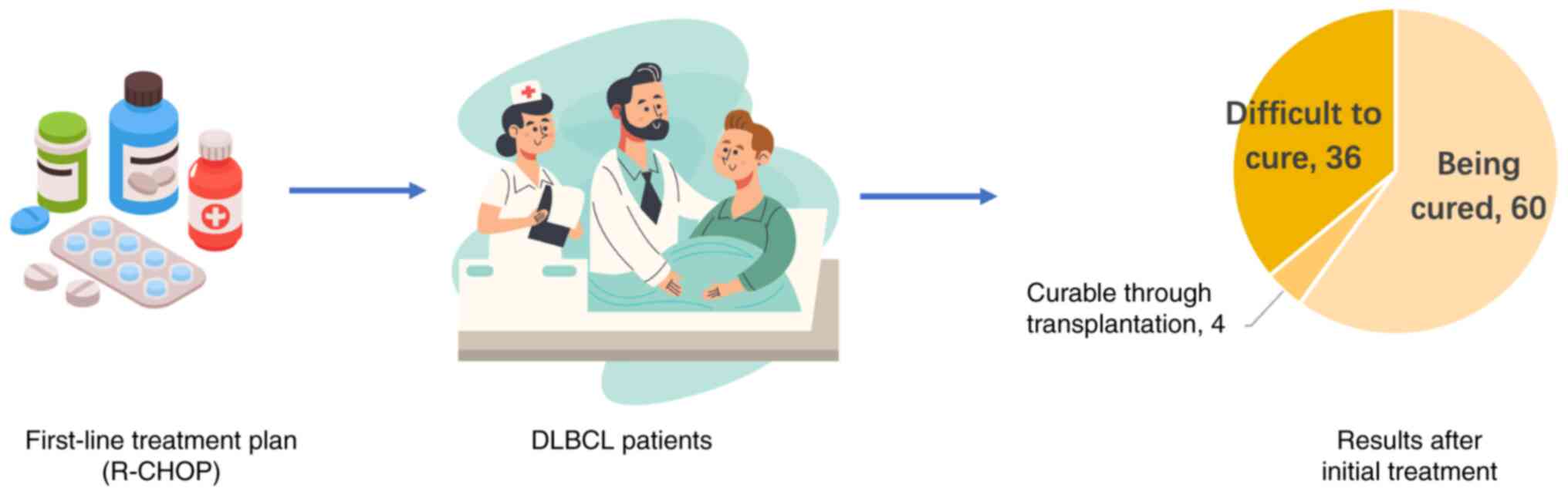
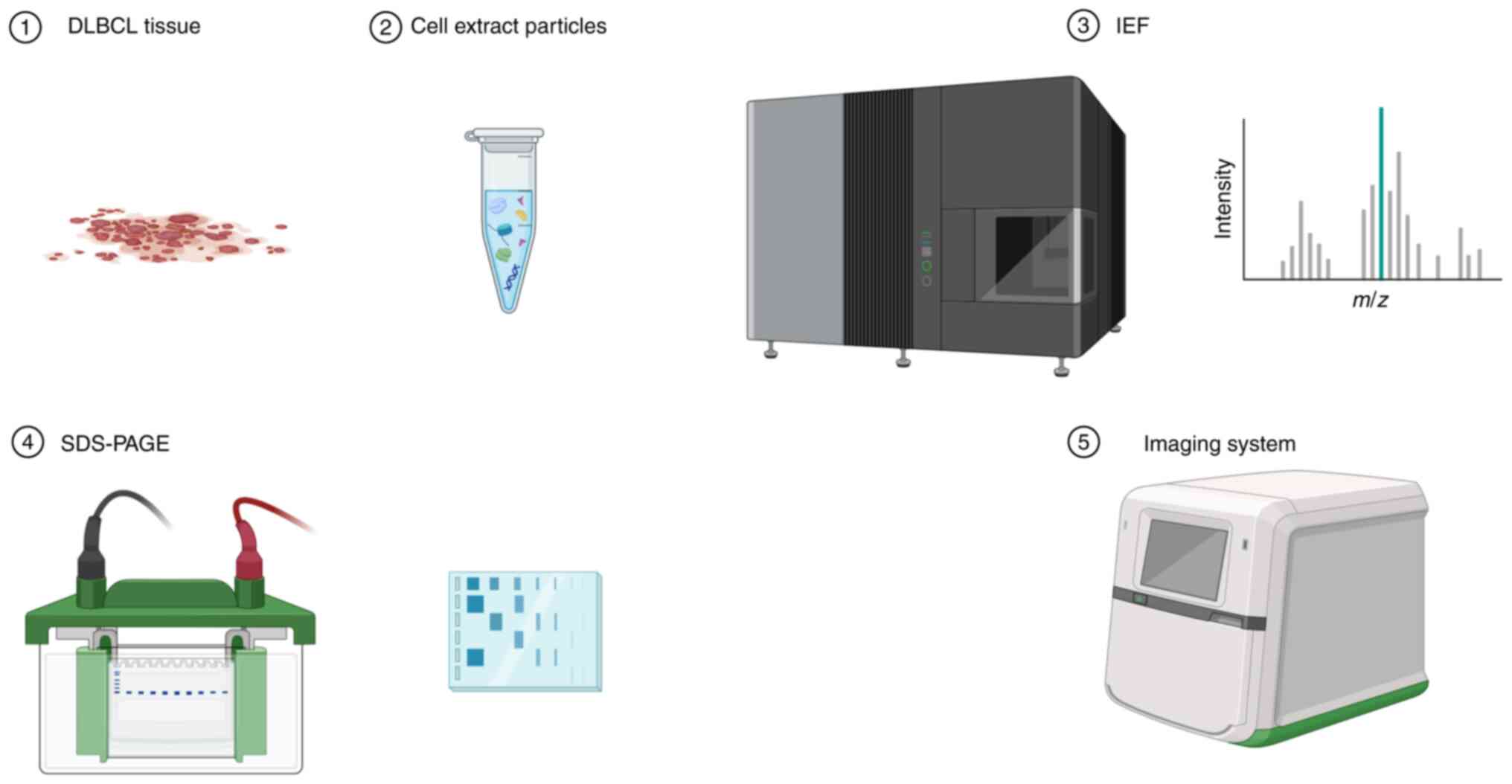
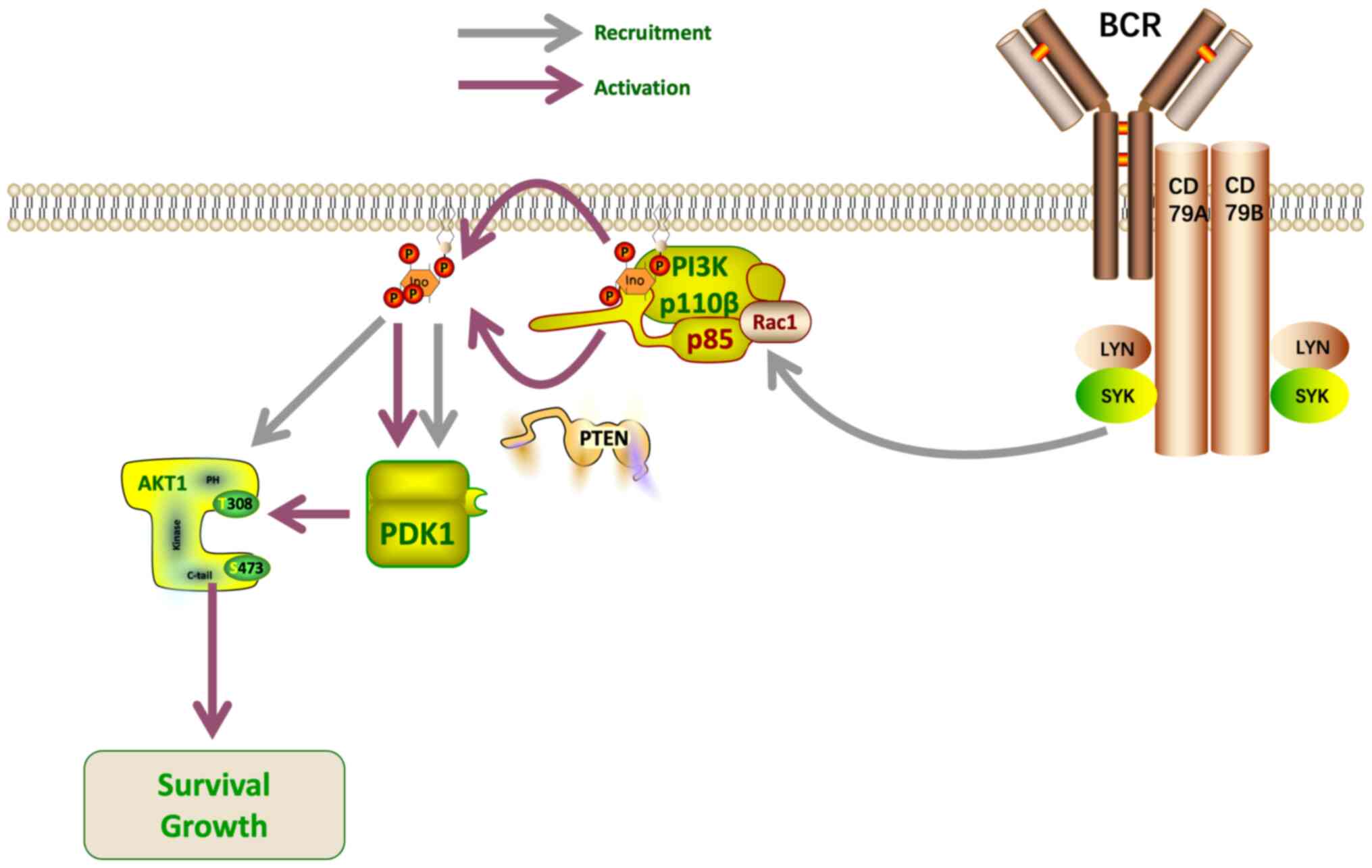
|
1
|
Thandra KC, Barsouk A, Saginala K, Padala
SA, Barsouk A and Rawla P: Epidemiology of non-hodgkin's lymphoma.
Med Sci (Basel). 9:52021.PubMed/NCBI
|
|
2
|
de Leval L and Jaffe ES: Lymphoma
classification. Cancer. 26:176–185. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harrington F, Greenslade M, Talaulikar D
and Corboy G: Genomic characterisation of diffuse large B-cell
lymphoma. Pathology. 53:367–376. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Opinto G, Vegliante MC, Negri A, Skrypets
T, Loseto G, Pileri SA, Guarini A and Ciavarella S: The tumor
microenvironment of DLBCL in the computational era. Front Oncol.
10:3512020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McCarthy L, Bentley-DeSousa A, Denoncourt
A, Tseng YC, Gabriel M and Downey M: Proteins required for vacuolar
function are targets of lysine polyphosphorylation in yeast. FEBS
Lett. 594:21–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kanduc D: The role of proteomics in
defining autoimmunity. Expert Rev Proteomics. 18:177–184. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liang XJ, Song XY, Wu JL, Liu D, Lin BY,
Zhou HS and Wang L: Advances in multi-omics study of prognostic
biomarkers of diffuse large B-cell lymphoma. Int J Biol Sci.
18:1313–1327. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stegemann M, Denker S and Schmitt CA:
DLBCL 1L-what to expect beyond R-CHOP? Cancers (Basel).
14:14532022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McArdle AJ and Menikou S: What is
proteomics? Arch Dis Child Educ Pract Ed. 106:178–181. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Punetha A and Kotiya D: Advancements in
oncoproteomics technologies: Treading toward translation into
clinical practice. Proteomes. 11:22023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang Z, Ma L, Huang C, Li Q and Nice EC:
Proteomic profiling of human plasma for cancer biomarker discovery.
Proteomics. 17:2017. View Article : Google Scholar
|
|
12
|
Kothalawala WJ, Barták BK, Nagy ZB,
Zsigrai S, Szigeti KA, Valcz G, Takács I, Kalmár A and Molnár B: A
detailed overview about the single-cell analyses of solid tumors
focusing on colorectal cancer. Pathol Oncol Res. 28:16103422022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao HX, Li SJ, Niu J, Ma ZP, Nuerlan A,
Xue J, Wang MB, Cui WL, Abulajiang G, Sang W, et al: TCL1 as a hub
protein associated with the PI3K/AKT signaling pathway in diffuse
large B-cell lymphoma based on proteomics methods. Pathol Res
Pract. 216:1527992020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bingham GC, Lee F, Naba A and Barker TH:
Spatial-omics: Novel approaches to probe cell heterogeneity and
extracellular matrix biology. Matrix Biol. 91-92:152–166. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ednersson SB, Stern M, Fagman H,
Nilsson-Ehle H, Hasselblom S, Thorsell A and Andersson PO:
Proteomic analysis in diffuse large B-cell lymphoma identifies
dysregulated tumor microenvironment proteins in non-GCB/ABC subtype
patients. Leuk Lymphoma. 62:2360–2373. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhuang K, Zhang Y, Mo P, Deng L, Jiang Y,
Yu L, Mei F, Huang S, Chen X, Yan Y, et al: Plasma proteomic
analysis reveals altered protein abundances in HIV-infected
patients with or without non-Hodgkin lymphoma. J Med Virol.
94:3876–3889. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ysebaert L, Quillet-Mary A, Tosolini M,
Pont F, Laurent C and Fournié JJ: Lymphoma heterogeneity unraveled
by single-cell transcriptomics. Front Immunol. 12:5976512021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang M, Bennani NN and Feldman AL:
Lymphoma classification update: T-cell lymphomas, Hodgkin
lymphomas, and histiocytic/dendritic cell neoplasms. Expert Rev
Hematol. 10:239–249. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang J, Gu Y and Chen B: Drug-resistance
mechanism and new targeted drugs and treatments of relapse and
refractory DLBCL. Cancer Manag Res. 15:245–225. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Zeng L, Zhang S, Zeng S, Huang J,
Tang Y and Zhong M: Identification of differentially expressed
proteins in chemotherapy-sensitive and chemotherapy-resistant
diffuse large B cell lymphoma by proteomic methods. Med Oncol.
30:5282013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xie M, Huang X, Ye X and Qian W:
Prognostic and clinicopathological significance of PD-1/PD-L1
expression in the tumor microenvironment and neoplastic cells for
lymphoma. Int Immunopharmacol. 77:1059992019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Steen CB, Luca BA, Esfahani MS, Azizi A,
Sworder BJ, Nabet BY, Kurtz DM, Liu CL, Khameneh F, Advani RH, et
al: The landscape of tumor cell states and ecosystems in diffuse
large B cell lymphoma. Cancer Cell. 39:1422–37.e10. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cioroianu AI, Stinga PI, Sticlaru L,
Cioplea MD, Nichita L, Popp C and Staniceanu F: Tumor
microenvironment in diffuse large B-cell lymphoma: role and
prognosis. Anal Cell Pathol (Amst). 2019:85863542019.PubMed/NCBI
|
|
24
|
Ceccato J, Piazza M, Pizzi M, Manni S,
Piazza F, Caputo I, Cinetto F, Pisoni L, Trojan D, Scarpa R, et al:
A bone-based 3D scaffold as an in-vitro model of
microenvironment-DLBCL lymphoma cell interaction. Front Oncol.
12:9478232022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
de Groot FA, de Groen RAL, van den Berg A,
Jansen PM, Lam KH, Mutsaers PGNJ, van Noesel CJM, Chamuleau MED,
Stevens WBC, Plaça JR, et al: Biological and clinical implications
of gene-expression profiling in diffuse large B-cell lymphoma: A
proposal for a targeted BLYM-777 consortium panel as part of a
multilayered analytical approach. Cancers (Basel). 14:18572022.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takahara T, Nakamura S, Tsuzuki T and
Satou A: The immunology of DLBCL. Cancers (Basel). 15:8352023.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ofori K, Bhagat G and Rai AJ: Exosomes and
extracellular vesicles as liquid biopsy biomarkers in diffuse large
B-cell lymphoma: Current state of the art and unmet clinical needs.
Brit J Clin Pharmaco. 87:284–294. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu X, Zhao X, Yang J, Wang H, Piao Y and
Wang L: High expression of AP2M1 correlates with worse prognosis by
regulating immune microenvironment and drug resistance to R-CHOP in
diffuse large B cell lymphoma. Eur J Haematol. 110:198–208. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ejtehadifar M, Zahedi S, Gameiro P,
Cabeçadas J, da Silva MG, Beck HC, Carvalho AS and Matthiesen R:
Meta-analysis of MS-based proteomics studies indicates interferon
regulatory factor 4 and nucleobindin1 as potential prognostic and
drug resistance biomarkers in diffuse large B cell lymphoma. Cells.
12:1962023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ma J, Pang X, Li J, Zhang W and Cui W: The
immune checkpoint expression in the tumor immune microenvironment
of DLBCL: Clinicopathologic features and prognosis. Front Oncol.
12:10693782022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kotlov N, Bagaev A, Revuelta MV, Phillip
JM, Cacciapuoti MT, Antysheva Z, Svekolkin V, Tikhonova E,
Miheecheva N, Kuzkina N, et al: Clinical and biological subtypes of
B-cell lymphoma revealed by microenvironmental signatures. Cancer
Discov. 11:1468–1489. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bouwstra R, He Y, de Boer J, Kooistra H,
Cendrowicz E, Fehrmann RSN, Ammatuna E, Zu Eulenburg C, Nijland M,
Huls G, et al: CD47 Expression defines efficacy of rituximab with
CHOP in non-germinal center B-cell (non-GCB) diffuse large B-cell
lymphoma patients (DLBCL), but not in GCB DLBCL. Cancer Immunol
Res. 7:1663–1671. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu-Monette ZY, Wei L, Fang X, Au Q, Nunns
H, Nagy M, Tzankov A, Zhu F, Visco C, Bhagat G, et al: Genetic
subtyping and phenotypic characterization of the immune
microenvironment and MYC/BCL2 double expression reveal
heterogeneity in diffuse large B-cell lymphoma. Clin Cancer Res.
28:972–983. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feng Y, Zhong M, Tang Y, Liu X, Liu Y,
Wang L and Zhou H: The role and underlying mechanism of exosomal
CA1 in chemotherapy resistance in diffuse large B cell lymphoma.
Mol Ther Nucleic Acids. 21:452–463. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Klein C, Jamois C and Nielsen T: Anti-CD20
treatment for B-cell malignancies: Current status and future
directions. Expert Opin Biol Ther. 21:161–181. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Poletto S, Novo M, Paruzzo L, Frascione
PMM and Vitolo U: Treatment strategies for patients with diffuse
large B-cell lymphoma. Cancer Treat Rev. 110:1024432022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Susanibar-Adaniya S and Barta SK: 2021
Update on diffuse large B cell lymphoma: A review of current data
and potential applications on risk stratification and management.
Am J Hematol. 96:617–629. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Roider T, Seufert J, Uvarovskii A,
Frauhammer F, Bordas M, Abedpour N, Stolarczyk M, Mallm JP, Herbst
SA, Bruch PM, et al: Dissecting intratumour heterogeneity of nodal
B-cell lymphomas at the transcriptional, genetic and drug-response
levels. Nat Cell Biol. 22:896–906. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ferreri AJM, Doorduijn JK, Re A, Cabras
MG, Smith J, Ilariucci F, Luppi M, Calimeri T, Cattaneo C, Khwaja
J, et al: MATRix-RICE therapy and autologous haematopoietic
stem-cell transplantation in diffuse large B-cell lymphoma with
secondary CNS involvement (MARIETTA): An international, single-arm,
phase 2 trial. Lancet Haematol. 8:e110–e121. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yan J, Yuan W, Zhang J, Li L, Zhang L,
Zhang X and Zhang M: Identification and validation of a prognostic
prediction model in diffuse large B-cell lymphoma. Front Endocrinol
(Lausanne). 13:8463572022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Stanwood SR, Chong LC, Steidl C and
Jefferies WA: Distinct gene expression patterns of calcium channels
and related signaling pathways discovered in lymphomas. Front
Pharmacol. 13:7951762022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Frontzek F, Karsten I, Schmitz N and Lenz
G: Current options and future perspectives in the treatment of
patients with relapsed/refractory diffuse large B-cell lymphoma.
Ther Adv Hematol. 13:204062072211033212022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li S, Young KH and Medeiros LJ: Diffuse
large B-cell lymphoma. Pathology. 50:74–87. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gao HX, Nuerlan A, Abulajiang G, Cui WL,
Xue J, Sang W, Li SJ, Niu J, Ma ZP, Zhang W and Li XX: Quantitative
proteomics analysis of differentially expressed proteins in
activated B-cell-like diffuse large B-cell lymphoma using
quantitative proteomics. Pathol Res Pract. 215:1525282019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Robotti E, Calà E and Marengo E:
Two-dimensional gel electrophoresis image analysis. Methods Mol
Biol. 2361:3–13. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rotello RJ and Veenstra TD: Mass
spectrometry techniques: Principles and practices for quantitative
proteomics. Curr Protein Pept Sci. 22:121–133. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang J, Li Y, Zhang Y, Fang X, Chen N,
Zhou X and Wang X: Sirt6 promotes tumorigenesis and drug resistance
of diffuse large B-cell lymphoma by mediating PI3K/Akt signaling. J
Exp Clin Cancer Res. 39:1422020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang X, Duan YT, Wang Y, Zhao XD, Sun YM,
Lin DZ, Chen Y, Wang YX, Zhou ZW, Liu YX, et al: SAF-248, a novel
PI3Kδ-selective inhibitor, potently suppresses the growth of
diffuse large B-cell lymphoma. Acta Pharmacol Sin. 43:209–219.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen CC, Hsu CC, Chen SL, Lin PH, Chen JP,
Pan YR, Huang CE, Chen YJ, Chen YY, Wu YY and Yang MH: RAS mediates
BET inhibitor-endued repression of lymphoma migration and
prognosticates a novel proteomics-based subgroup of DLBCL through
its negative regulator IQGAP3. Cancers (Basel). 13:50242021.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang N, Wu R, Tang D and Kang R: The BET
family in immunity and disease. Signal Transduct Target Ther.
6:232021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sun F, Fang X and Wang X: Signal pathways
and therapeutic prospects of diffuse large B cell lymphoma.
Anticancer Agents Med Chem. 19:2047–2059. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xu W, Berning P and Lenz G: Targeting
B-cell receptor and PI3K signaling in diffuse large B-cell
lymphoma. Blood. 138:1110–1119. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Dunleavy K, Erdmann T and Lenz G:
Targeting the B-cell receptor pathway in diffuse large B-cell
lymphoma. Cancer Treat Rev. 65:41–46. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bisserier M and Wajapeyee N: Mechanisms of
resistance to EZH2 inhibitors in diffuse large B-cell lymphomas.
Blood. 131:2125–2137. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Coronado BNL, da Cunha FBS, de Toledo
Nobrega O and Martins AMA: The impact of mass spectrometry
application to screen new proteomics biomarkers in ophthalmology.
Int Ophthalmol. 41:2619–2633. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dallavalasa S, Beeraka NM, Basavaraju CG,
Tulimilli SV, Sadhu SP, Rajesh K, Aliev G and Madhunapantula SV:
The role of tumor associated macrophages (TAMs) in cancer
progression, chemoresistance, angiogenesis and metastasis-current
status. Curr Med Chem. 28:8203–8236. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kelly RT: Single-cell proteomics: Progress
and prospects. Mol Cell Proteomics. 19:1739–1748. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hasin Y, Seldin M and Lusis A: Multi-omics
approaches to disease. Genome Biol. 18:832017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang L, Li LR and Young KH: New agents and
regimens for diffuse large B cell lymphoma. J Hematol Oncol.
13:1752020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xiong J, Cui BW, Wang N, Dai YT, Zhang H,
Wang CF, Zhong HJ, Cheng S, Ou-Yang BS, Hu Y, et al: Genomic and
transcriptomic characterization of natural killer T cell lymphoma.
Cancer Cell. 37:403–419.e6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
van der Meeren LE, Kluiver J, Rutgers B,
Alsagoor Y, Kluin PM, van den Berg A and Visser L: A super-SILAC
based proteomics analysis of diffuse large B-cell lymphoma-NOS
patient samples to identify new proteins that discriminate GCB and
non-GCB lymphomas. PLoS One. 14:e02232602019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang P and Zhang M: Epigenetic
alterations and advancement of treatment in peripheral T-cell
lymphoma. Clin Epigenetics. 12:1692020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jiang H, Li A, Ji Z, Tian M and Zhang H:
Role of radiomics-based baseline PET/CT imaging in lymphoma:
Diagnosis, prognosis, and response assessment. Mol Imaging Biol.
24:537–549. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Fornecker LM, Muller L, Bertrand F, Paul
N, Pichot A, Herbrecht R, Chenard MP, Mauvieux L, Vallat L, Bahram
S, et al: Multi-omics dataset to decipher the complexity of drug
resistance in diffuse large B-cell lymphoma. Sci Rep. 9:8952019.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bresnick AR, Weber DJ and Zimmer DB: S100
proteins in cancer. Nat Rev Cancer. 15:96–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ye X, Wang L, Nie M, Wang Y, Dong S, Ren
W, Li G, Li ZM, Wu K and Pan-Hammarström Q: A single-cell atlas of
diffuse large B cell lymphoma. Cell Rep. 39:1107132022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang N, Li X, Wang R and Ding Z: Spatial
transcriptomics and proteomics technologies for deconvoluting the
tumor microenvironment. Biotechnol J. 16:e21000412021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cumming IA, Degorce SL, Aagaard A,
Braybrooke EL, Davies NL, Diène CR, Eatherton AJ, Felstead HR,
Groombridge SD, Lenz EM, et al: Identification and optimisation of
a pyrimidopyridone series of IRAK4 inhibitors. Bioorg Med Chem.
63:1167292022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yoon SB, Hong H, Lim HJ, Choi JH, Choi YP,
Seo SW, Lee HW, Chae CH, Park WK, Kim HY, et al: A novel IRAK4/PIM1
inhibitor ameliorates rheumatoid arthritis and lymphoid malignancy
by blocking the TLR/MYD88-mediated NF-κB pathway. Acta Pharm Sin B.
13:1093–1109. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang J, Fu L, Shen B, Liu Y, Wang W, Cai
X, Kong L, Yan Y, Meng R, Zhang Z, et al: Assessing IRAK4 functions
in ABC DLBCL by IRAK4 kinase inhibition and protein degradation.
Cell Chem Biol. 27:1500–1509.e13. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Boșoteanu M, Cristian M, Așchie M, Deacu
M, Mitroi AF, Brînzan CS and Bălțătescu GI: Proteomics and genomics
of a monomorphic epitheliotropic intestinal T-cell lymphoma: An
extremely rare case report and short review of literature. Medicine
(Baltimore). 101:e319512022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Coradduzza D, Ghironi A, Azara E, Culeddu
N, Cruciani S, Zinellu A, Maioli M, De Miglio MR, Medici S, Fozza C
and Carru C: Role of polyamines as biomarkers in lymphoma patients:
A pilot study. Diagnostics (Basel). 12:21512022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Cheson BD, Nowakowski G and Salles G:
Diffuse large B-cell lymphoma: New targets and novel therapies.
Blood Cancer J. 11:682021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Rolland DCM, Basrur V, Jeon YK,
McNeil-Schwalm C, Fermin D, Conlon KP, Zhou Y, Ng SY, Tsou CC,
Brown NA, et al: Functional proteogenomics reveals biomarkers and
therapeutic targets in lymphomas. Proc Natl Acad Sci USA.
114:6581–6586. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Huang L, Brunell D, Stephan C, Mancuso J,
Yu X, He B, Zinner R, Kim J, Davies P and Wong STC: Driver network
as a biomarker: systematic integration and network modeling of
multi-omics data to derive driver signaling pathways for drug
combination prediction. Bioinformatics. 35:3709–3717. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Chakraborty S, Hosen MI, Ahmed M and
Shekhar HU: Onco-multi-OMICS approach: A new frontier in cancer
research. Biomed Res Int. 2018:98362562018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gohil SH, Iorgulescu JB, Braun DA, Keskin
DB and Livak KJ: Applying high-dimensional single-cell technologies
to the analysis of cancer immunotherapy. Nat Rev Clin Oncol.
18:244–256. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yang D, Wang J, Hu M, Li F, Yang F, Zhao
Y, Xu Y and Zhang X, Tang L and Zhang X: Combined multiomics
analysis reveals the mechanism of CENPF overexpression-mediated
immune dysfunction in diffuse large B-cell lymphoma in vitro. Front
Genet. 13:10726892022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Landeira-Viñuela A, Diez P, Juanes-Velasco
P, Lécrevisse Q, Orfao A, De Las Rivas J and Fuentes M: Deepening
into intracellular signaling landscape through integrative spatial
proteomics and transcriptomics in a lymphoma model. Biomolecules.
11:17762021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Jamil MO and Mehta A: Diffuse large B-cell
lymphoma: Prognostic markers and their impact on therapy. Expert
Rev Hematol. 9:471–477. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Maurer MJ, Micallef INM, Cerhan JR,
Katzmann JA, Link BK, Colgan JP, Habermann TM, Inwards DJ, Markovic
SN, Ansell SM, et al: Elevated serum free light chains are
associated with event-free and overall survival in two independent
cohorts of patients with diffuse large B-cell lymphoma. J Clin
Oncol. 29:1620–1626. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Witzig TE, Maurer MJ, Stenson MJ, Allmer
C, Macon W, Link B, Katzmann JA and Gupta M: Elevated serum
monoclonal and polyclonal free light chains and interferon
inducible protein-10 predicts inferior prognosis in untreated
diffuse large B-cell lymphoma. Am J Hematol. 89:417–422. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Grünwald BT, Devisme A, Andrieux G, Vyas
F, Aliar K, McCloskey CW, Macklin A, Jang GH, Denroche R, Romero
JM, et al: Spatially confined sub-tumor microenvironments in
pancreatic cancer. Cell. 184:5577–5592.e18. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Akhtar M, Haider A, Rashid S and Al-Nabet
ADMH: Paget's ‘seed and soil’ theory of cancer metastasis: An idea
whose time has come. Adv Anat Pathol. 26:69–74. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Cords L, Tietscher S, Anzeneder T,
Langwieder C, Rees M, de Souza N and Bodenmiller B:
Cancer-associated fibroblast classification in single-cell and
spatial proteomics data. Nat Commun. 14:42942023. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Franciosa G, Kverneland AH, Jensen AWP,
Donia M and Olsen JV: Proteomics to study cancer immunity and
improve treatment. Semin Immunopathol. 45:241–251. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Gatto L, Breckels LM and Lilley KS:
Assessing sub-cellular resolution in spatial proteomics
experiments. Curr Opin Chem Biol. 48:123–149. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Liu X, Salokas K, Tamene F, Jiu Y,
Weldatsadik RG, Öhman T and Varjosalo M: An AP-MS- and
BioID-compatible MAC-tag enables comprehensive mapping of protein
interactions and subcellular localizations. Nat Commun. 9:11882018.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Pankow S, Martínez-Bartolomé S, Bamberger
C and Yates JR: Understanding molecular mechanisms of disease
through spatial proteomics. Curr Opin Chem Biol. 48:19–25. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Guilliams M, Bonnardel J, Haest B,
Vanderborght B, Wagner C, Remmerie A, Bujko A, Martens L, Thoné T,
Browaeys R, et al: Spatial proteogenomics reveals distinct and
evolutionarily conserved hepatic macrophage niches. Cell.
185:379–396.e38. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lee PY, Saraygord-Afshari N and Low TY:
The evolution of two-dimensional gel electrophoresis-from
proteomics to emerging alternative applications. J Chromatogr A.
1615:4607632020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Strohkamp S, Gemoll T and Habermann JK:
Possibilities and limitations of 2DE-based analyses for identifying
low-abundant tumor markers in human serum and plasma. Proteomics.
16:2519–2532. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lin TT, Zhang T, Kitata RB, Liu T, Smith
RD, Qian WJ and Shi T: Mass spectrometry-based targeted proteomics
for analysis of protein mutations. Mass Spectrom Rev. 42:796–821.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Noor Z, Ahn SB, Baker MS, Ranganathan S
and Mohamedali A: Mass spectrometry-based protein identification in
proteomics-a review. Brief Bioinform. 22:1620–1638. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Ren AH, Diamandis EP and Kulasingam V:
Uncovering the depths of the human proteome: Antibody-based
technologies for ultrasensitive multiplexed protein detection and
quantification. Mol Cell Proteomics. 20:1001552021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Syu GD, Dunn J and Zhu H: Developments and
applications of functional protein microarrays. Mol Cell
Proteomics. 19:916–927. 2020. View Article : Google Scholar : PubMed/NCBI
|