Introduction
Colorectal cancer (CRC) has become a worldwide
public health problem with a high incidence and mortality (1). By 2030, an extra 2.2 million new cases
of CRC and 1.1 million cancer-related deaths are expected,
representing a 60% increased burden of CRC.
The early symptoms of CRC are not obvious. Notably,
>85% of patients with CRC are diagnosed at an advanced stage.
However, when the optimal treatment window is missed, the survival
time and quality of life for patients with advanced CRC are
significantly reduced, resulting in a 5-year survival rate of
<40% (2). By contrast, the
5-year survival rate of patients with CRC with early-stage disease
after treatment is as high as 95%.
The prognosis of patients with CRC largely depends
on the stage of the disease at first diagnosis. In most cases, CRC
cases are sporadic and transform from adenomas (3), and the transition from adenoma to CRC
typically spans several years. Detecting and removing adenomas at
an early stage can effectively impede their progression to CRC,
thus reducing the incidence of the disease (4,5).
Moreover, accurate, evidence-based screening could significantly
decrease the morbidity and mortality of CRC. Furthermore, early
screening can improve the clinical outcomes of patients, avoid
treatment delays, and reduce CRC mortality (6).
The U.S. Preventive Services Task Force (USPSTF)
strongly advocates for CRC screening for precise diagnosis
(7). Among the available methods,
colonoscopy stands out as the primary screening approach due to its
widespread use and high accuracy. In addition, the USPSTF
recommends that colonoscopy be performed promptly, as colonoscopy
can significantly reduce the incidence and mortality of CRC
(8). However, it is imperative to
acknowledge that colonoscopy does have certain limitations. One
such limitation is the reliance on the skill level of the
endoscopic physicians for diagnostic accuracy, which can vary among
practitioners. Ensuring that each patient undergoes an examination
by a highly skilled endoscopist can be challenging.
Artificial intelligence (AI) refers to the ability
of machines to imitate human cognitive functions and perform tasks
at or above the human level using a clever combination of computer
science, algorithms, machine learning (ML), and data science. In
recent years, advances in AI have permeated medicine, rapidly
changing the way cancer research is conducted. Research has shown
that AI-aided colonoscopy can enhance screening accuracy,
efficiency, and quality (9). The
availability of high-dimensional datasets, continuous advances in
high-performance computing power, and innovative deep-learning
architectures have all led to a rapidly emerging role for AI in CRC
screening.
The combination of AI technology and colonoscopy
holds great promise for controlling the morbidity and mortality of
CRC. Therefore, the aim of the present review was to examine the
advantages and limitations of colonoscopy while focusing on the
application of AI-aided colonoscopy, providing a theoretical
foundation for developing precise CRC screening.
Premalignant lesions of CRC and screening
modalities
Colorectal polyps are protrusions occurring in the
colorectal lumen, which can be divided into neoplastic and
non-neoplastic polyps (Table I).
Pathologically, neoplastic polyps can be classified as adenomatous
and serrated polyps, and non-neoplastic polyps include
inflammatory-associated polyps, hamartomatous polyps and
hyperplastic polyps (10,11). Adenomatous polyps include three
histological types: Tubular, tubulovillous, and villous (11). Conversely, serrated class lesions
are a heterogeneous group of lesions that can be further classified
into three categories: Hyperplastic polyps (HPs), sessile serrated
lesions (SSPs), and traditional serrated adenomas (TSAs) (Table I). The carcinogenesis process of CRC
involves four pathways: Adenoma-carcinoma, serrated neoplastic,
inflammatory, and de novo (12,13).
The first two pathways account for the vast majority of cases and
arise from colorectal polyps. The conventional adenoma-carcinoma
pathway leads to 70% of sporadic CRC cases (14), whereas the serrated neoplastic
pathway accounts for 15–30% of CRC cases.
 | Table I.Colorectal polyps. |
Table I.
Colorectal polyps.
| Neoplastic
polyps | Adenomatous
polyps | Tubular
adenoma |
|
|
| Tubulovillous
adenoma |
|
|
| Villous
adenoma |
|
| Serrated class
lesions | Hyperplastic
polyp |
|
|
| Sessile serrated
lesions |
|
|
| Traditional
serrated adenomas |
| Non-neoplastic
polyps |
Inflammatory-associated polyps | Inflammatory
polyps |
|
|
| Lymphoid
polyps |
|
|
| Schistosoma
polyps |
|
| Hamartomatous
polyp | Juvenile
polyps |
|
|
| Peutz-Jeghers
polyps |
|
|
| Cowden
syndrome-associated polyps |
|
|
| Canada-Cronkhite
syndrome-associated polyps |
|
| Hyperplastic
polyps | N/A |
High-sensitivity Guaiac fecal occult blood test,
fecal immunochemical test, multi-target fecal DNA, computed
tomography colonography (CTC), colonoscopy, and other methods are
currently the CRC screening methods recommended by the USPSTF
(7,8,15). The
diagnostic accuracy and effectiveness of visual screening (CTC,
flexible sigmoidoscopy, and colonoscopy) are far higher than those
of stool-based screening due to their ability to directly observe
lesions (6,16–25).
Compared to stool-based CRC screening, a colonoscopy may have lower
in-patient compliance and frequency, yet it remains significantly
more accurate in detecting colorectal lesions (26). This procedure allows for direct
detection, biopsy, and removal of polyps during visual assessment
of the entire colon. Colonoscopy has several benefits, including
high sensitivity and specificity, and enables direct biopsy or
excision of suspected polyps. As a result, the USPSTF has indicated
that colonoscopy has the highest validity and popularity among CRC
screening methods (8).
Colonoscopy in CRC screening
Colonoscopy is the most reliable form of CRC
screening. According to a large, prospective observational study
that included nearly 89,000 nurses and other health professionals,
the CRC mortality rate was lower in people who self-reported at
least one screening colonoscopy than in those who had never
undergone a colonoscopy (27).
Furthermore, the USPSTF included four studies (n=4,821) evaluating
the accuracy of colonoscopy in 2021, demonstrating that for
adenomas ≥10 mm, colonoscopy had a sensitivity of 89–95% and a
specificity of 89%. Additionally, for adenomas ≥6 mm, colonoscopy
had a sensitivity of 75–93% and a specificity of 94% (8). These results further support the high
accuracy and specificity of colonoscopy.
Although colonoscopy is considered the ‘gold
standard’ screening test, it does have its limitations (28). The challenge of endoscopic
procedures lies in the real-time interpretation of endoscopic
imagery, which is complex and sensitive to human error.
Consequently, subtle, and early premalignant lesions in the colon
and rectum can easily be missed by endoscopists. A systematic
review (29) showed that the rate
of missed adenomas on colonoscopy was 26%, and this was 9% for
advanced adenomas and up to 27% for serrated polyps. A prospective
study of individuals who underwent screening colonoscopy within a
National Colorectal Cancer Screening Program associated an
increased adenoma detection rate (ADR) with a reduced risk of
post-colonoscopy colorectal cancer and colorectal cancer death.
Notably, ADRs are negatively associated with CRC incidence, with
each 1% increase in ADR associated with a 3–6% reduction in the
risk of colorectal cancer (30). By
contrast, a higher rate of missed adenoma detection inevitably
increases the risk of colorectal cancer.
It is important to acknowledge that colonoscopy does
not always detect colorectal cancer, and some patients may develop
CRC even after receiving a negative examination result. When this
occurs before the next recommended screening or surveillance
examination, it is called interval cancer. However, the term
‘interval cancer’ is considered too restrictive to encompass all
aspects necessary for colonoscopy quality assurance purposes. To
address this, Rabeneck and Paszat introduced the term
‘post-colonoscopy colorectal cancer’ (PCCRC) in 2010 (31), defined as colorectal cancer not
detected by screening or surveillance examinations and occurring
before the recommended next examination date (32).
Colonoscopy, despite being a valuable screening
tool, is not infallible and can potentially miss early or advanced
non-characteristic lesions, leading to the risk of PCCRC (32). Studies have indicated a prevalence
of PCCRC ranging from 3.7 to 8.6% following colonoscopy screening
(33,34). However, failure to detect colorectal
neoplasia remains the most relevant cause of PCCRC (35). It can be observed from research that
an improvement in the ADR during screening colonoscopy, achieved
through a comprehensive quality assurance program, translates into
reduced risks of post-colonoscopy colorectal cancer. Specifically,
an ADR of a suboptimal endoscopist has been associated with a
substantial increase in the risk of post-colonoscopy colorectal
cancer incidence, whereas an ADR increase was effective in
reversing this detrimental effect (36). Undoubtedly, high-quality colonoscopy
will improve the diagnostic accuracy of adenoma and CRC lesions,
which is crucial for re-sectioning precancerous lesions and
preventing CRC (37).
AI has the potential to identify colorectal polyps
or CRC lesions that have gone undetected due to perceptual errors.
A multicenter and multi-county randomized crossover trial showed
that AI resulted in an ~50% reduction in the miss rate of
colorectal neoplasia. This finding highlights the potential of AI
in mitigating perceptual errors associated with small and subtle
lesions during standard colonoscopy (38). Consequently, combining colonoscopy
and AI may be a potential future development direction.
Artificial intelligence applications in
colonoscopy screening
AI is a generic term broadly referring to utilizing
computers to model intelligent behavior with minimal human
intervention (39). ML is a
subfield of AI that is capable of analyzing data through algorithms
to take particular actions in response to specific inputs and
improve (‘learn’) themselves as more data becomes available, i.e.,
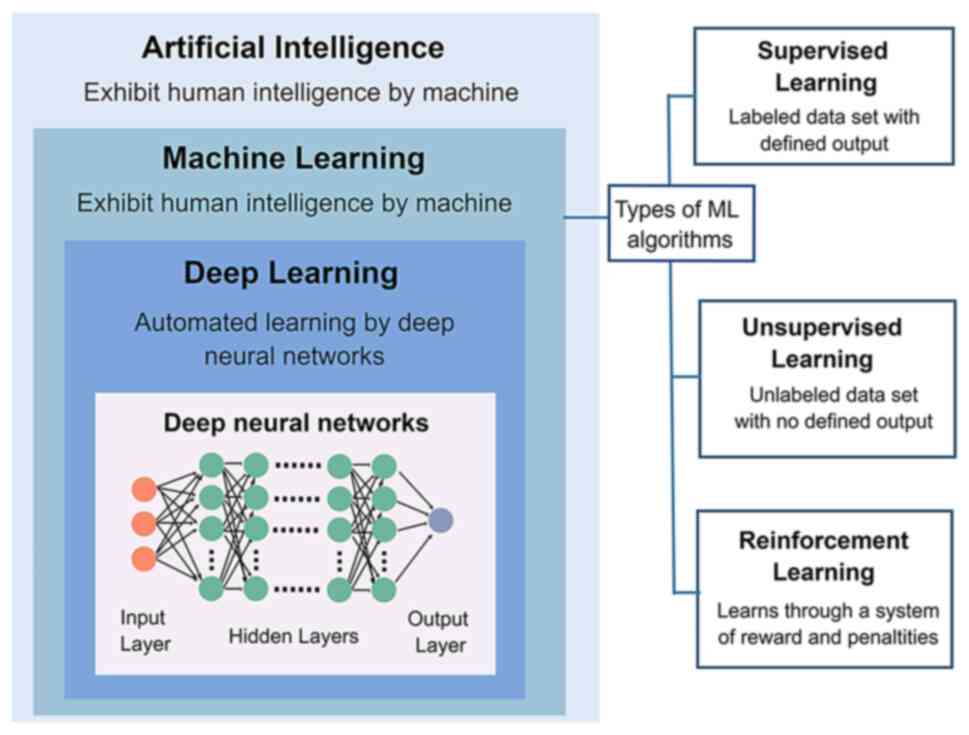
‘train’ (40). Supervised,
unsupervised, and reinforcement learning are three machine-learning
algorithm categories. Deep learning (DL) is an essential subfield
of ML that ‘learns’ from large data sets of raw images, leading to
higher accuracy and faster processing speeds when performing image
recognition, as this process does not require ‘instructions’ to
find specific image features (Fig.
1). Convolutional neural networks (CNNs), a classical branch of
DL, are frequently employed in medical image analysis (41). Thus far, these methods have
gradually penetrated the medical field with substantial success
(42).
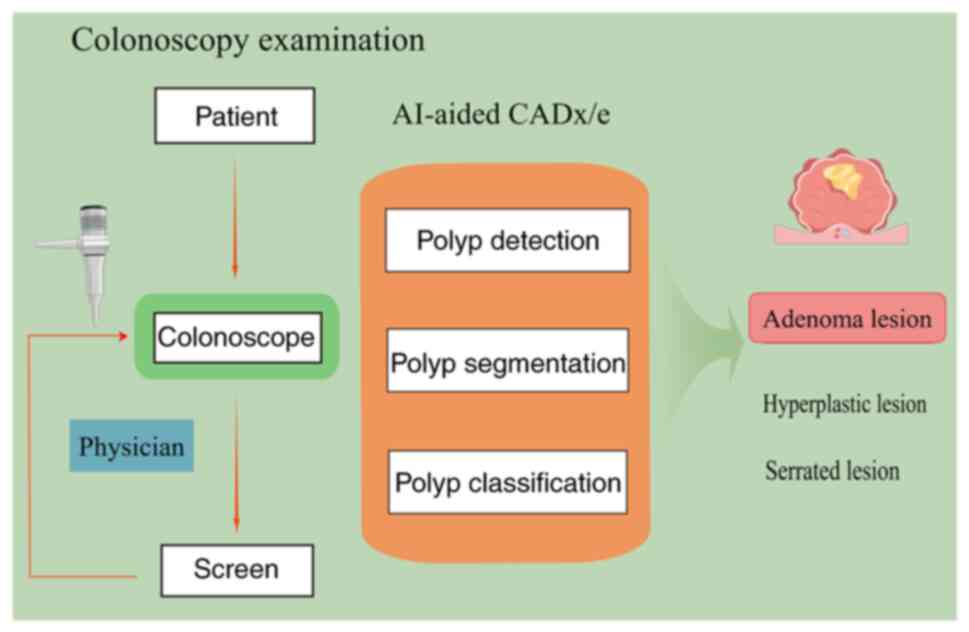
Numerous studies combining clinical and DL have
emerged in oncology screening in recent years. These studies have
utilized detection images and videos of colonoscopies or
pathological images of tumor tissues as input data to train models
with the help of ML (43). The aim
was to assist in the diagnosis and efficacy determination of
clinical tumors with the assistance of models to achieve efficient
precision medicine (Fig. 2)
(44). The combination of
colonoscopy and AI has also been realized using computer
algorithms.
Colonoscopy has established itself as the preferred
diagnostic modality for CRC due to its promising clinical results
and wide range. Unfortunately, due to challenges such as
inter-observer variability in lesion detection, time-consuming
biopsy protocols, and biopsy sampling errors (45), a substantial fluctuation in the ADR
remains present (7–53%) (46), and
the adenoma miss rate may be as high as 26% (29). Numerous studies have indicated that
endoscopists may achieve improved discrimination between
premalignant lesions and hyperplastic polyps through a combination
of AI and colonoscopy. The integration of both has contributed to
an elevated ADR (9) and markedly
reduced CRC morbidity and mortality (47–50).
Therefore, this review focuses on AI-aided colonoscopy, the most
promising and efficient CRC screening method in clinical
settings.
Computer-aided detection (CADe)
model
The concept of a CADe model was established in 2003
(51). This system supports the
diagnosis of CRC and detection of premalignant polyps by processing
endoscopic images or video frame sequences obtained during
colonoscopy (52–58) (Table
II). Karkanis et al (52) designed a CADe model based on color
and texture analysis of the intestinal mucosal surface. This model
had excellent sensitivity up to 99.3±0.3%, and specificity up to
93.6±0.8% in detecting abnormal colon regions associated with
adenomas. However, the CADe model identifies polyps based on static
colonoscopy images rather than real-time analysis of each image
frame in the colonoscopy video, limiting its clinical practicality.
To address this, AI models for automatically detecting polyps using
a series of different imaging feature quantities (such as edge
detection, texture analysis, and energy mapping) have been under
investigation (52). Nevertheless,
none of these methods have achieved a reliable detection rate of
≥90%, and real-time diagnosis has been hindered by computational
power limitations (59). It was not
until the advent of neural networks (NNs) that significant
improvements in this situation began to unfold.
 | Table II.Summary of studies on colonoscopy
combined with CADe. |
Table II.
Summary of studies on colonoscopy
combined with CADe.
| Authors, year | Study design | CADe model | Image type | Conclusion | (Refs.) |
|---|
| Karkanis et
al, 2003 | Retrospective
study | CADe model for
color and texture analysis of mucosal surfaces based on CWC
features | Static images | Sensitivity,
99.3±0.3% | (52) |
| Misawa et
al, 2018 | Retrospective
study | CNN-based CADe
model | Colonoscopy
video | Sensitivity,
90.0% | (53) |
| Urban et al,
2018 | Retrospective
study | CNN-based CADe
model | Colonoscopy
video | Sensitivity,
93.0% | (58) |
| Yamada et
al, 2019 | Retrospective
study | DNN-based CADe
model | Colonoscopy
video | Sensitivity,
97.3% | (54) |
| Wang et al,
2019 | Unblinded
prospective randomized controlled trial | DL-based CADe
model | Colonoscopy
video | ADR in CADe group
vs. standard colonoscopy group: 29.1 vs. 20.3f | (55) |
| Wang et al,
2020 | Double-blind
prospective randomized | DL-based CADe
model | Colonoscopy
video | ADR in CADe group
vs. control group: 34.1 vs. 28% | (56) |
|
| controlled
trial | ENDOANGEL
system | Colonoscopy | ADR in
ENDOANGEL | (57) |
| Gong et al,
2020 | Single-blind
prospective randomized controlled trial | based on DNN and
perceptual hash algorithm | video | group vs. standard
colonoscopy group: 16.34 vs. 7.74% |
|
NN algorithms have emerged and been proven to detect
and localize polyps automatically (60), thus improving the accuracy and
sensitivity of the CADe model for diagnosing polyps and CRC
lesions. In 2018, Misawa et al (53) developed a convolutional 3D NN
algorithm based on the CADe model, reporting that the model had a
sensitivity of 90.0% and a specificity of 63.3% for screening
polyps. In 2019, Yamada et al (54) developed a CADe model based on deep
neural networks (DNNs) and validated it using 705 static images
containing cancerous lesions and 4,135 static images of normal
tissues from 752 patients with CRC. The results were highly
promising, with the CADe model exhibiting exceptional diagnostic
accuracy for CRC, with a sensitivity of 97.3% and a specificity of
99.0%.
Previous prospective studies focusing on the
real-time performance of CADe models have been limited. However, in
2019, Wang et al conducted the first prospective unblinded
randomized controlled trial to investigate the impact of DL-based
CADe models on the accuracy of colorectal screening for polyps and
adenomas (55). A statistically
significant increase in the ADR was observed with the aid of CADe
compared to colorectal screening alone (29.1 vs. 20.3%;
P<0.001). Furthermore, colonoscopy detected more diminutive
adenomas using the CADe model than using colonoscopy alone (185 vs.
102; P<0.001). However, the study was unable to control the
subjective bias of the operating physicians since they were not
blinded to the CADe system. This could have influenced their
vigilance or reliance on the CADe system, potentially
overestimating or underestimating its effectiveness. To address
this issue, Wang et al conducted a double-blind, randomized
controlled trial (56) in 2020,
using a ‘dummy system’ that completely mimicked the false alarm of
the AI system without suggesting true polyps. The operating
physicians were double-blinded, allowing for a more rigorous
assessment of the effectiveness of the CADe system in improving the
detection rate of colonic adenomas and polyps. This study
demonstrated a significant increase of 23.4% in the ADR, from 27.6
to 34.1%, and a considerable increase in polyp detection rate (PDR)
in the CADe group compared to the control group. The study also
confirmed that a high-performance, real-time CADe model could
effectively enhance the detection rate of adenomas and colorectal
polyps, which may contribute to a lower prevalence of CRC. In
addition, the present study revealed a marked improvement in the
number of hyperplastic polyps detected in the CADe group compared
with the control group (114 vs. 52; P<0.001). This finding may
contribute to clinically reducing unnecessary polyp removal, thus
avoiding additional treatment risks such as perforation and massive
bleeding (61).
Negligence by endoscopists is responsible for a
significant percentage (71–86%) of interstitial colorectal cancers
(7,62). One of the main contributing factors
to this negligence is the challenge faced by physicians in
maintaining a standardized withdrawal time during long procedures
under high work pressure. In order to address this issue, Gong
et al (57) developed a CADe
system based on DNNs and perceptual hashing algorithms. By
performing real-time monitoring of the withdrawal speed, recording
of the withdrawal time, and alerting when the colonoscope slips,
the system provided normative feedback to the endoscopist. However,
in contrast to other AI-assisted systems, this system does not
improve the ADR by automatically examining polyps; instead, its
primary objective is to enhance technical elements of the procedure
to achieve improvement. The results revealed that the CADe group
had a prolonged mean negative withdrawal time (6.38 vs. 4.76 min)
and an ~100% enhancement in the ADR (16.34 vs. 7.74%) compared to
the colonoscopy-only group. These findings surpass previous reports
and indicate the practicality of improving the PDR and ADR by
standardizing endoscopist practices through the implementation of
the CADe system.
An AI-aided polyp detection system has shown a
significant increase in the detection rate of lesions, and the
ability of AI to detect lesions is not significantly affected by
factors such as size, location, and shape (63). Real-time AI-aided colonoscopy has
the potential to improved ADR even for experienced endoscopists
(64). Therefore, high-quality
clinical data are urgently required to demonstrate the
effectiveness and accuracy of AI-assisted endoscopy.
Recently, a review highlighted the approval of the
first AI-guided polyp detection system by the U.S. Food and Drug
Administration (65). The GI
Genius™ (Medtronic, Ltd.) system is a CADe system that integrates
existing endoscopy systems and improves adenoma detection during
colonoscopy. However, while the system shows promise in improving
adenoma detection, it is essential to assess its actual impact on
colorectal cancer prevention through large-scale population-based
studies. A study called COLO-DETECT will be the first multi-center
randomized controlled trial evaluting the GI Genius™ in real-world
colonoscopy practice and will be unique in evaluating its clinical
and cost effectiveness (66). The
results will significantly impact the future adoption of this novel
technology.
Computer-aided diagnosis (CADx)
model
Recently, the European Society of Gastrointestinal
Endoscopy (ESGE) published an official position statement aiming to
define simple, safe, and easy-to-measure competence standards for
endoscopists and AI systems performing optical diagnosis of
diminutive colorectal polyps (67).
In this regard, CADx has shown great potential in improving the
accuracy of colorectal polyp characterization (61,68–74)
(Table III). CADx systems may
improve the accuracy of colorectal polyp optical diagnosis, leading
to a reduction in the unnecessary removal of hyperplastic polyps.
Moreover, CADx could help implement cost-saving strategies in
colonoscopy by reducing the burden of polypectomy and pathology. In
other words, its application facilitates the implementation of
resect-and-discard (when polyps are resected and discarded without
histological evaluation) and ‘leave-in-situ’ (when non-neoplastic
lesions located in the rectum and sigmoid are left in situ
without resection, as they have no malignant potential) strategies
(75,76). Furthermore, the study conducted by
Hassan et al confirmed that a real-time CADx system has the
potential to reduce all polypectomies and related costs by 44.4% in
the study population (77),
highlighting the significant cost-saving benefits of this
technology.
 | Table III.Summary of studies on colonoscopy
combined with CADx. |
Table III.
Summary of studies on colonoscopy
combined with CADx.
| Authors, year | Study design | Research
objectives | Imaging
modality | CADx model | CADx model
performance | (Refs.) |
|---|
| Tischendorf et
al, 2010 | Prospective pilot
study | Differentiation of
neoplastic and non-neoplastic colorectal polyps | Magnifying NBI | SVM-based CADx | CADx: Sensitivity,
90%; Specificity, 70%; Accuracy, 85.3% | (71) |
| Gross et al,
2011 | Prospective
study | Differentiation
between neoplastic and non-neoplastic colorectal polyps (≤10
mm) | Magnifying NBI | SVM-based CADx | -CADx group:
Sensitivity, 93.4%; Specificity, 91.8%; Accuracy; 92.7% -Expert
panel: Sensitivity, 93.8%; Specificity, 85.7%; Accuracy, 91.9%
-Non-expert group: Sensitivity, 86%; Specificity, 87.8%; Accuracy,
86.8% | (72) |
| Aihara et
al, 2013 | Prospective
study | Differentiation of
neoplastic and non-neoplastic colorectal polyps | AFI | CADx based on AFE
numerical olor analysis | Sensitivity, 83.3%;
Specificity, 70.1%; PPV, 78.4%; NPV, 82.6% | (70) |
| Mori et al,
2018 | Prospective
study | Differentiation of
small (≤5 mm) neoplastic and non-neoplastic colorectal polyps | Magnifying NBI | SVM-based CADx | Sensitivity, 92.7%;
Specificity, 89.8%; PPV, 93.7%; NPV, 88.3% | (73) |
| Chen et al,
2018 | Prospective
study | Differentiation of
small (≤5 mm) neoplastic and non-neoplastic colorectal polyps | Magnifying NBI | DL-based CADx | Sensitivity, 96.3%;
Specificity, 78.1%; Accuracy, 90.1%; PPV, 89.6%; NPV, 91.5% | (61) |
| Min et al,
2019 | Prospective
study | Differentiation of
adenomatous and non-adenomatous polyps | LCI | GMM-based CADx | Sensitivity, 83.3%;
Specificity, 70.1%; Accuracy, 78.4%; PPV, 82.6%; NPV, 71.2% | (69) |
| Byrne et al,
2019 | Retrospective
study | Differentiation of
small (≤5 mm) neoplastic and non-neoplastic colorectal polyps | Magnifying NBI | DCNN-based
CADx | Sensitivity, 98%;
Specificity, 83%; Accuracy, 94%; PPV, 90%; NPV, 97% | (68) |
In contrast to CADe, in which only observation under
normal white light is possible, CADx is available not only with
white-light endoscopy (77,78) but also in combination with a variety
of other optical imaging techniques, including magnifying
narrow-band imaging (NBI) (68),
linked-color imaging (LCI) (69),
blue-light imaging (BLI) (79), and
autofluorescence imaging (AFI) (70). Among these, studies on CADx and NBI
are the most extensive. As an advanced endoscopic imaging method,
NBI provides excellent visualization, can evaluate mucosal surfaces
and microvascular structures, and is an excellent tool that can
differentiate between neoplastic and non-neoplastic lesions
(80). In 2010, Tischendorf et
al (71) developed a CADx model
that applied NBI and was capable of aiding the classification of
colorectal polyps based on three vascular structural features: Mean
vessel length, vessel circumference, and mean brightness as
observed using NBI. However, the diagnostic accuracy of this model
(85.3%) was markedly lower than that of endoscopic experts and
barely meets the clinical needs of experts. In 2011, Gross et
al (72) developed a CADx model
to assist in classifying colorectal polyps through the analytical
categorization of nine vessel characteristics (e.g., circumference
and brightness). With this model, the sensitivity, specificity, and
accuracy were 95, 90.3 and 93.1%, respectively. In addition, the
diagnostic performance of this model was comparable to that of an
endoscopic expert panel (93.4, 91.8 and 92.7% for sensitivity,
specificity, and accuracy, respectively) and significantly better
than that of a non-expert panel (86, 87.8 and 86.8% for
sensitivity, specificity, and accuracy, respectively). However,
these models lacked real-time diagnostic capabilities, highlighting
the importance of incorporating real-time diagnosis into CADx
technology for its practical application in clinical settings.
By constructing the CADx algorithm using real-time
decision outputs from a support vector machine (SVM), the CADx
algorithm has made significant progress in achieving real-time
diagnosis capabilities (81–83).
In 2018, Mori et al (73)
provided further evidence supporting the use of an SVM-based CADx
model for real-time assisted diagnosis in the NBI mode of
diminutive colorectal polyps. Low- and high-grade adenomas are
classified as neoplastic polyps; hyperplastic polyps, inflammatory
polyps, juvenile polyps, and benign lymphoid polyps are considered
non-neoplastic polyps (11).
Therefore, with a sensitivity of 92.7% and specificity of 89.8%,
this CADx model has sufficient potential to help endoscopists
differentiate between neoplastic and non-neoplastic polyps during
colonoscopy and to achieve the level of performance required for a
‘leave-in-situ’ strategy for patients with non-neoplastic
polyps.
A CADx model has the capability to assist the
endoscopist in differentiating between neoplastic and
non-neoplastic polyps, allowing for the implementation of a
‘leave-in-situ’ strategy for non-neoplastic polyps. Additionally,
it provides support in accurately grading neoplastic polyps for a
‘resect-and-discard’ approach. Min et al (68) designed a CADx model for predicting
the pathological outcome (adenomatous vs. non-adenomatous) of
colorectal polyps based on the results of image color assessments
performed using LCI. Subsequently, it assisted the endoscopist in
the selective resection of neoplastic colorectal polyps. The model
exhibited a sensitivity of 83.3%, specificity of 70.1%, and
accuracy of 78.4% in efficiently differentiating adenomatous from
non-adenomatous polyps. These results are comparable to the
accuracy achieved by endoscopic specialists (78.4 vs. 79.6%).
As a result of their technical shortcomings,
traditional ML methods (such as SVMs) perform poorly when
converting endoscopic images and video features into numerical
data, leading to severe limitations in the development of CADx.
However, the advent of DL has simplified the numerical conversion
process of these features and substantially reduced their
developmental hindrance. In 2018, Chen et al (61) developed a CADx model based on a DL
algorithm that accurately classified small colorectal polyps
(tumors or hyperplastic lesions). A total of 284 magnified NBI
image samples of small colorectal polyps obtained from 193 patients
were used to assess the diagnostic accuracy of this CADx model. The
results showed that the model had a disease sensitivity of 96.3%,
specificity of 78.1%, and accuracy of 90.1%. In addition, the
algorithm enabled the discrimination between tumor and hyperplastic
lesions in a shorter time than the time required by endoscopists
and trainee endoscopists (0.45±0.07 vs. 1.54±1.30 vs. 1.77±1.37
sec), demonstrating its feasibility in clinical practice.
The CADx companion diagnostic results offer
standardized and objective assessments independent of the expertise
and experience of the endoscopist, reducing variations between
beginners and experts. By utilizing CADx, the endoscopist can more
easily make a qualitative diagnosis of the lesion and assess
disease activity while maintaining a higher level of accuracy.
However, the available data for most commercially available AI
tools for lesion characterization remain inconclusive rather than
definitive. The performance of CADx systems should be further
evaluated in prospective randomized controlled trials conducted
among both expert endoscopists and trainees to establish reliable
data and evidence (84).
Future prospects
AI has garnered significant interest in healthcare,
and its potential applications extend to various areas, including
endoscopy. AI-aided colonoscopy has demonstrated promising accuracy
in laboratory settings, and the performance of AI-aided diagnostic
systems has been validated in prospective randomized controlled
trials conducted in diverse healthcare settings, involving
endoscopists with varying levels of experience (75,85).
While the success of AI has been evident in small-sample trials,
the challenge lies in its widespread implementation in clinical
practice. Several issues need addressing before AI can be
effectively integrated into daily practice.
Although several computer-aided colorectal polyp
detection and diagnosis systems have been proposed for clinical
applications, numerous remain susceptible to interference problems
such as low image clarity, unevenness, and low accuracy in the
analysis of dynamic images. These drawbacks affect the robustness
and practicality of these systems (86). In this regard, an intraprocedural AI
alert system for colonoscopy examination has been proposed using
feature extraction and classification alongside a CNN model
(87). This system can identify
blurred images, instances of inadequate bowel cleansing, and
instances of insufficient air insufflation during colonoscopy.
Nevertheless, further clinical trials are required to verify
whether this system can improve the detection rate of colorectal
adenomas. Considering that the data of the study only comes from a
single medical center (87), a
large-scale prospective multicenter clinical trial is required to
validate the efficacy of the proposed system in increasing the
colon polyp detection rate.
In addition to the aforementioned challenges, the
development, integration, and widespread implementation of AI
models in clinical practice require significant investments in
terms of time, resources, and expertise (88,89).
Future studies should carefully consider the potential effects of
these factors. For instance, constructing AI models requires
entering numerous training and validation samples. Nevertheless,
high-quality labeled samples are difficult to obtain in clinical
settings, as these samples often contain numerous labeling errors,
referred to as labeling noise or ‘noisy labels’ (90), which markedly decreases the accuracy
of the model. In addition, AI training involves powerful computer
configurations and long training times, and post-maintenance can be
cumbersome. Clinicians, who are mostly non-specialists, can only
assist in diagnosis based on predefined functions during clinical
work. Therefore, it becomes difficult for doctors to update the
database and algorithm when encountering new cases in clinics.
These factors have greatly hindered the popularity and optimization
of AI systems. Fortunately, these restrictions are gradually being
overcome as a result of advances in computing power, increases in
the number of digitally-stored medical images, and improvements in
deep network (DPN) architecture. Future research should consider
establishing an open data-sharing platform across multiple
institutions to overcome these barriers. Appropriate data sharing
would not only reduce competition among agencies but also alleviate
the difficulties and costs associated with data access while
enhancing data quality (91).
The current laws and regulations for newly developed
AI tools by regulatory agencies are inadequate at this stage.
Nonetheless, the situation is changing rapidly. In January 2021,
the U.S. FDA released the first AI/ML-Based Software as a Medical
Device (SaMD) Action Plan of the agency, which details several
guidelines for AI implementation (92). However, refining the original
legislation may not be sufficient to regulate AI in healthcare. It
is crucial for lawmakers to engage in a collaborative process with
computer scientists, clinicians, patients, professional
associations, and health technology companies to establish a robust
regulatory and legal framework for AI-based tools. This
collaborative effort aims to ensure that AI tools meet acceptable
standards of quality and safety.
From a clinical perspective, establishing trust in
the clinical system is of utmost importance in AI-assisted
decision-making (93). One crucial
aspect in this regard is the stability of AI models. For clinical
application, the AI model must withstand multiple fluctuations in
the input data, such as operator-operator and laboratory-laboratory
differences in data quality, resolution, intensity, and disease
characteristics. However, most AI models have not demonstrated
sufficient stability in the face of such fluctuations, which makes
rigorous quality control necessary. Both the passage of time and
changes in the patient population may lead to deviations in AI
model performance; therefore, AI models applied in clinical
settings must undergo regular quality monitoring and maintenance to
maintain a stable clinical performance (88). The development of standards and
guidelines for testing AI models could systematically assess the
performance of AI-based tools and obtain precise and uniform
measurements. This is the key component in future attempts to
overcome distrust in the clinical system.
The combination of AI and colonoscopy holds
practical and feasible potential, offering promising prospects for
the future (94). Genetic testing
and immune typing, coupled with AI technologies such as DL, have
shown promise in CRC research, providing insights into tumor
pathogenesis at the molecular level and offering theoretical
support for CRC diagnosis and treatment. Numerous studies have
supported that using AI to detect genetic mutations in CRC is a
reliable method to offer a new treatment option for targeted
therapy (94,95). Mutations in KRAS and BRAF genes are
the main predictive biomarkers for the response to anti-EGFR
monoclonal antibody-targeted therapy in metastatic colorectal
cancer (96). Some scholars have
used the DL method based on a residual NN and ML-based CT texture
analysis to achieve the noninvasive prediction of the KRAS mutation
status in CRC (97,98), while others have used a random
forest classifier (RFC) model to predict the V600E mutation in the
BRAF (97). This integration of AI
with genetic testing and immune typing has the potential to enhance
the accuracy and effectiveness of colonoscopy screening and
diagnosis. However, relevant literature was reviewed and it was
determined that there is no research currently on their use in
colonoscopy.
To summarize, the combination of AI and colonoscopy
is practical and feasible, and the future is bright; however,
further exploration and innovation are still expected.
Conclusion
CRC is one of the most common tumors worldwide,
accounting for 10% of all tumors. It is estimated that 608,000
people succumb to CRC annually (~8% of all cancer-related deaths).
In addition, CRC incidence and mortality rates among adults under
50 years of age have been consistently increasing at an annual rate
of 1.5% (2014–2018) and 1.2% (2005–2019), in recent years.
Moreover, the global CRC disease burden is continuously increasing,
with a trend toward younger incidence (1,99).
Although colonoscopy is valuable in decreasing the
mortality or morbidity of CRC, its diagnostic accuracy still falls
short of clinical needs, particularly for premalignant lesions or
early-stage CRC. The introduction of AI in colonoscopy may
potentially improve these deficiencies. For instance, various
studies have shown that AI-based high-level auxiliary diagnostic
systems can significantly improve the readability of medical images
and help clinicians make accurate diagnostic and therapeutic
decisions. In addition, CNNs can aid in the interpretation of
histopathological tissue images, reducing inter-observer
variability among doctors. Furthermore, CADe systems can
significantly improve polyp and ADRs during early colonoscopy
screenings, enhancing the differential diagnosis of non-neoplastic
vs. neoplastic polyps and adenomatous vs. non-adenomatous polyps,
thereby decreasing the possibility of mutating into CRC.
Additionally, AI has the potential to contribute to cost-saving
strategies by minimizing the need for unnecessary polypectomies and
pathology examinations. Overall, the key findings of this review
are that AI-aided colonoscopy could facilitate the efficiency and
accuracy of CRC screening and diagnosis and ameliorate patient
clinical outcomes and prognosis.
Preliminary data on AI-assisted systems are
promising; however, the lack of high-quality clinical studies
prevents reliable conclusions. It is essential to conduct
higher-quality research using modern trial designs to improve the
understanding of this field. Special attention should be given to
utilizing larger datasets and prospectively validating AI systems
in clinical settings. Moreover, these systems must provide quality
assurance within a robust ethical and legal framework before
clinicians and patients fully embrace them.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural Science
Foundation of China (grant nos. 82074214 and 81973598) and the Key
project of Administration of Traditional Chinese Medicine of
Zhejiang province (grant no. 2022ZZ014).
Availability of data and materials
Not applicable.
Authors' contributions
SZ and GC conceived and designed the review. MD and
JY collected and reviewed the literature as well as drafted the
manuscript. SZ, MD and JY edited and revised the manuscript. All
authors read and approved the final manuscript. Data authentication
is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
USPSTF
|
U.S. Preventive Services Taskforce
|
|
AI
|
artificial intelligence
|
|
ADR
|
adenoma detection rate
|
|
ML
|
machine learning
|
|
DL
|
deep learning
|
|
CADe
|
computer-aided detection
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Trepanier M, Minnella EM, Paradis T,
Awasthi R, Kaneva P, Schwartzman K, Carli F, Fried GM, Feldman LS
and Lee L: Improved Disease-free survival after prehabilitation for
colorectal cancer surgery. Ann Surg. 270:493–501. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Click B, Pinsky PF, Hickey T, Doroudi M
and Schoen RE: Association of colonoscopy adenoma findings with
Long-term colorectal cancer incidence. JAMA. 319:2021–2031. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shinya H and Wolff WI: Morphology,
anatomic distribution and cancer potential of colonic polyps. Ann
Surg. 190:679–683. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ladabaum U, Dominitz JA, Kahi C and Schoen
RE: Strategies for colorectal cancer screening. Gastroenterology.
158:418–432. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
US Preventive Services Task Force, .
Davidson KW, Barry MJ, Mangione CM, Cabana M, Caughey AB, Davis EM,
Donahue KE, Doubeni CA, Krist AH, et al: Screening for colorectal
cancer: US Preventive services task force recommendation statement.
JAMA. 325:1965–1977. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin JS, Perdue LA, Henrikson NB, Bean SI
and Blasi PR: Screening for colorectal cancer: Updated evidence
report and systematic review for the US preventive services task
force. JAMA. 325:1978–1998. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barua I, Vinsard DG, Jodal HC, Loberg M,
Kalager M, Holme O, Holme Ø, Misawa M, Bretthauer M and Mori Y:
Artificial intelligence for polyp detection during colonoscopy: A
systematic review and meta-analysis. Endoscopy. 53:277–284. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao P, Zhou K, Su W, Yu J and Zhou P:
Endoscopic management of colorectal polyps. Gastroenterol Rep
(Oxf). 11:goad0272023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meseeha M and Attia M: Colon Polyps.
StatPearls. Treasure Island (FL) ineligible companies. Disclosure:
Maximos Attia declares no relevant financial relationships with
ineligible companies. 2023.
|
|
12
|
Kamaradova K: Non-conventional types of
dysplastic changes in gastrointestinal tract mucosa-review of
morphological features of individual subtypes. Cesk Patol.
58:38–51. 2022.PubMed/NCBI
|
|
13
|
Keum N and Giovannucci E: Global burden of
colorectal cancer: Emerging trends, risk factors and prevention
strategies. Nat Rev Gastroenterol Hepatol. 16:713–732. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Crockett SD and Nagtegaal ID: Terminology,
molecular features, epidemiology, and management of serrated
colorectal neoplasia. Gastroenterology. 157:949–66.e4. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Haghighat S, Sussman DA and Deshpande A:
US preventive services task force recommendation statement on
screening for colorectal cancer. JAMA. 326:13282021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Carethers JM: Fecal DNA testing for
colorectal cancer screening. Annu Rev Med. 71:59–69. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mandel JS, Church TR, Bond JH, Ederer F,
Geisser MS, Mongin SJ, Snover DC and Schuman LM: The effect of
fecal occult-blood screening on the incidence of colorectal cancer.
N Engl J Med. 343:1603–1607. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Faivre J, Dancourt V, Lejeune C, Tazi MA,
Lamour J, Gerard D, Dassonville F and Bonithon-Kopp C: Reduction in
colorectal cancer mortality by fecal occult blood screening in a
French controlled study. Gastroenterology. 126:1674–1680. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kronborg O, Jorgensen OD, Fenger C and
Rasmussen M: Randomized study of biennial screening with a faecal
occult blood test: Results after nine screening rounds. Scand J
Gastroenterol. 39:846–851. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Scholefield JH, Moss SM, Mangham CM,
Whynes DK and Hardcastle JD: Nottingham trial of faecal occult
blood testing for colorectal cancer: A 20-year follow-up. Gut.
61:1036–1040. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shaukat A, Mongin SJ, Geisser MS, Lederle
FA, Bond JH, Mandel JS and Church TR: Long-term mortality after
screening for colorectal cancer. N Engl J Med. 369:1106–1114. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chiu HM, Chen SL, Yen AM, Chiu SY, Fann
JC, Lee YC, Pan SL, Wu MS, Liao CS, Chen HH, et al: Effectiveness
of fecal immunochemical testing in reducing colorectal cancer
mortality from the One Million Taiwanese Screening Program. Cancer.
121:3221–3229. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zorzi M, Fedeli U, Schievano E, Bovo E,
Guzzinati S, Baracco S, Fedato C, Saugo M and Dei Tos AP: Impact on
colorectal cancer mortality of screening programmes based on the
faecal immunochemical test. Gut. 64:784–790. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Redwood DG, Dinh TA, Kisiel JB, Borah BJ,
Moriarty JP, Provost EM, Sacco FD, Tiesinga JJ and Ahlquist DA:
Cost-Effectiveness of multitarget stool DNA testing vs colonoscopy
or fecal immunochemical testing for colorectal cancer screening in
alaska native people. Mayo Clin Proc. 96:1203–1217. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Atkin W, Wooldrage K, Parkin DM,
Kralj-Hans I, MacRae E, Shah U, Duffy S and Cross AJ: Long term
effects of once-only flexible sigmoidoscopy screening after 17
years of follow-up: The UK Flexible Sigmoidoscopy Screening
randomised controlled trial. Lancet. 389:1299–1311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wolf AMD, Fontham ETH, Church TR, Flowers
CR, Guerra CE, LaMonte SJ, Etzioni R, McKenna MT, Oeffinger KC and
Shih YT: Colorectal cancer screening for average-risk adults: 2018
guideline update from the American Cancer Society. CA Cancer J
Clin. 68:250–281. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nishihara R, Wu K, Lochhead P, Morikawa T,
Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, et al:
Long-term colorectal-cancer incidence and mortality after lower
endoscopy. N Engl J Med. 369:1095–1105. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Calderwood AH and Jacobson BC: Colonoscopy
quality: Metrics and implementation. Gastroenterol Clin North Am.
42:599–618. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao S, Wang S, Pan P, Xia T, Chang X,
Yang X, Guo L, Meng Q, Yang F, Qian W, et al: Magnitude, risk
factors, and factors associated with adenoma miss rate of tandem
colonoscopy: A Systematic review and meta-analysis.
Gastroenterology. 156:1661–1674.e11. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kaminski MF, Wieszczy P, Rupinski M,
Wojciechowska U, Didkowska J, Kraszewska E, Kobiela J, Franczyk R,
Rupinska M, Kocot B, et al: Increased rate of adenoma detection
associates with reduced risk of colorectal cancer and death.
Gastroenterology. 153:98–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rabeneck L and Paszat LF: Circumstances in
which colonoscopy misses cancer. Frontline Gastroenterol. 1:52–58.
2010.PubMed/NCBI
|
|
32
|
Rutter MD, Beintaris I, Valori R, Chiu HM,
Corley DA, Cuatrecasas M, Dekker E, Forsberg A, Gore-Booth J, Haug
U, et al: World endoscopy organization consensus statements on
Post-Colonoscopy and Post-Imaging colorectal cancer.
Gastroenterology. 155:909–25.e3. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kyu HH, Bachman VF, Alexander LT, Mumford
JE, Afshin A, Estep K, Veerman JL, Delwiche K, Iannarone ML, Moyer
ML, et al: Physical activity and risk of breast cancer, colon
cancer, diabetes, ischemic heart disease, and ischemic stroke
events: Systematic review and dose-response meta-analysis for the
Global Burden of Disease Study 2013. BMJ. 354:i38572016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Morris EJ, Rutter MD, Finan PJ, Thomas JD
and Valori R: Post-colonoscopy colorectal cancer (PCCRC) rates vary
considerably depending on the method used to calculate them: A
retrospective observational population-based study of PCCRC in the
English National Health Service. Gut. 64:1248–1256. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Anderson R, Burr NE and Valori R: Causes
of Post-colonoscopy colorectal cancers based on world endoscopy
organization system of analysis. Gastroenterology.
158:1287–1299.e2. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hassan C, Piovani D, Spadaccini M, Parigi
T, Khalaf K, Facciorusso A, Fugazza A, Rösch T, Bretthauer M, Mori
Y, et al: Variability in adenoma detection rate in control groups
of randomized colonoscopy trials: A systematic review and
meta-analysis. Gastrointest Endosc. 97:212–225.e7. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Burr N and Valori R: National
post-colonoscopy colorectal cancer data challenge services to
improve quality of colonoscopy. Endosc Int Open. 7:E728–E729. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wallace MB, Sharma P, Bhandari P, East J,
Antonelli G, Lorenzetti R, Vieth M, Speranza I, Spadaccini M, Desai
M, et al: Impact of artificial intelligence on miss rate of
colorectal neoplasia. Gastroenterology. 163:295–304.e5. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hamet P and Tremblay J: Artificial
intelligence in medicine. Metabolism. 69S:S36–S40. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bishop C: Pattern Recognition and Machine
Learning (Information Science and Statistics). Springer; April
6–2011, ISBN-10: 03873107382011. 2011.
|
|
41
|
LeCun Y, Bengio Y and Hinton G: Deep
learning. Nature. 521:436–444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Esteva A, Robicquet A, Ramsundar B,
Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S and Dean
J: A guide to deep learning in healthcare. Nat Med. 25:24–29. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Krenzer A, Makowski K, Hekalo A, Fitting
D, Troya J, Zoller WG, Hann A and Puppe F: Fast machine learning
annotation in the medical domain: A semi-automated video annotation
tool for gastroenterologists. Biomed Eng Online. 21:332022.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bera K, Schalper KA, Rimm DL, Velcheti V
and Madabhushi A: Artificial intelligence in digital pathology-new
tools for diagnosis and precision oncology. Nat Rev Clin Oncol.
16:703–715. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Corley DA, Jensen CD, Marks AR, Zhao WK,
Lee JK, Doubeni CA, Zauber AG, de Boer J, Fireman BH, Schottinger
JE, et al: Adenoma detection rate and risk of colorectal cancer and
death. N Engl J Med. 370:1298–1306. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Greenspan M, Rajan KB, Baig A, Beck T,
Mobarhan S and Melson J: Advanced adenoma detection rate is
independent of nonadvanced adenoma detection rate. Am J
Gastroenterol. 108:1286–1292. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Singh H, Turner D, Xue L, Targownik LE and
Bernstein CN: Risk of developing colorectal cancer following a
negative colonoscopy examination: Evidence for a 10-year interval
between colonoscopies. JAMA. 295:2366–2373. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Brenner H, Chang-Claude J, Seiler CM,
Rickert A and Hoffmeister M: Protection from colorectal cancer
after colonoscopy: A population-based, case-control study. Ann
Intern Med. 154:22–30. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Baxter NN, Goldwasser MA, Paszat LF,
Saskin R, Urbach DR and Rabeneck L: Association of colonoscopy and
death from colorectal cancer. Ann Intern Med. 150:1–8. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kahi CJ, Imperiale TF, Juliar BE and Rex
DK: Effect of screening colonoscopy on colorectal cancer incidence
and mortality. Clin Gastroenterol Hepatol. 7:770–775; quiz 11.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Maroulis DE, Iakovidis DK, Karkanis SA and
Karras DA: CoLD: A versatile detection system for colorectal
lesions in endoscopy video-frames. Comput Methods Programs Biomed.
70:151–166. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Karkanis SA, Iakovidis DK, Maroulis DE,
Karras DA and Tzivras M: Computer-aided tumor detection in
endoscopic video using color wavelet features. IEEE Trans Inf
Technol Biomed. 7:141–152. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Misawa M, Kudo SE, Mori Y, Cho T, Kataoka
S, Yamauchi A, Ogawa Y, Maeda Y, Takeda K, Ichimasa K, et al:
Artificial Intelligence-Assisted polyp detection for colonoscopy:
Initial experience. Gastroenterology. 154:2027–2029.e3. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yamada M, Saito Y, Imaoka H, Saiko M,
Yamada S, Kondo H, Takamaru H, Sakamoto T, Sese J, Kuchiba A, et
al: Development of a real-time endoscopic image diagnosis support
system using deep learning technology in colonoscopy. Sci Rep.
9:144652019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang P, Berzin TM, Glissen Brown JR,
Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, et al:
Real-time automatic detection system increases colonoscopic polyp
and adenoma detection rates: A prospective randomised controlled
study. Gut. 68:1813–1819. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang P, Liu X, Berzin TM, Glissen Brown
JR, Liu P, Zhou C, Lei L, Li L, Guo Z, Lei S, et al: Effect of a
deep-learning computer-aided detection system on adenoma detection
during colonoscopy (CADe-DB trial): A double-blind randomised
study. Lancet Gastroenterol Hepatol. 5:343–351. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gong D, Wu L, Zhang J, Mu G, Shen L, Liu
J, Wang Z, Zhou W, An P, Huang X, et al: Detection of colorectal
adenomas with a real-time computer-aided system (ENDOANGEL): A
randomised controlled study. Lancet Gastroenterol Hepatol.
5:352–361. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Urban G, Tripathi P, Alkayali T, Mittal M,
Jalali F, Karnes W and Baldi P: Deep learning localizes and
identifies polyps in real time with 96% accuracy in screening
colonoscopy. Gastroenterology. 155:1069–1078.e8. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kamitani Y, Nonaka K and Isomoto H:
Current status and future perspectives of artificial intelligence
in colonoscopy. J Clin Med. 11:29232022. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gonzalez-Bueno Puyal J, Brandao P, Ahmad
OF, Bhatia KK, Toth D, Kader R, Lovat L, Mountney P and Stoyanov D:
Polyp detection on video colonoscopy using a hybrid 2D/3D CNN. Med
Image Anal. 82:1026252022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH and
Tseng VS: Accurate classification of diminutive colorectal polyps
using computer-aided analysis. Gastroenterology. 154:568–575. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ng K, May FP and Schrag D: US preventive
services task force recommendations for colorectal cancer
screening: Forty-five is the new fifty. JAMA. 325:1943–1945. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Huang D, Shen J, Hong J, Zhang Y, Dai S,
Du N, Zhang M and Guo D: Effect of artificial intelligence-aided
colonoscopy for adenoma and polyp detection: A meta-analysis of
randomized clinical trials. Int J Colorectal Dis. 37:495–506. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Koh FH, Ladlad J, Centre SKHE, Teo EK, Lin
CL and Foo FJ: Real-time artificial intelligence (AI)-aided
endoscopy improves adenoma detection rates even in experienced
endoscopists: A cohort study in Singapore. Surg Endosc. 37:165–171.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Spadaccini M, Marco A, Franchellucci G,
Sharma P, Hassan C and Repici A: Discovering the first US
FDA-approved computer-aided polyp detection system. Future Oncol.
18:1405–1412. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Seager A, Sharp L, Hampton JS, Neilson LJ,
Lee TJW, Brand A, Evans R, Vale L, Whelpton J and Rees CJ: Trial
protocol for COLO-DETECT: A randomized controlled trial of lesion
detection comparing colonoscopy assisted by the GI Genius
artificial intelligence endoscopy module with standard colonoscopy.
Colorectal Dis. 24:1227–1237. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Houwen B, Hassan C, Coupe VMH, Greuter
MJE, Hazewinkel Y, Vleugels JLA, Antonelli G, Bustamante-Balén M,
Coron E, Cortas GA, et al: Definition of competence standards for
optical diagnosis of diminutive colorectal polyps: European Society
of Gastrointestinal Endoscopy (ESGE) Position Statement. Endoscopy.
54:88–99. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Byrne MF, Chapados N, Soudan F, Oertel C,
Linares Perez M, Kelly R, Iqbal N, Chandelier F and Rex DK:
Real-time differentiation of adenomatous and hyperplastic
diminutive colorectal polyps during analysis of unaltered videos of
standard colonoscopy using a deep learning model. Gut. 68:94–100.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Min M, Su S, He W, Bi Y, Ma Z and Liu Y:
Computer-aided diagnosis of colorectal polyps using linked color
imaging colonoscopy to predict histology. Sci Rep. 9:28812019.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Aihara H, Saito S, Inomata H, Ide D, Tamai
N, Ohya TR, Kato T, Amitani S and Tajiri H: Computer-aided
diagnosis of neoplastic colorectal lesions using ‘real-time’
numerical color analysis during autofluorescence endoscopy. Eur J
Gastroenterol Hepatol. 25:488–494. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tischendorf JJ, Gross S, Winograd R,
Hecker H, Auer R, Behrens A, Trautwein C, Aach T and Stehle T:
Computer-aided classification of colorectal polyps based on
vascular patterns: A pilot study. Endoscopy. 42:203–207. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Gross S, Trautwein C, Behrens A, Winograd
R, Palm S, Lutz HH, Schirin-Sokhan R, Hecker H, Aach T and
Tischendorf JJ: Computer-based classification of small colorectal
polyps by using narrow-band imaging with optical magnification.
Gastrointest Endosc. 74:1354–1359. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Mori Y, Kudo SE, Misawa M, Saito Y,
Ikematsu H, Hotta K, Ohtsuka K, Urushibara F, Kataoka S, Ogawa Y,
et al: Real-time use of artificial intelligence in identification
of diminutive polyps during colonoscopy: A prospective study. Ann
Intern Med. 169:357–366. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Vinsard DG, Mori Y, Misawa M, Kudo SE,
Rastogi A, Bagci U, Rex DK and Wallace MB: Quality assurance of
computer-aided detection and diagnosis in colonoscopy. Gastrointest
Endosc. 90:55–63. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Spadaccini M, Massimi D, Mori Y, Alfarone
L, Fugazza A, Maselli R, Sharma P, Facciorusso A, Hassan C and
Repici A: Artificial intelligence-aided endoscopy and colorectal
cancer screening. Diagnostics (Basel). 13:11022023. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Hassan C, Pickhardt PJ and Rex DK: A
resect and discard strategy would improve cost-effectiveness of
colorectal cancer screening. Clin Gastroenterol Hepatol.
8:865–869.e1-e3. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Hassan C, Balsamo G, Lorenzetti R, Zullo A
and Antonelli G: Artificial intelligence allows leaving-in-situ
colorectal polyps. Clin Gastroenterol Hepatol. 20:2505–2513.e4.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Sánchez-Montes C, Sánchez FJ, Bernal J,
Córdova H, López-Cerón M, Cuatrecasas M, Rodríguez de Miguel C,
García-Rodríguez A, Garcés-Durán R, Pellisé M, et al:
Computer-aided prediction of polyp histology on white light
colonoscopy using surface pattern analysis. Endoscopy. 51:261–265.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Yoshida N, Inoue K, Tomita Y, Kobayashi R,
Hashimoto H, Sugino S, Hirose R, Dohi O, Yasuda H, Morinaga Y, et
al: An analysis about the function of a new artificial
intelligence, CAD EYE with the lesion recognition and diagnosis for
colorectal polyps in clinical practice. Int J Colorectal Dis.
36:2237–2245. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Barbeiro S, Libanio D, Castro R,
Dinis-Ribeiro M and Pimentel-Nunes P: Narrow-band imaging: Clinical
application in gastrointestinal endoscopy. GE Port J Gastroenterol.
26:40–53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Tamaki T, Yoshimuta J, Kawakami M,
Raytchev B, Kaneda K, Yoshida S, Takemura Y, Onji K, Miyaki R and
Tanaka S: Computer-aided colorectal tumor classification in NBI
endoscopy using local features. Med Image Anal. 17:78–100. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wimmer G, Tamaki T, Tischendorf JJ, Hafner
M, Yoshida S, Tanaka S and Uhl A: Directional wavelet based
features for colonic polyp classification. Med Image Anal.
31:16–36. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Hafner M, Tamaki T, Tanaka S, Uhl A,
Wimmer G and Yoshida S: Local fractal dimension based approaches
for colonic polyp classification. Med Image Anal. 26:92–107. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Mori Y, Neumann H, Misawa M, Kudo SE and
Bretthauer M: Artificial intelligence in colonoscopy-Now on the
market. What's next? J Gastroenterol Hepatol. 36:7–11. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Nazarian S, Glover B, Ashrafian H, Darzi A
and Teare J: Diagnostic accuracy of artificial intelligence and
computer-aided diagnosis for the detection and characterization of
colorectal polyps: Systematic review and Meta-analysis. J Med
Internet Res. 23:e273702021. View
Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hassan C, Badalamenti M, Maselli R,
Correale L, Iannone A, Radaelli F, Rondonotti E, Ferrara E,
Spadaccini M, Alkandari A, et al: Computer-aided detection-assisted
colonoscopy: Classification and relevance of false positives.
Gastrointest Endosc. 92:900–904.e4. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Hsu CM, Hsu CC, Hsu ZM, Chen TH and Kuo T:
Intraprocedure artificial intelligence alert system for colonoscopy
examination. Sensors (Basel). 23:12112023. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Elemento O, Leslie C, Lundin J and
Tourassi G: Artificial intelligence in cancer research, diagnosis
and therapy. Nat Rev Cancer. 21:747–752. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Wei JW, Suriawinata AA, Vaickus LJ, Ren B,
Liu X, Lisovsky M, Tomita N, Abdollahi B, Kim AS, Snover DC, et al:
Evaluation of a deep neural network for automated classification of
colorectal polyps on histopathologic slides. JAMA Netw Open.
3:e2033982020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Karimi D, Dou H, Warfield SK and Gholipour
A: Deep learning with noisy labels: Exploring techniques and
remedies in medical image analysis. Med Image Anal. 65:1017592020.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Huang P, Feng Z, Shu X, Wu A, Wang Z, Hu
T, Cao Y, Tu Y and Li Z: A bibliometric and visual analysis of
publications on artificial intelligence in colorectal cancer
(2002–2022). Front Oncol. 13:10775392023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Clark P, Kim J and Aphinyanaphongs Y:
Marketing and US food and drug administration clearance of
artificial intelligence and machine learning enabled software in
and as medical devices: A systematic review. JAMA Netw Open.
6:e23217922023. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Bhinder B, Gilvary C, Madhukar NS and
Elemento O: Artificial intelligence in cancer research and
precision medicine. Cancer Discov. 11:900–915. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yin Z, Yao C, Zhang L and Qi S:
Application of artificial intelligence in diagnosis and treatment
of colorectal cancer: A novel Prospect. Front Med (Lausanne).
10:11280842023. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Sorokin M, Zolotovskaia M, Nikitin D,
Suntsova M, Poddubskaya E, Glusker A, Garazha A, Moisseev A, Li X,
Sekacheva M, et al: Personalized targeted therapy prescription in
colorectal cancer using algorithmic analysis of RNA sequencing
data. BMC Cancer. 22:11132022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Sanchez-Ibarra HE, Jiang X,
Gallegos-Gonzalez EY, Cavazos-Gonzalez AC, Chen Y, Morcos F and
Barrera-Saldaña HA: KRAS, NRAS, and BRAF mutation prevalence,
clinicopathological association, and their application in a
predictive model in Mexican patients with metastatic colorectal
cancer: A retrospective cohort study. PLoS One. 15:e02354902020.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
He K, Liu X, Li M, Li X, Yang H and Zhang
H: Noninvasive KRAS mutation estimation in colorectal cancer using
a deep learning method based on CT imaging. BMC Med Imaging.
20:592020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Taguchi N, Oda S, Yokota Y, Yamamura S,
Imuta M, Tsuchigame T, Nagayama Y, Kidoh M, Nakaura T, Shiraishi S,
et al: CT texture analysis for the prediction of KRAS mutation
status in colorectal cancer via a machine learning approach. Eur J
Radiol. 118:38–43. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Spaander MCW, Zauber AG, Syngal S, Blaser
MJ, Sung JJ, You YN and Kuipers EJ: Young-onset colorectal cancer.
Nat Rev Dis Primers. 9:212023. View Article : Google Scholar : PubMed/NCBI
|