Introduction
Cervical cancer is the fourth most frequently
occurring cancer and the fourth leading cause of cancer-associated
mortality among women, with an estimated 604,000 new cases and
342,000 related deaths worldwide in 2020 (1). Human papillomavirus (HPV) infection is
a necessary, but not sufficient cause of cervical cancer. Over 100
genotypes of HPV have been identified (2), and persistent infection with high-risk
HPV types (HR-HPVs) is the causal factor in >99% of cervical
cancer cases and the main drivers of HR-HPV-associated oncogenesis
are the oncoproteins E5, E6 and E7 (3). To date, the thinprep cytologic test
(TCT) or Papanicolaou (pap) test together with HPV genotype
screening programs have been applied successful and decrease the
morbidity of cervical cancer in a number of countries in Europe,
Oceania and North American (4,5). In
addition, the HPV vaccination programs reduce the long-term future
burden of cervical cancer (6,7).
MicroRNAs (miRNAs/miRs) are the most abundant class
of small non-coding RNAs. They are transcribed from genomic DNA and
generally processed with two endonucleases, Drosha and Dicer.
miRNAs regulate gene expression at the post-transcriptional level
in a sequence-dependent manner. Dysregulated miRNA expression
levels have been identified in various types of cancer (8) including cervical cancer (9,10).
Previously, some dysregulated miRNAs were subjected to in-depth
investigations and their downstream functional mechanisms were
elucidated. For example, the authors have previously performed
various biochemical approaches, such as CLIP-Chip and PAR-CLIP, and
identified the different targets of miR-205 and miR-944 in cervical
cancer cells, respectively (11,12).
Some studies have also explored the reasons for the abnormal
expression of miRNAs in tumor tissues from different aspects. For
example, the loss of chromosome 13q14 linked the reduced miR-15a
and miR-16-1 expression in chronic lymphocytic leukemia (13). Another study demonstrated that the
key transcription factor, c-Myc, resulted in the upregulated
expression of the miR-17/92 cluster members due to its
amplification and overexpression in multiple cancer types (14). Although studies have reported the
transcriptional regulation of miRNAs from transcription factors and
epigenetic regulations (15–17),
the exact regulatory mechanisms for the specific miRNA
dysregulation remain to be fully elucidated.
Hsa-miR-27a-3p is located in the minus strand of
human 19p13.12 and is transcribed from the same primary transcript
together with miR-23a-3p/5p and miR-24-2-3p/5p, the other members
of the miR-23a/27a/24-2 cluster. The abnormal expression of miR-27a
has been reported in multiple cancer types and its involvement in
different signaling pathways plays dual roles based on the
different cell or tissue types. Previous studies have demonstrated
that miR-27a-3p is involved in the PI3K/Akt pathway (18), TGF-β signaling pathway (19) and Wnt/β-catenin pathway (20,21).
Recently, several long non-coding RNAs were reported to be involved
in the regulatory effects of miR-27a and its mRNA target (22–24).
Moreover, previous studies have linked miR-27a with the aggressive
phenotypes of cervical cancer cells (25,26);
however, the presented results are still controversial due to the
different cell lines or the different materials used, and thus,
further studies are warranted.
TGF-β activated kinase 1 binding protein 3 (TAB3) is
a protein which functions in the NF-κB signaling transduction
pathway (27). Together with TAB1/2
and TAK1 forming the ternary complex, TAB3 responds to the
stimulation of pro-inflammatory cytokines, such as TNF-α or IL-1β,
and subsequently triggers the activation of the NF-κB signaling
pathway (28). TAB3 promotes the
proliferation and/or invasion of various cancer cells via the
activation of NF-κB signaling (29–32).
For example, the TAB3/TAK1 complex directly activates STAT3
signaling via the phosphorylation of STAT3 and promotes colorectal
cancer growth (29). The expression
levels of TAB3 and proliferating cell nuclear antigen have been
found to be gradually increased in ovarian cancer tissues and cell
lines, suggesting the novel oncogenic functions of TAB3 in ovarian
cancer (31). Ding et al
(33) applied a genome-wide
screening strategy and revealed that miR-195 exerted its
tumor-suppressive effects via the inhibition of TAB3 and IKKα
expression in hepatocellular carcinoma. However, the exact
expression and functions of TAB3 in cervical cancer have not yet
been reported, at least to the best of our knowledge; thus, further
investigations are required.
Based on previously unpublished screening data by
the authors, miR-27a-3p was one of the differently expressed miRNAs
identified by silencing NFKB1. The present study thus focused on
miR-27a-3p and investigated its upstream regulatory and downstream
functions. The transcription factor, NF-κB subunit p65, was
identified to activate the transcription of the miR-23a/27a/24-2
cluster by directly binding to the promoter region of
pri-miR-23a/27a/24-2 and contributed to the upregulated expression
of miR-27a-3p. Functionally, miR-27a-3p promoted the malignant
potential of cervical cancer cells by increasing the colony
formation rates, the migratory and invasive abilities, epithelial
mesenchymal transition (EMT) and inhibiting apoptosis.
Mechanistically, miR-27a-3p directly bound to the 3′UTR of TAB3 and
enhanced its expression, followed by the subsequent activation of
NF-κB signaling. To the best of our knowledge, this is the first
functional report of TAB3 in cervical cancer cells, linking TAB3
with miR-27a-3p, and demonstrates the possible post-transcriptional
regulation for TAB3 expression. The data presented herein suggest
that the upregulation of miR-27a-3p expression by p65 further
enhances TAB3 expression at the post-transcriptional level and
sequentially mediates the activation of the NF-κB signaling pathway
via TAB3 upregulation, composing of a positive feedback regulatory
loop, namely the p65/miR-27a-3p/TAB3/NF-κB axis. These findings may
provide novel insight into the mechanisms underlying the
carcinogenesis of cervical cancer and may provide potential novel
biomarkers for its management.
Materials and methods
Cells, cell culture and
transfection
The human cervical cancer cell lines, HeLa (CCL-2)
and C33A (HTB-31), were originally purchased from the American Type
Culture Collection (ATCC) and cultured in the tumor, virus and
ncRNA group. The (HeLa and C33A) cells were cultured in DMEM
(VivaCell Biosciences), supplemented with 10% fetal bovine serum
(FBS; Biological Industries), 100 µg/ml streptomycin and 100 IU/ml
penicillin (cat. no. P1400, Beijing Solarbio Science &
Technology Co., Ltd.). All cells were cultured at 37°C with 5%
CO2.
All transfection experiments were carried out using
Lipofectamine 2000® reagent (Thermo Fisher Scientific,
Inc.) following the manufacturer's instructions. Plasmids (2 µg per
well in a six-well plate or 500 ng per well in 24-well plate) or
antisense oligonucleotides (ASO-NC/ASO-miR-27a-3p, 50 nM) were
transfected into cervical cancer cells. After adding the complex
(mixture of transfection reagent and DNA or ASO) to the wells, the
cells were incubated at 37°C for 24–48 h for further analyses.
Plasmid construction
The strand-specific miR-27a-3p expression vector was
constructed following a previously described strategy (34). In brief, the mature miR-27a-3p
sequence with its first 19 nucleotides complementary sequence and
the loop sequence were synthesized and inserted into the
pcDNA3-U6M2 vector (modified from the pcDNA3 vector, replacing the
CMV promoter with U6 promoter, abbreviated as U6M2; a gift from Dr
Per Johansson, Karolinska Institutet, Sweden) between the
BglII/KpnI sites. For miR-27a-3p inhibition, the
synthesized 2′-O-methyl-modified antisense oligonucleotides of
miR-27a-3p (ASO-miR-27a-3p) and the scramble control
oligonucleotides (ASO-NC) were purchased from Shanghai GenePharma
Co., Ltd.
To construct the overexpression vector for TAB3, the
full coding sequence of TAB3 from the cDNA of HeLa cells was
amplified and cloned into the pcDNA3-3×Flag vector (a modified
vector, named as pcDNA3/Flag-KBE, abbreviated as KBE, since the
3×Flag sequences inserted between the NcoI/KpnI
sites, the cloned fragment can only be inserted downstream of the
KpnI site) between the KpnI/EcoRI sites,
respectively. The overexpression vectors for p65 (KBE-p65), p50
(KBE-p50) were provided by Dr Weiying Liu (Tianjin Medical
University), the shRNA vector targeting NFKB1 and the empty vector
control pSilencer2.1-U6 neo were provided by Dr Qi Sun (Tianjin
Medical University). The shRNA vectors targeting TAB3, NF-κB/p65
and Agonaute (AGO)2 (shR-TAB3, shR-p65, shR-AGO2) were constructed
by annealing the synthesized oligonucleotides into pcDNA3-U6M2
vector between the BglII/KpnI sites.
The luciferase reporter vectors for the miR-27a
promoter were constructed by amplifying the respected sequences
from the genomic DNA of HeLa cells and cloned into the
pGL3-enhancer vector (Promega Corporation), between the
XhoI/HindIII sites (pGL3-P1 and pGL3-P2). The series
truncated promoter reporter vectors (R175, R259, R509, F259 and
F509) were constructed using the same strategy by amplifying the
related fragments from the pGL3-P2 plasmid. The mutated reporter
vector with the p65 binding site deletion (F509-Del546) was
synthesized by Sangon Biotech Co., Ltd.
The wild-type and the mutated form of the 3′UTR EGFP
reporter vectors for TAB3, mitogen-activated protein kinase kinase
kinase 14 (MAP3K14) and TNF receptor associated factor 3 (TRAF3)
were constructed by annealing the synthesized oligonucleotides
(containing the predicted miR-27a-3p binding sites, wild-type and
mutated form) and cloning into the pcDNA3/EGFP vector between the
BamHI/EcoRI sites. All constructed vectors were
verified using Sanger sequencing at GENEWIZ. All primers used for
PCR amplification and all oligonucleotides for preparing constructs
were synthesized by Synbio Technologies or General Biol, and their
sequences are listed in Supplementary Table SI.
Bioinformatics analyses
The online miRNA target prediction tool, TargetScan
(https://www.targetscan.org/), was used
to predict the potential targets of miR-27a-3p. An online analyses
tool (Assistant for Clinical Bioinformatics, http://www.aclbi.com) based on The Cancer Genome Atlas
(TCGA)-Cervical Squamous Cell Carcinoma and Endocervical
Adenocarcinoma (CESC) dataset (http://portal.gdc.cancer.gov) was used to analyze the
expression levels of miRNAs and the selected potential targets. The
NCBI GEO datasets (GSE86100 and GSE20592) were also selected to
analyze the expression levels of miR-27a-3p and the
miR-23a/27a/24-2 cluster members. GEPIA2 (http://gepia2.cancer-pku.cn/#index) was used to
analyze the overall and disease-free survival based on the
expression levels of TAB3, MAP3K14 and TRAF3 from the TCGA-CESC
dataset, and the online tool HIPLOT (https://hiplot.com.cn/cloud-tool/drawing-tool/detail/122)
was applied to draw the survival curves based on miR-27a-3p
expression from the TCGA-CESC dataset.
For promoter prediction, with the help of FANTOM
(https://fantom.gsc.riken.jp/) and
Promoter 2.0 (https://services.healthtech.dtu.dk/service.php?Promoter-2.0)
predictions, combining with CpG island analysis, the upstream (~8
kb) sequences of miR-27a-3p were analyzed.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the cultured cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's recommendations. The
concentration of the RNA was measured using a NanoDrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.) and stored at
−80°C. cDNA was synthesized using the HiScript II Q Select RT
SuperMix for qPCR kit (cat. no. R233-01, Nanjing Vazyme Biotech
Co., Ltd.). A total of 1 µg RNA was first treated by genome DNA
wiper mix at 42°C for 2 min and followed by 50°C/15 min plus 85°C/5
sec for cDNA synthesis. The expression levels of miRNAs and mRNAs
were quantified by qPCR using 2X Universal SYBR-Green fast qPCR Mix
(ABclonal Biotech Co., Ltd.). β-actin and U6 snRNA were used as the
endogenous controls for mRNA and miRNA quantification,
respectively, reported as 2−∆CCq (35) and further normalized to their
respective controls. All primers used for RT-qPCR were synthesized
by Synbio Technologies and are listed in Supplementary Table SI.
Western blot analyses
Cells were collected after 48 h post-transfection
and lysed in RIPA buffer (cat. no. R0010, Beijing Solarbio Science
& Technology Co., Ltd.). Proteins (50–100 µg per lane,
determined using the BCA protein assay kit, Beyotime Institute of
Biotechnology, Inc.), were separated by 10% SDS-PAGE and
transferred to nitrocellulose membranes. After blocking with 5%
non-fat milk in 1×TBST (cat. no. BF-0113, Beijing Dingguo
Biotechnology Co. Ltd.) at room temperature for 2 h, followed by
incubation with the indicated dilutions of the primary antibodies
overnight at 4°C. GAPDH served as the endogenous loading control
for the whole cell lysates and the cytoplasmic sub-fractions, and
Lamin B1 was selected for the internal reference index for the
nuclear fractions. After washing three times (5 min for each) with
TBST, the secondary antibodies (anti-mouse or anti-rabbit IgG-HRP)
were added and incubated for a further 1 h at room temperature.
Detection was performed using ECL HRP chemiluminescent substrate
reagent (cat. no. BL520B, Biosharp Life Sciences). The protein
expression levels were quantified on the blots using ImageJ
software (version 1.52v, National Institutes of Health). All
antibodies used in the present study are listed in Table I.
 | Table I.The detailed information for all the
antibodies used in the present study. |
Table I.
The detailed information for all the
antibodies used in the present study.
| Primary
antibodies |
|---|
|
|---|
| Antibody | Manufacturer | Cat. no. | Species | Dilution |
|---|
| TAB3 | Tianjin Saier
Biotechnology Co., Ltd. | SRP08919 | Rabbit | 1:500 (WB) |
| Cleaved PARP | Beyotime Institute
of Biotechnology | AF1567 | Rabbit | 1: 1,000 WB) |
| Vimentin | Beyotime Institute
of Biotechnology | AF0318 | Mouse | 1:3,000 (WB) |
| EpCAM | Chengdu Zen
Bioscience Co., Ltd. | R24219 | Rabbit | 1:1,000 (WB) |
| Flag | Beyotime Institute
of Biotechnology | AF519 | Mouse | 1:1,000 (WB) |
| Agonaute 2 | Chengdu Zen
Bioscience Co., Ltd. | R23523 | Rabbit | 1:1,000 (WB) |
| MAP3K14 | Tianjin Saier
Biotechnology Co., Ltd. | SRP06501 | Rabbit | 1:500 (WB) |
| TRAF3 | Wanleibio Co.,
Ltd. | WL04574 | Rabbit | 1:250 (WB) |
| GAPDH | Beyotime Institute
of Biotechnology | AF1186 | Rabbit | 1:10,000 (WB) |
| Lamin B1 | Tianjin Saier
Biotechnology Co., Ltd. | SRP13156 | Rabbit | 1:3,000 (WB) |
| NF-κB/p65 | Beyotime Institute
of Biotechnology | AF1234 | Rabbit | 1:1,000 (WB) |
| NF-κB/p65 | Beyotime Institute
of Biotechnology | AF1234 | Rabbit | 1:200 (IF) |
|
| Secondary
antibodies used for western blot analysis and
immunofluorescence |
|
| Goat anti-mouse
IgG-HRP | Boster Biological
Technology Co., Ltd. | BA1050 | Goat | 1:80,000 (WB) |
| Goat anti-rabblit
IgG-HRP | Boster Biological
Technology Co., Ltd. | BA1054 | Goat | 1:80,000 (WB) |
| Goat anti-rabbit
IgG-FITC | Tianjin Sungene
Biotech Co., Ltd. | AJ1027 | Goat | 1:200 (IF) |
Luciferase promoter reporter and EGFP
3′UTR reporter analyses
For promoter luciferase reporter assays, the HeLa
cells were seeded in 96-well plates and transfected with the
indicated series reporter vectors (100 ng/well) with/without the
modulation of NF-κB/p65 or NFKB1/p50. Each well was co-transfected
with the pRL-CMV vector (10 ng/well) for normalization. The
activities of luciferases (Firefly and Renilla, FL and RL)
were measured at 30 h post-transfection using the Dual-Luciferase
Assay kit (cat. no. E1910, Promega Corporation) following the
recommendations of the manufacturer. The relative Firefly
activities were normalized to the Renilla luciferase
activities in each well.
For 3′UTR EGFP fluorescence reporter assays, the
HeLa cells were seeded in 24-well plates and transfected with the
wild-type or mutated form of the reporter vectors (500 ng/well)
with/without the overexpression of miR-27a-3p. pDsRed2-N1 (100
ng/well, Clontech; Takara Bio USA) co-transfected into each well
for normalization. At 48 h following transfection, the cells were
lysed with RIPA buffer and the fluorescence intensities of EGFP and
RFP were determined using a microplate reader (Tecan Spark, Tecan
Group, Ltd.).
Colony formation assays
Following 24 h of transfection, the HeLa and C33A
cells were re-suspended and seeded in a 24-well plate (300
cells/well) or 12-well plate (500 cells/well) and incubated for a
further 10–14 days a 37°C in a 5% CO2 incubator. During
the incubation period, fresh culture medium was changed every 3–4
days. The cells were fixed with 4% paraformaldehyde (cat. no.
P1110; Beijing Solarbio Science & Technology Co., Ltd.) for 30
min and stained with 0.5% crystal violet (cat. no. C0121, Beyotime
Institute of Biotechnology) for 10 min at room temperature, and the
colonies including >50 cells were counted using a light
microscope (ECLIPSE TS100, Nikon Corporation).
Transwell migration and invasion
assays
Cell migration and invasion were analyzed using
24-well Boyden chambers with an 8-µm pore size polycarbonate
membrane. The insert chambers were placed in a 24-well plate,
containing 800 µl culture medium with 30% FBS. For the invasion
assays, the Matrigel (cat. no. 356234, Corning Inc.) was 1:7
diluted with cold serum-free medium, placed onto the chamber
membrane in advance and incubated at 37°C for at least 2 h. In
brief, following 24 h of transfection, 6×105 cells (for
migration) or 8×105 cells (for invasion) were
re-suspended in serum-free culture medium and seeded in the upper
chamber and incubated at 37°C for a further 36 h. At the end of the
incubation period, the non-migrated or non-invaded cells on the top
surface of membrane were removed using cotton swabs, and the
migrated or invaded cells on the bottom surface of membrane were
fixed with 4% paraformaldehyde for 30 min and stained with crystal
violet for 10 min at room temperature. For quantification, the
stained migrated or invaded cells were dissolved using 33% acetic
acid reagent and the optical densities at 570 nm were determined
using an MQX200 microplate reader (BioTek Instruments, Inc.).
Immunofluorescence analysis
The HeLa cells were seeded in a 12-well plate with
coverslips inside. Following transfection with 1 µg plasmids
(miR-27a-3p, TAB3 and their respective vector controls) for 48 h,
the cells were fixed with 4% paraformaldehyde for 10 min at room
temperature, permeabilized with 0.01% Triton X-100 (v/v, in 1X PBS)
for 2 min, and blocked in 10% donkey serum (cat. no. S9100, Beijing
Solarbio Science & Technology Co., Ltd.) at room temperature
for 30 min. Following overnight incubation with anti-p65 antibody
(1:200; cat no. AF1234, Beyotime Institute of Biotechnology) at
4°C, the FITC-labeled secondary antibody (1:200; cat. no. AJ1027,
Tianjin Sungene Biotech Co., Ltd.) were added and incubated for 2 h
at 4°C in the dark. The cells were mounted with DAPI Fluoromount-G
and images were captured using a TS2 fluorescence microscope (Nikon
Corporation). ImageJ software (version 1.52v, National Institutes
of Health) was used to analyze the mean fluorescence intensities
from at least three views, including ~15-25 individual cells, and
the quantification was normalized to their respective control
groups.
Cell cytosolic and nuclear fraction
extraction
At 48 h post-transfection, the cells (in a six-well
plate) were trypsinized, harvested with cold PBS and lysed with 150
µl Buffer I [0.4% NP-40 (cat. no. DH218 (Guangzhou Dingguo Biology)
in PBS with 1X Protease Inhibitor Cocktail, cat. no. P6730, Beijing
Solarbio Science & Technology Co., Ltd.] by vortexing.
Following incubation for 5 min at 4°C, the lysates were centrifuged
at 5,000 × g for 10 min at 4°C. The supernatant (i.e., the
cytosolic extracts) was carefully collected and stored at −20°C for
further analyses. The precipitates were re-suspended in 100 µl
Buffer II (0.1% NP-40 in PBS with 1X Protease Inhibitor Cocktail)
by gently pipetting up and down, followed by centrifugation at
5,000 × g for 10 min at 4°C. The supernatants were discarded and
the precipitates were dissolved in 100 µl RIPA buffer (cat. no.
R0010, Beijing Solarbio Science & Technology Co., Ltd.) and
incubated at −80°C for 30 min; these were the nuclear extracts used
for further analyses.
Statistical analyses
All experiments were performed at least in
triplicate independently. All data were expressed as the mean ± SEM
and were analyzed using GraphPad Prism 7 (GraphPad Software, Inc.).
The significance of the differences between two or multiple groups
was evaluated using an unpaired Student's t-test or one-way ANOVA
followed by a Tukey's post hoc test. The paired Student's t-test
was only applied with the GSE20592 dataset. P≤0.05 was considered
to indicate a statistically significant difference. Pearson's
correlation test was applied to analyze the correlation between the
miR-27a-3p and TAB3 expression levels.
Results
The high expression of miR-27a-3p
facilitates the malignant properties and enhances the nuclear
translocation of NF-κB/p65 in cervical cancer cells
To investigate the expression levels of miR-27a-3p
and the other members from miR-23a/27a/24-2 cluster in cervical
cancer tissues, expression analyses were performed based on the
GSE86100, GSE20592 and TCGA-CESC datasets. As demonstrated in
Figs. 1A and S1, the expression of miR-27a-3p was
significantly higher in cervical cancer tissues compared with
normal tissues, while the expression levels of the other miRNAs
were not consistent. Further RT-qPCR analyses were first applied to
investigate the effectiveness of the constructed miR-27a-3p strand
specific expression vector and the synthesized ASO-miR-27a-3p. As
shown in Fig. 1B, the
stand-specific miR-27a-3p construct enhanced and ASO-miR-27a-3p
decreased the expression of miR-27a-3p in HeLa cells. Subsequently,
gain- and loss-of-function experiments were performed to
investigate its multiple effects on cell malignancies. The
overexpression of miR-27a-3p significantly enhanced the colony
formation rates of cervical cancer cells, while the inhibition of
miR-27a-3p resulted in decreased colony formation rates in both the
HeLa and C33A cells (Fig. 1C).
Moreover, miR-27a-3p overexpression inhibited the apoptosis of
cervical cancer cells, since the expression of cleaved PARP (89
kDa) increased upon miR-27a-3p inhibition and decreased with the
overexpression of miR-27a-3p (Fig.
1D).
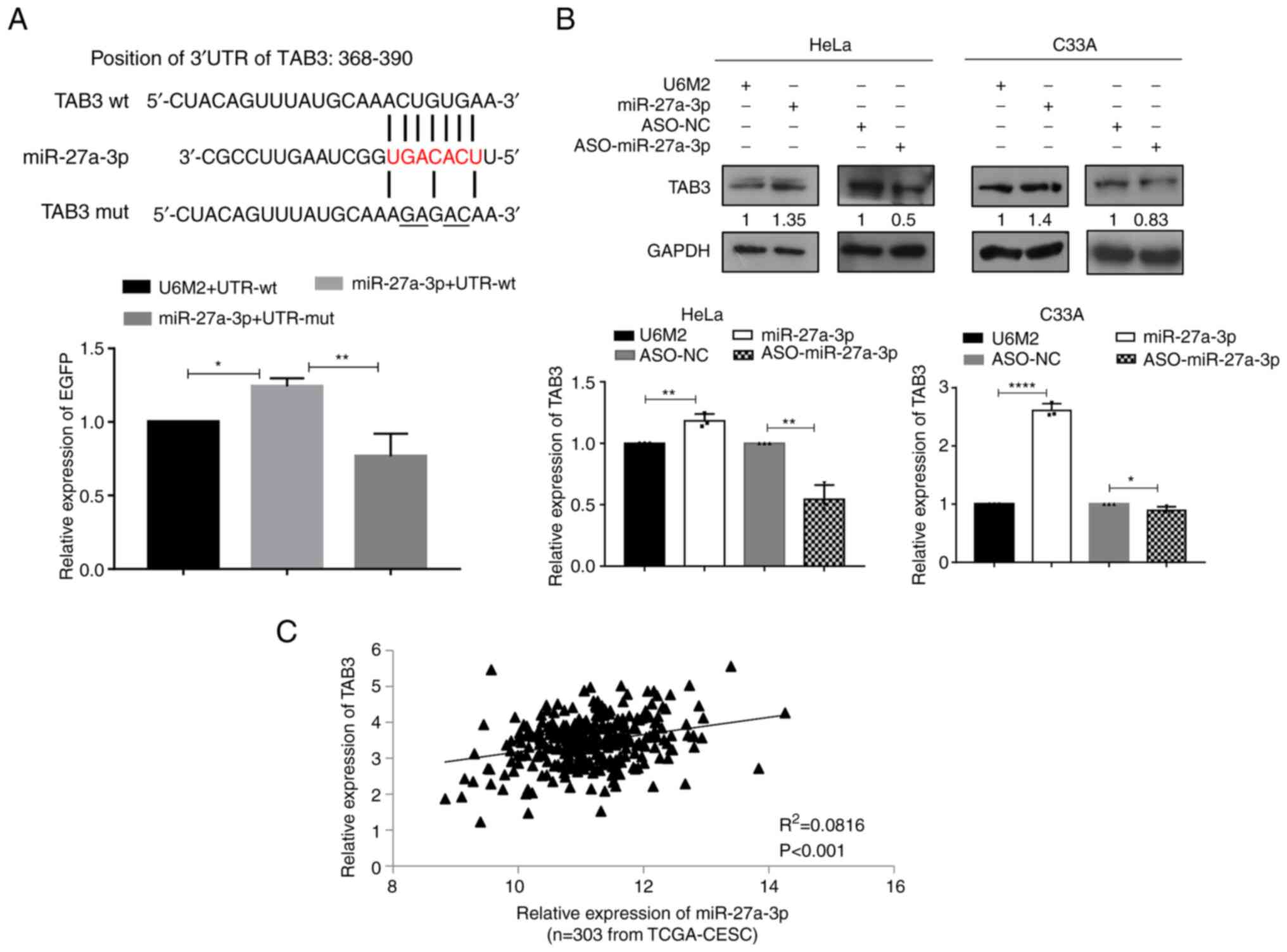
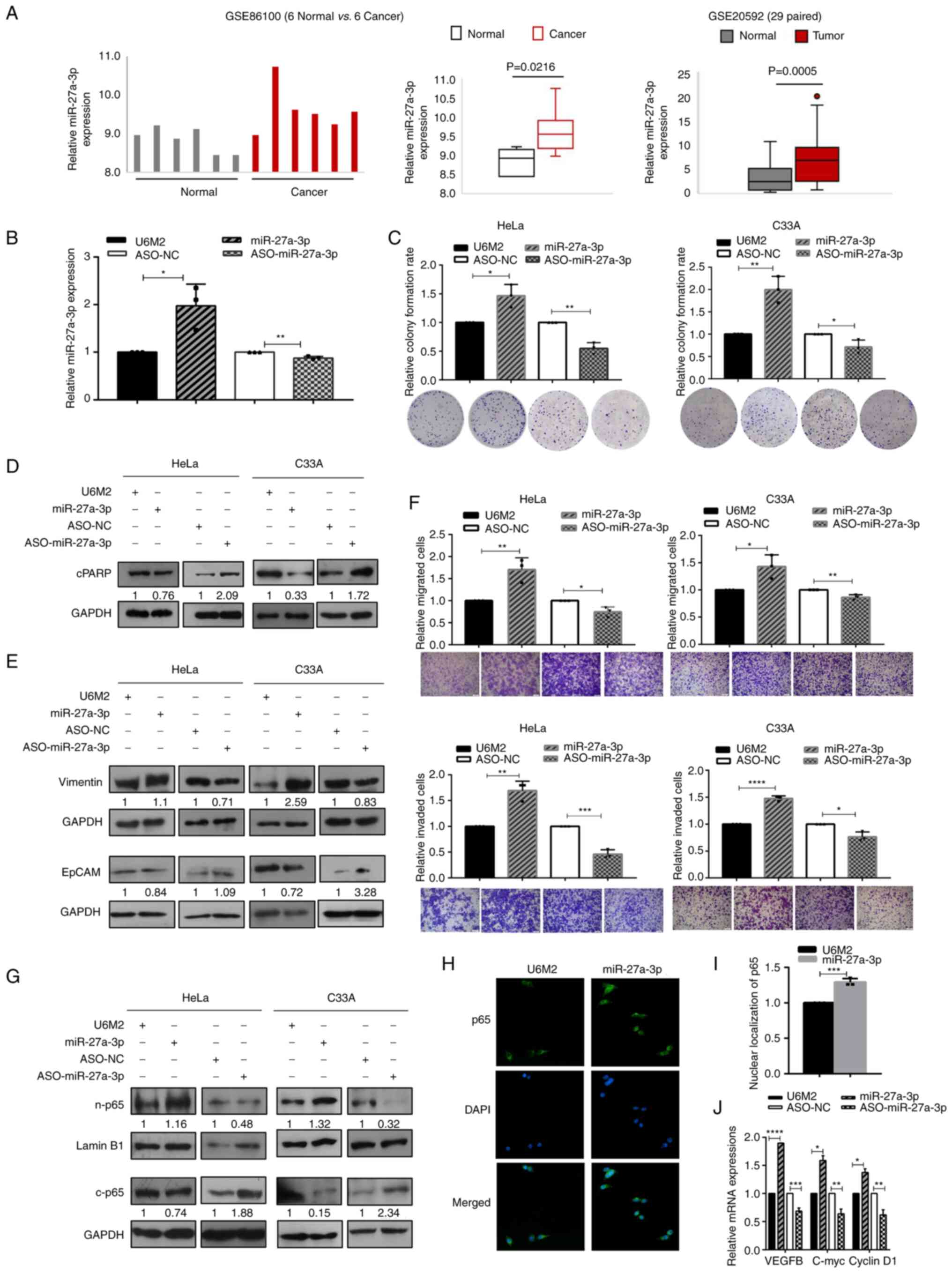 | Figure 1.The high expression of miR-27a-3p
facilitates the malignant properties and enhances the nuclear
translocation of NF-κB/p65 in cervical cancer cells. (A) miR-27a-3p
was highly expressed in cervical cancer tissues compared with
normal tissues based on the GSE86100 and GSE20592 datasets. (B)
RT-qPCR assays were used to determine the effectiveness of
miR-27a-3p overexpression and inhibition in HeLa cells. (C) The
effects of miR-27a-3p on cell growth were evaluated using colony
formation assays in cervical cancer cells. (D) The expression of
cleaved PARP (as an indicator of cell apoptosis) was examined using
western blot analysis upon the modulation of miR-27a-3p expression
in cervical cancer cells. (E) Western blot analysis was performed
to determine the specific biomarkers (Vimentin for mesenchymal
cells, and EpCAM for epithelial cells) in the epithelial
mesenchymal transition process in cervical cancer cells upon
miR-27a-3p overexpression and inhibition. (F) The effects of
miR-27a-3p on migration (upper panel) and invasion (lower panel)
were determined using Transwell assays in cervical cancer cells;
scale bars, 100 µm. (G) Western blot analysis of sub-fraction
extracts presented the nuclear and cytoplasmic p65 expression upon
the modulation of miR-27a-3p expression in cervical cancer cells.
(H) Representative images of immunofluorescence assays indicated
the enhanced nuclear translocation of p65 upon miR-27a-3p
overexpression in HeLa cells (magnification, ×400). (I) The
quantification of nuclear p65 expression based on
immunofluorescence assays. (J) RT-qPCR assays were performed to
determine the expression of NF-κB dependent genes upon the
modulation of miR-27a-3p expression. (C and F) The raw
representative graphs of the stained cells are presented under the
quantitative histograms. (D, E and G) The numbers under the western
blot bands indicate the relative quantifications, as normalized to
their endogenous loading control, respectively. *P<0.05,
**P<0.01, ***P<0.001 and ****P<0.0001, vs. their
respective control group. RT-qPCR, reverse
transcription-quantitative PCR. |
Furthermore, the expression levels of different
biomarkers in the epithelial-mesenchymal transition (EMT) processes
were examined using western blot analyses. As shown in Fig. 1E, the expression of the epithelial
cell marker, EpCAM, decreased and that of the mesenchymal cell
marker, Vimentin, increased with the overexpression of miR-27a-3p,
and vice versa. Transwell migration and invasion assays were used
to evaluate the effects of miR-27a-3p in cervical cancer cells. The
ectopic expression of miR-27a-3p significantly enhanced the
migratory (upper panel) and invasive (lower panel) abilities of the
HeLa and C33A cells, and the inhibition of miR-27a-3p significantly
decreased the migration and invasion of cervical cancer cells
(Fig. 1F). Taken together, these
results indicate the cancer-promoting effects of miR-27a-3p in
cervical cancer cells.
Since miR-27a-3p was identified as one of the
differentially expressed miRNAs associated with NF-κB silencing in
previous unpublished screening data by the authors, in the present
study, western blot and immunofluorescence analyses were performed
to examine the effects of miR-27a-3p on nuclear p65 expression and
the potential activation of NF-κB signaling. The sub-fraction
extracts examined using western blot analysis revealed the
increased nuclear p65 and decreased cytoplasmic p65 expression
levels upon the overexpression of miR-27a-3p in the HeLa and C33A
cells, and the opposite results were obtained following the
inhibition of miR-27a-3p expression (Fig. 1G). The enhanced nuclear p65 signals
upon miR-27a-3p overexpression were also observed in the HeLa
cells, as evaluated using immunofluorescence analyses (Fig. 1H and I). Further RT-qPCR assays were
performed to evaluate the expression levels of NF-κB-dependent
genes, such as VEGFB, c-Myc and cyclin D1 upon miR-27a-3p
modulation. As shown in Fig. 1J,
the mRNA expression levels of VEGFB, c-Myc and cyclin D1 were
significantly enhanced/decreased by miR-27a-3p
overexpression/inhibition in HeLa cells. Taken together, these data
suggest that the enhanced p65 nuclear translocation and the
activation of NF-κB signaling are induced by miR-27a-3p
overexpression.
TAB3 is upregulated by miR-27a-3p via
directly binding to its 3′UTR
To further investigate the regulatory mechanisms of
miR-27a-3p and its possible connections with the NF-κB signaling
pathway, bioinformatics analyses were first performed to search for
its potential targets and focused on the molecules involved in
NF-κB signaling. TAB3, MAP3K14 and TRAF3 were first selected for
verification using the 3′UTR EGFP fluorescence reporter systems. As
shown in Fig. 2A, there was a
putative miR-27a-3p binding site in the position 368–390 of 3′UTR
of TAB3. When the miR-27a-3p expression vector and the wild-type
3′UTR reporter vector were co-transfected, the normalized EGFP
intensities were significantly increased, while the mutant 3′UTR
reporter vector co-transfection abolished the enhancement of EGFP
intensities induced by miR-27a-3p, suggesting the direct binding of
miR-27a-3p with the 3′UTR of TAB3. Further western blot and RT-qPCR
analyses revealed the increased TAB3 expression at both the protein
and mRNA level upon miR-27a-3p overexpression, and the decreased
TAB3 expression upon miR-27a-3p inhibition using ASO-miR-27a-3p in
HeLa and C33A cells (Fig. 2B).
Further correlation analyses based on the available TCGA-CESC
dataset including 303 cervical cancer tissues revealed a
significant positive correlation between miR-27a-3p and TAB3
expression levels (Fig. 2C).
Similarly, bioinformatics analyses also revealed the
putative miR-27a-3p binding sites present in the 3′UTRs of MAP3K14
(position:1245-1267) and TRAF3 (position:1613-1635), as presented
in Fig. S2A. The 3′UTR EGFP
fluorescence reporter analyses obtained the similar results as
TAB3, i.e., the increased EGFP intensities were induced by
miR-27a-3p overexpression, and this enhancement was abolished by
co-transfection with the mutated 3′UTR reporter vectors (Fig. S2B). The MAP3K14 and TRAF3
expression levels were also analyzed upon the modulation of
miR-27a-3p expression in cervical cancer cells. As shown in
Fig. S2C, the mRNA expression
levels of MAP3K14 and TRAF3 were significantly increased following
miR-27a-3p overexpression and decreased upon miR-27a-3p inhibition
in both cervical cancer cells, and enhanced protein expression
levels were also observed upon the ectopic expression of miR-27a-3p
in HeLa cells (Fig. S2D). Further
survival analyses based on the expression levels of TAB3, MAP3K14,
TRAF3 and miR-27a-3p from the TCGA-CESC dataset were conducted
using online tools. As illustrated in Fig. S3, only MAP3K14 expression was
associated with significant changes in the overall survival of
patients; i.e., patients with a higher MAP3K14 expression exhibited
an improved survival rate; however, no significant associations
were found between TAB3, TRAF3 and miR-27a-3p expression and
patient survival.
To further investigate the mechanisms responsible
for the promoting effects on TAB3 expression induced by miR-27a-3p,
the present study first determined whether AGO2 protein
participated in this process. By constructing shRNA vectors for
AGO2 (shR1-AGO2 and shR2-AGO2) and determining their efficiencies
(Fig. S4A), miR-27a-3p was
overexpressed in C33A cells with/without shR-AGO2 and the protein
expression levels of TAB3 were re-evaluated. As demonstrated in
Fig. S4B, a similar upregulated
TAB3 expression was observed upon miR-27a-3p overexpression with
the inhibition of AGO2, indicating that AGO2 was involved in the
upregulation of TAB3 induced by miR-27a-3p. Notably, the globally
decreased TAB3 protein expression was also observed upon the
silencing of AGO2 compared with the control group, suggesting the
potential regulation of TAB3 by other AGO2-associated mechanisms or
other AGO2-dependent miRNA regulations.
TAB3 contributes to the malignant
phenotypes of human cervical cancer cells
Overexpression and knockdown vectors were
constructed for TAB3 (KBE-TAB3, shR1-TAB3 and shR2-TAB3) for the
further investigation of its biological roles. Firstly, their
efficiencies were evaluated using western blot analyses with
anti-TAB3 and anti-Flag. As shown in Fig. 3A, the KBE-TAB3 vector efficiently
enhanced TAB3 expression, and both the shR1-TAB3 and shR2-TAB3
vectors inhibited TAB3 expression. The shR2-TAB3 vector was
selected for performing the further loss-of-function experiments,
since its silencing effect was more prominent than that of
shR1-TAB3. By gain- and loss-of-function transit transfection, the
series functional effects of TAB3 on the colony formation,
apoptosis, migration and invasion of cervical cancer cells, as well
as the expression levels of EMT-associated biomarkers were
determined. The results revealed increased colony formation rates
upon TAB3 overexpression in cervical cancer cells, and TAB3
inhibition using shRNA significantly decreased the colony formation
abilities (Fig. 3B). The decreased
expression of cleaved PARP upon TAB3 overexpression and the
enhanced expression of cleaved PARP following the silencing of TAB3
expression were observed using western blot analyses in the
cervical cancer cells (Fig.
3C).
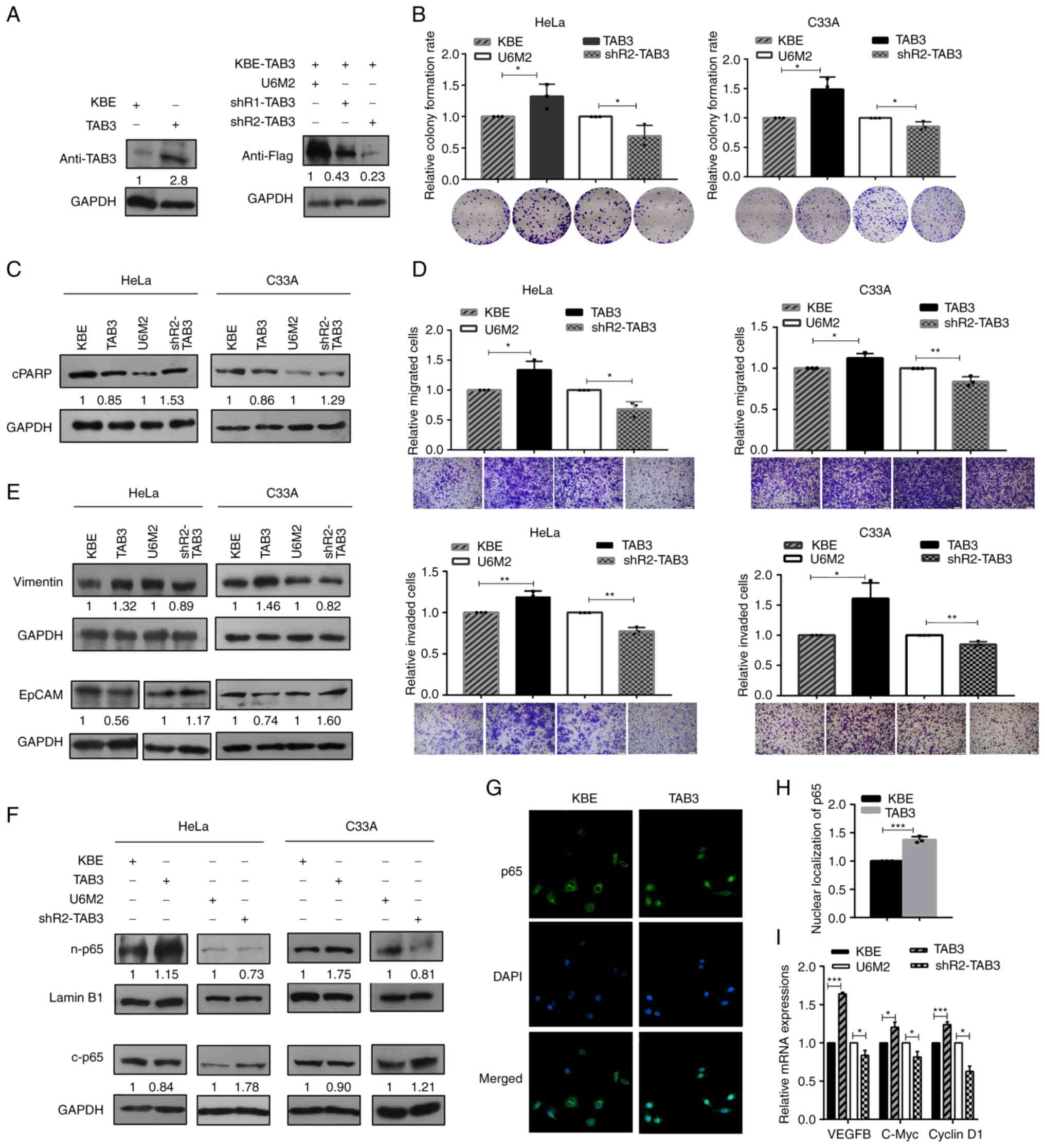 | Figure 3.TAB3 contributes to the malignant
phenotypes of human cervical cancer cells. (A) The efficiencies of
the constructed TAB3 overexpression and inhibition vectors
(KBE-TAB3, abbreviated as TAB3, and shRNA vectors, indicated as
shR1-TAB3 and shR2-TAB3) were evaluated using western blot
analysis. (B) Colony formation assays were performed to evaluate
the effects of TAB3 modulation on cell growth. (C) Representative
western blots of cleaved PARP illustrating the effects of TAB3
overexpression and inhibition on apoptosis. (D) Transwell migration
(upper) and invasion (lower) assays were applied to determine the
effects of the modulation of TAB3 expression; scale bars, 100 µm.
(E) The specific biomarkers (Vimentin and EpCAM) of the
epithelial-mesenchymal transition process were determined using
western blot analysis to investigate the effects of TAB3
overexpression and inhibition. (F) Western blot analysis indicated
the increased nuclear p65 and decreased cytoplasmic p65 expression
upon the ectopic expression of TAB3 in cervical cancer cells, and
vice versa. (G) Representative images of immunofluorescence assays
indicated the enhanced nuclear translocation of p65 upon TAB3
overexpression in HeLa cells (magnification, ×400). (H) The
quantification of nuclear p65 expression based on
immunofluorescence assays. (I) Reverse transcription-quantitative
PCR assays were performed to determine the expression of
NF-κB-dependent genes (VEGFB, C-Myc and cyclin D1) upon the
modulation of TAB3 expression. (B and D) The raw representative
graphs of the stained cells are presented under the quantitative
histograms. (A, C, E and F) The numbers under the western blot
bands indicate the relative quantifications, as normalized to their
endogenous loading control, respectively. All experiments, apart
from those in (A) (n=2) were performed at least three times
independently. *P<0.05, **P<0.01 and ***P<0.001, vs. their
respective control group. TAB3, TGF-β activated kinase 1 binding
protein 3. |
Transwell migration and invasion assays revealed the
elevated migration (upper panel) and invasion (lower panel)
properties following the ectopic expression of TAB3, while the
significantly decreased migratory and invasive capacities were
observed in the TAB3 inhibition groups (Fig. 3D). Moreover, the increased
expression of Vimentin, one of the mesenchymal cell markers, and
the reduced expression of the epithelial cell marker, EpCAM, were
observed following TAB3 overexpression in the cervical cancer
cells. The opposite results, i.e., the decreased expression of
Vimentin and the increased expression of EpCAM were observed upon
the inhibition of TAB3, suggesting that TAB3 promoted the EMT
processes (Fig. 3E). Furthermore,
the analyses of p65 expression based on the sub-fraction extracts
revealed the increased nuclear p65 and decreased cytoplasmic p65
expression following TAB3 overexpression; opposite results were
obtained by the silencing of TAB3 expression in both HeLa and C33A
cells (Fig. 3F). The
immunofluorescence assays also revealed the enhanced p65 nuclear
signals upon TAB3 overexpression in HeLa cells (Fig. 3G and H). RT-qPCR analyses to
determine the expression of NF-κB-dependent genes (VEGFB, c-Myc and
cyclin D1) revealed their significantly increased expression levels
upon TAB3 overexpression, while the silencing of TAB3 decreased
their expression levels in HeLa cells (Fig. 3I), suggesting the enhanced nuclear
p65 expression and the activation of NF-κB signaling by TAB3
overexpression in cervical cancer cells.
Inhibition of TAB3 abolishes the
effects promoted by miR-27a-3p in cervical cancer cells
Since it was observed that miR-27a-3p and TAB3
exerted similar effects on the aggressive phenotypes of cervical
cancer cells and that miR-27a-3p promoted the expression of TAB3,
the present study therefore determined whether the promoting
effects of miR-27a-3p on the malignant potential of cervical cancer
cells were achieved due to its promoting effects on TAB3
expression. A series of functional rescue experiments was conducted
by co-transfection with miR-27a-3p + shR-TAB3 and ASO-miR-27a-3p +
TAB3. As was expected, the elevated colony formation rates induced
by miR-27a-3p overexpression were counteracted by the silencing of
TAB3 expression, and the suppressed colony formation properties
induced by ASO-miR-27a-3p were rescued by TAB3 overexpression in
both HeLa and C33A cells (Fig. 4A).
The expression levels of EMT-associated specific molecules, i.e.,
the decreased EpCAM and increased Vimentin expression induced by
miR-27a-3p were restored to their normal levels by co-transfection
with shR2-TAB3, and vice versa (Fig.
4B). Furthermore, the knockdown of TAB3 mostly abolished the
enhanced migratory (Fig. 4C) and
invasive (Fig. 4D) abilities of the
cervical cancer cells induced by miR-27a-3p overexpression; in
addition, the suppressed migratory and invasive properties induced
by ASO-miR-27a-3p transfection were counteracted by the ectopic
expression of TAB3. The decreased/increased expression of cleaved
PARP induced by miR-27a-3p/ASO-miR-27a-3p was also neutralized by
the silencing/overexpressing TAB3 expression (Fig. 4E).
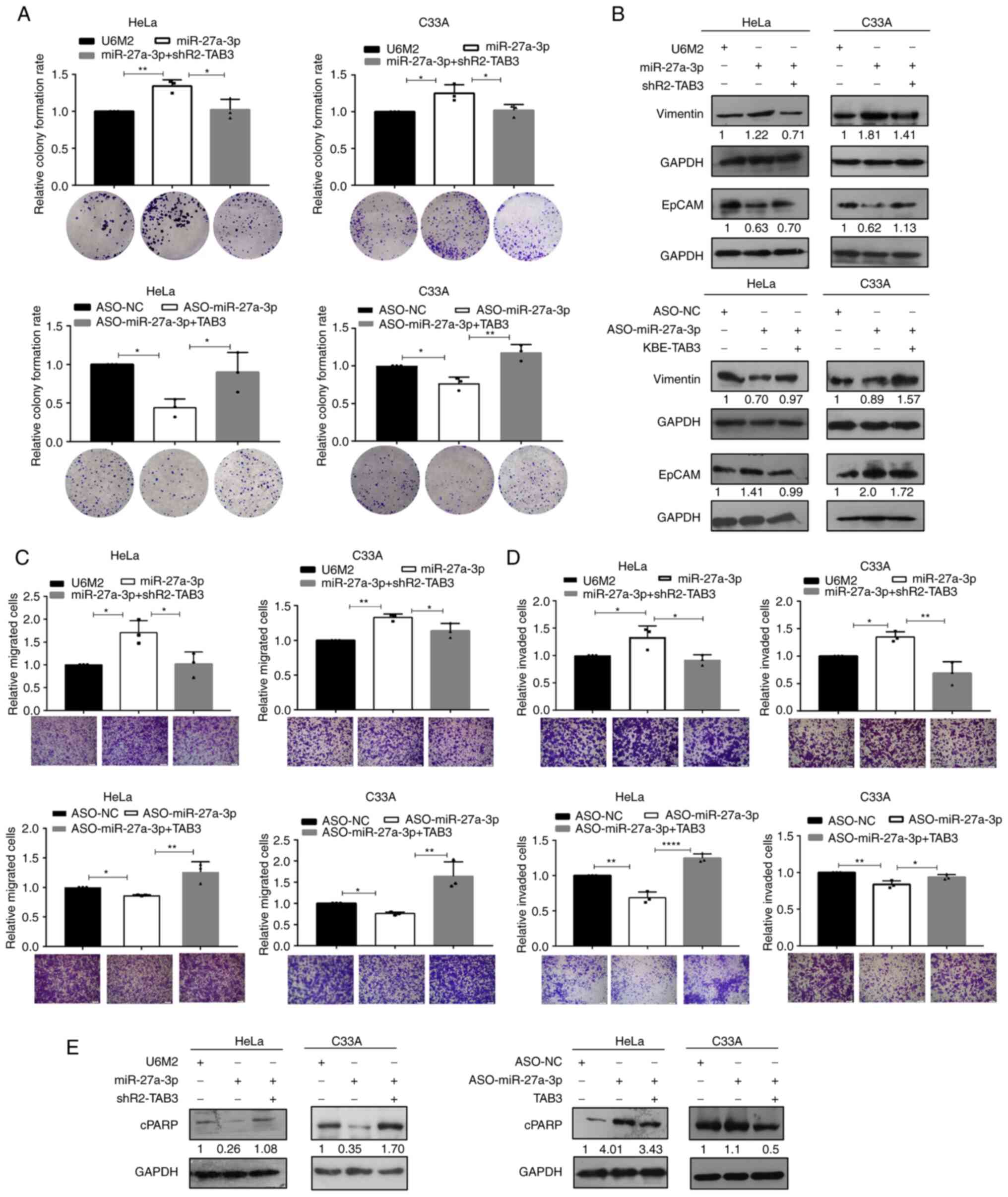 | Figure 4.Inhibition of TAB3 abolishes the
effects induced by miR-27a-3p in cervical cancer cells. The
functional consequences were determined using (A) colony formation
assay, (B) western blot analysis for the epithelial-mesenchymal
transition specific biomarkers, (C) migration, (D) invasion and (E)
apoptosis assays following co-transfection with miR-27a-3p +
shR2-TAB3 and ASO-miR-27a-3p + TAB3 to evaluate the functional
rescue effects of the miR-27a-3p-induced TAB3 upregulation. (A, C
and D) The raw representative graphs of the stained cells are
presented under the quantitative histograms; scale bars, 100 µm. (B
and E) The numbers under the western blot bands indicate the
relative quantifications, as normalized to their endogenous loading
control, respectively. *P<0.05, **P<0.01 and ****P<0.0001,
vs. their respective control group. TAB3, TGF-β activated kinase 1
binding protein 3. |
Taken together, the findings demonstrated that the
suppression of TAB3 at least partially eliminated the effects
generated by overexpression miR-27a-3p, and the ectopic expression
of TAB3 abolished the effects induced by the inhibition of
miR-27a-3p expression, indicating that TAB3 served as a direct
functional target gene of miR-27a-3p, and that the enhanced
malignant effects induced by miR-27a-3p overexpression were
mediated by its promoting effects on TAB3 expression in cervical
cancer cells.
The transcription factor p65 directly
binds to the promoter region of miR-27a-3p and activates its
transcription
Previous studies have reported the dysregulated
expression of miR-27a-3p in cervical cancer samples (25,26).
In an unpublished screening study by the authors, miR-27a-3p was
identified as one of the dysregulated miRNAs by silencing NFKB1 in
human cancer cells. The present study thus further focused on the
transcriptional regulation of miR-27a-3p, as well as the
miR-23a/27a/24-2 cluster. Using bioinformatics analyses, two
putative transcription start sites (TSSs) were identified in the
analyzed fragment and two promoter reporter vectors containing the
two predicted TSSs were constructed, referred to as pGL3-P1 and
pGL3-P2, respectively (Fig. 5A). As
demonstrated in Fig. S5A, the
relative luciferase activity of the fragment P2 was ~4-fold higher
than that of the fragment P1, suggesting that the fragment P2
contained the functional promoter sequences. Further analyses of
the fragment P2 sequence identified two potential p65 binding sites
presented (Fig. 5A, positions:
214–223 and 546–555 in this 773 bp full length) and a series of
truncated promoter reporter vectors with none or either one p65
binding site and flanking sequence were constructed, respectively.
Of note, the truncated fragments F259 and F509 had higher promoter
activities compared with the full-length fragment P2, and both of
these only contained the predicted second p65 binding site
(position 546–555), suggesting a critical positive regulatory role
for this site, while the other one (position 214–223) appeared to
have negative effects (Fig. 5B).
Following the verification of the efficiencies of the shR-p65 and
KBE-p65 vectors (Fig. S5B), the
enhanced/decreased promoter activities of fragment P2 and its
truncated form F509 were observed upon the modulation of p65
expression (Fig. 5C). Furthermore,
this p65 binding site was deleted in fragment F509 and the promoter
activities were re-evaluated using dual luciferase reporter
systems. As illustrated in Figs. 5C
and S5C and D, the significantly
decreased promoter activities induced by the silencing of p65 and
the silencing of NFKB1 were abolished, and the increased promoter
activities induced by the ectopic expression of p65 or NFKB1/p50
were partially decreased, although these were still significantly
higher compared with the wild-type sequence of F509 which included
the p65 binding site, suggesting the importance of this binding
site.
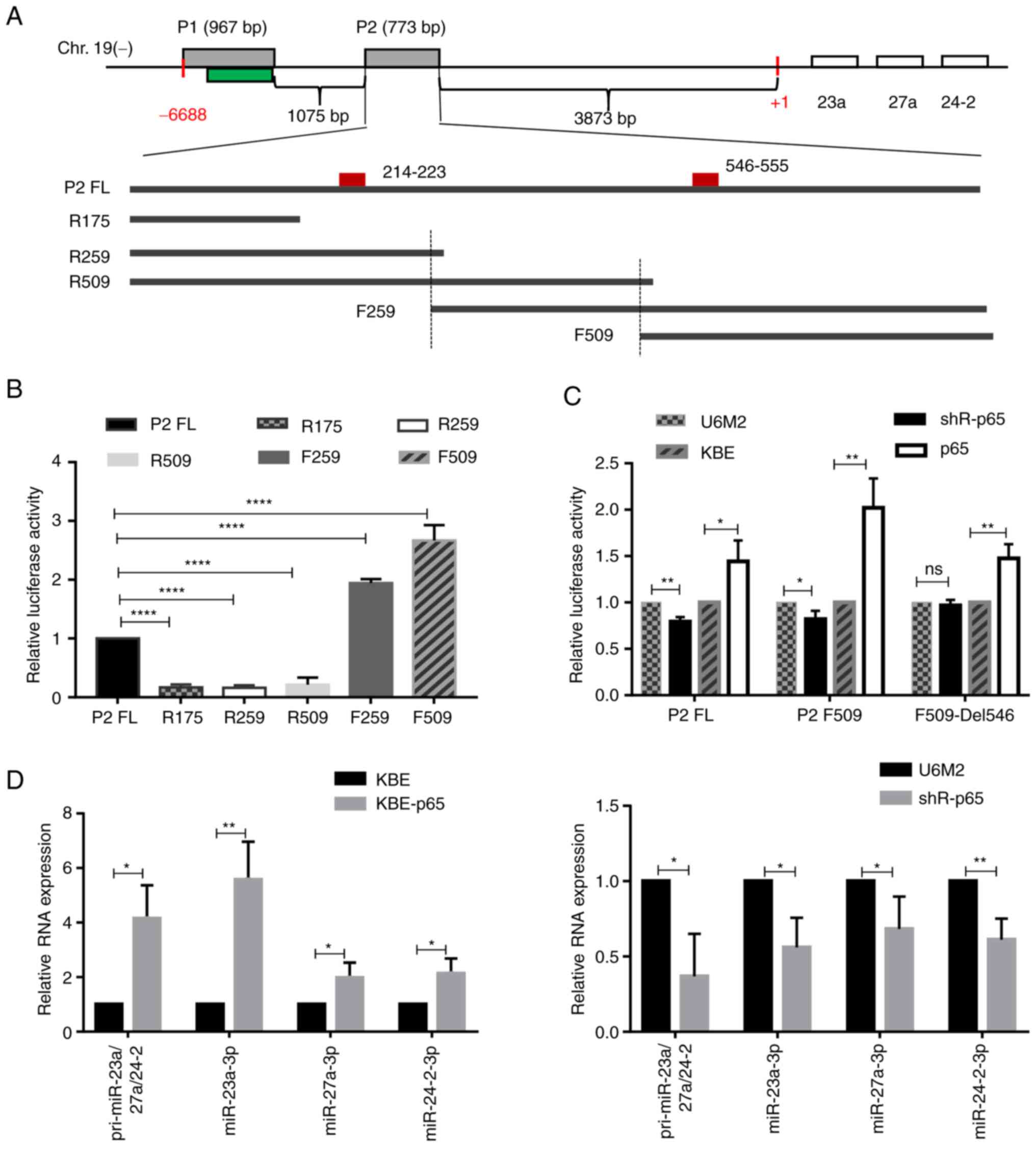 | Figure 5.p65 directly binds to the promoter
region of miR-27a and activates its transcription. (A) The graph
illustrates the location of miR-23a/27a/24-2 cluster and miR-27a-3p
in the minus strand of human chromosome 19, as well as the
constructed series truncated promoter reporter vectors (P2 full
length abbreviated as P2 FL, and the truncated ones, as indicated
R175, R259, R509, F259 and F509). The two grey boxes indicate the
two amplified fragments (P1 and P2) and the green one indicates the
predicted CpG island region. The small red box in the P2 fragment
indicates the predicted p65 binding site and their relative
positions are labelled with numbers. (B) The relative luciferase
activities of the series P2 truncated promoter reporter vectors
were determined using dual luciferase reporter systems. (C) The
effects of the modulation of p65 expression on the promoter
activities of the full length P2, the truncated F509 and the p65
binding site deleted one (F509-Del546). (D) Reverse
transcription-quantitative PCR assays were performed to determine
the effects of the modulation of p65 expression on the expression
levels of pri-miR-23a/27a/24-2, miR-27a-3p, miR-23a-3p and
miR-24-2-3p. *P<0.05, **P<0.01 and ****P<0.0001, vs. (B)
the P2 Full length group, or (C and D) vs. the respective control
group. ns, not significant. |
Furthermore, RT-qPCR assays revealed the increased
or decreased primary transcript for this cluster (miR-23a/27a/24-2)
and miR-27a-3p expression by p65 overexpression or inhibition, as
well as the similar expression patterns for miR-23a-3p and
miR-24-2-3p (Fig. 5D), the other
two abundant members from the same miRNA cluster, suggesting the
members from the same miRNA cluster transcript together. Taken
together, these data suggested that the direct binding of p65 to
this specific site contributed to the upregulated expression of
miR-27a-3p by enhancing its transcription.
Discussion
NF-κB is a transcription factor firstly identified
in mature B cells in 1988 (36,37).
The mammalian NF-κB family is composed of five members, including
RELA/p65, RelB, c-Rel, NFKB1/p50 and NFKB2/p52. Generally, the
heterodimeric complex p50-p65 is the most abundant form of NF-κB in
the majority of cells (38).
Increasing evidence has indicated that NF-κB is highly activated in
a variety of cancer types (39,40).
miRNAs not only regulate NF-κB expression directly, but also up- or
downregulate NF-κB activity via the upstream or downstream
signaling pathways of NF-κB. For example, miR-146a/b can inhibit
TRAF6 and IRAK1 expression and further inhibit the NF-κB activity
in breast cancer cells (41).
miR-1290 was previously found to be upregulated by NF-κB in colon
cancer tissues compared with normal adjacent tissues and to
directly suppress NF-κB repressing factor, which in turn activated
the Akt and NF-κB pathways (42).
The present study first investigated the functional effects of
miR-27a-3p, one of the identified dysregulated miRNAs by silencing
NFKB1, and further investigated its regulatory roles in cervical
cancer cells. Using bioinformatics analyses and experimental
validation, it was found that miR-27a-3p upregulated TAB3
expression by binding to its 3′UTR and subsequently triggered the
activation of NF-κB signaling. Functionally, it was demonstrated
that miR-27a-3p promoted the malignant potential of cervical cancer
cells, and rescue experiments suggested that the enhanced malignant
potential induced by miR-27a-3p overexpression was directly
mediated by its upregulation of TAB3.
As a class of non-coding RNA molecules, the
functions of miRNAs have been extensively studied in various cancer
types over the past decade, while the mechanisms responsible for
their dysregulated expression remain to be fully elucidated. It has
been reported that the miR-203 locus is highly methylated and
epigenetically silenced by DNA methylation in several metastatic
tumor cells (16,17). Ma et al (43) demonstrated a positive feedback loop
composed of KLF3, miR-23a/27a, β-like globin gene and the
miR-23a/27a/24-2 cluster during erythropoiesis. In addition,
another study demonstrated that c-Fos transcriptionally activated
miR-27a expression and miR-27a regulated the translocation of
apoptosis-inducing factor from the mitochondria to the nucleus by
targeting ATAD3a and contributed to myocardial ischemia-reperfusion
injury (44). Tao et al
(45) concluded that the
transcription factor p53 significantly decreased the expression of
miR-27a by binding to the specific sites of the promoter region in
mouse ovarian granulosa cells. The present study identified one
specific p65 binding site in the promoter region of the
miR-23a/27a/24-2 cluster, and p65 transcriptionally enhanced the
expression levels of primary miRNA transcript and mature miRNAs,
while the silencing of p65 significantly decreased their expression
levels in cervical cancer cells. In particular, it was found that
NFKB1/p50 had similar effects as p65, although no p50 binding site
was present in the reporter construct, which was reasonable and
explainable, since the p50-p65 heterodimer was the most abundant
form of the NF-κB transcription factor. Moreover, as an upstream
activator of the NF-κB pathway, TAB3 was also upregulated by
miR-27a-3p and enhanced the nuclear translocation of the p65-p50
heterodimer, suggesting the positive feedback loop among
miR-27a-3p, TAB3 and the NF-κB signaling pathway.
In recent years, several studies have presented
controversial functional results on miR-27a in cervical cancer
cells, and indicated its oncogenic or tumor suppressor roles in
different situations. For example, as previously demonstrated,
miR-27a-3p plays an oncogenic functional role by promoting the
proliferation, migration and invasion of HeLa cells, mediated by
the inhibition of BTG2 and FBXW7 expression using miR-27a-3p mimic
individually (24,26). However, Fang et al (25) reported that miR-27a-3p
overexpression using Agomir (chemically modified miRNA mimic)
inhibited the proliferation, migration and invasion of HeLa cells
by downregulating TGF-βRI. The present study investigated the
functional effects of miR-27a-3p using the strand specific
expression vector in both the cervical adenocarcinoma cell line,
HeLa, and the cervical squamous cell carcinoma cell line, C33A.
Compared with the traditional construction method for miRNA
overexpression, in which both −3p and −5p are expressed together,
the current strand specific expression construct can only produce
the indicated miR-27a-3p molecule. The results suggested the
oncogenic roles of miR-27a-3p in both HeLa and C33A cells by
promoting the colony formation, migratory and invasive abilities
upon the overexpression of miR-27a-3p.
Typically, miRNAs negatively regulate multiple genes
and lead to mRNA degradation or translation suppression via binding
to the 3′UTRs (46). Some studies
have also shown the positive regulation of gene expression by
miRNAs. For example, a previous study demonstrated that miR-744
upregulated CCNB1 expression by promoting the enrichment of RNA
polymerase II and the tri-methylation of histone 3 at lysine 4
(H3K4me3) at its transcription start site in mouse NIH/3T3 cells
(47). Another study demonstrated
that miR-122 enhanced hepatitis C virus (HCV) replication by
targeting its 5′-noncoding elements in the HCV genome (48). In addition, it was previously found
that miR-1 exerted inhibitory effects in the cytoplasm, while it
stimulated the translation of mitochondrial genome-encoded ND1 and
COX1 when GW182 was silenced in the cytoplasm (49). Song et al (50) reported that miR-346 bound to the
3′UTR of hTERT and upregulated its expression by recruiting the
mRNA to the ribosome and promoted its translation in an
AGO2-independent manner, which was mediated by G-rich RNA sequence
binding factor 1 (GRSF1). In the present study, the enhanced
expression of TAB3 was observed following the overexpression of
miR-27a-3p and this enhancement may be at least partially mediated
in an AGO2-dependent manner, since similar upregulated TAB3
expression patterns (miR-27a-3p vs. control) were observed
following the silencing of AGO2. However, whether other proteins,
such as GRSF1 are also involved in the promoting effects remains
unclear. The transfected HeLa cells were further treated with
cycloheximide and actinomycin D individually, and the stabilities
of TAB3 were determined; no significant differences were observed
in the presence of altering miR-27a-3p expression, which indicated
that the upregulated TAB3 expression induced by miR-27a-3p was not
due to the increased protein stability or the cessative
transcription activity (data not shown). It was thus hypothesized
that this upregulation may probably resulted from the enhanced
synthesis of transcripts. However, further mechanistic analyses are
required for further clarifications.
Of note, the globally decreased endogenous
expression of TAB3 was observed following the silencing of AGO2
expression with/without miR-27a-3p overexpression in cervical
cancer cells. AGO2 may be involved in the inhibition of translation
initiation either by binding to the 7-methylguanosine cap (51) or via the interaction with eIF6 and
preventing the assembly of ribosome (52), further resulting in mRNA degradation
in the processing bodies (P-bodies). This may be a reasonable
explanation for the globally decreased TAB3 protein expression by
inhibiting AGO2. On the other hand, the AGO2 protein is required
for RNA-mediated gene silencing by the RNA-induced silencing
complex. According to the miRTarBase prediction, 18 miRNAs were
predicted to target TAB3 in total, and some miRNAs were verified in
experimental systems (53,54). Thus, it can be hypothesized that the
silencing of AGO2 may affect the miRNA-mediated regulatory effects
on TAB3, which may also result in the globally reduced expression
of TAB3. However, further experimental evidence is required to
answer this question precisely and this may represent another
challenging research topic for the future.
Moreover, TRAF3 and MAP3K14 were also identified and
verified as direct targets of miR-27a-3p using EGFP 3′UTR
reporters, RT-qPCR and western blot analyses in the present study.
Among these, MAP3K14 could bind to TRAF2 and stimulate NF-κB
activity, while TRAF3 functioned as a constitutive negative
regulator of the alternative NF-κB pathway. Taken together, it can
be hypothesized that the effects induced by miR-27a-3p on NF-κB
activities not only derived from its upregulation of TAB3, but may
also be attributed to other molecules involved in the NF-κB
signaling pathway, such as MAP3K14 and TRAF3 presented herein. It
may be of interest to formulate the miR-27a-3p and NF-κB signaling
pathway-associated regulatory network.
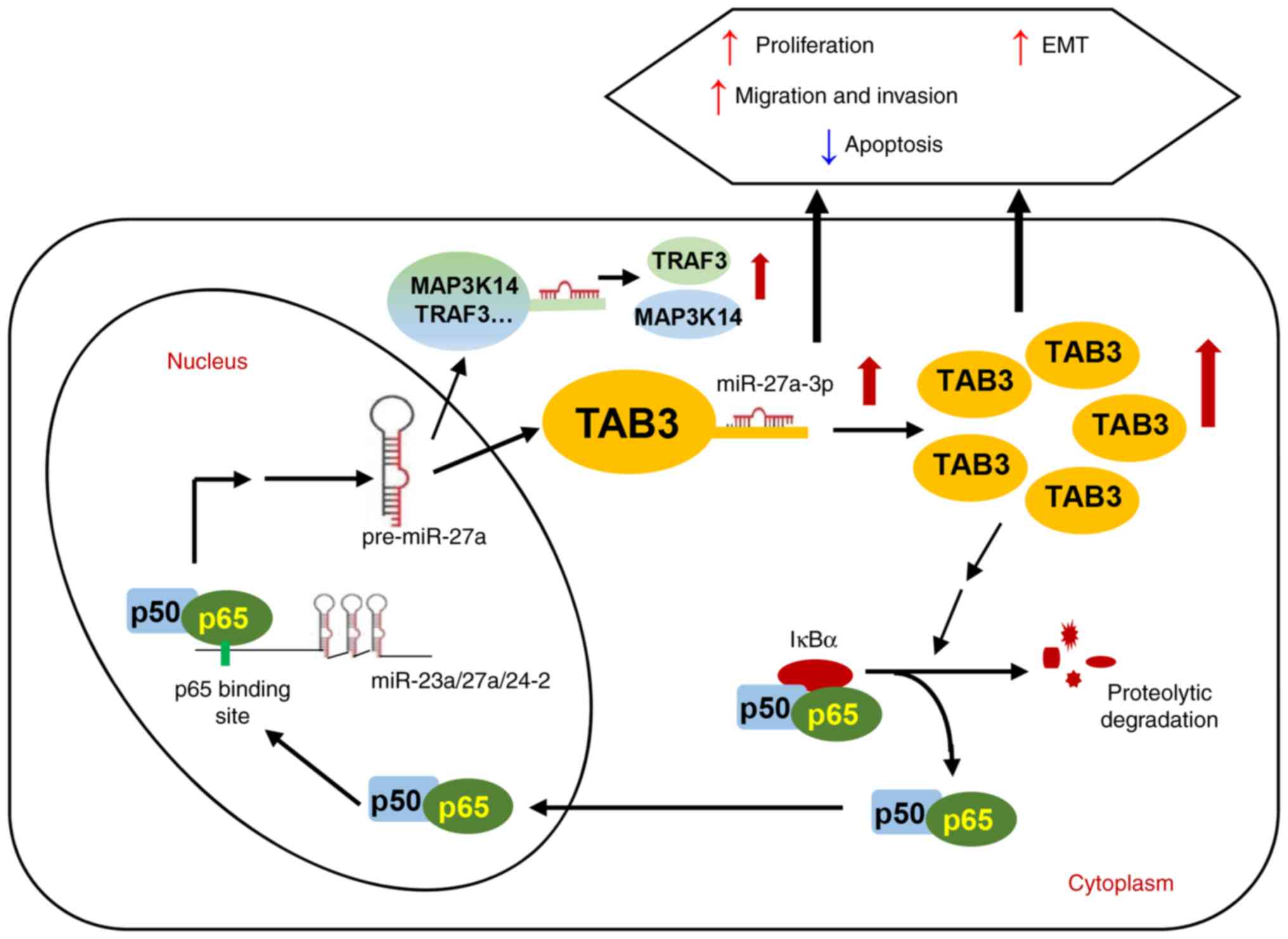
In conclusion, the present study revealed that
NF-κB/p65 transcriptionally activated miR-27a-3p and miR-27a-3p
promoted TAB3 expression by binding to its 3′UTR and subsequently
activated the NF-κB signaling pathway, which contributed to
cervical tumorigenesis. Functionally, it was demonstrated that
miR-27a-3p enhanced the colony formation, migratory and invasive
abilities of cervical cancer cells and promoted the EMT process.
Moreover, TAB3 played an oncogenic role and functional rescue
experiments indicated that the oncogenic functions induced by
miR-27a-3p were mediated by its upregulation of TAB3 expression in
cervical cancer cells. In addition, the miR-27a-3p upstream
transcription factor, p65, was identified to promote the
transcription and expression of the miR-23a/27a/24-2 cluster
members. Taken together, the present study demonstrated a positive
feedback regulatory loop composed of p65, miR-27a-3p, TAB3 and the
NF-κB pathway in the progression of cervical cancer (Fig. 6), suggesting a novel miRNA-mediated
mechanism between pro-inflammation and tumor transformation by the
continuous activation of the NF-κB pathway. A limitation of the
present study was lack of in vivo evaluations, while studies
from other groups have also indicated the oncogenic roles of
miR-27a-3p in cervical cancer cells (21,23),
which may function as a supplement to the in vitro
experiments herein. The present study highlighted the oncogenic
roles of TAB3 and miR-27a-3p in cervical cancer cells, which may
provide new insight into cervical cancer malignancies, and may
provide potential valuable novel biomarkers for clinical
applications.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Weiying Liu and
Dr Qi Sun from Tianjin Medical University for providing the
KBE-p65, KBE-p50 vectors and shR-NFKB1 vector, and Dr Per Johansson
from Karolinska Institutet for providing the pcDNA3-U6M2
vector.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 81773002 and 31971100) and the
Natural Science Foundation of Tianjin City (grant no.
16JCYBJC42400).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HX conceived, designed and supervised the study. HX,
ML, ZG and SW performed the experiments. HX, ML, ZG, SW and YZ
analyzed and interpreted the data. ML, YZ and HX wrote the
manuscript. ML and HX confirm the authenticity of all the raw data.
All authors provided comments, and have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
zur Hausen H: Papillomaviruses in the
causation of human cancers-a brief historical account. Virology.
384:260–265. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Scarth JA, Patterson MR, Morgan EL and
Macdonald A: The human papillomavirus oncoproteins: A review of the
host pathways targeted on the road to transformation. J Gen Virol.
102:0015402021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liang LA, Einzmann T, Franzen A, Schwarzer
K, Schauberger G, Schriefer D, Radde K, Zeissig SR, Ikenberg H,
Meijer CJLM, et al: Cervical cancer screening: Comparison of
conventional pap smear test, liquid-based cytology, and human
papillomavirus testing as stand-alone or Cotesting strategies.
Cancer Epidemiol Biomarkers Prev. 30:474–484. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Perkins RB, Guido RL, Saraiya M, Sawaya
GF, Wentzensen N, Schiffman M and Feldman S: Summary of current
guidelines for cervical cancer screening and management of abnormal
test results: 2016–2020. J Womens Health (Larchmt). 30:5–13. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lei J, Ploner A, Elfström KM, Wang J, Roth
A, Fang F, Sundström K, Dillner J and Sparén P: HPV Vaccination and
the risk of invasive cervical cancer. N Engl J Med. 383:1340–1348.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Palmer T, Wallace L, Pollock KG, Cuschieri
K, Robertson C, Kavanagh K and Cruickshank M: Prevalence of
cervical disease at age 20 after immunisation with bivalent HPV
vaccine at age 12–13 in Scotland: Retrospective population study.
BMJ. 365:l11612019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in Human Cancer. MicroRNA Cancer Regulation. pp1–20.
2013. View Article : Google Scholar
|
|
9
|
He Y, Lin J, Ding Y, Liu G, Luo Y, Huang
M, Xu C, Kim TK, Etheridge A, Lin M, et al: A systematic study on
dysregulated microRNAs in cervical cancer development. Int J
Cancer. 138:1312–1327. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lui WO, Pourmand N, Patterson BK and Fire
A: Patterns of known and novel small rnas in human cervical cancer.
Cancer Res. 67:6031–6043. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie H, Zhao YG, Caramuta S, Larsson C and
Lui WO: miR-205 expression promotes cell proliferation and
migration of human cervical cancer cells. PLoS One. 7:e469902012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie H, Lee L, Scicluna P, Kavak E, Larsson
C, Sandberg R and Lui WO: Novel functions and targets of miR-944 in
human cervical cancer cells. Int J Cancer. 136:E230–E241. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jin HY, Oda H, Lai M, Skalsky RL, Bethel
K, Shepherd J, Kang SG, Liu WH, Sabouri-Ghomi M, Cullen BR, et al:
MicroRNA-17~92 plays a causative role in lymphomagenesis by
coordinating multiple oncogenic pathways. EMBO J. 32:2377–2391.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Feng Y, Zhou S, Li G, Hu C, Zou W, Zhang H
and Sun L: Nuclear factor-κB-dependent microRNA-130a upregulation
promotes cervical cancer cell growth by targeting phosphatase and
tensin homolog. Arch Biochem Biophys. 598:57–65. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bueno MJ, Pérez de Castro I, Gómez de
Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM,
Fernández-Piqueras J and Malumbres M: Genetic and epigenetic
silencing of MicroRNA-203 enhances ABL1 and BCR-ABL1 oncogene
expression. Cancer Cell. 13:496–506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cui X, Chen X, Wang W, Chang A, Yang L,
Liu C, Peng H, Wei Y, Liang W, Li S, et al: Epigenetic silencing of
miR-203 in Kazakh patients with esophageal squamous cell carcinoma
by MassARRAY spectrometry. Epigenetics. 12:698–707. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang Y, Yang Z, Zhang R, Jia C, Mao R,
Mahati S, Zhang Y, Wu G, Sun YN, Jia XY, et al: MiR-27a-3p enhances
the cisplatin sensitivity in hepatocellular carcinoma cells through
inhibiting PI3K/Akt pathway. Biosci Rep. 41:BSR201920072021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chae DK, Ban E, Yoo YS, Kim EE, Baik JH
and Song EJ: MIR-27a regulates the TGF-signaling pathway by
targeting SMAD2 and SMAD4 in lung cancer. Mol Carcinogen.
56:1992–1998. 2017. View Article : Google Scholar
|
|
20
|
Kong LY, Xue M, Zhang QC and Su CF: In
vivo and in vitro effects of microRNA-27a on proliferation,
migration and invasion of breast cancer cells through targeting of
SFRP1 gene via Wnt/β-catenin signaling pathway. Oncotarget.
8:15507–15519. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liang JT, Tang JM, Shi HJ, Li H, Zhen T,
Duan J, Kang L, Zhang F, Dong Y and Han A: miR-27a-3p targeting RXR
α promotes colorectal cancer progression by activating
Wnt/β-catenin pathway. Oncotarget. 8:82991–83008. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
You J, Li J, Ke C, Xiao Y, Lu C, Huang F,
Mi Y, Xia R and Li Q: Oncogenic long intervening noncoding RNA
Linc00284 promotes c-Met expression by sponging miR-27a in
colorectal cancer. Oncogene. 40:4151–4166. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi J, Yang C, An J, Hao D, Liu C, Liu J,
Sun J and Jiang J: KLF5-induced BBOX1-AS1 contributes to cell
malignant phenotypes in non-small cell lung cancer via sponging
miR-27a-5p to up-regulate MELK and activate FAK signaling pathway.
J Exp Clin Cancer Res. 40:1482021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li S, Han Y, Liang X and Zhao M: LINC01089
inhibits the progression of cervical cancer via inhibiting
miR-27a-3p and increasing BTG2. J Gene Med. 23:e32802020.PubMed/NCBI
|
|
25
|
Fang F, Huang B, Sun S, Xiao M, Guo J, Yi
X, Cai J and Wang Z: miR-27a inhibits cervical adenocarcinoma
progression by downregulating the TGF-βRI signaling pathway. Cell
Death Dis. 9:3952018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ben W, Zhang G, Huang Y and Sun Y:
MiR-27a-3p regulated the aggressive phenotypes of cervical cancer
by targeting FBXW7. Cancer Manag Res. 12:2925–2935. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jin G, Klika A, Callahan M, Faga B, Danzig
J, Jiang Z, Li X, Stark GR, Harrington J and Sherf B:
Identification of a human NF-kappa B-activating protein, TAB3. Proc
Natl Acad Sci USA. 101:2028–2033. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheung PCF, Nebreda AR and Cohen P: TAB3,
a new binding partner of the protein kinase TAK1. Biochem J.
378:27–34. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Q, Chen L, Luo C, ChenYan, Ge J, Zhu Z,
Wang K, Yu X, Lei J, Liu T, et al: TAB3 upregulates PIM1 expression
by directly activating the TAK1-STAT3 complex to promote colorectal
cancer growth. Exp Cell Res. 391:1119752020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao J, Gai L, Gao Y, Xia W, Shen D, Lin
Q, Mao W, Wang F, Liu P, Chen J, et al: TAB3 promotes human
esophageal squamous cell carcinoma proliferation and invasion via
the NF-κB pathway. Oncol Rep. 40:2876–2885. 2018.PubMed/NCBI
|
|
31
|
Chen Y, Wang X, Duan C, Chen J, Su M, Jin
Y, Deng Y, Wang D, Chen C, Zhou L, et al: Loss of TAB3 expression
by shRNA exhibits suppressive bioactivity and increased chemical
sensitivity of ovarian cancer cell lines via the NF-B pathway. Cell
Prolif. 49:657–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Luo C, Yuan R, Chen L, Zhou W, Shen W, Qiu
Y and Shao J, Yan J and Shao J: TAB3 upregulates Survivin
expression to promote colorectal cancer invasion and metastasis by
binding to the TAK1-TRAF6 complex. Oncotarget. 8:106565–106576.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ding J, Huang S, Wang Y, Tian Q, Zha R,
Shi H, Wang Q, Ge C, Chen T, Zhao Y, et al: Genome-wide screening
reveals that miR-195 targets the TNF-α/NF-κB pathway by
down-regulating IκB kinase alpha and TAB3 in hepatocellular
carcinoma. Hepatology. 58:654–666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kumar S, Xie H, Shi H, Gao J, Juhlin CC,
Björnhagen V, Höög A, Lee L, Larsson C and Lui WO: Merkel cell
polyomavirus oncoproteins induce microRNAs that suppress multiple
autophagy genes. Int J Cancer. 146:1652–1666. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Method. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Baeuerle PA and Baltimore D: Activation of
DNA-binding activity in an apparently cytoplasmic precursor of the
NF-kappa B transcription factor. Cell. 53:211–217. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Baeuerle PA and Baltimore D: I kappa B: A
Specific Inhibitor of the NF-kappa B Transcription Factor. Science.
242:540–546. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Karin M, Yamamoto Y and Wang QM: The IKK
NF-kappaB system: A treasure trove for drug development. Nat Rev
Drug Discov. 3:17–26. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Biswas DK, Shi Q, Baily S, Strickland I,
Ghosh S, Pardee AB and Iglehart JD: NF-kappa B activation in human
breast cancer specimens and its role in cell proliferation and
apoptosis. Proc Natl Acad Sci USA. 101:10137–10142. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sakamoto K, Maeda S, Hikiba Y, Nakagawa H,
Hayakawa Y, Shibata W, Yanai A, Ogura K and Omata M: Constitutive
NF-kappa B activation in colorectal carcinoma plays a key role in
angiogenesis, promoting tumor growth. Clin Cancer Res.
15:2248–2258. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bhaumik D, Scott GK, Schokrpur S, Patil
CK, Campisi J and Benz CC: Expression of microRNA-146 suppresses
NF-kappaB activity with reduction of metastatic potential in breast
cancer cells. Oncogene. 27:5643–5647. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu J, Ji X, Zhu L, Jiang Q, Wen Z, Xu S,
Shao W, Cai J, Du Q, Zhu Y and Mao J: Up-regulation of
microRNA-1290 impairs cytokinesis and affects the reprogramming of
colon cancer cells. Cancer Lett. 329:155–163. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ma Y, Wang B, Jiang F, Wang D, Liu H, Yan
Y, Dong H, Wang F, Gong B, Zhu Y, et al: A feedback loop consisting
of MicroRNA 23a/27a and the β-Like globin suppressors KLF3 and SP1
regulates globin gene expression. Mol Cell Biol. 33:3994–4007.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bao YD, Qiao Y, Yu H, Zhang Z, Yang H, Xin
X, Chen Y, Guo Y, Wu N and Jia D: miRNA-27a transcription activated
by c-Fos regulates myocardial ischemia-reperfusion injury by
targeting ATAD3a. Oxid Med Cell Longev. 2021:25149472021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tao H, Xiong Q, Ji Z, Zhang F, Liu Y and
Chen M: NFAT5 is Regulated by p53/miR-27a Signal axis and promotes
mouse ovarian granulosa cells proliferation. Int J Biol Sci.
15:287–297. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Meijer HA, Kong YW, Lu WT, Wilczynska A,
Spriggs RV, Robinson SW, Godfrey JD, Willis AE and Bushell M:
Translational repression and eIF4A2 activity are critical for
MicroRNA-Mediated gene regulation. Science. 340:82–85. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Huang V, Place RF, Portnoy V, Wang J, Qi
Z, Jia Z, Yu A, Shuman M, Yu J and Li LC: Upregulation of Cyclin B1
by miRNA and its implications in cancer. Nucleic Acids Res.
40:1695–1707. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Roberts APE, Lewis AP and Jopling CL:
miR-122 activates hepatitis C virus translation by a specialized
mechanism requiring particular RNA components. Nucleic Acids Res.
39:7716–7729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou
Y, Huang J, Zhao X, Zhou J, Yan Y, et al: MicroRNA directly
enhances mitochondrial translation during muscle differentiation.
Cell. 158:607–619. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Song G, Wang R, Guo J, Liu X, Wang F, Qi
Y, Wan H, Liu M, Li X and Tang H: miR-346 and miR-138 competitively
regulate hTERT in GRSF1- and AGO2-dependent manners, respectively.
Sci Rep. 5:157932015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kiriakidou M, Tan GS, Lamprinaki S, De
Planell-Saguer M, Nelson PT and Mourelatos Z: An mRNA m7G cap
binding-like motif within human Ago2 represses translation. Cell.
129:1141–1151. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chendrimada TP, Finn KJ, Ji XJ, Baillat D,
Gregory RI, Liebhaber SA, Pasquinelli AE and Shiekhattar R:
MicroRNA silencing through RISC recruitment of eIF6. Nature.
447:823–821. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jiang L, Yu L, Zhang X, Lei F, Wang L, Liu
X, Wu S, Zhu J, Wu G, Cao L, et al: miR-892b silencing activates
NF-κB and promotes aggressiveness in breast cancer. Cancer Res.
76:1101–1111. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhou X, Chen JJ, Zhang HJ, Chen X and Shao
GH: MicroRNA-23b attenuates the H2O2-induced
injury of microglial cells via TAB3/NF-κB signaling pathway. Int J
Clin Exp Pathol. 11:5765–5773. 2018.PubMed/NCBI
|