|
1
|
Weiderpass E and Stewart BW: World cancer
report: Cancer research for cancer prevention. International Agency
for Research on Cancer; Lyon: 2020
|
|
2
|
Feng Y, Spezia M, Huang S, Yuan C, Zeng Z,
Zhang L, Ji X, Liu W, Huang B, Luo W, et al: Breast cancer
development and progression: risk factors, cancer stem cells,
signaling pathways, genomics, and molecular pathogenesis. Genes
Dis. 5:77–106. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Laissue P: The forkhead-box family of
transcription factors: Key molecular players in colorectal cancer
pathogenesis. Mol Cancer. 18:52019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bach DH, Long NP, Luu TT, Anh NH, Kwon SW
and Lee SK: The dominant role of forkhead box proteins in cancer.
Int J Mol Sci. 19:32792018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weigel D, Jürgens G, Küttner F, Seifert E
and Jäckle H: The homeotic gene fork head encodes a nuclear protein
and is expressed in the terminal regions of the Drosophila embryo.
Cell. 57:645–658. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vaidya HJ, Briones Leon A and Blackburn
CC: FOXN1 in thymus organogenesis and development. Eur J Immunol.
46:1826–1837. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lam EW and Gomes AR: Forkhead box
transcription factors in cancer initiation, progression and
chemotherapeutic drug response. Front Oncol. 4:3052014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li C, Zhang K, Chen J, Chen L, Wang R and
Chu X: MicroRNAs as regulators and mediators of forkhead box
transcription factors function in human cancers. Oncotarget.
8:12433–12450. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Seachrist DD, Anstine LJ and Keri RA:
FOXA1: A pioneer of nuclear receptor action in breast cancer.
Cancers (Basel). 13:52052021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Czerny CC, Borschel A, Cai M, Otto M and
Hoyer-Fender S: FOXA1 is a transcriptional activator of
Odf2/Cenexin and regulates primary ciliation. Sci Rep.
12:214682022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cirillo LA and Zaret KS: Specific
interactions of the wing domains of FOXA1 transcription factor with
DNA. J Mol Biol. 366:720–724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bernardo GM and Keri RA: FOXA1: A
transcription factor with parallel functions in development and
cancer. Biosci Rep. 32:113–130. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dai X, Cheng H, Bai Z and Li J: Breast
cancer cell line classification and its relevance with breast tumor
subtyping. J Cancer. 8:3131–3141. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y, Zhao Y, Skerry B, Wang X,
Colin-Cassin C, Radisky DC, Kaestner KH and Li Z: Foxa1 is
essential for mammary duct formation. Genesis. 54:277–285. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brisken C and O'Malley B: Hormone action
in the mammary gland. Cold Spring Harb Perspect Biol.
2:a0031782010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Robinson JL, Macarthur S, Ross-Innes CS,
Tilley WD, Neal DE, Mills IG and Carroll JS: Androgen receptor
driven transcription in molecular apocrine breast cancer is
mediated by FoxA1. EMBO J. 30:3019–3027. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang YA, Zhao JC, Fong KW, Kim J, Li S,
Song C, Song B, Zheng B, He C and Yu J: FOXA1 potentiates
lineage-specific enhancer activation through modulating TET1
expression and function. Nucleic Acids Res. 44:8153–8164. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bernardo GM, Lozada KL, Miedler JD,
Harburg G, Hewitt SC, Mosley JD, Godwin AK, Korach KS, Visvader JE,
Kaestner KH, et al: FOXA1 is an essential determinant of ERalpha
expression and mammary ductal morphogenesis. Development.
137:2045–2054. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takaku M, Grimm SA, De Kumar B, Bennett BD
and Wade PA: Cancer-specific mutation of GATA3 disrupts the
transcriptional regulatory network governed by Estrogen Receptor
alpha, FOXA1 and GATA3. Nucleic Acids Res. 48:4756–4768. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ghosh S, Gu F, Wang CM, Lin CL, Liu J,
Wang H, Ravdin P, Hu Y, Huang TH and Li R: Genome-wide DNA
methylation profiling reveals parity-associated hypermethylation of
FOXA1. Breast Cancer Res Treat. 147:653–659. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
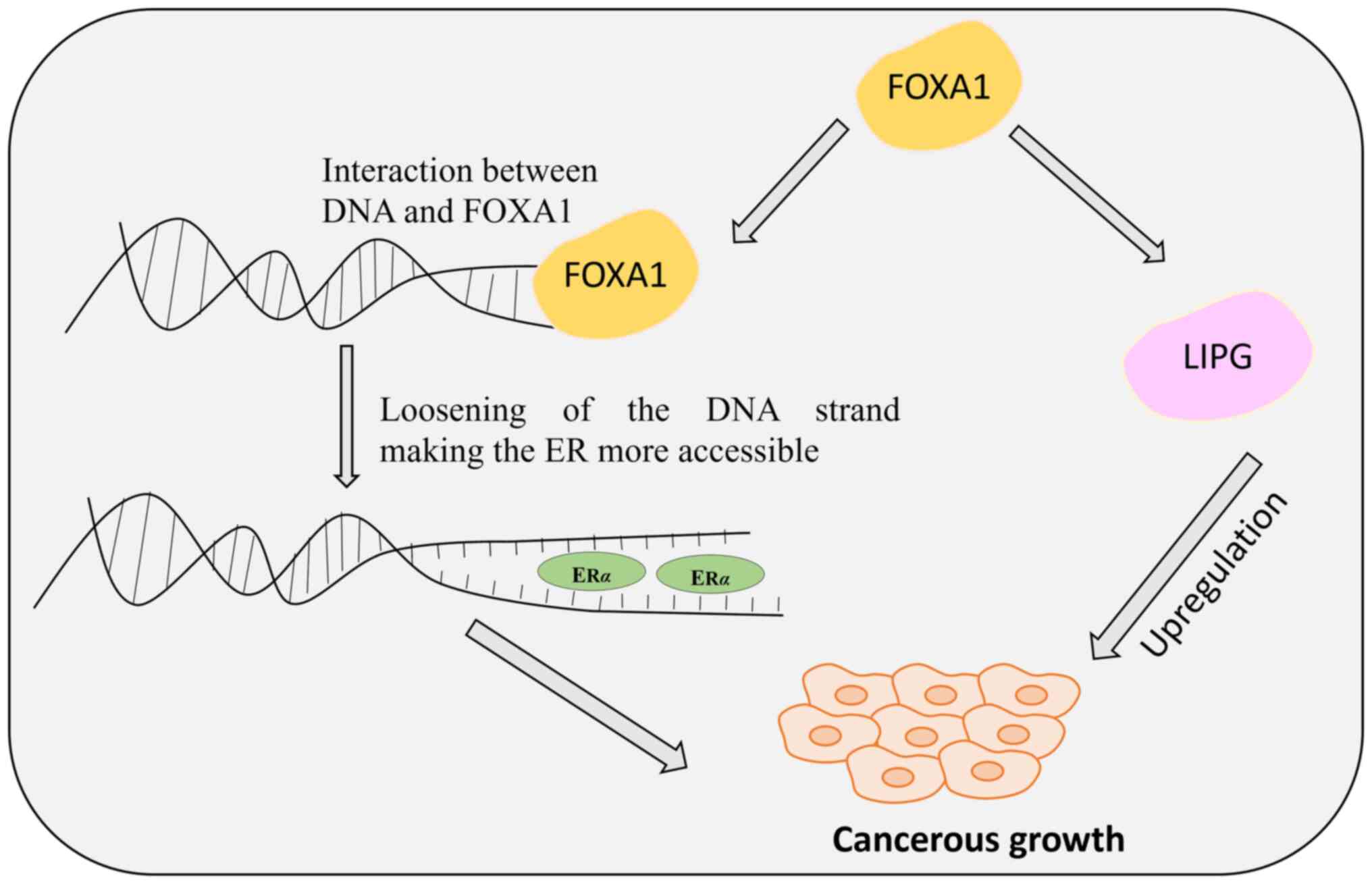
Slebe F, Rojo F, Vinaixa M, García-Rocha
M, Testoni G, Guiu M, Planet E, Samino S, Arenas EJ, Beltran A, et
al: FoxA and LIPG endothelial lipase control the uptake of
extracellular lipids for breast cancer growth. Nat Commun.
7:111992016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Anzai E, Hirata K, Shibazaki M, Yamada C,
Morii M, Honda T and Yamaguchi N and Yamaguchi N: FOXA1 induces
E-cadherin expression at the protein level via suppression of slug
in epithelial breast cancer cells. Biol Pharm Bull. 40:1483–1489.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ambrosone CB and Higgins MJ: Relationships
between breast feeding and breast cancer subtypes: Lessons learned
from studies in humans and in mice. Cancer Res. 80:4871–4877. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xia K, Huang W, Zhao X, Huang X, Chen Y,
Yu L and Tan Y: Increased FOXA1 levels induce apoptosis and inhibit
proliferation in FOXA1-low expressing basal breast cancer cells. Am
J Cancer Res. 12:2641–2658. 2022.PubMed/NCBI
|
|
26
|
Cantor JR and Sabatini DM: Cancer cell
metabolism: One hallmark, many faces. Cancer Discov. 2:881–898.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Zhou Y and Graves DT: FOXO
transcription factors: Their clinical significance and regulation.
Biomed Res Int. 2014:9253502014.PubMed/NCBI
|
|
28
|
Jiramongkol Y and Lam EW: FOXO
transcription factor family in cancer and metastasis. Cancer
Metastasis Rev. 39:681–709. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dumont SN, Lazar AJ, Bridge JA, Benjamin
RS and Trent JC: PAX3/7-FOXO1 fusion status in older
rhabdomyosarcoma patient population by fluorescent in situ
hybridization. J Cancer Res Clin Oncol. 138:213–220. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Parry P, Wei Y and Evans G: Cloning and
characterization of the t(X;11) breakpoint from a leukemic cell
line identify a new member of the forkhead gene family. Genes
Chromosomes Cancer. 11:79–84. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guttilla IK and White BA: Coordinate
regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast
cancer cells. J Biol Chem. 284:23204–23216. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bullock M: FOXO factors and breast cancer:
Outfoxing endocrine resistance. Endocr Relat Cancer. 23:R113–R130.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Farhan M, Wang H, Gaur U, Little PJ, Xu J
and Zheng W: FOXO signaling pathways as therapeutic targets in
cancer. Int J Biol Sci. 13:815–827. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Di Blasio L, Gagliardi PA, Puliafito A and
Primo L: Serine/threonine kinase 3-phosphoinositide-dependent
protein Kinase-1 (PDK1) as a key regulator of cell migration and
cancer dissemination. Cancers (Basel). 9:252017. View Article : Google Scholar : PubMed/NCBI
|
|
35
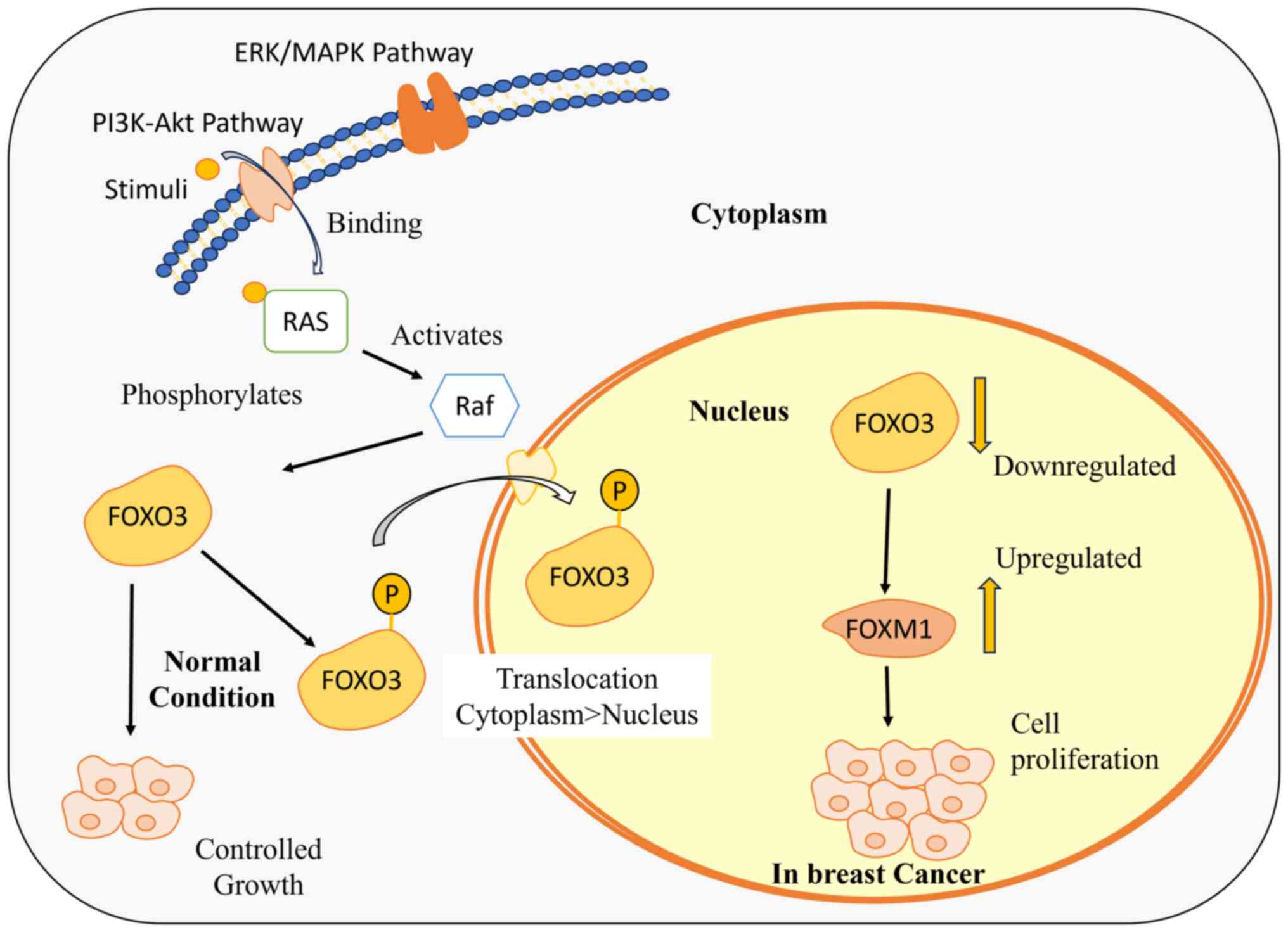
|
Shaw RJ and Cantley LC: Ras, PI(3)K and
mTOR signalling controls tumour cell growth. Nature. 441:424–430.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
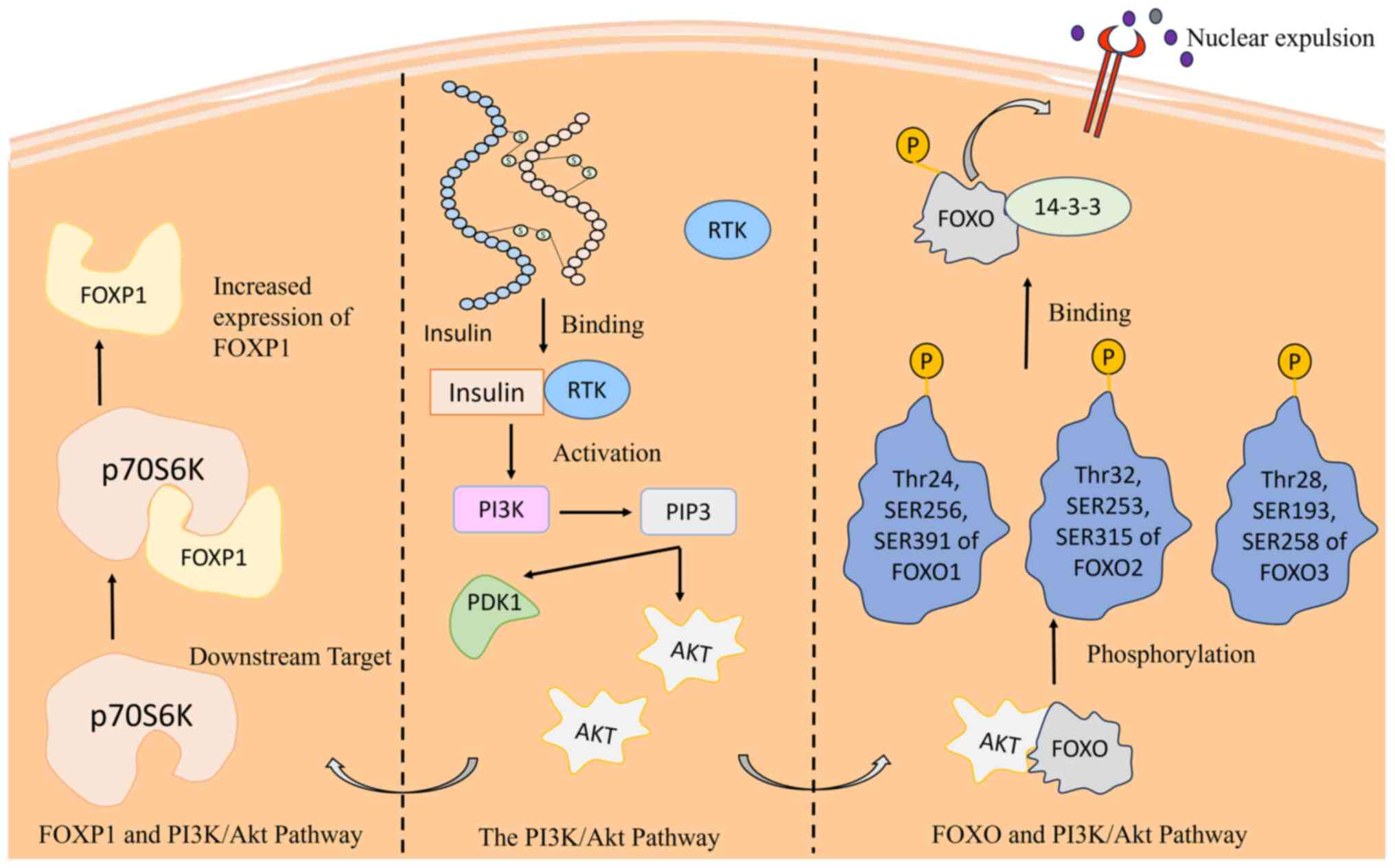
Tzivion G, Dobson M and Ramakrishnan G:
FoxO transcription factors; regulation by AKT and 14-3-3 proteins.
Biochim Biophys Acta. 1813:1938–1945. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim S, Kim Y, Lee J and Chung J:
Regulation of FOXO1 by TAK1-Nemo-like kinase pathway. J Biol Chem.
285:8122–8129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu H, Liu K and Dong Z: The role of
p21-activated kinases in cancer and beyond: Where are we heading?
Front Cell Dev Biol. 9:6413812021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Khan MA, Massey S, Ahmad I, Sada f, Akhter
N, Habib M, Mustafa S, Deo SVS and Husain SA: FOXO1 gene
downregulation and promoter methylation exhibits significant
correlation with clinical parameters in Indian breast cancer
patients. Front Genet. 13:8429432022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Peck B, Chen CY, Ho KK, Di Fruscia P,
Myatt SS, Coombes RC, Fuchter MJ, Hsiao CD and Lam EW: SIRT
inhibitors induce cell death and p53 acetylation through targeting
both SIRT1 and SIRT2. Mol Cancer Ther. 9:844–855. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gong C, Yao S, Gomes AR, Man EP, Lee HJ,
Gong G, Chang S, Kim SB, Fujino K, Kim SW, et al: BRCA1 positively
regulates FOXO3 expression by restricting FOXO3 gene methylation
and epigenetic silencing through targeting EZH2 in breast cancer.
Oncogenesis. 5:e2142016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu H, Song Y, Qiu H, Liu Y, Luo K, Yi Y,
Jiang G, Lu M, Zhang Z, Yin J, et al: Downregulation of FOXO3a by
DNMT1 promotes breast cancer stem cell properties and
tumorigenesis. Cell Death Differ. 27:966–983. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sanders DA, Gormally MV, Marsico G,
Beraldi D, Tannahill D and Balasubramanian S: FOXM1 binds directly
to non-consensus sequences in the human genome. Genome Biol.
16:1302015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Korver W, Roose J, Heinen K, Weghuis DO,
de Bruijn D, van Kessel AG and Clevers H: The human
TRIDENT/HFH-11/FKHL16 gene: Structure, localization, and promoter
characterization. Genomics. 46:435–442. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kalathil D, John S and Nair AS: FOXM1 and
cancer: Faulty cellular signaling derails homeostasis. Front Oncol.
10:6268362021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ye H, Kelly TF, Samadani U, Lim L, Rubio
S, Overdier DG, Roebuck KA and Costa RH: Hepatocyte nuclear factor
3/fork head homolog 11 is expressed in proliferating epithelial and
mesenchymal cells of embryonic and adult tissues. Mol Cell Biol.
17:1626–1641. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Halasi M and Gartel AL: FOX(M1) news-it is
cancer. Mol Cancer Ther. 12:245–254. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
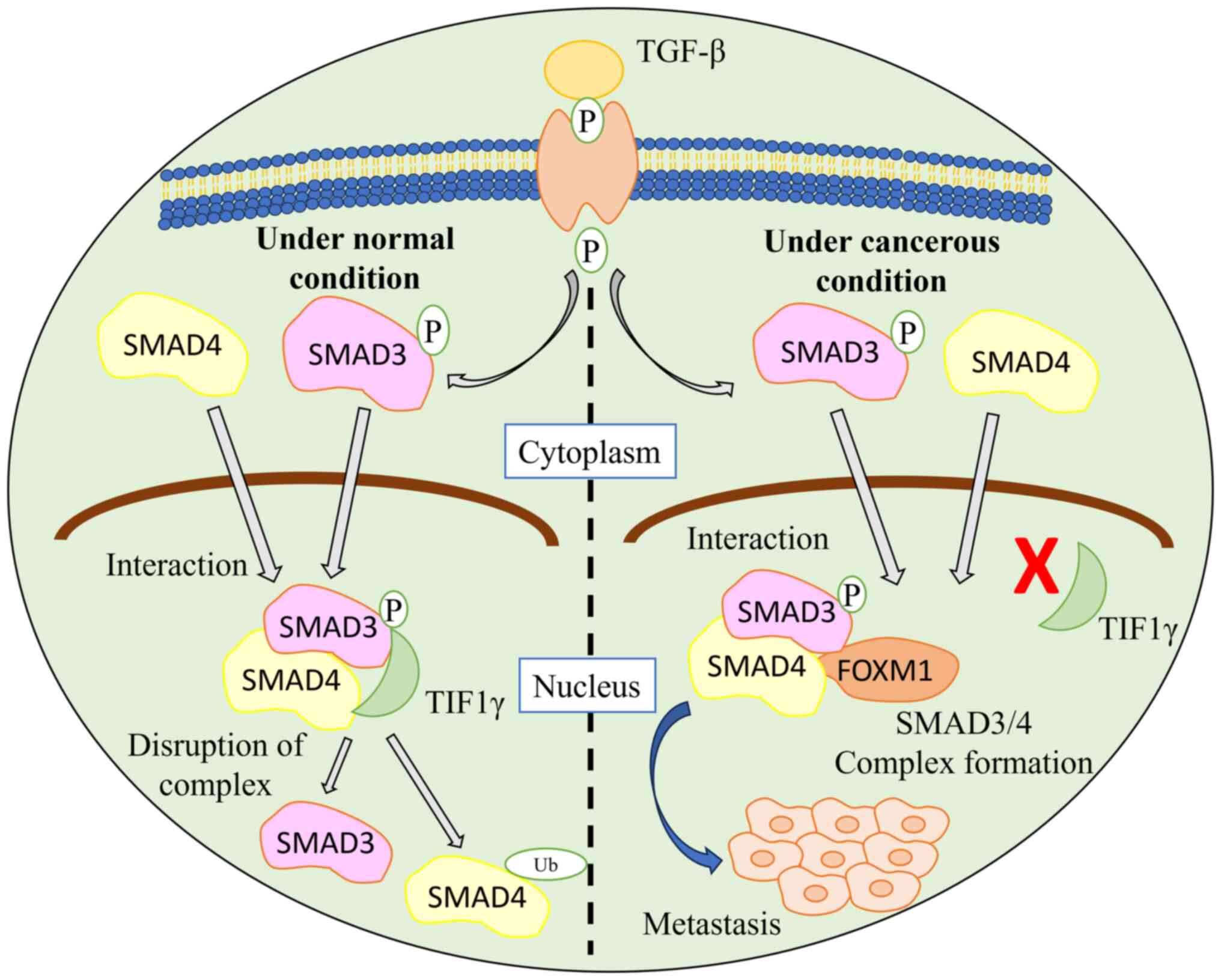
Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu
M, Feng XH, Sawaya R, Medema RH, Hung MC and Huang S: Sustained
activation of SMAD3/SMAD4 by FOXM1 promotes TGF-β-dependent cancer
metastasis. J Clin Invest. 124:564–579. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Speirs V and Walker RA: New perspectives
into the biological and clinical relevance of oestrogen receptors
in the human breast. J Pathol. 211:499–506. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Iqbal N and Iqbal N: Human epidermal
growth factor Receptor 2 (HER2) in cancers: Overexpression and
therapeutic implications. Mol Biol Int. 2014:8527482014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Francis RE, Myatt SS, Krol J, Hartman J,
Peck B, McGovern UB, Wang J, Guest SK, Filipovic A, Gojis O, et al:
FoxM1 is a downstream target and marker of HER2 overexpression in
breast cancer. Int J Oncol. 35:57–68. 2009.PubMed/NCBI
|
|
52
|
Chen X, Wei H, Li J, Liang X, Dai S, Jiang
L, Guo M, Qu L, Chen Z, Chen L and Chen Y: Structural basis for DNA
recognition by FOXC2. Nucleic Acids Res. 47:3752–3764. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pierrou S, Enerbäck S and Carlsson P:
Selection of high-affinity binding sites for sequence-specific, DNA
binding proteins from random sequence oligonucleotides. Anal
Biochem. 229:99–105. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yin L, Duan JJ, Bian XW and Yu SC:
Triple-negative breast cancer molecular subtyping and treatment
progress. Breast Cancer Res. 22:612020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Han B, Bhowmick N, Qu Y, Chung S, Giuliano
AE and Cui X: FOXC1: An emerging marker and therapeutic target for
cancer. Oncogene. 36:3957–3963. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang J, Ray PS, Sim MS, Zhou XZ, Lu KP,
Lee AV, Lin X, Bagaria SP, Giuliano AE and Cui X: FOXC1 regulates
the functions of human basal-like breast cancer cells by activating
NF-kappaB signaling. Oncogene. 31:4798–4802. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Nieto MA: Epithelial plasticity: A common
theme in embryonic and cancer cells. Science. 342:12348502013.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Bloushtain-Qimron N, Yao J, Snyder EL,
Shipitsin M, Campbell LL, Mani SA, Hu M, Chen H, Ustyansky V,
Antosiewicz JE, et al: Cell type-specific DNA methylation patterns
in the human breast. Proc Natl Acad Sci USA. 105:14076–14081. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Powell AA, Talasaz AH, Zhang H, Coram MA,
Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, et
al: Single cell profiling of circulating tumor cells:
Transcriptional heterogeneity and diversity from breast cancer cell
lines. PLoS One. 7:e337882012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lindley LE and Briegel KJ: Molecular
characterization of TGFbeta-induced epithelial-mesenchymal
transition in normal finite lifespan human mammary epithelial
cells. Biochem Biophys Res Commun. 399:659–664. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Clark KL, Halay ED, Lai E and Burley SK:
Co-crystal structure of the HNF-3/fork head DNA-recognition motif
resembles histone H5. Nature. 364:412–420. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Perumal K, Dirr HW and Fanucchi S: A
single amino acid in the hinge loop region of the FOXP forkhead
domain is significant for dimerisation. Protein J. 34:111–121.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Stroud JC, Wu Y, Bates DL, Han A, Nowick
K, Paabo S, Tong H and Chen L: Structure of the forkhead domain of
FOXP2 bound to DNA. Structure. 14:159–166. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shigekawa T, Ijichi N, Ikeda K,
Horie-Inoue K, Shimizu C, Saji S, Aogi K, Tsuda H, Osaki A, Saeki T
and Inoue S: FOXP1, an estrogen-inducible transcription factor,
modulates cell proliferation in breast cancer cells and 5-year
recurrence-free survival of patients with tamoxifen-treated breast
cancer. Horm Cancer. 2:286–297. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Beelen K, Hoefnagel LD, Opdam M, Wesseling
J, Sanders J, Vincent AD, van Diest PJ and Linn SC: PI3K/AKT/mTOR
pathway activation in primary and corresponding metastatic breast
tumors after adjuvant endocrine therapy. Int J Cancer.
135:1257–1263. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Banham AH, Beasley N, Campo E, Fernandez
PL, Fidler C, Gatter K, Jones M, Mason DY, Prime JE, Trougouboff P,
et al: The FOXP1 winged helix transcription factor is a novel
candidate tumor suppressor gene on chromosome 3p. Cancer Res.
61:8820–8829. 2001.PubMed/NCBI
|
|
68
|
Liu Y, Chen T, Guo M, Li Y, Zhang Q, Tan
G, Yu L and Tan Y: FOXA2-interacting FOXP2 prevents
epithelial-mesenchymal transition of breast cancer cells by
stimulating E-cadherin and PHF2 transcription. Front Oncol.
11:6050252021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Sada f, Akhter N, Alharbi RA, Sindi AAA,
Najm MZ, Alhumaydhi FA, Khan MA, Deo SVS and Husain SA: Epigenetic
alteration and its association with downregulated FOXP3 gene in
indian breast cancer patients. Front Genet. 12:7814002021.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu C, Han J, Li X, Huang T, Gao Y, Wang
B, Zhang K, Wang S, Zhang W, Li W, et al: FOXP3 inhibits the
metastasis of breast cancer by downregulating the expression of
MTA1. Front Oncol. 11:6561902021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ma B, Miao W, Xiao J, Chen X, Xu J and Li
Y: The role of FOXP3 on tumor metastasis and its interaction with
traditional Chinese medicine. Molecules. 27:67062022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Dai X, Cheng H, Chen X, Li T, Zhang J, Jin
G, Cai D and Huang Z: FOXA1 is prognostic of triple negative breast
cancers by transcriptionally suppressing SOD2 and IL6. Int J Biol
Sci. 15:1030–1041. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Cao J, Wang X, Wang D, Ma R, Li X, Feng H,
Wang J, Liu S and Wang L: PGC-1β cooperating with FOXA2 inhibits
proliferation and migration of breast cancer cells. Cancer Cell
Int. 19:932019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Song Y, Zeng S, Zheng G, Chen D, Li P,
Yang M, Luo K, Yin J, Gu Y, Zhang Z, et al: FOXO3a-driven miRNA
signatures suppresses VEGF-A/NRP1 signaling and breast cancer
metastasis. Oncogene. 40:777–790. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wei G, Yang X, Lu H, Zhang L, Wei Y, Li H,
Zhu M and Zhou X: Prognostic value and immunological role of FOXM1
in human solid tumors. Aging (Albany NY). 14:9128–9148. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Broughton JP, Lovci MT, Huang JL, Yeo GW
and Pasquinelli AE: Pairing beyond the seed supports microRNA
targeting specificity. Mol Cell. 64:320–333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Arora T, Kausar MA, Aboelnaga SM, Anwar S,
Hussain MA, Sadaf S, Kaur S, Eisa AA, Shingatgeri VMM, Najm MZ and
Aloliqi AA: miRNAs and the Hippo pathway in cancer: Exploring the
therapeutic potential (Review). Oncol Rep. 48:1352022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Denli AM, Tops BB, Plasterk RH, Ketting RF
and Hannon GJ: Processing of primary microRNAs by the
microprocessor complex. Nature. 432:231–235. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Alarcón CR, Lee H, Goodarzi H, Halberg N
and Tavazoie SF: N6-methyladenosine marks primary microRNAs for
processing. Nature. 519:482–485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Okada C, Yamashita E, Lee SJ, Shibata S,
Katahira J, Nakagawa A, Yoneda Y and Tsukihara T: A high-resolution
structure of the pre-microRNA nuclear export machinery. Science.
326:1275–1279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang H, Kolb FA, Jaskiewicz L, Westhof E
and Filipowicz W: Single processing center models for human Dicer
and bacterial RNase III. Cell. 118:57–68. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wang W and Luo YP: MicroRNAs in breast
cancer: Oncogene and tumor suppressors with clinical potential. J
Zhejiang Univ Sci B. 16:18–31. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Corcoran C, Friel AM, Duffy MJ, Crown J
and O'Driscoll L: Intracellular and extracellular microRNAs in
breast cancer. Clin Chem. 57:18–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Guo X, Connick MC, Vanderhoof J, Ishak MA
and Hartley RS: MicroRNA-16 modulates HuR regulation of cyclin E1
in breast cancer cells. Int J Mol Sci. 16:7112–7132. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Jin T, Suk Kim H, Ki Choi S, Hye Hwang E,
Woo J, Suk Ryu H, Kim K, Moon A and Kyung Moon W: microRNA-200c/141
upregulates SerpinB2 to promote breast cancer cell metastasis and
reduce patient survival. Oncotarget. 8:32769–32782. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Chen S, Wang Y, Ni C, Meng G and Sheng X:
HLF/miR-132/TTK axis regulates cell proliferation, metastasis and
radiosensitivity of glioma cells. Biomed Pharmacother. 83:898–904.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang D, Ren J, Ren H, Fu JL and Yu D:
MicroRNA-132 suppresses cell proliferation in human breast cancer
by directly targeting FOXA1. Acta Pharmacol Sin. 39:124–131. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Gao T, Zou M, Shen T and Duan S:
Dysfunction of miR-802 in tumors. J Clin Lab Anal. 35:e239892021.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yuan F and Wang W: MicroRNA-802 suppresses
breast cancer proliferation through downregulation of FoxM1. Mol
Med Rep. 12:4647–4651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wang Q, Ye B, Wang P, Yao F, Zhang C and
Yu G: Overview of microRNA-199a regulation in cancer. Cancer Manag
Res. 11:10327–10335. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Cuiffo BG, Campagne A, Bell GW, Lembo A,
Orso F, Lien EC, Bhasin MK, Raimo M, Hanson SE, Marusyk A, et al:
MSC-regulated microRNAs converge on the transcription factor FOXP2
and promote breast cancer metastasis. Cell Stem Cell. 15:762–774.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu
C, Li J, Wang X and Song L: Unregulated miR-96 induces cell
proliferation in human breast cancer by downregulating
transcriptional factor FOXO3a. PLoS One. 5:e157972010. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yin Z, Wang W, Qu G, Wang L, Wang X and
Pan Q: MiRNA-96-5p impacts the progression of breast cancer through
targeting FOXO3. Thorac Cancer. 11:956–963. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Tan X, Li Z, Ren S, Rezaei K, Pan Q,
Goldstein AT, Macri CJ, Cao D, Brem RF and Fu SW: Dynamically
decreased miR-671-5p expression is associated with oncogenic
transformation and radiochemoresistance in breast cancer. Breast
Cancer Res. 21:892019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kumar U, Hu Y, Masrour N,
Castellanos-Uribe M, Harrod A, May ST, Ali S, Speirs V, Coombes RC
and Yagüe E: MicroRNA-495/TGF-β/FOXC1 axis regulates multidrug
resistance in metaplastic breast cancer cells. Biochem Pharmacol.
192:1146922021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Badve S, Turbin D, Thorat MA, Morimiya A,
Nielsen TO, Perou CM, Dunn S, Huntsman DG and Nakshatri H: FOXA1
expression in breast cancer-correlation with luminal subtype A and
survival. Clin Cancer Res. 13:4415–4421. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Fu X, Jeselsohn R, Pereira R,
Hollingsworth EF, Creighton CJ, Li F, Shea M, Nardone A, De Angelis
C, Heiser LM, et al: FOXA1 overexpression mediates endocrine
resistance by altering the ER transcriptome and IL-8 expression in
ER-positive breast cancer. Proc Natl Acad Sci USA. 113:E6600–E6609.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Hurtado A, Holmes KA, Ross-Innes CS,
Schmidt D and Carroll JS: FOXA1 is a key determinant of estrogen
receptor function and endocrine response. Nat Genet. 43:27–33.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
100
|
Yamaguchi N, Nakayama Y and Yamaguchi N:
Down-regulation of Forkhead box protein A1 (FOXA1) leads to cancer
stem cell-like properties in tamoxifen-resistant breast cancer
cells through induction of interleukin-6. J Biol Chem.
292:8136–8148. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Karunarathna U, Kongsema M, Zona S, Gong
C, Cabrera E, Gomes AR, Man EP, Khongkow P, Tsang JW, Khoo US, et
al: OTUB1 inhibits the ubiquitination and degradation of FOXM1 in
breast cancer and epirubicin resistance. Oncogene. 35:1433–1444.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Nestal de Moraes G, Delbue D, Silva KL,
Robaina MC, Khongkow P, Gomes AR, Zona S, Crocamo S, Mencalha AL,
Magalhães LM, et al: FOXM1 targets XIAP and Survivin to modulate
breast cancer survival and chemoresistance. Cell Signal.
27:2496–2505. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Nestal de Moraes G, Bella L, Zona S,
Burton MJ and Lam EW: Insights into a critical role of the
FOXO3a-FOXM1 axis in DNA damage response and genotoxic drug
resistance. Curr Drug Targets. 17:164–177. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Di Fruscia P, Zacharioudakis E, Liu C,
Moniot S, Laohasinnarong S, Khongkow M, Harrison IF, Koltsida K,
Reynolds CR, Schmidtkunz K, et al: The discovery of a highly
selective
5,6,7,8-tetrahydrobenzo[4,5]thieno[2,3-d]pyrimidin-4(3H)-one SIRT2
inhibitor that is neuroprotective in an in vitro Parkinson's
disease model. ChemMedChem. 10:69–82. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Saba R, Alsayed A, Zacny JP and Dudek AZ:
The role of forkhead box protein M1 in breast cancer progression
and resistance to therapy. Int Breast Cancer.
2016:97681832016.PubMed/NCBI
|
|
106
|
Ji X, Tian X, Feng S, Zhang L, Wang J, Guo
R, Zhu Y, Yu X, Zhang Y, Du H, et al: Intermittent F-actin
perturbations by magnetic fields inhibit breast cancer metastasis.
Research (Wash DC). 6:00802023.PubMed/NCBI
|
|
107
|
Halasi M, Hitchinson B, Shah BN, Váraljai
R, Khan I, Benevolenskaya EV, Gaponenko V, Arbiser JL and Gartel
AL: Honokiol is a FOXM1 antagonist. Cell Death Dis. 9:842018.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Rajamanickam S, Panneerdoss S, Gorthi A,
Timilsina S, Onyeagucha B, Kovalskyy D, Ivanov D, Hanes MA,
Vadlamudi RK, Chen Y, et al: Inhibition of FoxM1-mediated DNA
repair by imipramine blue suppresses breast cancer growth and
metastasis. Clin Cancer Res. 22:3524–3536. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Lopez JS and Banerji U: Combine and
conquer: Challenges for targeted therapy combinations in early
phase trials. Nat Rev Clin Oncol. 14:57–66. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Martel S, Bruzzone M, Ceppi M, Maurer C,
Ponde NF, Ferreira AR, Viglietti G, Del Mastro L, Prady C, de
Azambuja E and Lambertini M: Risk of adverse events with the
addition of targeted agents to endocrine therapy in patients with
hormone receptor-positive metastatic breast cancer: A systematic
review and meta-analysis. Cancer Treat Rev. 62:123–132. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Pegram MD, Konecny GE, O'Callaghan C,
Beryt M, Pietras R and Slamon DJ: Rational combinations of
trastuzumab with chemotherapeutic drugs used in the treatment of
breast cancer. J Natl Cancer Inst. 96:739–749. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Guillen VS, Ziegler Y, Gopinath C, Kumar
S, Dey P, Plotner BN, Dawson NZ, Kim SH, Katzenellenbogen JA and
Katzenellenbogen BS: Effective combination treatments for breast
cancer inhibition by FOXM1 inhibitors with other targeted cancer
drugs. Breast Cancer Res Treat. 198:607–621. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Lin Z, Huang W, He Q, Li D, Wang Z, Feng
Y, Liu D, Zhang T, Wang Y, Xie M, et al: FOXC1 promotes HCC
proliferation and metastasis by Upregulating DNMT3B to induce DNA
Hypermethylation of CTH promoter. J Exp Clin Cancer Res. 40:502021.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Bader AG and Lammers P: The therapeutic
potential of microRNAs. Innov Pharm Technol. 52–55. 2011.
|
|
116
|
Broderick JA and Zamore PD: MicroRNA
therapeutics. Gene Ther. 18:1104–1110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Tate CR, Rhodes LV, Segar HC, Driver JL,
Pounder FN, Burow ME and Collins-Burow BM: Targeting
triple-negative breast cancer cells with the histone deacetylase
inhibitor panobinostat. Breast Cancer Res. 14:R792012. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Linares A, Dalenc F, Balaguer P, Boulle N
and Cavailles V: Manipulating protein acetylation in breast cancer:
A promising approach in combination with hormonal therapies? J
Biomed Biotechnol. 2011:8569852011. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Khafaga AF, Shamma RN, Abdeen A, Barakat
AM, Noreldin AE, Elzoghby AO and Sallam MA: Celecoxib repurposing
in cancer therapy: Molecular mechanisms and nanomedicine-based
delivery technologies. Nanomedicine (Lond). 16:1691–1712. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
National Library of Medicine (NIH), .
Study to Compare Alisertib with Paclitaxel vs Paclitaxel Alone in
Metastatic or Locally Recurrent Breast Cancer. Clinical trial:
NCT02187991. NIH; Bethesda, MD: 2022, https://www.mycancergenome.org/content/clinical_trials/NCT02187991/December
1–2022
|
|
121
|
National Library of Medicine (NIH), .
First Time in Human Study of AZD8701 With or Without Durvalumab in
Participants with Advanced Solid Tumours. Clinical trial:
NCT04504669. NIH; Bethesda, MD: 2022, https://www.mycancergenome.org/content/clinical_trials/NCT04504669/December
1–2022
|
|
122
|
National Library of Medicine (NIH), .
Pre-op Pembro + Radiation Therapy in Breast Cancer (P-RAD).
Clinical trial: NCT04443348. NIH; Bethesda, MD: 2022, https://www.mycancergenome.org/content/clinical_trials/NCT04443348/December
1–2022
|
|
123
|
National Library of Medicine (NIH), . A
Study of PDR001 in Combination with LCL161, Everolimus or
Panobinostat. Clinical trial: NCT02890069. NIH; Bethesda, MD: 2022,
https://www.mycancergenome.org/content/clinical_trials/NCT02890069/December
1–2022
|
|
124
|
National Library of Medicine (NIH), .
Phase I, Dose Study to Look at the Safety and Pharmacokinetics of
AZD8835 in Patients with Advanced Solid Tumours. Clinical trial:
NCT02260661. NIH; Bethesda, MD: 2022, https://www.mycancergenome.org/content/clinical_trials/NCT02260661/December
1–2022
|