J, Jiang Z, Wang C, Chen X, Li R, Sun
N, Liu X, Wang H, Hong L, Zheng K, et al: Inhibition of Aurora-A
promotes CD8+ T-cell infiltration by mediating IL10
production in cancer cells. Mol Cancer Res. 18:1589–1602. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yin T, Zhao ZB, Guo J, Wang T, Yang JB,
Wang C, Long J, Ma S, Huang Q, Zhang K, et al: Aurora A inhibition
eliminates myeloid cell-mediated immunosuppression and enhances the
efficacy of anti-PD-L1 therapy in breast cancer. Cancer Res.
79:3431–3444. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Punt S, Malu S, McKenzie JA, Manrique SZ,
Doorduijn EM, Mbofung RM, Williams L, Silverman DA, Ashkin EL,
Dominguez AL, et al: Aurora kinase inhibition sensitizes melanoma
cells to T-cell-mediated cytotoxicity. Cancer Immunol Immunother.
70:1101–1113. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Basu A, Ramamoorthi G, Albert G, Gallen C,
Beyer A, Snyder C, Koski G, Disis ML, Czerniecki BJ and Kodumudi K:
Differentiation and regulation of TH cells: A balancing
act for cancer immunotherapy. Front Immunol. 12:6694742021.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sammarco G, Varricchi G, Ferraro V,
Ammendola M, De Fazio M, Altomare DF, Luposella M, Maltese L, Currò
G, Marone G, et al: Mast cells, angiogenesis and lymphangiogenesis
in human gastric cancer. Int J Mol Sci. 20:21062019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Majorini MT, Colombo MP and Lecis D: Few,
but efficient: The role of mast cells in breast cancer and other
solid tumors. Cancer Res. 82:1439–1447. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang B, Li L, Liao Y, Li J, Yu X, Zhang Y,
Xu J, Rao H, Chen S, Zhang L and Zheng L: Mast cells expressing
interleukin 17 in the muscularis propria predict a favorable
prognosis in esophageal squamous cell carcinoma. Cancer Immunol
Immunother. 62:1575–1585. 2013. View Article : Google Scholar : PubMed/NCBI
|