Lymphomas are malignant tumors in which lymphocytes
in the human body undergo different stages of development and
differentiation (17). Lymphomas
exhibit high levels of heterogeneity and a complex pathological
classification, with different treatment responses among different
pathological types. In addition, treatment responses may differ
between patients with the same pathological type. Numerous factors,
including cell proteomics and molecular features may impact the
prognosis of patients (18). In
clinical practice, patients with DLBCL often develop drug
resistance (19), which is a
barrier to treatment within clinical practice. Liu et al
(20) examined samples from 14
patients with untreated DLBCL using mass spectrometry and
two-dimensional (2D) gel electrophoresis, and quantitatively
identified differentially expressed proteins between patients who
were susceptible to CHOP treatment and those who were resistant.
This approach allowed the comprehensive characterization of the
proteomic landscape associated with chemotherapy response in DLBCL;
thus, providing valuable insights into potential biomarkers and
therapeutic targets for improving treatment outcomes. Results of
the previous study demonstrated that the protein expression levels
of histone H2A.2, S100A9, Ezrin and Pleckstrin were significantly
increased. In addition, the protein expression levels of 61 kD
protein, collagen alpha 1 (VI), glutathione S-transferase pi-1 and
heat shock protein beta 1 were significantly lower in patients who
were susceptible to CHOP treatment, compared with those that were
resistant. Analyzing the protein network associated with resistance
to CHOP chemotherapy may aid in identifying patients with DLBCL
with CHOP resistance; thus, providing a novel theoretical basis for
the identification of therapeutic targets.
Tumors form dynamic, complex and heterogeneous
environments with various cells and surrounding components, known
as the TME (21). The heterogeneity
of DLBCL is associated with the types of cells in the TME (22), including matrix components,
dendritic cells, macrophages, monocytes, fibroblasts and T cells
(23). Notably, the extracellular
matrix interacts with lymphoma cells (24), and high numbers of M2 macrophages,
natural killer cells and plasma cells are associated with lower
survival rates in patients with DLBCL (25). The TME plays a significant role in
the initiation, development and treatment resistance of DLBCL, and
these factors ultimately impact the prognosis of patients (19). In total, ~75% of patients with DLBCL
possess aberrations in genes associated with immune escape
(26), and the TME includes
numerous inhibitory immune detection points (27). Liu et al (28) suggested that adaptor-related protein
complex 2 subunit mu1 subunit may contribute to the resistance of
DLBCL to chemotherapy and targeted medications through controlling
the TME. Notably, results of previous studies highlighted that
multiple components in the TME may impact the occurrence and
development of DLBCL. Spatial proteomics analysis may provide
location information for cells in the tissue (29), and this method may be used to
explore the interaction between DLBCL cells and the TME (30).
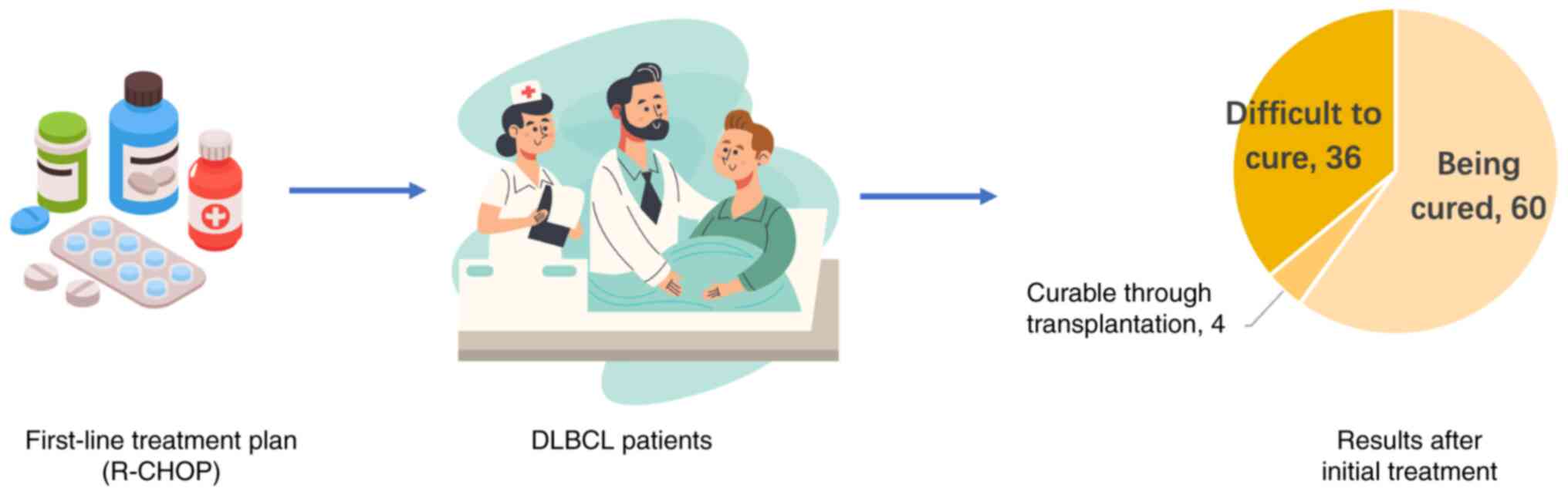
The discovery and application of anti-CD20
monoclonal antibodies in the early 20th century led to a new era of
DLBCL treatment (35). At present,
global treatment guidelines recommend first-line therapy with
R-CHOP, comprising rituximab with cyclophosphamide, doxorubicin,
vincristine and prednisone (36),
leading to a cure in ~60% of patients (37). However, a small number of patients
continue to exhibit refractory disease or relapse following
complete remission (38), and
traditional salvage immunochemotherapy combined with autologous
hematopoietic stem cell transplantation only achieves a cure in
~10% of these patients (39). Thus,
the remaining 90% of patients exhibit poor treatment outcomes
(Fig. 1). Thus, improving the
prognosis of these patients is complex (40), and further proteomic analyses are
required to determine the signaling pathways associated with the
onset and progression of DLBCL (41). Novel developments in proteomics
technology have led to the discovery of multiple drug resistance
mechanisms in lymphoma (42); thus,
strategies and methods that eliminate the drug resistance of
lymphoma cells and improve the therapeutic effects are also
required (43).
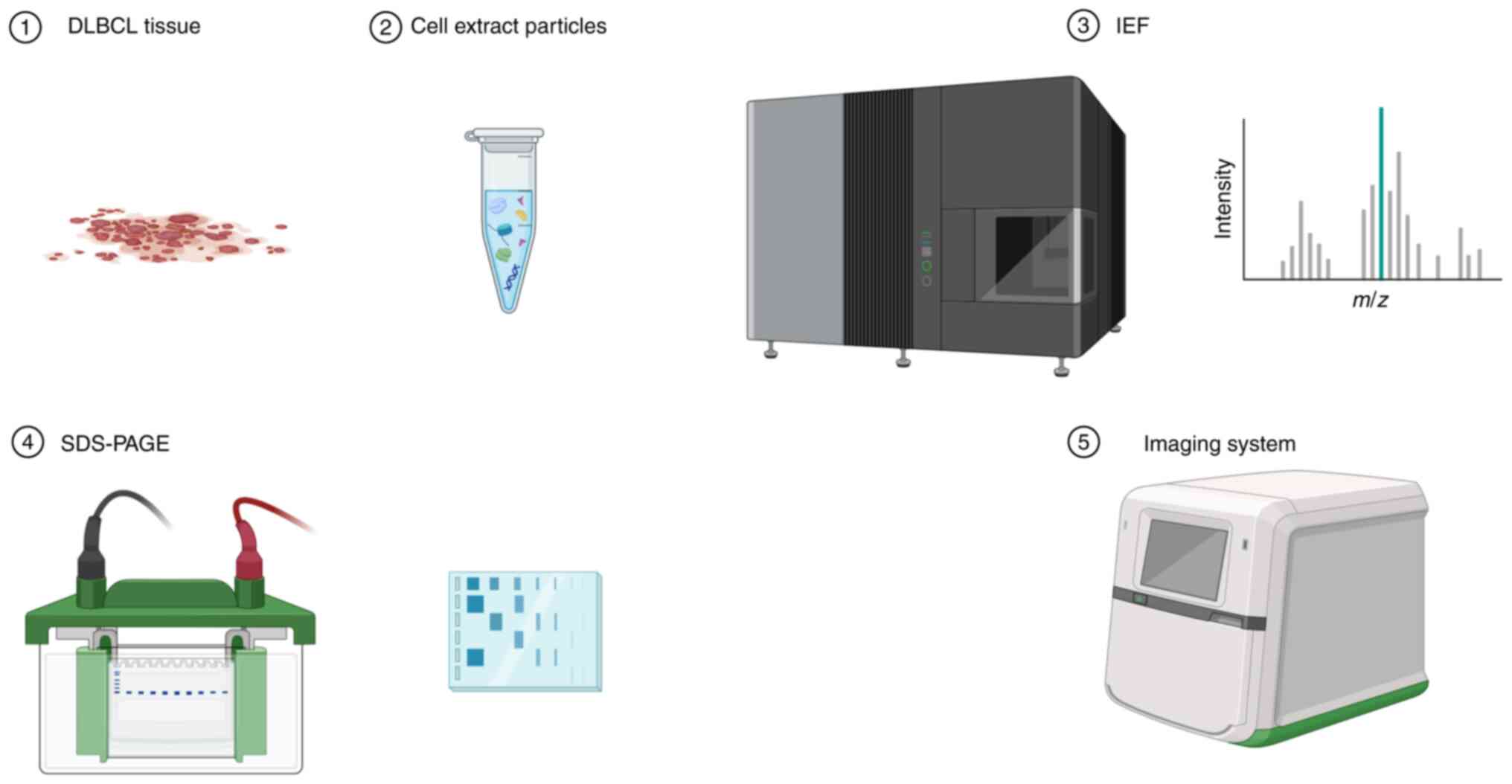
Proteomics includes the identification of
differentially expressed proteins in DLBCL tissues (44), obtaining 2D electrophoresis profiles
(45), and the use of mass
spectrometry to identify associated proteins (46) (Fig.
2). Bioinformatics analysis is also used to identify
differentially expressed proteins for further validation at the
tissue level, which may provide a theoretical basis for subsequent
experiments (47). In addition,
further investigations are required to determine the specific
mechanisms of DLBCL resistance and verify the feasibility of
differentially expressed proteins as drug-resistance-related
targets (48). Chen et al
(49) carried out mRNA/protein
analysis of clinicopathological samples, and the results
demonstrated that inhibitors of bromodomain and extraterminal (BET)
protein inhibited the progression of DLBCL. BET inhibition led to
upregulation of GTPase regulatory protein (IQGAP3), which inhibited
RAS protein activity in DLBCL cells, indicating that patients with
DLBCL with low IQGAP3 expression levels exhibited a poor prognosis.
In addition, BET inhibitors effectively controlled the progression
of DLBCL. Collectively, these results provided a theoretical basis
for targeting the BET protein (50)
as a potential treatment strategy for DLBCL.
Advances in proteomics-associated technologies have
demonstrated that the emergence of DLBCL chemoresistance is closely
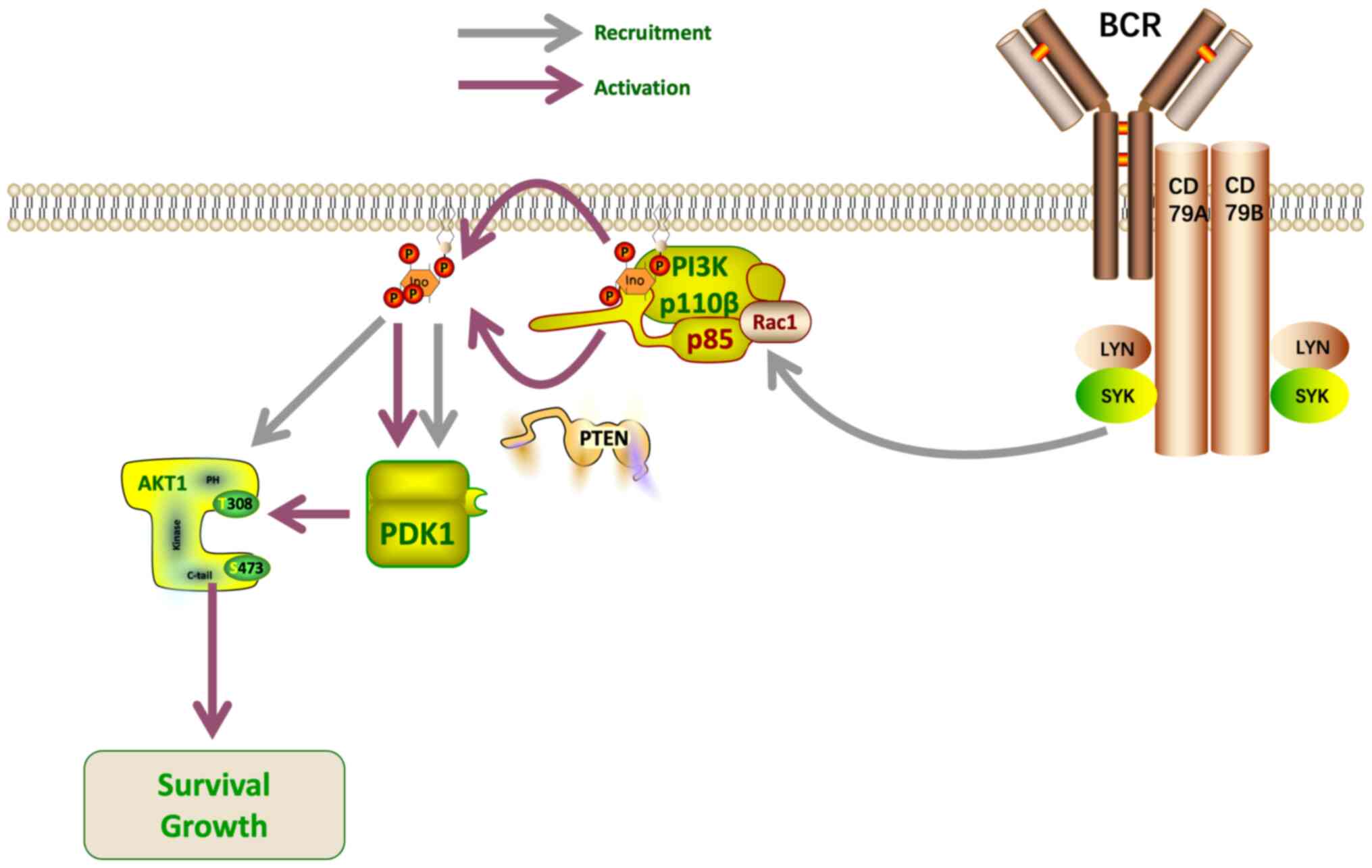
associated with signaling pathways (51), including the PI3K/Akt pathway. Akt
promotes cell survival and proliferation (52), as well as dysregulation of key
effectors controlling cell metabolism (53). A proteomics analysis conducted by Xu
et al (52) revealed that
removal of the PI3K/Akt signaling pathway antagonist, PTEN, led to
inactivation of the PI3K/Akt pathway in GCB DLBCL. Thus, PI3K/Akt
activation may play a key role in the development of GCB DLBCL, and
these findings demonstrate the potential value of PTEN as a
therapeutic target. PTEN acts as a lipoprotein phosphatase,
dephosphorylating the 3′ position of phosphatidylinositol
triphosphate, thereby reducing Akt activation (Fig. 3). Measurement of phosphorylated Akt
levels indicated that PTEN expression was negatively correlated
with PI3K/Akt activation in both a GCB DLBCL model and primary
DLBCL samples (52). Bisserier and
Wajapeyee (54) demonstrated that
DLBCL cells resistant to Enhancer of Zeste Homolog 2 inhibitors
exhibited activation of insulin-like growth factor I receptor,
PI3K, and mitogen-activated protein kinase pathways. Feng et
al (34) used proteomics
technology to demonstrate the increased expression levels of
exocrine carbonic anhydrase (CA)1, and the role of this protein as
a biomarker for the prognosis of DLBCL. Notably, CA1 expression
levels were also associated with an increased resistance to
chemotherapy via the signal transducer and activator of
transcription 3 signaling pathways and nuclear factor-κB.
Collectively, these results indicated that proteomic
techniques exhibit potential in the differential and enrichment
analyses of DLBCL-associated proteins for the subsequent discovery
of novel therapeutic targets. Specific signaling pathway inhibitors
also exhibit potential in highlighting the molecular mechanisms
underlying drug resistance; thus, leading to the development of
novel therapeutic options.
Proteomics technologies have improved the current
understanding of the molecular changes associated with DLBCL
(55,56). Storage of large amounts of
proteomics data (57) is
challenging; however, these often contain biologically significant
results (7). Data storage may be
aided through a combination of multiple omics techniques (58), and numerous biological techniques
are used to examine lymphomas (59), including genomics (60), proteomics (61), epigenetics (62) and radiomics (63).
Moreover, results of previous studies revealed a
regulatory role of interleukin-1 receptor-associated kinase (IRAK4)
in lymphoma cell proliferation and inflammation through proteome
and phosphorylation modifications (68). A series of targeted degradation
agents of IRAK4 were used to study the effects of impaired IRAK4
function on the phosphorylation levels of downstream signaling
proteins, and the results demonstrated that IRAK4 only partially
participated in the regulation of ABC DLBCL cell proliferation and
inflammatory signals (69). The
survival of ABC DLBCL cells was not solely dependent on the
function of IRAK4; thus, highlighting a requirement for the
development of other drug targets in ABC DLBCL (70).
The integration of multi-omics technologies exhibits
potential in the treatment of DLBCL (76). Proteomics may also be used in
conjunction with other omics techniques, such as transcriptomics,
metabolomics and genomics, to further the current understanding of
the molecular landscape and mechanisms underlying DLBCL (77). This integrative approach exhibits
potential in the discovery of novel biomarkers, therapeutic targets
and personalized treatment strategies for patients with DLBCL.
In conclusion, proteomics techniques are widely
established in the study of DLBCL (78), and proteomics have been used in
investigating the pathogenesis, drug resistance and mechanisms of
lymphoma, the evaluation of prognosis, and guiding treatment plans
(79). Further developments in
proteomics-associated technologies are required for the
identification of novel drugs and drug targets for the treatment of
DLBCL (80). For example, Maurer
et al (81) found that DLBCL
patients with elevated serum free light chain (sFLC) had a
relatively poor prognosis using FREELITE analysis. Then, Witzig
et al (82) conducted a
6-year monitoring of FLC concentrations in patients with DLBCL and
found that patients with DLBCL belonged to the FLC monoclonal and
polyclonal groups; and the results revealed that elevated FLC was
an adverse factor in the poor prognosis of DLBCL patients, and the
aforementioned study provides new ideas for the treatment of DLBCL
(7). Spatial proteomics, also known
as spatiomics technology, is advancing (83). This technique is used to examine
biological components, such as RNA and proteins, and adds
‘location’ dimensional information to further the current
understanding of the microenvironment (84). Spatial proteomics has been used in
breast cancer research and treatment; Cords et al (85) used highly multiplexed imaging mass
cytometry on breast cancer samples matched to single-cell RNA
sequencing datasets to confirm their cancer-associated fibroblast
phenotypes defined at the protein level, and used spatial
proteomics to analyze their spatial distributions in tumors, which
provided a new strategy for this treatment. Notably, spatial
proteomics is being used in lymphoma research (86), and may exhibit potential in the
treatment of DLBCL. Spatial proteomics involves analysis of the
subcellular localization of proteins in a systematic and
high-throughput manner (87), where
proteins simultaneously exist in different subcellular locations
(88) and travel between them
(89). For example, spatial
proteomics may be used to demonstrate the spatial profile of
proteins in the liver of patients with obesity, and these results
are compared with healthy individuals to determine the localization
of hepatocytes. Thus, spatial proteomics may aid in the treatment
of patients with liver disease (90). In an era of rapid advances in
medical technology, the use of spatial proteomics for the analysis
and precise treatment of DLBCL can help to develop personalized
treatment plans for patients and improve the cure and survival
rates of DLBCL patients. However, due to the limitations of
research methods and research data, there is still a lot of space
for the wide application of spatial proteomics.
Not applicable.
The present study was supported by the Natural Science
Foundation of Jilin (grant no. YDZJ202201ZYTS117).
Not applicable.
ZG and CW authored or reviewed drafts of the
manuscript, and approved the final draft. XS, ZW, JT and JM
provided figures and helped with proofreading of draft. LB prepared
tables and approved the final draft. All authors read and approved
the final manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Thandra KC, Barsouk A, Saginala K, Padala
SA, Barsouk A and Rawla P: Epidemiology of non-hodgkin's lymphoma.
Med Sci (Basel). 9:52021.PubMed/NCBI
|
|
2
|
de Leval L and Jaffe ES: Lymphoma
classification. Cancer. 26:176–185. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harrington F, Greenslade M, Talaulikar D
and Corboy G: Genomic characterisation of diffuse large B-cell
lymphoma. Pathology. 53:367–376. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Opinto G, Vegliante MC, Negri A, Skrypets
T, Loseto G, Pileri SA, Guarini A and Ciavarella S: The tumor
microenvironment of DLBCL in the computational era. Front Oncol.
10:3512020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McCarthy L, Bentley-DeSousa A, Denoncourt
A, Tseng YC, Gabriel M and Downey M: Proteins required for vacuolar
function are targets of lysine polyphosphorylation in yeast. FEBS
Lett. 594:21–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kanduc D: The role of proteomics in
defining autoimmunity. Expert Rev Proteomics. 18:177–184. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liang XJ, Song XY, Wu JL, Liu D, Lin BY,
Zhou HS and Wang L: Advances in multi-omics study of prognostic
biomarkers of diffuse large B-cell lymphoma. Int J Biol Sci.
18:1313–1327. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stegemann M, Denker S and Schmitt CA:
DLBCL 1L-what to expect beyond R-CHOP? Cancers (Basel).
14:14532022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McArdle AJ and Menikou S: What is
proteomics? Arch Dis Child Educ Pract Ed. 106:178–181. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Punetha A and Kotiya D: Advancements in
oncoproteomics technologies: Treading toward translation into
clinical practice. Proteomes. 11:22023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang Z, Ma L, Huang C, Li Q and Nice EC:
Proteomic profiling of human plasma for cancer biomarker discovery.
Proteomics. 17:2017. View Article : Google Scholar
|
|
12
|
Kothalawala WJ, Barták BK, Nagy ZB,
Zsigrai S, Szigeti KA, Valcz G, Takács I, Kalmár A and Molnár B: A
detailed overview about the single-cell analyses of solid tumors
focusing on colorectal cancer. Pathol Oncol Res. 28:16103422022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao HX, Li SJ, Niu J, Ma ZP, Nuerlan A,
Xue J, Wang MB, Cui WL, Abulajiang G, Sang W, et al: TCL1 as a hub
protein associated with the PI3K/AKT signaling pathway in diffuse
large B-cell lymphoma based on proteomics methods. Pathol Res
Pract. 216:1527992020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bingham GC, Lee F, Naba A and Barker TH:
Spatial-omics: Novel approaches to probe cell heterogeneity and
extracellular matrix biology. Matrix Biol. 91-92:152–166. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ednersson SB, Stern M, Fagman H,
Nilsson-Ehle H, Hasselblom S, Thorsell A and Andersson PO:
Proteomic analysis in diffuse large B-cell lymphoma identifies
dysregulated tumor microenvironment proteins in non-GCB/ABC subtype
patients. Leuk Lymphoma. 62:2360–2373. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhuang K, Zhang Y, Mo P, Deng L, Jiang Y,
Yu L, Mei F, Huang S, Chen X, Yan Y, et al: Plasma proteomic
analysis reveals altered protein abundances in HIV-infected
patients with or without non-Hodgkin lymphoma. J Med Virol.
94:3876–3889. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ysebaert L, Quillet-Mary A, Tosolini M,
Pont F, Laurent C and Fournié JJ: Lymphoma heterogeneity unraveled
by single-cell transcriptomics. Front Immunol. 12:5976512021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang M, Bennani NN and Feldman AL:
Lymphoma classification update: T-cell lymphomas, Hodgkin
lymphomas, and histiocytic/dendritic cell neoplasms. Expert Rev
Hematol. 10:239–249. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang J, Gu Y and Chen B: Drug-resistance
mechanism and new targeted drugs and treatments of relapse and
refractory DLBCL. Cancer Manag Res. 15:245–225. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Zeng L, Zhang S, Zeng S, Huang J,
Tang Y and Zhong M: Identification of differentially expressed
proteins in chemotherapy-sensitive and chemotherapy-resistant
diffuse large B cell lymphoma by proteomic methods. Med Oncol.
30:5282013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xie M, Huang X, Ye X and Qian W:
Prognostic and clinicopathological significance of PD-1/PD-L1
expression in the tumor microenvironment and neoplastic cells for
lymphoma. Int Immunopharmacol. 77:1059992019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Steen CB, Luca BA, Esfahani MS, Azizi A,
Sworder BJ, Nabet BY, Kurtz DM, Liu CL, Khameneh F, Advani RH, et
al: The landscape of tumor cell states and ecosystems in diffuse
large B cell lymphoma. Cancer Cell. 39:1422–37.e10. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cioroianu AI, Stinga PI, Sticlaru L,
Cioplea MD, Nichita L, Popp C and Staniceanu F: Tumor
microenvironment in diffuse large B-cell lymphoma: role and
prognosis. Anal Cell Pathol (Amst). 2019:85863542019.PubMed/NCBI
|
|
24
|
Ceccato J, Piazza M, Pizzi M, Manni S,
Piazza F, Caputo I, Cinetto F, Pisoni L, Trojan D, Scarpa R, et al:
A bone-based 3D scaffold as an in-vitro model of
microenvironment-DLBCL lymphoma cell interaction. Front Oncol.
12:9478232022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
de Groot FA, de Groen RAL, van den Berg A,
Jansen PM, Lam KH, Mutsaers PGNJ, van Noesel CJM, Chamuleau MED,
Stevens WBC, Plaça JR, et al: Biological and clinical implications
of gene-expression profiling in diffuse large B-cell lymphoma: A
proposal for a targeted BLYM-777 consortium panel as part of a
multilayered analytical approach. Cancers (Basel). 14:18572022.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takahara T, Nakamura S, Tsuzuki T and
Satou A: The immunology of DLBCL. Cancers (Basel). 15:8352023.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ofori K, Bhagat G and Rai AJ: Exosomes and
extracellular vesicles as liquid biopsy biomarkers in diffuse large
B-cell lymphoma: Current state of the art and unmet clinical needs.
Brit J Clin Pharmaco. 87:284–294. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu X, Zhao X, Yang J, Wang H, Piao Y and
Wang L: High expression of AP2M1 correlates with worse prognosis by
regulating immune microenvironment and drug resistance to R-CHOP in
diffuse large B cell lymphoma. Eur J Haematol. 110:198–208. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ejtehadifar M, Zahedi S, Gameiro P,
Cabeçadas J, da Silva MG, Beck HC, Carvalho AS and Matthiesen R:
Meta-analysis of MS-based proteomics studies indicates interferon
regulatory factor 4 and nucleobindin1 as potential prognostic and
drug resistance biomarkers in diffuse large B cell lymphoma. Cells.
12:1962023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ma J, Pang X, Li J, Zhang W and Cui W: The
immune checkpoint expression in the tumor immune microenvironment
of DLBCL: Clinicopathologic features and prognosis. Front Oncol.
12:10693782022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kotlov N, Bagaev A, Revuelta MV, Phillip
JM, Cacciapuoti MT, Antysheva Z, Svekolkin V, Tikhonova E,
Miheecheva N, Kuzkina N, et al: Clinical and biological subtypes of
B-cell lymphoma revealed by microenvironmental signatures. Cancer
Discov. 11:1468–1489. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bouwstra R, He Y, de Boer J, Kooistra H,
Cendrowicz E, Fehrmann RSN, Ammatuna E, Zu Eulenburg C, Nijland M,
Huls G, et al: CD47 Expression defines efficacy of rituximab with
CHOP in non-germinal center B-cell (non-GCB) diffuse large B-cell
lymphoma patients (DLBCL), but not in GCB DLBCL. Cancer Immunol
Res. 7:1663–1671. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu-Monette ZY, Wei L, Fang X, Au Q, Nunns
H, Nagy M, Tzankov A, Zhu F, Visco C, Bhagat G, et al: Genetic
subtyping and phenotypic characterization of the immune
microenvironment and MYC/BCL2 double expression reveal
heterogeneity in diffuse large B-cell lymphoma. Clin Cancer Res.
28:972–983. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feng Y, Zhong M, Tang Y, Liu X, Liu Y,
Wang L and Zhou H: The role and underlying mechanism of exosomal
CA1 in chemotherapy resistance in diffuse large B cell lymphoma.
Mol Ther Nucleic Acids. 21:452–463. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Klein C, Jamois C and Nielsen T: Anti-CD20
treatment for B-cell malignancies: Current status and future
directions. Expert Opin Biol Ther. 21:161–181. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Poletto S, Novo M, Paruzzo L, Frascione
PMM and Vitolo U: Treatment strategies for patients with diffuse
large B-cell lymphoma. Cancer Treat Rev. 110:1024432022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Susanibar-Adaniya S and Barta SK: 2021
Update on diffuse large B cell lymphoma: A review of current data
and potential applications on risk stratification and management.
Am J Hematol. 96:617–629. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Roider T, Seufert J, Uvarovskii A,
Frauhammer F, Bordas M, Abedpour N, Stolarczyk M, Mallm JP, Herbst
SA, Bruch PM, et al: Dissecting intratumour heterogeneity of nodal
B-cell lymphomas at the transcriptional, genetic and drug-response
levels. Nat Cell Biol. 22:896–906. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ferreri AJM, Doorduijn JK, Re A, Cabras
MG, Smith J, Ilariucci F, Luppi M, Calimeri T, Cattaneo C, Khwaja
J, et al: MATRix-RICE therapy and autologous haematopoietic
stem-cell transplantation in diffuse large B-cell lymphoma with
secondary CNS involvement (MARIETTA): An international, single-arm,
phase 2 trial. Lancet Haematol. 8:e110–e121. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yan J, Yuan W, Zhang J, Li L, Zhang L,
Zhang X and Zhang M: Identification and validation of a prognostic
prediction model in diffuse large B-cell lymphoma. Front Endocrinol
(Lausanne). 13:8463572022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Stanwood SR, Chong LC, Steidl C and
Jefferies WA: Distinct gene expression patterns of calcium channels
and related signaling pathways discovered in lymphomas. Front
Pharmacol. 13:7951762022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Frontzek F, Karsten I, Schmitz N and Lenz
G: Current options and future perspectives in the treatment of
patients with relapsed/refractory diffuse large B-cell lymphoma.
Ther Adv Hematol. 13:204062072211033212022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li S, Young KH and Medeiros LJ: Diffuse
large B-cell lymphoma. Pathology. 50:74–87. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gao HX, Nuerlan A, Abulajiang G, Cui WL,
Xue J, Sang W, Li SJ, Niu J, Ma ZP, Zhang W and Li XX: Quantitative
proteomics analysis of differentially expressed proteins in
activated B-cell-like diffuse large B-cell lymphoma using
quantitative proteomics. Pathol Res Pract. 215:1525282019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Robotti E, Calà E and Marengo E:
Two-dimensional gel electrophoresis image analysis. Methods Mol
Biol. 2361:3–13. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rotello RJ and Veenstra TD: Mass
spectrometry techniques: Principles and practices for quantitative
proteomics. Curr Protein Pept Sci. 22:121–133. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang J, Li Y, Zhang Y, Fang X, Chen N,
Zhou X and Wang X: Sirt6 promotes tumorigenesis and drug resistance
of diffuse large B-cell lymphoma by mediating PI3K/Akt signaling. J
Exp Clin Cancer Res. 39:1422020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang X, Duan YT, Wang Y, Zhao XD, Sun YM,
Lin DZ, Chen Y, Wang YX, Zhou ZW, Liu YX, et al: SAF-248, a novel
PI3Kδ-selective inhibitor, potently suppresses the growth of
diffuse large B-cell lymphoma. Acta Pharmacol Sin. 43:209–219.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen CC, Hsu CC, Chen SL, Lin PH, Chen JP,
Pan YR, Huang CE, Chen YJ, Chen YY, Wu YY and Yang MH: RAS mediates
BET inhibitor-endued repression of lymphoma migration and
prognosticates a novel proteomics-based subgroup of DLBCL through
its negative regulator IQGAP3. Cancers (Basel). 13:50242021.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang N, Wu R, Tang D and Kang R: The BET
family in immunity and disease. Signal Transduct Target Ther.
6:232021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sun F, Fang X and Wang X: Signal pathways
and therapeutic prospects of diffuse large B cell lymphoma.
Anticancer Agents Med Chem. 19:2047–2059. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xu W, Berning P and Lenz G: Targeting
B-cell receptor and PI3K signaling in diffuse large B-cell
lymphoma. Blood. 138:1110–1119. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Dunleavy K, Erdmann T and Lenz G:
Targeting the B-cell receptor pathway in diffuse large B-cell
lymphoma. Cancer Treat Rev. 65:41–46. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bisserier M and Wajapeyee N: Mechanisms of
resistance to EZH2 inhibitors in diffuse large B-cell lymphomas.
Blood. 131:2125–2137. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Coronado BNL, da Cunha FBS, de Toledo
Nobrega O and Martins AMA: The impact of mass spectrometry
application to screen new proteomics biomarkers in ophthalmology.
Int Ophthalmol. 41:2619–2633. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dallavalasa S, Beeraka NM, Basavaraju CG,
Tulimilli SV, Sadhu SP, Rajesh K, Aliev G and Madhunapantula SV:
The role of tumor associated macrophages (TAMs) in cancer
progression, chemoresistance, angiogenesis and metastasis-current
status. Curr Med Chem. 28:8203–8236. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kelly RT: Single-cell proteomics: Progress
and prospects. Mol Cell Proteomics. 19:1739–1748. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hasin Y, Seldin M and Lusis A: Multi-omics
approaches to disease. Genome Biol. 18:832017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang L, Li LR and Young KH: New agents and
regimens for diffuse large B cell lymphoma. J Hematol Oncol.
13:1752020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xiong J, Cui BW, Wang N, Dai YT, Zhang H,
Wang CF, Zhong HJ, Cheng S, Ou-Yang BS, Hu Y, et al: Genomic and
transcriptomic characterization of natural killer T cell lymphoma.
Cancer Cell. 37:403–419.e6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
van der Meeren LE, Kluiver J, Rutgers B,
Alsagoor Y, Kluin PM, van den Berg A and Visser L: A super-SILAC
based proteomics analysis of diffuse large B-cell lymphoma-NOS
patient samples to identify new proteins that discriminate GCB and
non-GCB lymphomas. PLoS One. 14:e02232602019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang P and Zhang M: Epigenetic
alterations and advancement of treatment in peripheral T-cell
lymphoma. Clin Epigenetics. 12:1692020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jiang H, Li A, Ji Z, Tian M and Zhang H:
Role of radiomics-based baseline PET/CT imaging in lymphoma:
Diagnosis, prognosis, and response assessment. Mol Imaging Biol.
24:537–549. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Fornecker LM, Muller L, Bertrand F, Paul
N, Pichot A, Herbrecht R, Chenard MP, Mauvieux L, Vallat L, Bahram
S, et al: Multi-omics dataset to decipher the complexity of drug
resistance in diffuse large B-cell lymphoma. Sci Rep. 9:8952019.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bresnick AR, Weber DJ and Zimmer DB: S100
proteins in cancer. Nat Rev Cancer. 15:96–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ye X, Wang L, Nie M, Wang Y, Dong S, Ren
W, Li G, Li ZM, Wu K and Pan-Hammarström Q: A single-cell atlas of
diffuse large B cell lymphoma. Cell Rep. 39:1107132022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang N, Li X, Wang R and Ding Z: Spatial
transcriptomics and proteomics technologies for deconvoluting the
tumor microenvironment. Biotechnol J. 16:e21000412021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cumming IA, Degorce SL, Aagaard A,
Braybrooke EL, Davies NL, Diène CR, Eatherton AJ, Felstead HR,
Groombridge SD, Lenz EM, et al: Identification and optimisation of
a pyrimidopyridone series of IRAK4 inhibitors. Bioorg Med Chem.
63:1167292022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yoon SB, Hong H, Lim HJ, Choi JH, Choi YP,
Seo SW, Lee HW, Chae CH, Park WK, Kim HY, et al: A novel IRAK4/PIM1
inhibitor ameliorates rheumatoid arthritis and lymphoid malignancy
by blocking the TLR/MYD88-mediated NF-κB pathway. Acta Pharm Sin B.
13:1093–1109. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang J, Fu L, Shen B, Liu Y, Wang W, Cai
X, Kong L, Yan Y, Meng R, Zhang Z, et al: Assessing IRAK4 functions
in ABC DLBCL by IRAK4 kinase inhibition and protein degradation.
Cell Chem Biol. 27:1500–1509.e13. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Boșoteanu M, Cristian M, Așchie M, Deacu
M, Mitroi AF, Brînzan CS and Bălțătescu GI: Proteomics and genomics
of a monomorphic epitheliotropic intestinal T-cell lymphoma: An
extremely rare case report and short review of literature. Medicine
(Baltimore). 101:e319512022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Coradduzza D, Ghironi A, Azara E, Culeddu
N, Cruciani S, Zinellu A, Maioli M, De Miglio MR, Medici S, Fozza C
and Carru C: Role of polyamines as biomarkers in lymphoma patients:
A pilot study. Diagnostics (Basel). 12:21512022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Cheson BD, Nowakowski G and Salles G:
Diffuse large B-cell lymphoma: New targets and novel therapies.
Blood Cancer J. 11:682021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Rolland DCM, Basrur V, Jeon YK,
McNeil-Schwalm C, Fermin D, Conlon KP, Zhou Y, Ng SY, Tsou CC,
Brown NA, et al: Functional proteogenomics reveals biomarkers and
therapeutic targets in lymphomas. Proc Natl Acad Sci USA.
114:6581–6586. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Huang L, Brunell D, Stephan C, Mancuso J,
Yu X, He B, Zinner R, Kim J, Davies P and Wong STC: Driver network
as a biomarker: systematic integration and network modeling of
multi-omics data to derive driver signaling pathways for drug
combination prediction. Bioinformatics. 35:3709–3717. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Chakraborty S, Hosen MI, Ahmed M and
Shekhar HU: Onco-multi-OMICS approach: A new frontier in cancer
research. Biomed Res Int. 2018:98362562018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gohil SH, Iorgulescu JB, Braun DA, Keskin
DB and Livak KJ: Applying high-dimensional single-cell technologies
to the analysis of cancer immunotherapy. Nat Rev Clin Oncol.
18:244–256. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yang D, Wang J, Hu M, Li F, Yang F, Zhao
Y, Xu Y and Zhang X, Tang L and Zhang X: Combined multiomics
analysis reveals the mechanism of CENPF overexpression-mediated
immune dysfunction in diffuse large B-cell lymphoma in vitro. Front
Genet. 13:10726892022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Landeira-Viñuela A, Diez P, Juanes-Velasco
P, Lécrevisse Q, Orfao A, De Las Rivas J and Fuentes M: Deepening
into intracellular signaling landscape through integrative spatial
proteomics and transcriptomics in a lymphoma model. Biomolecules.
11:17762021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Jamil MO and Mehta A: Diffuse large B-cell
lymphoma: Prognostic markers and their impact on therapy. Expert
Rev Hematol. 9:471–477. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Maurer MJ, Micallef INM, Cerhan JR,
Katzmann JA, Link BK, Colgan JP, Habermann TM, Inwards DJ, Markovic
SN, Ansell SM, et al: Elevated serum free light chains are
associated with event-free and overall survival in two independent
cohorts of patients with diffuse large B-cell lymphoma. J Clin
Oncol. 29:1620–1626. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Witzig TE, Maurer MJ, Stenson MJ, Allmer
C, Macon W, Link B, Katzmann JA and Gupta M: Elevated serum
monoclonal and polyclonal free light chains and interferon
inducible protein-10 predicts inferior prognosis in untreated
diffuse large B-cell lymphoma. Am J Hematol. 89:417–422. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Grünwald BT, Devisme A, Andrieux G, Vyas
F, Aliar K, McCloskey CW, Macklin A, Jang GH, Denroche R, Romero
JM, et al: Spatially confined sub-tumor microenvironments in
pancreatic cancer. Cell. 184:5577–5592.e18. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Akhtar M, Haider A, Rashid S and Al-Nabet
ADMH: Paget's ‘seed and soil’ theory of cancer metastasis: An idea
whose time has come. Adv Anat Pathol. 26:69–74. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Cords L, Tietscher S, Anzeneder T,
Langwieder C, Rees M, de Souza N and Bodenmiller B:
Cancer-associated fibroblast classification in single-cell and
spatial proteomics data. Nat Commun. 14:42942023. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Franciosa G, Kverneland AH, Jensen AWP,
Donia M and Olsen JV: Proteomics to study cancer immunity and
improve treatment. Semin Immunopathol. 45:241–251. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Gatto L, Breckels LM and Lilley KS:
Assessing sub-cellular resolution in spatial proteomics
experiments. Curr Opin Chem Biol. 48:123–149. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Liu X, Salokas K, Tamene F, Jiu Y,
Weldatsadik RG, Öhman T and Varjosalo M: An AP-MS- and
BioID-compatible MAC-tag enables comprehensive mapping of protein
interactions and subcellular localizations. Nat Commun. 9:11882018.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Pankow S, Martínez-Bartolomé S, Bamberger
C and Yates JR: Understanding molecular mechanisms of disease
through spatial proteomics. Curr Opin Chem Biol. 48:19–25. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Guilliams M, Bonnardel J, Haest B,
Vanderborght B, Wagner C, Remmerie A, Bujko A, Martens L, Thoné T,
Browaeys R, et al: Spatial proteogenomics reveals distinct and
evolutionarily conserved hepatic macrophage niches. Cell.
185:379–396.e38. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lee PY, Saraygord-Afshari N and Low TY:
The evolution of two-dimensional gel electrophoresis-from
proteomics to emerging alternative applications. J Chromatogr A.
1615:4607632020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Strohkamp S, Gemoll T and Habermann JK:
Possibilities and limitations of 2DE-based analyses for identifying
low-abundant tumor markers in human serum and plasma. Proteomics.
16:2519–2532. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lin TT, Zhang T, Kitata RB, Liu T, Smith
RD, Qian WJ and Shi T: Mass spectrometry-based targeted proteomics
for analysis of protein mutations. Mass Spectrom Rev. 42:796–821.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Noor Z, Ahn SB, Baker MS, Ranganathan S
and Mohamedali A: Mass spectrometry-based protein identification in
proteomics-a review. Brief Bioinform. 22:1620–1638. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Ren AH, Diamandis EP and Kulasingam V:
Uncovering the depths of the human proteome: Antibody-based
technologies for ultrasensitive multiplexed protein detection and
quantification. Mol Cell Proteomics. 20:1001552021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Syu GD, Dunn J and Zhu H: Developments and
applications of functional protein microarrays. Mol Cell
Proteomics. 19:916–927. 2020. View Article : Google Scholar : PubMed/NCBI
|