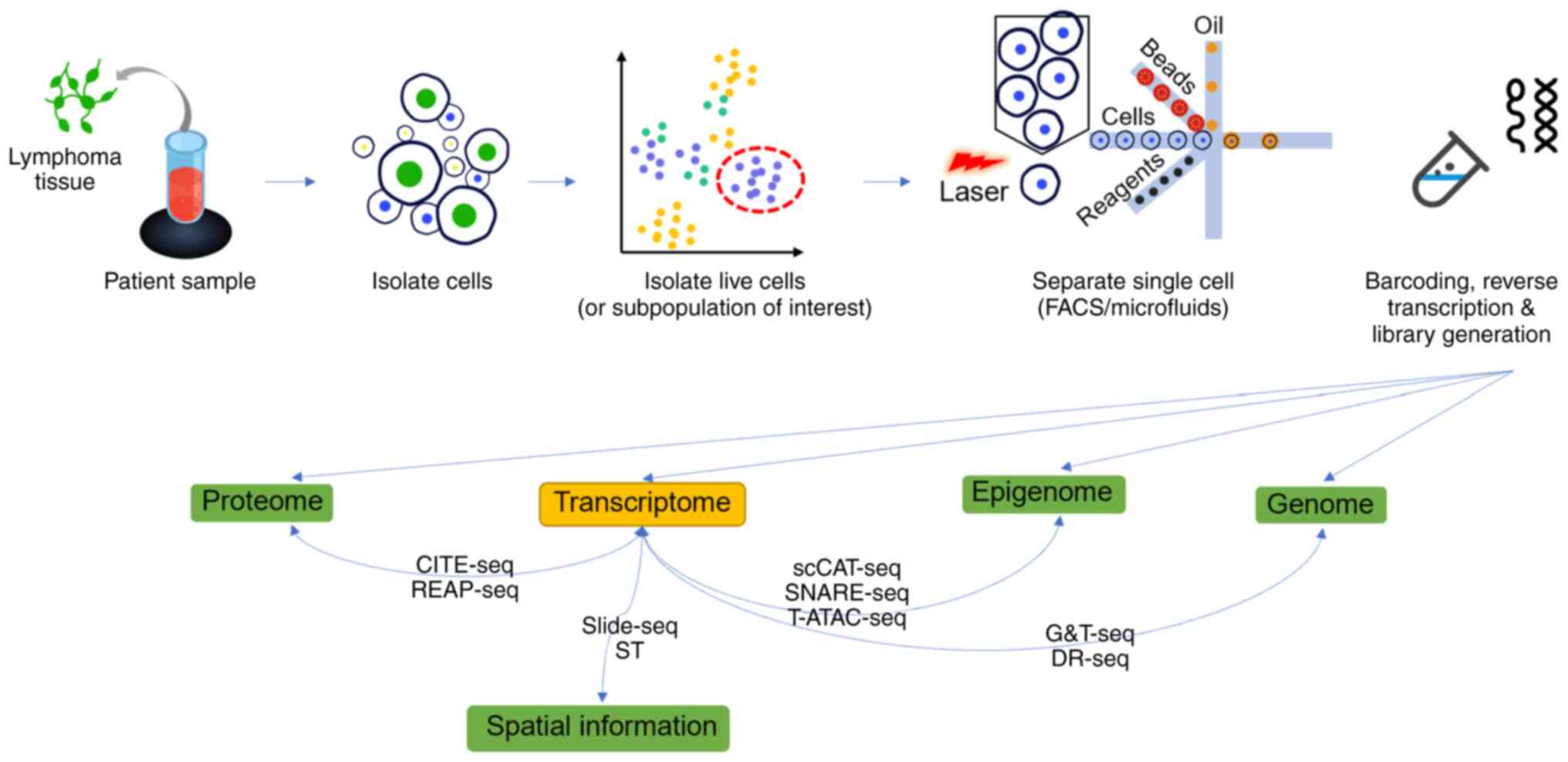
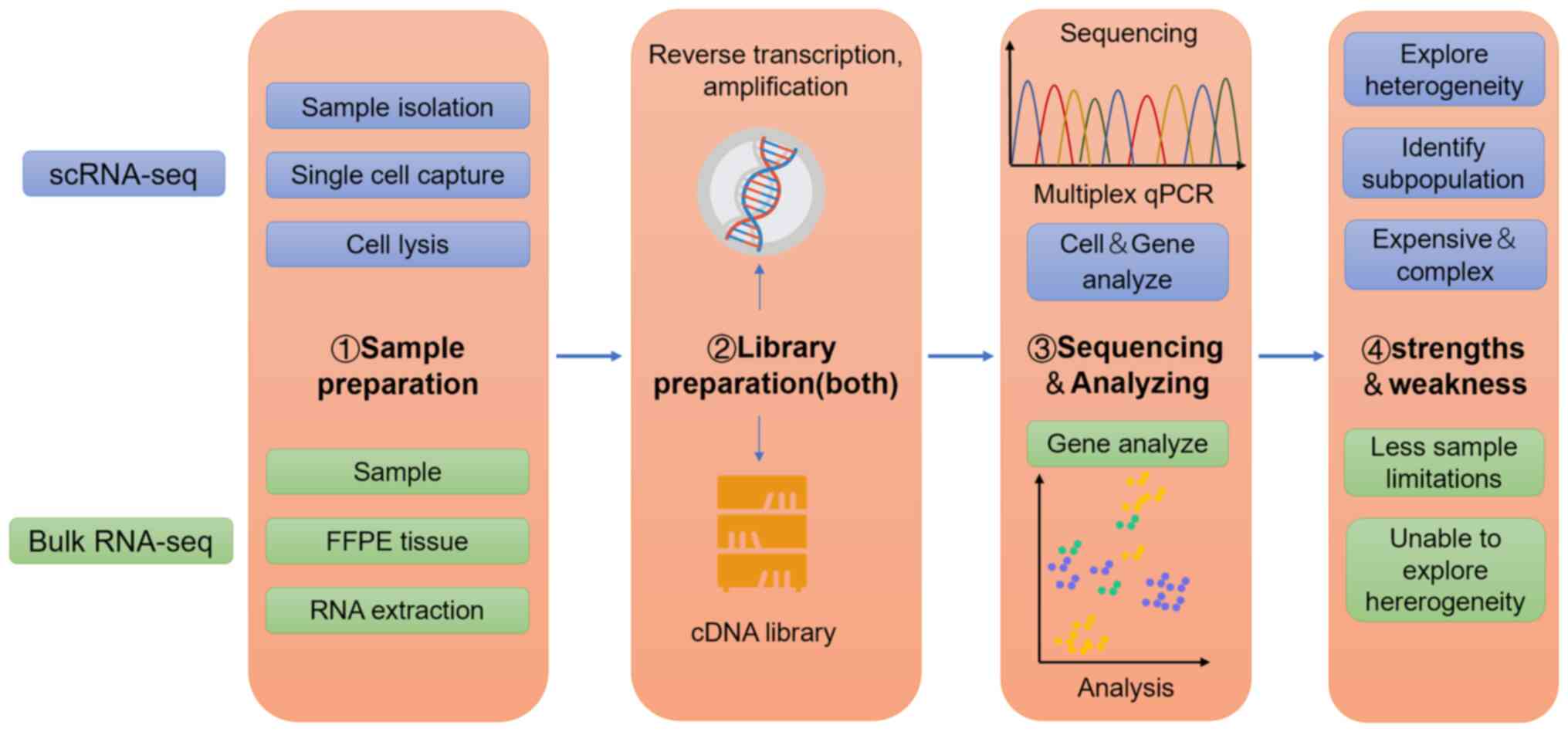
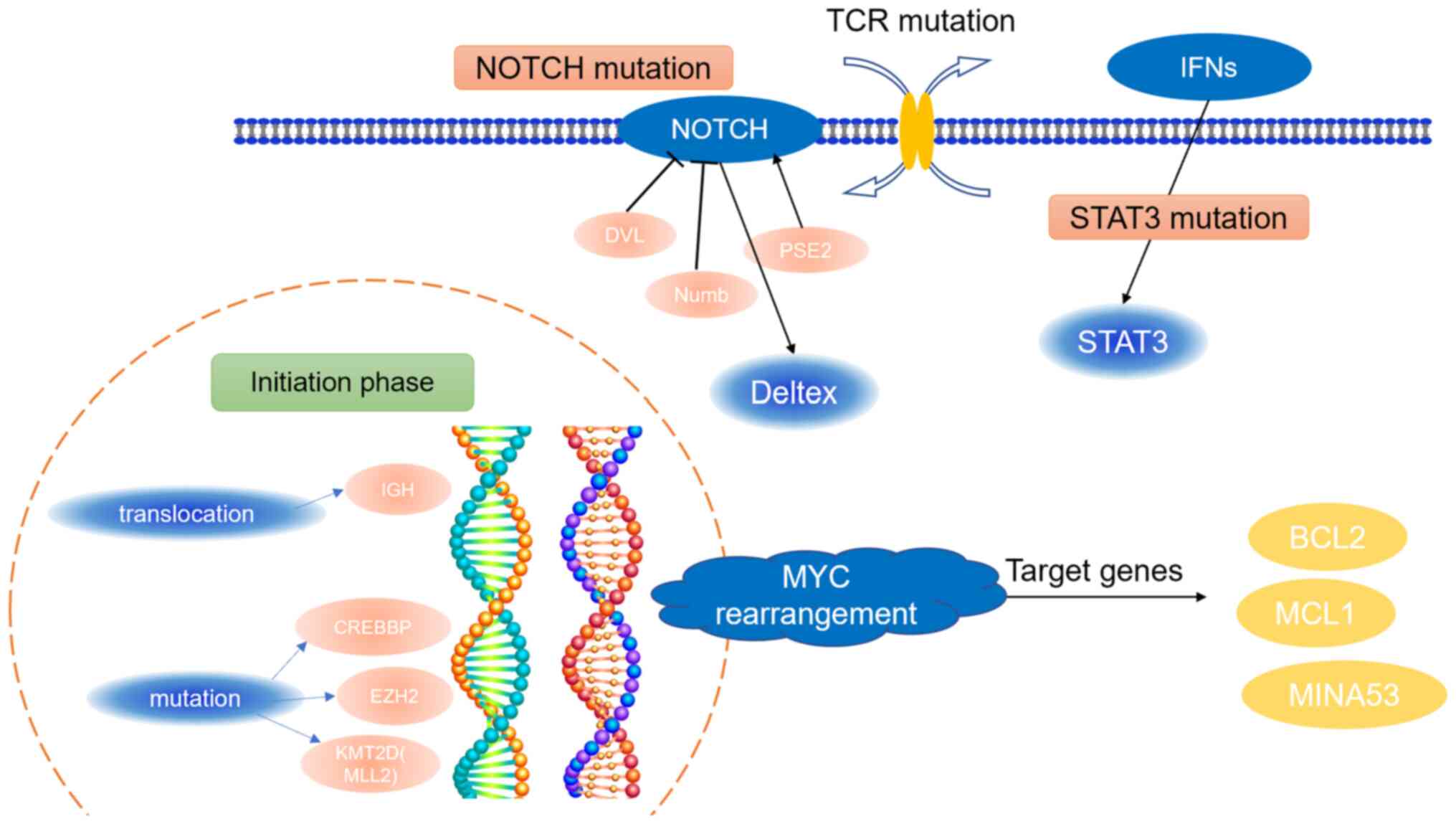
|
1
|
Ysebaert L, Quillet-Mary A, Tosolini M,
Pont F, Laurent C and Fournié JJ: Lymphoma heterogeneity unraveled
by single-cell transcriptomics. Front Immunol. 12:5976512021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bocci F, Gearhart-Serna L, Boareto M,
Ribeiro M, Ben-Jacob E, Devi GR, Levine H, Onuchic JN and Jolly MK:
Toward understanding cancer stem cell heterogeneity in the tumor
microenvironment. Proc Natl Acad Sci USA. 116:148–157. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lei Y, Tang R, Xu J, Wang W, Zhang B, Liu
J, Yu X and Shi S: Applications of single-cell sequencing in cancer
research: Progress and perspectives. J Hematol Oncol. 14:912021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alizadeh AA, Eisen MB, Davis RE, Ma C,
Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al:
Distinct types of diffuse large B-cell lymphoma identified by gene
expression profiling. Nature. 403:503–511. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gentles AJ, Newman AM, Liu CL, Bratman SV,
Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al: The
prognostic landscape of genes and infiltrating immune cells across
human cancers. Nat Med. 21:938–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kothalawala WJ, Barták BK, Nagy ZB,
Zsigrai S, Szigeti KA, Valcz G, Takács I, Kalmár A and Molnár B: A
detailed overview about the single-cell analyses of solid tumors
focusing on colorectal cancer. Pathol Oncol Res. 28:16103422022.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bingham GC, Lee F, Naba A and Barker TH:
Spatial-omics: Novel approaches to probe cell heterogeneity and
extracellular matrix biology. Matrix Biol. 91–92. 152–166.
2020.
|
|
10
|
Borcherding N, Voigt AP, Liu V, Link BK,
Zhang W and Jabbari A: Single-Cell profiling of cutaneous T-Cell
lymphoma reveals underlying heterogeneity associated with disease
progression. Clin Cancer Res. 25:2996–3005. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gaydosik AM, Tabib T, Geskin LJ, Bayan CA,
Conway JF, Lafyatis R and Fuschiotti P: Single-Cell lymphocyte
heterogeneity in advanced cutaneous T-cell lymphoma skin tumors.
Clin Cancer Res. 25:4443–4454. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang W, Yang B, Weng L, Li J, Bai J, Wang
T, Wang J, Ye J, Jing H, Jiao Y, et al: Single cell sequencing
reveals cell populations that predict primary resistance to
imatinib in chronic myeloid leukemia. Aging (Albany NY).
12:25337–25355. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ren J, Qu R, Rahman NT, Lewis JM, King
ALO, Liao X, Mirza FN, Carlson KR, Huang Y, Gigante S, et al:
Integrated transcriptome and trajectory analysis of cutaneous
T-cell lymphoma identifies putative precancer populations. Blood
Adv. 7:445–457. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamagishi M, Kubokawa M, Kuze Y, Suzuki A,
Yokomizo A, Kobayashi S, Nakashima M, Makiyama J, Iwanaga M, Fukuda
T, et al: Chronological genome and single-cell transcriptome
integration characterizes the evolutionary process of adult T cell
leukemia-lymphoma. Nat Commun. 12:48212021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Haebe S, Shree T, Sathe A, Day G,
Czerwinski DK, Grimes SM, Lee H, Binkley MS, Long SR, Martin B, et
al: Single-cell analysis can define distinct evolution of tumor
sites in follicular lymphoma. Blood. 137:2869–2880. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Borcherding N, Severson KJ, Henderson N,
Ortolan LS, Rosenthal AC, Bellizzi AM, Liu V, Link BK, Mangold AR
and Jabbari A: Single-cell analysis of Sézary syndrome reveals
novel markers and shifting gene profiles associated with treatment.
Blood Adv. 7:321–335. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Valentin Hansen S, Høy Hansen M, Cédile O,
Møller MB, Haaber J, Abildgaard N and Guldborg Nyvold C: Detailed
characterization of the transcriptome of single B cells in mantle
cell lymphoma suggesting a potential use for SOX4. Sci Rep.
11:190922021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pritchett JC, Yang ZZ, Kim HJ, Villasboas
JC, Tang X, Jalali S, Cerhan JR, Feldman AL and Ansell SM:
High-dimensional and single-cell transcriptome analysis of the
tumor microenvironment in angioimmunoblastic T cell lymphoma
(AITL). Leukemia. 36:165–176. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wei B, Liu Z, Fan Y, Wang S, Dong C, Rao
W, Yang F, Cheng G and Zhang J: Analysis of cellular heterogeneity
in immune microenvironment of primary central nervous system
lymphoma by single-cell sequencing. Front Oncol. 11:6830072021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Macosko EZ, Basu A, Satija R, Nemesh J,
Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck
EM, et al: Highly parallel genome-wide expression profiling of
individual cells using nanoliter droplets. Cell. 161:1202–1214.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Klein AM, Mazutis L, Akartuna I,
Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA and Kirschner
MW: Droplet barcoding for single-cell transcriptomics applied to
embryonic stem cells. Cell. 161:1187–1201. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng GX, Terry JM, Belgrader P, Ryvkin P,
Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et
al: Massively parallel digital transcriptional profiling of single
cells. Nat Commun. 8:140492017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gong H, Do D and Ramakrishnan R:
Single-Cell mRNA-Seq using the fluidigm C1 system and integrated
fluidics circuits. Methods Mol Biol. 1783:193–207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Han X, Chen H, Huang D, Chen H, Fei L,
Cheng C, Huang H, Yuan GC and Guo G: Mapping human pluripotent stem
cell differentiation pathways using high throughput single-cell
RNA-sequencing. Genome Biol. 19:472018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ziegenhain C, Vieth B, Parekh S, Reinius
B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I
and Enard W: Comparative analysis of single-cell RNA sequencing
methods. Mol Cell. 65:631–643.e4. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xin Y, Kim J, Ni M, Wei Y, Okamoto H, Lee
J, Adler C, Cavino K, Murphy AJ, Yancopoulos GD, et al: Use of the
Fluidigm C1 platform for RNA sequencing of single mouse pancreatic
islet cells. Proc Natl Acad Sci USA. 113:3293–3298. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gierahn TM, Wadsworth MH II, Hughes TK,
Bryson BD, Butler A, Satija R, Fortune S, Love JC and Shalek AK:
Seq-Well: Portable, low-cost RNA sequencing of single cells at high
throughput. Nat Methods. 14:395–398. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aicher TP, Carroll S, Raddi G, Gierahn T,
Wadsworth MH II, Hughes TK, Love C and Shalek AK: Seq-Well: A
sample-efficient, portable picowell platform for massively parallel
single-cell RNA sequencing. Methods Mol Biol. 1979:111–132. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han X, Wang R, Zhou Y, Fei L, Sun H, Lai
S, Saadatpour A, Zhou Z, Chen H, Ye F, et al: Mapping the mouse
cell atlas by microwell-seq. Cell. 173:13072018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lai S, Huang W, Xu Y, Jiang M, Chen H,
Cheng C, Lu Y, Huang H, Guo G and Han X: Comparative transcriptomic
analysis of hematopoietic system between human and mouse by
Microwell-seq. Cell Discov. 4:342018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Briggs JA, Weinreb C, Wagner DE, Megason
S, Peshkin L, Kirschner MW and Klein AM: The dynamics of gene
expression in vertebrate embryogenesis at single-cell resolution.
Science. 360:eaar57802018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tosches MA, Yamawaki TM, Naumann RK,
Jacobi AA, Tushev G and Laurent G: Evolution of pallium,
hippocampus, and cortical cell types revealed by single-cell
transcriptomics in reptiles. Science. 360:881–888. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jaitin DA, Kenigsberg E, Keren-Shaul H,
Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A
and Amit I: Massively parallel single-cell RNA-seq for marker-free
decomposition of tissues into cell types. Science. 343:776–779.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Stoeckius M, Hafemeister C, Stephenson W,
Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R and Smibert
P: Simultaneous epitope and transcriptome measurement in single
cells. Nat Methods. 14:865–868. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mimitou EP, Cheng A, Montalbano A, Hao S,
Stoeckius M, Legut M, Roush T, Herrera A, Papalexi E, Ouyang Z, et
al: Multiplexed detection of proteins, transcriptomes, clonotypes
and CRISPR perturbations in single cells. Nat Methods. 16:409–412.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Harris NL, Jaffe ES, Stein H, Banks PM,
Chan JK, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter
KC, et al: A revised European-American classification of lymphoid
neoplasms: A proposal from the International Lymphoma Study Group.
Blood. 84:1361–1392. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Barbui T, Thiele J, Gisslinger H,
Kvasnicka HM, Vannucchi AM, Guglielmelli P, Orazi A and Tefferi A:
The 2016 WHO classification and diagnostic criteria for
myeloproliferative neoplasms: Document summary and in-depth
discussion. Blood Cancer J. 8:152018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Glaser SL and Jarrett RF: The epidemiology
of Hodgkin's disease. Baillieres Clin Haematol. 9:401–416. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Marafioti T, Hummel M, Foss HD, Laumen H,
Korbjuhn P, Anagnostopoulos I, Lammert H, Demel G, Theil J, Wirth T
and Stein H: Hodgkin and reed-sternberg cells represent an
expansion of a single clone originating from a germinal center
B-cell with functional immunoglobulin gene rearrangements but
defective immunoglobulin transcription. Blood. 95:1443–1450. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kanzler H, Küppers R, Hansmann ML and
Rajewsky K: Hodgkin and Reed-Sternberg cells in Hodgkin's disease
represent the outgrowth of a dominant tumor clone derived from
(crippled) germinal center B cells. J Exp Med. 184:1495–1505. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Grimm KE and O'Malley DP: Aggressive B
cell lymphomas in the 2017 revised WHO classification of tumors of
hematopoietic and lymphoid tissues. Ann Diagn Pathol. 38:6–10.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Thandra KC, Barsouk A, Saginala K, Padala
SA, Barsouk A and Rawla P: Epidemiology of Non-Hodgkin's Lymphoma.
Med Sci (Basel). 9:52021.PubMed/NCBI
|
|
43
|
de Leval L and Jaffe ES: Lymphoma
Classification. Cancer J. 26:176–185. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
García-Sanz R and Jiménez C: Time to move
to the single-cell level: Applications of single-cell multi-omics
to hematological malignancies and Waldenström's Macroglobulinemia-A
particularly heterogeneous lymphoma. Cancers (Basel). 13:15412021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Glass DR, Tsai AG, Oliveria JP, Hartmann
FJ, Kimmey SC, Calderon AA, Borges L, Glass MC, Wagar LE, Davis MM
and Bendall SC: An Integrated Multi-omic Single-cell atlas of human
B cell identity. Immunity. 53:217–232.e5. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Crinier A, Dumas PY, Escalière B,
Piperoglou C, Gil L, Villacreces A, Vély F, Ivanovic Z, Milpied P,
Narni-Mancinelli É and Vivier É: Single-cell profiling reveals the
trajectories of natural killer cell differentiation in bone marrow
and a stress signature induced by acute myeloid leukemia. Cell Mol
Immunol. 18:1290–1304. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Aoki T, Chong LC, Takata K, Milne K, Hav
M, Colombo A, Chavez EA, Nissen M, Wang X, Miyata-Takata T, et al:
Single-Cell transcriptome analysis reveals disease-defining T-cell
subsets in the tumor microenvironment of classic Hodgkin Lymphoma.
Cancer Discov. 10:406–421. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pizzolato G, Kaminski H, Tosolini M,
Franchini DM, Pont F, Martins F, Valle C, Labourdette D, Cadot S,
Quillet-Mary A, et al: Single-cell RNA sequencing unveils the
shared and the distinct cytotoxic hallmarks of human TCRVδ1 and
TCRVδ2 γδ T lymphocytes. Proc Natl Acad Sci USA. 116:11906–11915.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Singh M, Al-Eryani G, Carswell S, Ferguson
JM, Blackburn J, Barton K, Roden D, Luciani F, Giang Phan T,
Junankar S, et al: High-throughput targeted long-read single cell
sequencing reveals the clonal and transcriptional landscape of
lymphocytes. Nat Commun. 10:31202019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Vitak SA, Torkenczy KA, Rosenkrantz JL,
Fields AJ, Christiansen L, Wong MH, Carbone L, Steemers FJ and Adey
A: Sequencing thousands of single-cell genomes with combinatorial
indexing. Nat Methods. 14:302–328. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xiong J, Cui BW, Wang N, Dai YT, Zhang H,
Wang CF, Zhong HJ, Cheng S, Ou-Yang BS, Hu Y, et al: Genomic and
transcriptomic characterization of natural killer T cell lymphoma.
Cancer Cell. 37:403–419.e6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li P, Chai J, Chen Z, Liu Y, Wei J, Liu Y,
Zhao D, Ma J, Wang K, Li X, et al: Genomic mutation profile of
primary gastrointestinal diffuse large B-Cell Lymphoma. Front
Oncol. 11:6226482021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Radke J, Ishaque N, Koll R, Gu Z, Schumann
E, Sieverling L, Uhrig S, Hübschmann D, Toprak UH, López C, et al:
The genomic and transcriptional landscape of primary central
nervous system lymphoma. Nat Commun. 13:25582022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yi S, Yan Y, Jin M, Bhattacharya S, Wang
Y, Wu Y, Yang L, Gine E, Clot G, Chen L, et al: Genomic and
transcriptomic profiling reveals distinct molecular subsets
associated with outcomes in mantle cell lymphoma. J Clin Invest.
132:e1532832022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nadeu F, Martin-Garcia D, Clot G,
Díaz-Navarro A, Duran-Ferrer M, Navarro A, Vilarrasa-Blasi R, Kulis
M, Royo R, Gutiérrez-Abril J, et al: Genomic and epigenomic
insights into the origin, pathogenesis, and clinical behavior of
mantle cell lymphoma subtypes. Blood. 136:1419–1432. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
De Bie J, Demeyer S, Alberti-Servera L,
Geerdens E, Segers H, Broux M, De Keersmaecker K, Michaux L,
Vandenberghe P, Voet T, et al: Single-cell sequencing reveals the
origin and the order of mutation acquisition in T-cell acute
lymphoblastic leukemia. Leukemia. 32:1358–1369. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
López C, Kleinheinz K, Aukema SM, Rohde M,
Bernhart SH, Hübschmann D, Wagener R, Toprak UH, Raimondi F, Kreuz
M, et al: Genomic and transcriptomic changes complement each other
in the pathogenesis of sporadic Burkitt lymphoma. Nat Commun.
10:14592019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Küçük C, Hu X, Gong Q, Jiang B, Cornish A,
Gaulard P, McKeithan T and Chan WC: Diagnostic and biological
significance of KIR EXPRESSION PROFILE DETErmined by RNA-Seq in
Natural Killer/T-Cell Lymphoma. Am J Pathol. 186:1435–1441. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Andor N, Simonds EF, Czerwinski DK, Chen
J, Grimes SM, Wood-Bouwens C, Zheng GXY, Kubit MA, Greer S, Weiss
WA, et al: Single-cell RNA-Seq of follicular lymphoma reveals
malignant B-cell types and coexpression of T-cell immune
checkpoints. Blood. 133:1119–1129. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Huang Z, Ma L, Huang C, Li Q and Nice EC:
Proteomic profiling of human plasma for cancer biomarker discovery.
Proteomics. 17((6))2017.
|
|
61
|
Kim MK, Song JY, Koh DI, Kim JY, Hatano M,
Jeon BN, Kim MY, Cho SY, Kim KS and Hur MW: Reciprocal negative
regulation between the tumor suppressor protein p53 and B cell
CLL/lymphoma 6 (BCL6) via control of caspase-1 expression. J Biol
Chem. 294:299–313. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Daniunaite K, Jarmalaite S and Kriukiene
E: Epigenomic technologies for deciphering circulating tumor DNA.
Curr Opin Biotechnol. 55:23–29. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ng SB, Yan J, Huang G, Selvarajan V, Tay
JL, Lin B, Bi C, Tan J, Kwong YL, Shimizu N, et al: Dysregulated
microRNAs affect pathways and targets of biologic relevance in
nasal-type natural killer/T-cell lymphoma. Blood. 118:4919–4929.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang X, Ji W, Huang R, Li L, Wang X, Li
L, Fu X, Sun Z, Li Z, Chen Q and Zhang M: MicroRNA-155 is a
potential molecular marker of natural killer/T-cell lymphoma.
Oncotarget. 7:53808–53819. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Yan J, Ng SB, Tay JL, Lin B, Koh TL, Tan
J, Selvarajan V, Liu SC, Bi C, Wang S, et al: EZH2 overexpression
in natural killer/T-cell lymphoma confers growth advantage
independently of histone methyltransferase activity. Blood.
121:4512–4520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liang L, Nong L, Zhang S, Zhao J, Ti H,
Dong Y, Zhang B and Li T: The downregulation of PRDM1/Blimp-1 is
associated with aberrant expression of miR-223 in extranodal
NK/T-cell lymphoma, nasal type. J Exp Clin Cancer Res. 33:72014.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Küçük C, Hu X, Jiang B, Klinkebiel D, Geng
H, Gong Q, Bouska A, Iqbal J, Gaulard P, McKeithan TW and Chan WC:
Global promoter methylation analysis reveals novel candidate tumor
suppressor genes in natural killer cell lymphoma. Clin Cancer Res.
21:1699–1711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chen YW, Guo T, Shen L, Wong KY, Tao Q,
Choi WW, Au-Yeung RK, Chan YP, Wong ML, Tang JC, et al:
Receptor-type tyrosine-protein phosphatase κ directly targets STAT3
activation for tumor suppression in nasal NK/T-cell lymphoma.
Blood. 125:1589–1600. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ranzoni AM, Tangherloni A, Berest I, Riva
SG, Myers B, Strzelecka PM, Xu J, Panada E, Mohorianu I, Zaugg JB
and Cvejic A: Integrative Single-Cell RNA-Seq and ATAC-seq analysis
of human developmental hematopoiesis. Cell Stem Cell.
28:472–487.e7. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yu J, Gemenetzis G, Kinny-Köster B, Habib
JR, Groot VP, Teinor J, Yin L, Pu N, Hasanain A, van Oosten F, et
al: Pancreatic circulating tumor cell detection by targeted
single-cell next-generation sequencing. Cancer Lett. 493:245–253.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhang L, Dong X, Lee M, Maslov AY, Wang T
and Vijg J: Single-cell whole-genome sequencing reveals the
functional landscape of somatic mutations in B lymphocytes across
the human lifespan. Proc Natl Acad Sci USA. 116:9014–9019. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Psatha K, Kollipara L, Voutyraki C,
Divanach P, Sickmann A, Rassidakis GZ, Drakos E and Aivaliotis M:
Deciphering lymphoma pathogenesis via state-of-the-art mass
spectrometry-based quantitative proteomics. J Chromatogr B Analyt
Technol Biomed Life Sci. 1047:2–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Bacher U, Haferlach T, Alpermann T, Kern
W, Schnittger S and Haferlach C: Several lymphoma-specific genetic
events in parallel can be found in mature B-cell neoplasms. Genes
Chromosomes Cancer. 50:43–50. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Okosun J, Bödör C, Wang J, Araf S, Yang
CY, Pan C, Boller S, Cittaro D, Bozek M, Iqbal S, et al: Integrated
genomic analysis identifies recurrent mutations and evolution
patterns driving the initiation and progression of follicular
lymphoma. Nat Genet. 46:176–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sewastianik T, Prochorec-Sobieszek M,
Chapuy B and Juszczyński P: MYC deregulation in lymphoid tumors:
Molecular mechanisms, clinical consequences and therapeutic
implications. Biochim Biophys Acta. 1846:457–467. 2014.PubMed/NCBI
|
|
76
|
Rosenthal A and Rimsza L: Genomics of
aggressive B-cell lymphoma. Hematology Am Soc Hematol Educ Program.
2018:69–74. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gawad C, Koh W and Quake SR: Dissecting
the clonal origins of childhood acute lymphoblastic leukemia by
single-cell genomics. Proc Natl Acad Sci USA. 111:17947–17952.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Snuderl M, Kolman OK, Chen YB, Hsu JJ,
Ackerman AM, Dal Cin P, Ferry JA, Harris NL, Hasserjian RP,
Zukerberg LR, et al: B-cell lymphomas with concurrent IGH-BCL2 and
MYC rearrangements are aggressive neoplasms with clinical and
pathologic features distinct from Burkitt lymphoma and diffuse
large B-cell lymphoma. Am J Surg Pathol. 34:327–340. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Huang W, Medeiros LJ, Lin P, Wang W, Tang
G, Khoury J, Konoplev S, Yin CC, Xu J, Oki Y and Li S:
MYC/BCL2/BCL6 triple hit lymphoma: A study of 40 patients with a
comparison to MYC/BCL2 and MYC/BCL6 double hit lymphomas. Mod
Pathol. 31:1470–1478. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Moore EM, Aggarwal N, Surti U and Swerdlow
SH: Further exploration of the complexities of large B-Cell
Lymphomas With MYC abnormalities and the importance of a blastoid
morphology. Am J Surg Pathol. 41:1155–1166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Jiang Y, Redmond D, Nie K, Eng KW, Clozel
T, Martin P, Tan LH, Melnick AM, Tam W and Elemento O: Deep
sequencing reveals clonal evolution patterns and mutation events
associated with relapse in B-cell lymphomas. Genome Biol.
15:4322014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Ding S, Chen X and Shen K: Single-cell RNA
sequencing in breast cancer: Understanding tumor heterogeneity and
paving roads to individualized therapy. Cancer Commun (Lond).
40:329–344. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kim E, Hurtz C, Koehrer S, Wang Z,
Balasubramanian S, Chang BY, Müschen M, Davis RE and Burger JA:
Ibrutinib inhibits pre-BCR(+) B-cell acute lymphoblastic leukemia
progression by targeting BTK and BLK. Blood. 129:1155–1165. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Wang L, Mo S, Li X, He Y and Yang J:
Single-cell RNA-seq reveals the immune escape and drug resistance
mechanisms of mantle cell lymphoma. Cancer Biol Med. 17:726–739.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Marcus R, Imrie K, Solal-Celigny P,
Catalano JV, Dmoszynska A, Raposo JC, Offner FC, Gomez-Codina J,
Belch A, Cunningham D, et al: Phase III study of R-CVP compared
with cyclophosphamide, vincristine, and prednisone alone in
patients with previously untreated advanced follicular lymphoma. J
Clin Oncol. 26:4579–4586. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lim SH, Vaughan AT, Ashton-Key M, Williams
EL, Dixon SV, Chan HT, Beers SA, French RR, Cox KL, Davies AJ, et
al: Fc gamma receptor IIb on target B cells promotes rituximab
internalization and reduces clinical efficacy. Blood.
118:2530–2540. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang J, Dominguez-Sola D, Hussein S, Lee
JE, Holmes AB, Bansal M, Vlasevska S, Mo T, Tang H, Basso K, et al:
Disruption of KMT2D perturbs germinal center B cell development and
promotes lymphomagenesis. Nat Med. 21:1190–1198. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Arber DA, Orazi A, Hasserjian R, Thiele J,
Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M and Vardiman JW:
The 2016 revision to the World Health Organization classification
of myeloid neoplasms and acute leukemia. Blood. 127:2391–2405.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Swerdlow SH and Cook JR: As the world
turns, evolving lymphoma classifications-past, present and future.
Hum Pathol. 95:55–77. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Takagi M: DNA damage response and
hematological malignancy. Int J Hematol. 106:345–356. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Flinders C, Lam L, Rubbi L, Ferrari R,
Fitz-Gibbon S, Chen PY, Thompson M, Christofk H, B Agus D, Ruderman
D, et al: Epigenetic changes mediated by polycomb repressive
complex 2 and E2a are associated with drug resistance in a mouse
model of lymphoma. Genome Med. 8:542016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Chiche J, Reverso-Meinietti J, Mouchotte
A, Rubio-Patiño C, Mhaidly R, Villa E, Bossowski JP, Proics E,
Grima-Reyes M, Paquet A, et al: GAPDH expression predicts the
response to R-CHOP, the tumor metabolic status, and the response of
DLBCL patients to metabolic inhibitors. Cell Metab.
29:1243–1257.e10. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Klener P and Klanova M: Drug Resistance in
Non-Hodgkin Lymphomas. Int J Mol Sci. 21:20812020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wang X, Li G, Zhang Y, Li L, Qiu L, Qian
Z, Zhou S, Wang X, Li Q and Zhang H: Pan-Cancer analysis reveals
genomic and clinical characteristics of TRPV Channel-related genes.
Front Oncol. 12:8131002022. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Russo M, Crisafulli G, Sogari A, Reilly
NM, Arena S, Lamba S, Bartolini A, Amodio V, Magrì A, Novara L, et
al: Adaptive mutability of colorectal cancers in response to
targeted therapies. Science. 366:1473–1480. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kater L, Kater B, Jakupec MA, Keppler BK
and Prokop A: KP772 overcomes multiple drug resistance in malignant
lymphoma and leukemia cells in vitro by inducing Bcl-2-independent
apoptosis and upregulation of Harakiri. J Biol Inorg Chem.
26:897–907. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Sciarrillo R, Wojtuszkiewicz A, Assaraf
YG, Jansen G, Kaspers GJL, Giovannetti E and Cloos J: The role of
alternative splicing in cancer: From oncogenesis to drug
resistance. Drug Resist Updat. 53:1007282020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Roider T, Seufert J, Uvarovskii A,
Frauhammer F, Bordas M, Abedpour N, Stolarczyk M, Mallm JP, Herbst
SA, Bruch PM, et al: Dissecting intratumour heterogeneity of nodal
B-cell lymphomas at the transcriptional, genetic and drug-response
levels. Nat Cell Biol. 22:896–906. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Wills QF, Livak KJ, Tipping AJ, Enver T,
Goldson AJ, Sexton DW and Holmes C: Single-cell gene expression
analysis reveals genetic associations masked in whole-tissue
experiments. Nat Biotechnol. 31:748–752. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Kotlov N, Bagaev A, Revuelta MV, Phillip
JM, Cacciapuoti MT, Antysheva Z, Svekolkin V, Tikhonova E,
Miheecheva N, Kuzkina N, et al: Clinical and Biological Subtypes of
B-cell lymphoma revealed by microenvironmental signatures. Cancer
Discov. 11:1468–1489. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Croci GA, Au-Yeung RKH, Reinke S, Staiger
AM, Koch K, Oschlies I, Richter J, Poeschel V, Held G, Loeffler M,
et al: SPARC-positive macrophages are the superior prognostic
factor in the microenvironment of diffuse large B-cell lymphoma and
independent of MYC rearrangement and double-/triple-hit status. Ann
Oncol. 32:1400–1409. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Abe Y, Sakata-Yanagimoto M, Fujisawa M,
Miyoshi H, Suehara Y, Hattori K, Kusakabe M, Sakamoto T, Nishikii
H, Nguyen TB, et al: A single-cell atlas of non-haematopoietic
cells in human lymph nodes and lymphoma reveals a landscape of
stromal remodelling. Nat Cell Biol. 24:565–578. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Ferreri AJ, Cwynarski K, Pulczynski E,
Ponzoni M, Deckert M, Politi LS, Torri V, Fox CP, Rosée PL, Schorb
E, et al: Chemoimmunotherapy with methotrexate, cytarabine,
thiotepa, and rituximab (MATRix regimen) in patients with primary
CNS lymphoma: Results of the first randomisation of the
International Extranodal Lymphoma Study Group-32 (IELSG32) phase 2
trial. Lancet Haematol. 3:e217–e227. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Bromberg JEC, Issa S, Bakunina K, Minnema
MC, Seute T, Durian M, Cull G, Schouten HC, Stevens WBC, Zijlstra
JM, et al: Rituximab in patients with primary CNS lymphoma (HOVON
105/ALLG NHL 24): A randomised, open-label, phase 3 intergroup
study. Lancet Oncol. 20:216–228. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Ruggieri S, Tamma R, Resta N, Albano F,
Coccaro N, Loconte D, Annese T, Errede M, Specchia G, Senetta R, et
al: Stat3-positive tumor cells contribute to vessels neoformation
in primary central nervous system lymphoma. Oncotarget.
8:31254–31269. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhou Y, Liu W, Xu Z, Zhu H, Xiao D, Su W,
Zeng R, Feng Y, Duan Y, Zhou J and Zhong M: Analysis of genomic
alteration in primary central nervous system lymphoma and the
expression of some related genes. Neoplasia. 20:1059–1069. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Ribatti D, Nico B, Ranieri G, Specchia G
and Vacca A: The role of angiogenesis in human non-Hodgkin
lymphomas. Neoplasia. 15:231–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Clozel T, Yang S, Elstrom RL, Tam W,
Martin P, Kormaksson M, Banerjee S, Vasanthakumar A, Culjkovic B,
Scott DW, et al: Mechanism-based epigenetic chemosensitization
therapy of diffuse large B-cell lymphoma. Cancer Discov.
3:1002–1019. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Hazar B, Paydas S, Zorludemir S, Sahin B
and Tuncer I: Prognostic significance of microvessel density and
vascular endothelial growth factor (VEGF) expression in
non-Hodgkin's lymphoma. Leuk Lymphoma. 44:2089–2093. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Carlo-Stella C and Santoro A:
Microenvironment-related biomarkers and novel targets in classical
Hodgkin's lymphoma. Biomark Med. 9:807–817. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Holmes AB, Corinaldesi C, Shen Q, Kumar R,
Compagno N, Wang Z, Nitzan M, Grunstein E, Pasqualucci L,
Dalla-Favera R and Basso K: Single-cell analysis of germinal-center
B cells informs on lymphoma cell of origin and outcome. J Exp Med.
217:e202004832020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Mintz MA and Cyster JG: T follicular
helper cells in germinal center B cell selection and
lymphomagenesis. Immunol Rev. 296:48–61. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Turqueti-Neves A, Otte M, Prazeres da
Costa O, Höpken UE, Lipp M, Buch T and Voehringer D:
B-cell-intrinsic STAT6 signaling controls germinal center
formation. Eur J Immunol. 44:2130–2138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Mintz MA, Felce JH, Chou MY, Mayya V, Xu
Y, Shui JW, An J, Li Z, Marson A, Okada T, et al: The HVEM-BTLA
Axis Restrains T cell help to germinal center B cells and functions
as a cell-extrinsic suppressor in lymphomagenesis. Immunity.
51:310–323.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Hashwah H, Schmid CA, Kasser S, Bertram K,
Stelling A, Manz MG and Müller A: Inactivation of CREBBP expands
the germinal center B cell compartment, down-regulates MHCII
expression and promotes DLBCL growth. Proc Natl Acad Sci USA.
114:9701–9706. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Deng Q, Han G, Puebla-Osorio N, Ma MCJ,
Strati P, Chasen B, Dai E, Dang M, Jain N, Yang H, et al:
Characteristics of anti-CD19 CAR T cell infusion products
associated with efficacy and toxicity in patients with large B cell
lymphomas. Nat Med. 26:1878–1887. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Parker KR, Migliorini D, Perkey E, Yost
KE, Bhaduri A, Bagga P, Haris M, Wilson NE, Liu F, Gabunia K, et
al: Single-Cell analyses identify brain mural cells expressing CD19
as potential off-tumor targets for CAR-T immunotherapies. Cell.
183:126–142.e17. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Shi Y, Ding W, Gu W, Shen Y, Li H, Zheng
Z, Zheng X, Liu Y and Ling Y: Single-cell phenotypic profiling to
identify a set of immune cell protein biomarkers for relapsed and
refractory diffuse large B cell lymphoma: A single-center study. J
Leukoc Biol. 112:1633–1648. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Shin D, Lee W, Lee JH and Bang D:
Multiplexed single-cell RNA-seq via transient barcoding for
simultaneous expression profiling of various drug perturbations.
Sci Adv. 5:eaav22492019. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Krieg C, Nowicka M, Guglietta S, Schindler
S, Hartmann FJ, Weber LM, Dummer R, Robinson MD, Levesque MP and
Becher B: High-dimensional single-cell analysis predicts response
to anti-PD-1 immunotherapy. Nat Med. 24:144–153. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Yost KE, Satpathy AT, Wells DK, Qi Y, Wang
C, Kageyama R, McNamara KL, Granja JM, Sarin KY, Brown RA, et al:
Clonal replacement of tumor-specific T cells following PD-1
blockade. Nat Med. 25:1251–1259. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Wu TD, Madireddi S, de Almeida PE,
Banchereau R, Chen YJ, Chitre AS, Chiang EY, Iftikhar H, O'Gorman
WE, Au-Yeung A, et al: Peripheral T cell expansion predicts tumour
infiltration and clinical response. Nature. 579:274–278. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
La Manno G, Soldatov R, Zeisel A, Braun E,
Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg
P, Furlan A, et al: RNA velocity of single cells. Nature.
560:494–498. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Simmons SK, Lithwick-Yanai G, Adiconis X,
Oberstrass F, Iremadze N, Geiger-Schuller K, Thakore PI, Frangieh
CJ, Barad O, Almogy G, et al: Mostly natural
sequencing-by-synthesis for scRNA-seq using Ultima sequencing. Nat
Biotechnol. 41:204–211. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Fan X, Zhang X, Wu X, Guo H, Hu Y, Tang F
and Huang Y: Single-cell RNA-seq transcriptome analysis of linear
and circular RNAs in mouse preimplantation embryos. Genome Biol.
16:1482015. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Macaulay IC, Haerty W, Kumar P, Li YI, Hu
TX, Teng MJ, Goolam M, Saurat N, Coupland P, Shirley LM, et al:
G&T-seq: Parallel sequencing of single-cell genomes and
transcriptomes. Nat Methods. 12:519–522. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P, Wu
X, Wen L, Tang F, Huang Y and Peng J: Single-cell triple omics
sequencing reveals genetic, epigenetic, and transcriptomic
heterogeneity in hepatocellular carcinomas. Cell Res. 26:304–319.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Katzenelenbogen Y, Sheban F, Yalin A, Yofe
I, Svetlichnyy D, Jaitin DA, Bornstein C, Moshe A, Keren-Shaul H,
Cohen M, et al: Coupled scRNA-Seq and intracellular protein
activity reveal an immunosuppressive role of TREM2 in cancer. Cell.
182:872–885.e19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Svensson V, Vento-Tormo R and Teichmann
SA: Exponential scaling of single-cell RNA-seq in the past decade.
Nat Protoc. 13:599–604. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Bai X, Li Y, Zeng X, Zhao Q and Zhang Z:
Single-cell sequencing technology in tumor research. Clin Chim
Acta. 518:101–109. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
McGinnis CS, Patterson DM, Winkler J,
Conrad DN, Hein MY, Srivastava V, Hu JL, Murrow LM, Weissman JS,
Werb Z, et al: MULTI-seq: Sample multiplexing for single-cell RNA
sequencing using lipid-tagged indices. Nat Methods. 16:619–626.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Kashima Y, Sakamoto Y, Kaneko K, Seki M,
Suzuki Y and Suzuki A: Single-cell sequencing techniques from
individual to multiomics analyses. Exp Mol Med. 52:1419–1427. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Ando Y, Kwon AT and Shin JW: An era of
single-cell genomics consortia. Exp Mol Med. 52:1409–1418. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Landeira-Viñuela A, Díez P, Juanes-Velasco
P, Lécrevisse Q, Orfao A, De Las Rivas J and Fuentes M: Deepening
into intracellular signaling landscape through integrative spatial
proteomics and transcriptomics in a lymphoma model. Biomolecules.
11:17762021. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Du J, Qiu C, Li WS, Wang B, Han XL, Lin
SW, Fu XH, Hou J and Huang ZF: Spatial transcriptomics analysis
reveals that CCL17 and CCL22 are robust indicators of a suppressive
immune environment in angioimmunoblastic T cell lymphoma (AITL).
Front Biosci (Landmark Ed). 27:2702022. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Tripodo C, Zanardi F, Iannelli F, Mazzara
S, Vegliante M, Morello G, Di Napoli A, Mangogna A, Facchetti F,
Sangaletti S, et al: A spatially resolved dark-versus light-zone
microenvironment signature subdivides germinal center-related
aggressive B cell lymphomas. iScience. 23:1015622020. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Colombo AR, Hav M, Singh M, Xu A, Gamboa
A, Lemos T, Gerdtsson E, Chen D, Houldsworth J, Shaknovich R, et
al: Single-cell spatial analysis of tumor immune architecture in
diffuse large B-cell lymphoma. Blood Adv. 6:4675–4690. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Efremova M, Vento-Tormo R, Park JE,
Teichmann SA and James KR: Immunology in the Era of single-cell
technologies. Annu Rev Immunol. 38:727–757. 2020. View Article : Google Scholar : PubMed/NCBI
|