Introduction
Chemotherapy, as a comprehensive treatment for
breast cancer (BRCA), has improved the survival rate of patients
with BRCA, to a certain extent (1).
Docetaxel (DTX), a semisynthetic derivative of paclitaxel, is a
second-generation paclitaxel anticancer drug and serves as the
cornerstone of various BRCA chemotherapy regimens (2). However, the occurrence of drug
resistance has limited the clinical efficacy of taxanes to some
extent (3). Congenital and acquired
chemoresistance constitute important reasons for long-term
treatment failure in patients with BRCA (4). The mechanisms of congenital resistance
mainly include decreased drug activation, abnormal expression of
membrane transporters such as ABC transporters (5), abnormal apoptosis or autophagy-induced
chemotherapy resistance (6),
changes in the expression and/or function of drug targets, reduced
efficiency of drug-target interactions (7), and enhanced DNA damage repair ability
(8). Acquired resistance is
influenced by genetic or environmental factors that promote the
development of resistant stem cells or induce mutations in enzymes
involved in related metabolic pathways (9). Currently, an increasing number of
studies have focused on the role of metabolic reprogramming in
tumor chemotherapy resistance (10).
Lysine methyltransferase 5A (KMT5A, also known as
SETD8; SET8; SET07; PR-Set7, and PR/SET07) is a member of the SET
domain-containing methyltransferase family that specifically
catalyses the addition of histone H4 lysine 20 (H4K20) me1
(11). The expression of this
protein methyltransferase fluctuates during the cell cycle and
peaks during the G2/M transition (12). KMT5A affects the development and
progression of a variety of cancers (13–15)
and causes a variety of tumors to develop chemotherapy resistance
(16). Fructose-1,6-bisphosphatase
1 (FBP1) acts as a key enzyme for gluconeogenesis and regulates
energy metabolism in tumor cells, and it is downregulated in a
variety of cancers, which affects the evolution of drug resistance
and the prognosis of cancers (17–19).
KMT5A participates in regulating the energy metabolism process of
malignant tumors and promotes tumor cell invasion and metastasis
(20). In the present study, the
expression of KMT5A and FBP1 in normal BRCA and paraneoplastic
tissues were analysed. Changes in DTX sensitivity in stable
KMT5A-knockdown BRCA cells were detected with a Cell Counting Kit-8
(CCK-8) kit. The mechanism through which KMT5A promotes DTX
resistance in BRCA was preliminarily explored by inhibiting FBP1 to
provide a theoretical basis for overcoming drug resistance.
Materials and methods
Cell lines and cell culture
The MCF-7 cell line was purchased from the Chinese
Academy of Sciences (Shanghai, China), the MDA-MB-231 cell line was
donated by Fudan University Cancer Hospital (Shanghai, China), and
the 293T cell line was purchased by Shanghai Sixth Hospital Central
Laboratory (Shanghai, China). The MCF-7 and 293T cell lines were
cultured in DMEM supplemented with 10% fetal bovine serum (FBS)
(Wuhan Servicebio Technology Co., Ltd.) at 37°C and 5%
CO2. MDA-MB-231 cell line was cultured in Leibovitz's
L-15 medium supplemented with 10% FBS without CO2 at
37°C. The cells were digested with 0.25% trypsin-EDTA (cat. no.
25200072) (Gibco; Thermo Fisher Scientific, Inc.) and passaged.
Plasmids
The full-length cDNAs of KMT5A and FBP1 were
obtained by PCR amplification from human tissue and directly
connected downstream of the CMV promoter of the lentiviral
expression vector after enzyme digestion through ligase reaction.
The overexpression plasmid was
Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin (GV358) (Shanghai GeneChem
Co., Ltd.), and GV358-Control and GV358-KMT5A (Shanghai GeneChem
Co., Ltd.) were constructed by sequencing and identification. The
vector with the correct expression sequence of the recombinant FBP1
gene was selected by gene sequencing, and the plasmid GV358-Control
and GV358-FBP1 (Shanghai GeneChem Co., Ltd.) were constructed by
sequencing. shRNA targets design software (https://rnaidesigner.thermofisher.com/rnaiexpress/sort.do)
from human KMT5A (NM_020382) was used. The synthetical oligos were
inserted into the lentivirus expression plasmid
hU6-MCS-Ubiquitin-EGFP-IRES-puromycin (GV248) (Shanghai GeneChem
Co., Ltd.) (additional vector information and the sequences of
shRNA are included in Tables SI
and SII). Empty vectors were used
as negative controls in the aforementioned plasmid construction
experiments of overexpression and knockdown expression.
Lentivirus production and
transduction
For virus packaging, 293T cells were seeded into a
10-cm culture dish with puromycin (2 µg/ml). When the cells reached
a density of 60–70%, KMT5A(GV358-KMT5A) (2.67 µg)/
shKMT5A(GV248-KMT5A) (4 µg)/FBP1(GV358-FBP1) (1.33 µg) plasmid or
control vector was transfected into 293T cells along with
lentiviral helper plasmid (ps-PAX2, pMD2G tagged in green
fluorescent protein) (Shanghai GeneChem Co., Ltd.) using
Lipofectamine® 3000 transfection kit (Invitrogen; Thermo
Fisher Scientific, Inc.) at 25°C. The proportions were as follows:
target plasmid: ps-PAX2: pMD2G=1:3:4. After transfection for 48 h,
the culture media containing recombinant lentivirus were collected,
filtered, concentrated, purified and stored at −80°C. The sequence
of shRNA was CGCAACAGAATCGCAAACTTA.
Lentiviral infection of human BRCA
cells
The MDA-MB-231 cells and MCF-7 cells were cultured
in 6-well plates. Then prepared recombinant lentivirus was added
into the culture medium with multiplicity of infection of 20. After
72 h infection, the GFP expression was observed under a
fluorescence microscope to evaluate lentivirus infection
efficiency. Total RNA and protein were extracted 72 h after
infection.
Reverse transcription-quantitative
(RT-q) PCR
The cells were collected (6-well plate with 80% cell
density) and centrifuged at 14,000 × g for 5 min at 25°C, after
which total RNA was extracted with TRIzol® reagent
(Shanghai Pufei Biotechnology Co., Ltd.). Total RNA (2.0 µg) was
used for complementary DNA (cDNA) synthesis via a Promega M-MLV kit
(cat no. M1701; Promega Corporation) following the manufacturer's
protocol. RT-qPCR was conducted with SYBR Premix Ex Taq (Takara
Bio, Inc.) via a two-step method, and a melting curve was prepared
for quantitative data analysis. The thermocycling conditions were
as follows: 95°C for 10 min, 95°C for 15 sec, 59°C for 40 sec, 72°C
for 45 sec, and the cycles were 40 times. The relative mRNA levels
were calculated using the comparative Ct method (2−ΔΔCq)
(21). The primer pairs were
designed with Primer Express 3.0 (ABI Inc.). The sequences of
primers (Biosune; http://www.biosune.com/) were as follows: KMT5A
forward, 5′-GAAGTCCGAGGAACAGAAG-3′ and reverse,
5′-ACAGGGTAGAAATCCGTAA-3′; FBP1 forward, 5′-TCTACCAACGTGACAGGTGA-3′
and reverse, 5′-ATCAAGGGGATCAAAACAGA-3′; and β-actin forward,
5′-GCGTGACATTAAGGAGAAGC-3′ and reverse,
5′-CCACGTCACACTTCATGATGG-3′.
In vitro chemosensitivity assay
The MTT assay was used to evaluate the cytotoxicity
of DTX against BRCA cells. First, the MDA-MB-231 cells were
incubated at 37°C after seeding into 96-well plates at
5×104 cells/well for 24 h. Then, the cells were treated
with gradient concentrations of DTX at equal concentrations. MTT
solution (20 µl, 5 mg/ml) (Shanghai Dingguo Biotechnology Co.,
Ltd.) was added to each well according to the design time. After 4
h of incubation at 37°C, the culture medium containing unreacted
MTT was completely removed from the wells, and dimethyl sulfoxide
(100 µl) was added to each well. After the samples were oscillated
for 3–5 min, the OD values were detected by a microplate reader at
490/570 nm. Finally, the data were analyzed.
Detection of drug sensitivity by Cell
Counting Kit-8 (CCK-8)
Cell proliferation and cell activities were measured
by a CCK-8 assay (Dojindo Laboratories, Inc.). All the MDA-MB-231
or MCF-7 cells were plated at 1×104 cells per well and
treated with different concentrations of DTX for 48 h. Then, CCK-8
solution (100 µl/well) was added to all the wells and incubated at
37°C for 4 h, the absorbance was read at 450 nm, and calculated the
percentage of cell viability.
BRCA cell lines were treated with KMT5A inhibitor
UNC0379 (0, 5 or 10 µM) (MedChemExpress) and DTX (20 µM) for 48 h.
A total of 10 µl of diluted CCK-8 was added into each well and
cells were incubated in a 5% CO2 incubator for 2 h at
37°C. Absorbance was measured at 450 nm.
Flow cytometry
When the cells cultured in 6-cm dishes reached to a
confluence rate of ~80% (the cells did not reach the growth plateau
stage), they were digested with pancreatic enzymes and collected,
and three wells were set in each group (to ensure a sufficient
number of cells on the machine, ≥106 cells were used).
The cells were centrifuged at 350 × g for 5 min at 4°C, washed with
precooled Dulbecco's phosphate-buffered saline (DPBS) (pH=7.2~7.4)
at 4°C and precipitated once. The cells were centrifuged at 350 × g
for 5 min at 4°C. The cell staining agent used was 40X propidium
iodide (PI) (2 mg/ml) (MilliporeSigma), 100X RNase (10 mg/ml)
(Thermo Fisher Scientific, Inc.), 1X DPBS and 25X Triton X-100
(MilliporeSigma) (25:10:1,000:40). The cell suspension was added to
a certain volume of cell staining solution (0.6–1 ml) to ensure
that the cell passage rate was 300 to 800 cells/sec. After machine
(CytoFLEX; Beckman Coulter, Inc.) detection, the data analysis was
performed by ModFit software (5.0) (Verity Software House,
Inc.).
Immunohistochemistry (IHC)
A total of 60 patients (age range, 31–82 years) with
BRCA admitted to Shanghai Sixth People's Hospital Affiliated to
Shanghai Jiao Tong University School of Medicine from August 2023
to December 2023 were selected as the present study's subjects. The
present study was approved (approval no. YS-2017-006) by the Ethics
Committee of the Sixth People's Hospital Affiliated to Shanghai
Jiao Tong University School of Medicine (Shanghai, China), and
written informed consent was obtained from each patient. The
inclusion criteria were as follows: i) Meeting the diagnostic
criteria for BRCA; ii) Surgical resection was performed, and
samples of BRCA tissues and adjacent tissues were obtained. The
diagnosis of BRCA was confirmed by pathological examination, iii)
for the first time, no anticancer treatment such as endocrine
therapy, radiotherapy and chemotherapy was taken before surgery,
iv) women, v) complete clinical data; and vi) signed informed
consent for the present study. Exclusion criteria were the
following: i) Combined with systemic infectious diseases,
cardiovascular and cerebrovascular diseases, liver and kidney
insufficiency; ii) combined with other types of tumors; iii)
combined with terminal disease; iv) tissue samples that cannot be
obtained; and v) ment0al system diseases and suicidal
tendencies.
Normal tumor and breast tissues were excised from
patients. These tissues were fixed in 10% neutral buffered
formalin, embedded in paraffin at 4°C for 12 h, sectioned (5 µm),
and stained with haematoxylin-eosin staining. Immunohistochemical
staining using KMT5A antibody (1:100; cat. no. PA5-102712; Thermo
Fisher Scientific, Inc.; 22°C for 1.5 h) and Rabbit IgG (H + L)
cross-Adsorbed Secondary Antibody (1:2,000; cat. no. A-11008;
Thermo Fisher Scientific, Inc.; 25°C for 30 min) was performed by
the Department of Pathology at Shanghai Sixth People's Hospital
Affiliated with Shanghai Jiao Tong University School of Medicine.
KMT5A immunohistochemical markers were assessed using light
microscopy. The immunostained slides were scored according to the
proportion of tumor cells that exhibited nuclear staining. KMT5A
expression was considered positive when >25% of the tumor cell
nuclei were stained (Table
SIII).
Western blotting
MCF-7 or MDA-MB-231 cells in 6-well plates were
collected and washed twice with phosphate- buffered saline (PBS)
(Wuhan Servicebio Technology Co., Ltd.). Protein was extracted from
MCF-7 or MDA-MB-231 cells by RIPA lysis buffer (Beyotime Institute
of Biotechnology). The concentration of the proteins was determined
with a BCA protein assay kit (Beyotime Institute of Biotechnology).
The protein lysates were separated by sodium dodecyl
sulphate-polyacrylamide gel electrophoresis. The separation gel
with 10% polyacrylamide was prepared, and 30 µl protein was loaded
to each lane and transferred onto polyvinylidene fluoride (PVDF)
membranes (MilliporeSigma). The PVDF membranes were blocked with
0.1% TBST solution (Wuhan Servicebio Technology Co., Ltd.)
containing 5% skim milk at 25°C for 1 h. After the membranes were
incubated with primary antibodies KMT5A, (1:1,000; cat. no.
14063-1-AP; Proteintech Group, Inc.; 37°C for 1 h;) and FBP1
(1:2,000; cat. no. 12842-1-AP; Proteintech Group, Inc.; 25°C for
1.5 h;) and secondary antibody Multi-rAb HRP-Goat Anti-Rabbit
Recombinant Secondary Antibody (1:5,000; cat. no. RGAR001;
Proteintech Group, Inc.; 25°C for 1 h;), an enhanced
chemiluminescence (ECL) kit (Thermo Fisher Scientific, Inc.) and
X-ray (Carestream Health, Inc.) were used to visualize the
membranes. The X-rays were removed, the samples were allowed to
dry, and the blots were analysed. Because the molecular weights of
KMT5A and actin were both 43 KD. At first, the target protein KMT5A
was incubated. After KMT5A was stripped, the internal reference
protein actin was incubated on the same membrane.
Dual-luciferase reporter assay
KMT5A 3′-UTR sequence was obtained from KMT5A and
twist family BHLH transcription factor 1 (TWIST1) full-length cDNA
vector (Shanghai GeneChem Co., Ltd.) and sub-cloned to
pcDNA3.1-control vector (Shanghai GeneChem Co., Ltd.) as wild-type
vector pcDNA3.1-KMT5A-3′-UTR-WT (KMT5A-WT) and
pcDNA3.1-TWIST1-3′-UTR respectively. The mutant vector
pcDNA3.1-KMT5A-3′-UTR-Mut (KMT5A-R295G) was obtained by
site-directed mutagenesis using QuikChange®
Site-Directed Mutagenesis Kit (Stratagene; Agilent). MDA-MB-231
cells were seeded in a 24-well culture plate in triplicate and were
transfected followed by pcDNA3.1-KMT5A-3′-UTR-WT or
pcDNA3.1-KMT5A-3′-UTR-Mut or pcDNA3.1-TWIST1-3′-UTR by ExFect
Transfection Reagent (Vazyme Biotech Co., Ltd.) according to the
manufacturer's procedure.
Cells were seeded in 24-well plates at a density of
500 µl/well and transfected with the target plasmid (Table SIV). After 48 h at 37°C, the
luciferase activity of each group was detected by a dual luciferase
system (cat. no. E1910; Promega Corporation). The cell lysate was
supplemented with firefly luciferase (100 µl), after which firefly
luciferase activity was measured (F value). After adding
Renilla luciferase (100 µl), the cell lysate was immediately
placed into the fluorescence detector to calculate the
Renilla luciferase activity (R-value). Based on the F and R
values of each group detected previously, the relative luciferase
activity was calculated to get promoter activity. The primers for
the FBP1 promoter were: forward, 5′-GACAGAAGGGCCAGGTGA-3′ and
reverse, 5′-GCCAGAGAGAAAGCTATGACTG-3′; transcription start site:
(967–979 bp) AAGCCAGATGA.
Determination of glucose uptake and
lactate production
To detect glucose consumption and lactate production
in BRCA cells, a Glucose Uptake Colorimetric Assay Kit (cat. no.
K676-100; BioVision, Inc.; Abcam) and a Colorimetric/Fluorometric
Assay Kit (cat. no. K607-100; BioVision, Inc.; Abcam) were used
according to the manufacturer's instructions. A total of 2,000
MDA-MB-231 cells were seeded in each well of a 96-well plate and
incubated for 48 h at 37°C. After that, based on the Glucose Uptake
Colorimetric Assay Kit protocol, absorbance was measured at 412 nm
in a microplate reader at 37°C every 5 min until the 100 pmol
standard reached 1.5–2.0 OD. An endpoint reading of all samples and
standards was taken. The 2-deoxy-D-glucose 6P standard curve was
plotted and the glucose intake was calculated. Based on the
Colorimetric/Fluorometric Assay Kit protocol, absorbance was
measured at 570 nm in a microplate reader. The lactate standard
curve was plotted and sample lactate concentration was calculated.
The glucose and lactate levels were calculated using a standard
calibration curve prepared under the same conditions.
TMT labelling and bioinformatics
analysis
The peptide mixture (100 µg) of each sample was
labelled by TMT reagent according to the manufacturer's
instructions (Thermo Fisher Scientific, Inc.). TMT-labelled
peptides (10 µl) were fractionated by peptide fractionation with
reversed phase chromatography using the Agilent 1260 infinity II
HPLC. Each fraction was injected for nano LC-MS/MS analysis. The
peptide mixture was loaded onto the C18-reversed phase analytical
column (Thermo Fisher Scientific, Inc.; Acclaim PepMap RSLC 50 µm
×15 cm, nano viper, P/N164943) in buffer A (0.1% formic acid) and
separated with a linear gradient of buffer B (80% acetonitrile and
0.1% formic acid) at a flow rate of 300 nl/min. LC-MS/MS analysis
was performed on a Q Exactive Plus mass spectrometer (Thermo Fisher
Scientific, Inc.) that was coupled to Easy nLC (Thermo Fisher
Scientific, Inc.) for 60 min. MS/MS raw files were processed using
MASCOT engine (version 2.6; Matrix Science, Ltd.) embedded into
Proteome Discoverer 2.2, and searched against the UniProt database,
downloaded from https://www.uniprot.org/, including
HomoSapiens_20367_20200226 sequences. Proteins with Fold change
>1.2 and adjusted. P<0.05 (Student's t-test) were considered
to be differentially expressed proteins. The annotation from gene
ontology (GO) terms to proteins was completed by Blast2GO Command
Line. After the elementary annotation, InterProScan (https://www.ebi.ac.uk/interpro/) was used to
search the EBI database (https://www.ebi.ac.uk/) by motif and then add the
functional information of motif to proteins to improve annotation.
Then further improvement of annotation and connection between GO
terms were carried out (Table SV).
Fisher's Exact Test was used to enrich GO terms by comparing the
number of differentially expressed proteins and total proteins
associated to GO terms. Pathway analysis was performed using Kyoto
Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/pathway.html). Fisher's
Exact Test was used to identify the significantly enriched pathways
by comparing the number of differentially expressed proteins and
total proteins associated to pathways (Table SVI Table VII, Table VIII, Table IX, Table SX) and protein structure (Table SXI).
Source databases
The partial immunohistochemistry images and
interaction network were obtained from The Human Protein Atlas
(HPA, http://www.proteinatlas.org/)
(22). The gene expression levels
in healthy breast and BRCA were obtained from The University of
Alabama at Birmingham database (UALCAN, http://ualcan.path.uab.edu/index.html) (23). Some data were downloaded from the
TCGA database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga).
The expression data of KMT5A gene in MDA-MB-231 and MCF-7 cell
lines was downloaded from the ‘Expression 23Q4 Public’ dataset of
the Cancer Cell Line Encyclopedia database (CCLE, http://www.broadinstitute.org/ccle) (24).
Ethics
The present study was approved (approval no.
YS-2017-006) by the Ethics Committee of the Sixth People's Hospital
Affiliated to Shanghai Jiao Tong University School of Medicine
(Shanghai, China), and written informed consent was obtained from
each patient.
Statistical analysis
SPSS software 26.0 (IBM Corp.) was used for Pearson
correlation statistical analyses. Most of the data were analyzed
and bar charts were generated with GraphPad Prism 9.5 software
(Dotmatics). Volcano map was expressed by R studio (4.3.2)
(RStudio, Inc.). KEGG and GO analysis were drawn by bioinformatics
(https://www.bioinformatics.com.cn).
Data are presented as the mean ± standard deviation (minimum three
repeats, depending of the experiments). Comparisons between groups
were analyzed using one-way ANOVA followed by Bonferroni's (for
comparing within treatment conditions) or Dunnett's (for comparing
between treatment conditions) or Multiple Comparison Test.
P<0.05 was considered to indicate a statistically significant
deference.
Results
KMT5A is upregulated in BRCA
tissues
The UALCAN database was searched (https://ualcan.path.uab.edu/index.html)
(23) to determine the expression
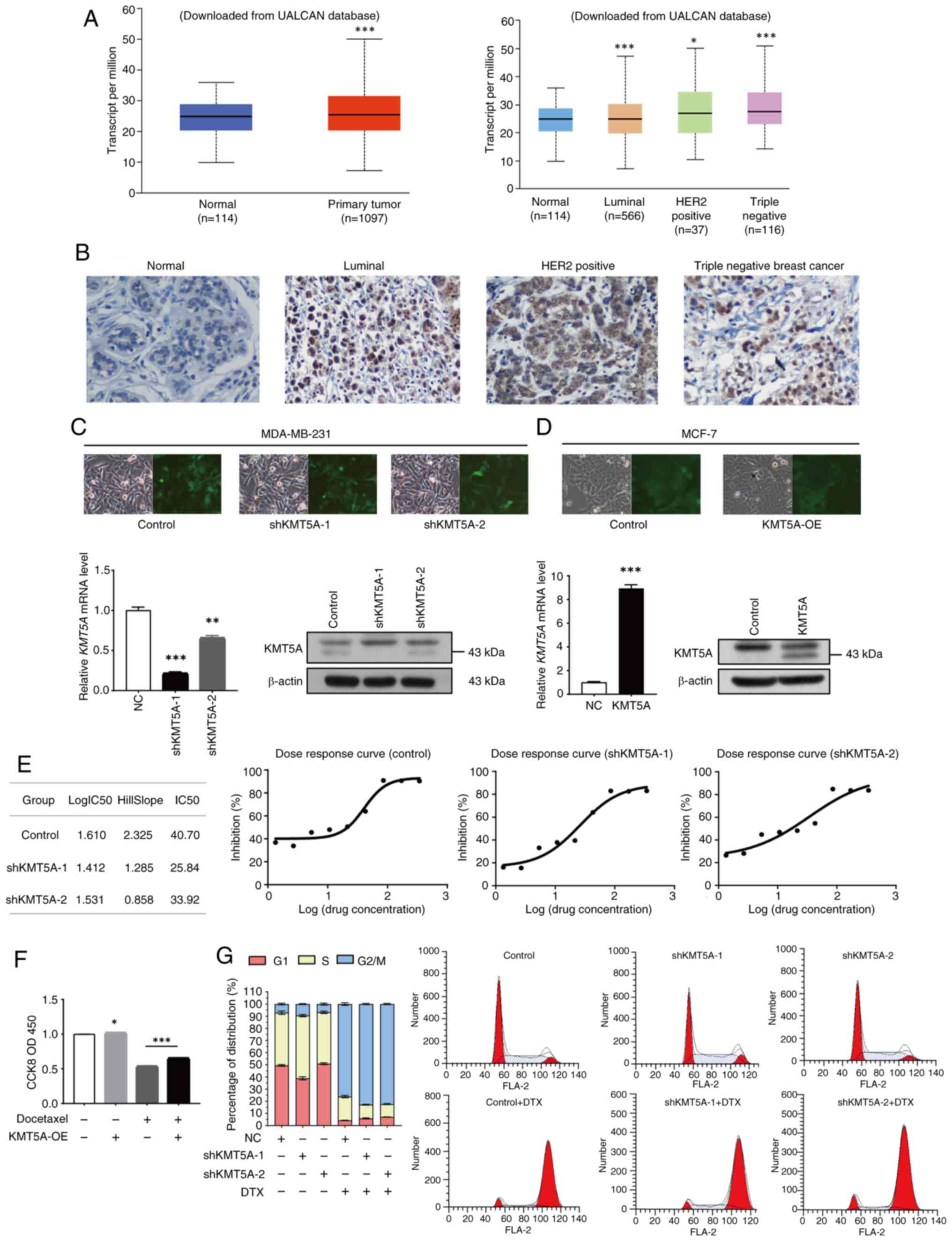
of KMT5A in BRCA. KMT5A was upregulated in BRCA (P<0.05). The
expression level of KMT5A was the highest in triple-negative BRCA
(P<0.05) and the lowest in the luminal subtype (P<0.05)
(Fig. 1A) (All data and
significance levels were from the UALCAN database). The
immunohistochemical results of 60 patients were included in the
analysis. Patients ranged in age from 31 to 82 years, with a median
age of 60 years. IHC also revealed that KMT5A was highly expressed
in BRCA tissues (Fig. 1A).
 | Figure 1.KMT5A mediates DTX resistance in BRCA
cells. (A) Expression of KMT5A in BRCA. (B) Representative images
of immunohistochemical staining of KMT5A in BRCA tumors. (C and D)
After 72-h infection, cells were observed, and images were captured
by fluorescence microscope. Magnification, ×100. Expression of
KMT5A in BRCA cells was detected by reverse
transcription-quantitative PCR and western blotting. (E) MTT assay
was performed to examine the cell viability and proliferation. (F)
The sensitivity of MCF-7 cell line overexpressing KMT5A to DTX was
detected by a CCK-8 assay. (G) Effect of KMT5A knockdown and/or DTX
on the cell cycle of the MDA-MB-231 cell line. *P<0.05;
**P<0.01 and ***P<0.001. KMT5A, lysine methyltransferase 5A;
DTX, docetaxel; BRCA, breast cancer; CCK-8, Cell Counting Kit-8;
UALCAN, the university of Alabama at Birmingham database; sh, short
hairpin; OE, overexpression; NC, negative control. |
Knockdown of KMT5A in MDA-MB-231 cells
and overexpression of KMT5A in MCF-7 cells
The expression of endogenous KMT5A was lower in
MCF-7 cell lines (5.35), however higher in MDA-MB-231 cell lines
(5.74) (Data was from ‘Expression 23Q4 Public’ dataset of the CCLE
database). Therefore, lentivirus-mediated KMT5A shRNA silenced
KMT5A expression in MDA-MB-231 cell line, and overexpressed KMT5A
mRNA in MCF-7 cell line. As demonstrated in Fig. 1B, the stably transfected MDA-MB-231
BRCA cell line was subsequently generated via lentivirus infection
and puromycin resistance screening and subsequently divided into
three groups for identification. The negative control (NC) group
was generated by stably transforming MDA-MB-231 cells with a
control lentiviral vector. The shKMT5A-1 and shKMT5A-2 group were
stably transformed into MDA-MB-231 cells via the
LVpFU-GW-007-mediated transfer of the lentiviral vector. KMT5A
knockdown was confirmed at the mRNA and protein levels. The
shKMT5A-1 and shKMT5A-2 groups exhibited significantly lower levels
of the KMT5A mRNA and protein (P<0.05) (Fig. 1C). Furthermore, the knockdown
efficiency of shKMT5A-1 was greater than that of shKMT5A-2.
Moreover, RT-qPCR and western blot analysis confirmed that KMT5A
was significantly highly expressed in MCF-7 cells transfected with
an overexpression lentiviral vector (Fig. 1D).
Effect of KMT5A on sensitivity to
chemical resistance and the cell cycle
To further confirm whether KMT5A knockdown affects
chemical resistance and the cell cycle, MDA-MB-231 cells were
treated with DTX, the half-maximal inhibitory concentration
(IC50) was measured, dose-response curves were
constructed for each group, and the proportions of cells in
different phases were evaluated. After KMT5A knockdown, the
IC50 of DTX decreased significantly (Fig. 1E). In the DTX positive KMT5A-OE
negative group, the proliferation rate of MCF-7 cells when DTX (20
µM) was added only was 53.9%. However, in the case of high
expression of KMT5A, the cell activity increased to 66.5% after DTX
addition compared with the DTX positive KMT5A-OE negative group. It
was revealed that high expression of KMT5A antagonized DTX and
promoted cell proliferation (Fig.
1F). KMT5A expression may mediate the development of DTX
resistance in BRCA cells.
In the control group, the proportion of G1-phase
cells was 49.51±0.71%, the proportion of S-phase cells was
43.43±1.38%, and the proportion of G2/M-phase cells was 7.06±0.67%.
The proportions of G1 phase cells, S phase cells, and G2/M phase
cells in the shKMT5A-1 group were 38.91±1.27, 51.60±0.93% and
9.49±0.37%, respectively. After adding DTX, the proportion of cells
in the G2/M phase increased, and the proportion of cells in the S
phase decreased significantly. In the control group, the proportion
of G2/M phase cells increased to 82.82±0.53%, and the proportion of
S phase cells decreased to 11.28±0.36%. The proportion of G2/M
phase cells in the shKMT5A-1 group was 76.15±1.04%, and the
proportion of S phase cells was 19.50±0.96%. After KMT5A knockdown,
the proportion of S-phase cells was markedly greater than that of
parental cells (Fig. 1G). First of
all, DTX is effective against various pathological types of BRCA,
including triple-negative BRCA, and remains the first-choice
chemotherapy drug for this type. The triple-negative BRCA cell line
(MDA-MB-231) is less sensitive to DTX than the hormone
receptor-positive MCF-7 cell line. Secondly, the mechanism of DTX
is to enhance the polymerization of tubulin and inhibit the
depolymerization of microtubules, leading to the formation of
stable non-functional microtubule bundles, thus disrupting the
mitosis of tumor cells. The present study revealed that MDA-MB-231
cell line was blocked in mitosis and remained in the G2/M phase,
which is consistent with its pharmacological effect and previous
findings (25).
The levels of monomethylated H4K20 and KMT5A changed
dynamically during different phases of the cell cycle, and these
dynamic changes regulated the progression of the cell cycle. KMT5A
and monomethylated H4K20 are expressed at low levels in the S
phase, and KMT5A and monomethylated H4K20 reach their peak
expression in the G2/M phase. The normal process of the S phase
requires KMT5A (26); therefore,
after KMT5A expression was knocked down, cells were arrested in the
S phase and could not enter the G2/M phase. However, DTX mainly
affects M-phase cells (27);
therefore, when DTX is added, the proportion of G2/M phase cells
was greater than that of parent cells (negative control without
DTX).
Bioinformatics analysis reveals that
KMT5A knockdown induces FBP1 upregulation
To identify the downstream proteins regulated by
KMT5A, protein expression levels were evaluated in the BRCA cell
line MDA-MB-231 transfected with NC lentivirus (negative control)
or KMT5A-targeting lentivirus (shKMT5A). A total of three samples
were randomly selected from NC and shKMT5A groups, and the protein
was quantitatively analyzed by TMT (Tandem Mass Tag) proteomics
technology and the differentially expressed protein was screened
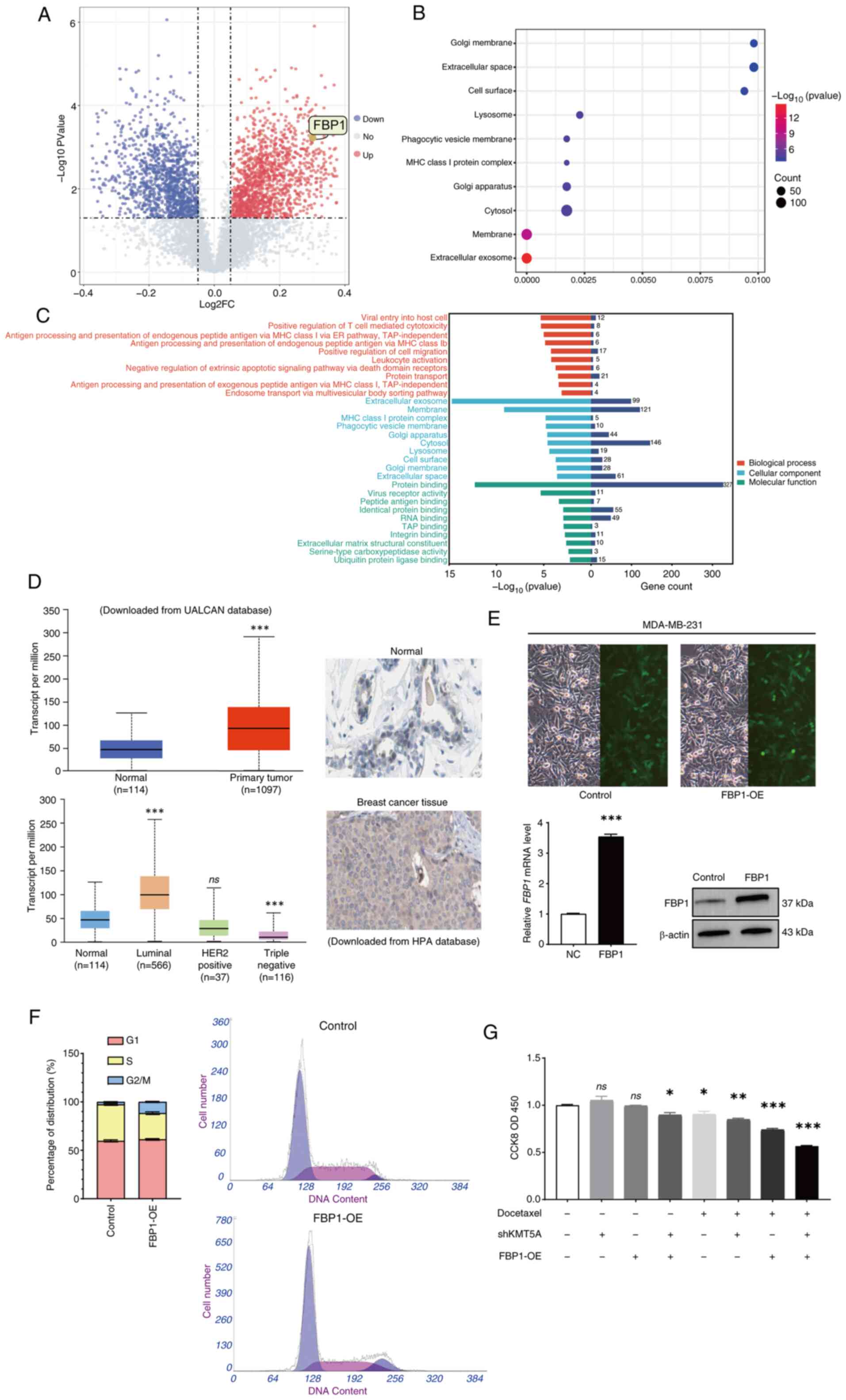
out. The analysis of differential gene expression revealed that the
expression levels of 261 proteins increased after KMT5A was knocked
down. Conversely, the expression of 153 proteins was downregulated
(Fig. 2A). As a key gluconeogenic
enzyme, FBP1 expression increased significantly (Fold change=1.23,
adjusted. P<0.05) after KMT5A was knocked down.
 | Figure 2.FBP1 is upregulated by KMT5A
knockdown. (A) Volcano map demonstrating 414 differentially
expressed proteins between control (n=3) and KMT5A-knockdown groups
(n=3). Red indicates upregulation, and blue indicates deregulation.
(B) Kyoto Encyclopedia of Genes and Genomes pathway enrichment
analysis of 414 target proteins. (C) GO analysis of 414 target
proteins. The colors represent GO functional categories (red:
Biological Process, green: Molecular Function, blue: Cellular
Component). The length of the bar chart to the right of the origin
indicates the number of different expressed proteins included in
the function. (D) Expression of FBP1 in breast cancer. (E) After 72
h of infection, cells were observed and images were captured by
fluorescence microscope. Magnification, ×100. Expression of FBP1 in
breast cancer cells was detected by reverse
transcription-quantitative PCR and western blotting. (F) The cell
cycle analysis of the FBP1-overexpressing group. (G) CCK-8 assay
was used to detect the cell viability and proliferation after the
addition of docetaxel. *P<0.05, **P<0.01 and ***P<0.001.
FBP1, fructose-1,6-bisphosphatase; KMT5A, lysine methyltransferase
5A; GO, Gene Ontology; CCK-8, Cell Counting Kit-8; DTX, docetaxel;
UALCAN, the university of Alabama at Birmingham database; HPA,
human protein atlas; sh-, short hairpin; NC, negative controls; OE,
overexpression; ns, no significance (P≥0.05). |
Pathway enrichment analysis demonstrated that KMT5A
regulates several pathways, including the extracellular exosome and
membrane pathways. In addition to these pathways being enriched, it
was revealed that they were tightly connected to chemoresistance
and glucose metabolism (Fig. 2B).
Moreover, the ‘extracellular exosome’ plate differs most
significantly among the cell components, and exosomes have been
revealed to lead to chemical resistance in various cancers and
regulate chemical resistance in several ways (Fig. 2C). Exosomes transport glycolytic
enzymes. According to the proteomic analysis results, exosomes lead
to metabolic reprogramming and promote alterations in both glucose
metabolism and lactate production.
Overexpressed FBP1 in MDA-MB-231
cells
UALCAN database was searched and was revealed that
the expression of FBP1was significantly increased in BRCA, however
there were significant differences among the different subtypes.
FBP1 was significantly increased in luminal BRCA (P<0.05) but
significantly decreased in triple-negative BRCA (P<0.05)
(Fig. 2D). IHC also demonstrated
that FBP1 was highly expressed in BRCA tissues (Fig. 2D). Inadequately, IHC cannot divide
into different subtypes. IHC results of FBP1 in BRCA and adjacent
normal breast tissue were obtained from the HPA database and could
not be differentiated into different subtypes for analysis.
MDA-MB-231 cells were transfected with the
lentivirus LVCON238 (negative control) or LVKL68505-2 (FBP1-OE).
RT-qPCR also revealed that the mRNA level of FBP1 was significantly
increased in cells transfected with the lentivirus LVKL68505-2
(FBP1-OE) (P<0.05) (Fig. 2E).
Western blot analysis revealed that FBP1 was markedly overexpressed
in MDA-MB-231 cells transfected with the lentivirus LVKL68505-2
(FBP1-OE) (Fig. 2E).
Effect of FBP1 on the cell cycle and
chemotherapy resistance
After overexpression of FBP1, the proportion of G2/M
phase cells increased significantly (Fig. 2F). DTX is a G2/M phase
cycle-specific drug, and overexpression of FBP1 may enhance the
proapoptotic effect of DTX on BRCA cells. Therefore, the effect of
overexpression of FBP1 was detected on cell proliferation using a
CCK-8 assay and MDA-MB-231 cells were subsequently treated with
DTX. After the addition of the DTX (20 µM), cell proliferation was
further inhibited in the FBP1 overexpression group (Fig. 2G).
FBP1 is downregulated by KMT5A
overexpression
TCGA-BRCA data were downloaded from the TCGA
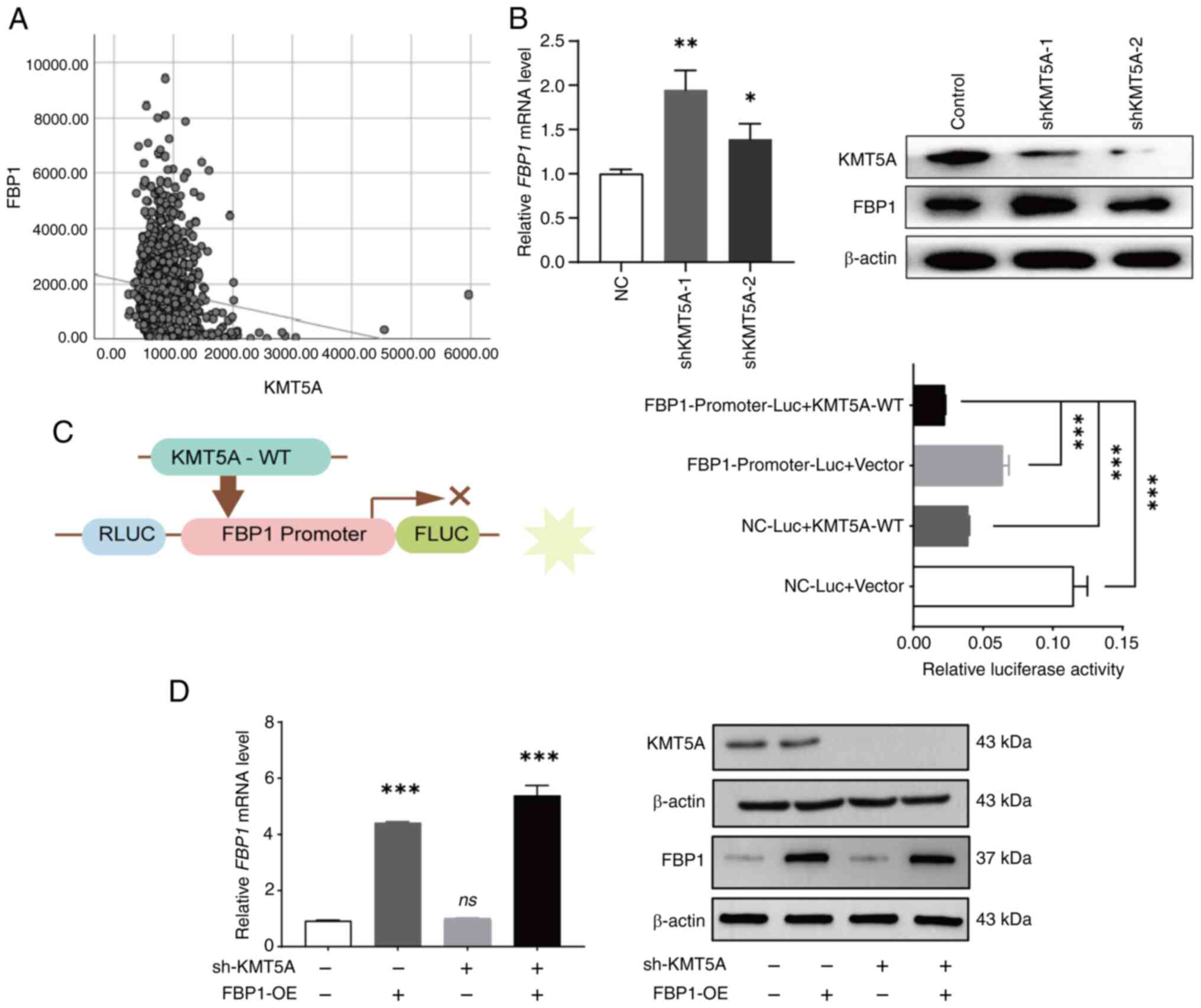
database. The expression of FBP1 was negatively correlated with
that of KMT5A [Pearson correlation coefficient (r=−0.131,
P<0.05] (Fig. 3A). To verify the
relationship between KMT5A and FBP1, the expression of FBP1 was
detected after knocking down KMT5A in MDA-MB-231 cell line. The
expression of FBP1 was increased in the shKMT5A group (Fig. 3B). A dual-luciferase reporter gene
assay was used to determine the regulatory relationship between
KMT5A and FBP1. The NC-Luc + KMT5A-WT group and the
FBP1-Promoter-Luc + vector group were designed to observe the
internal interaction between the reporter vector and the internal
reference vector to eliminate the background influence. The
luciferase activity of the FBP1-Promoter-Luc + KMT5A-WT group was
lower than that of the FBP1-Promoter-Luc+vector group and NC-Luc +
KMT5A-WT group. The results revealed that FBP1 demonstrated strong
promoter activity when KMT5A was not expressed. Thus, it was found
that KMT5A inhibits FBP1 after the exclusion of endogenous
influences (Fig. 3C).
 | Figure 3.KMT5A inhibits FBP1 expression. (A)
Correlation between the expression of KMT5A and FBP1 based on TCGA
cohort [Pearson correlation coefficient (r)=0.131, P<0.05]. (B)
The expression of FBP1 was detected by RT-qPCR and western blotting
after KMT5A was knocked down. (C) The relationship between KMT5A
and FBP1 was verified by dual-luciferase reporter gene assay. (D)
The effect of infection of shKMT5A and FBP1 lentivirus in the
breast cancer cells were detected by RT-qPCR and western blotting.
*P<0.05, **P<0.01 and ***P<0.001. KMT5A, lysine
methyltransferase 5A; FBP1, fructose-1,6-bisphosphatase; RT-qPCR,
reverse transcription-quantitative PCR; sh-, short hairpin; NC,
negative controls; WT, wild-type; RLUC, Renilla luciferase;
FLUC, firefly luciferase; Luc, luciferase; OE, overexpression; ns,
no significance (P≥0.05). |
FBP1 overexpression combined with
KMT5A knockdown inhibits MDA-MB-231 cell line proliferation
To determine whether FBP1 has a reverse regulatory
effect on KMT5A, a lentivirus was used to construct a NC group, an
FBP1 overexpression group (FBP1-OE), a KMT5A low-expression group
(shKMT5A) and an FBP1 overexpression + KMT5A low-expression group
(FBP1-OE + shKMT5A). Moreover, RT-qPCR and western blot analysis
successfully verified that FBP1 was overexpressed and KMT5A was
downregulated in the MDA-MB-231 cell line (Fig. 3D).
When DTX was not added, no matter whether KMT5A was
knocked down alone (shKMT5A) or FBP1 was overexpressed (FBP1-OE),
the proliferation inhibition rate of MDA-MB-231 cells was not
significantly different from that of the NC group. However, the
inhibitory rate of cell proliferation in the FBP1-OE + shKMT5A
group was 10.3±4.4%. After addition of DTX, the cell proliferation
inhibition rate was 26.1±2.6% in the FBP1-OE group alone, 15.2±2.7%
in the shKMT5A group alone, however increased to 43.4±1.3% in the
FBP1-OE + shKMT5A group. Neither the overexpression of FBP1 nor low
expression of KMT5A alone affected the proliferation of BRCA cells.
When KMT5A was simultaneously expressed at low levels and FBP1 was
overexpressed, the proliferation of BRCA cells was significantly
inhibited after DTX (20 µM) was added (Fig. 2G). It was also revealed that the
MDA-MB-231 cell activity decreased to ~55.8 and 41.0% respectively
after DTX addition without/with FBP1 overexpression. As
demonstrated in Fig. 2G, the
MDA-MB-231 cell activity decreased to ~91.4 and 73.9% respectively
after DTX addition without/with FBP1 overexpression. Therefore,
after the addition of DTX, overexpression of FBP1 or/and
downregulation of KMT5A jointly inhibited the proliferation of
MDA-MB-231 cells.
The effect of KMT5A on
chemotherapeutic drug resistance and glycolysis depends on
methylase activity
UNC0379 is a selective inhibitor of KMT5A, however
it can also target other histone methyltransferases apart from
KMT5A (28). The DTX groups contain
both DTX (20 µM) and DTX solvent. UNC0379 (0, 5 or 10 µM) was added
respectively the DTX solvent groups and DTX groups. As it can be
observed, UNC0379 can also cooperate with DTX to inhibit the
proliferation of BRCA cells. Moreover, the change in concentration
of UNC0379 can enhance the inhibitory effect on the proliferation
of BRCA cells. As the concentration UNC0379 was increased,
MDA-MB-231 cell proliferation was more strongly inhibited (Fig. 4A).
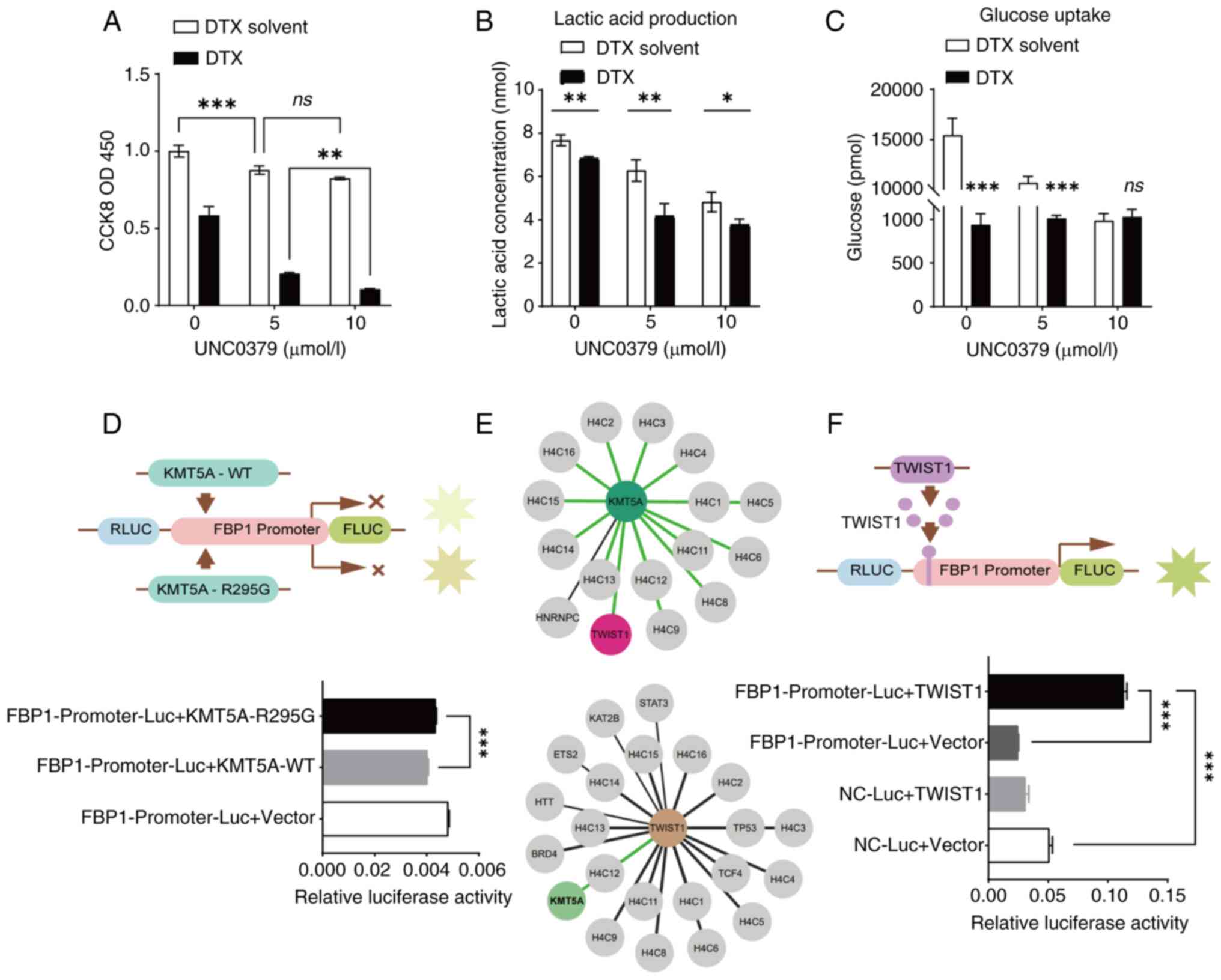 | Figure 4.KMT5A relies on methylase activity to
inhibit FBP1. (A) KMT5A inhibitor UNC0379 was used to treat breast
cancer cells to detect cell activity. (B and C) MDA-MB-231 cells
were treated with KMT5A inhibitor UNC0379 to detect lactic acid
production and glucose uptake. (D) The effect of KMT5A methylation
activity on the FBP1 promoter was detected by dual-luciferase
reporter gene assay. KMT5A-R295G is a mutant without methylase
activity. (E) The Human Protein Atlas database (https://www.proteinatlas.org/) revealed the
interaction networks of KMT5A and TWIST1. (F) Correlation between
TWIST1 and FBP1 promoter was detected by dual-luciferase reporter
gene assay. ns, P≥0.05; *P<0.05, **P<0.01 and ***P<0.001.
KMT5A, lysine methyltransferase 5A; FBP1,
fructose-1,6-bisphosphatase; TWIST1, twist family BHLH
transcription factor 1; DTX, docetaxel; WT, wild-type; RLUC,
Renilla luciferase; FLUC, firefly luciferase; Luc,
luciferase; ns, no significance (P≥0.05). |
As the concentration of the inhibitor UNC0379
increased, the amount of lactic acid produced by the BRCA cells
decreased (Fig. 4B). Moreover,
after the addition of the inhibitor UNC0379, the glucose uptake of
BRCA cells was also significantly reduced (Fig. 4C).
KMT5A inhibits FBP1 expression through
methylase activity
As revealed in Fig.
3C, FBP1 promoter activity decreased when KMT5A-WT was
overexpressed. To further verify whether KMT5A methylase activity
affects FBP1 transcription, a KMT5A mutant (KMT5A-R295G) was added
to the FBP1 promoter. Both KMT5A-WT and KMT5A-R295G inhibited FBP1
gene expression. Therefore, compared with the NC-Luc + Vector
(control) group, the relative activity of luciferase in both groups
decreased. The results also revealed that when KMT5A lost methylase
activity, FBP1 promoter activity increased compared with KMT5A-WT
group (Fig. 4D). This suggests that
KMT5A has an inhibitory effect on the activation and regulation of
FBP1 promoter depending on its methylase activity.
How does KMT5A affect FBP1 expression through
methylase activity? Yang et al (29) demonstrated that SET8 and TWIST
commonly promote epithelial-mesenchymal transformation and enhance
the invasive potential of BRCA cells. In BRCA, KMT5A (SETD8)
interacts with TWIST and synergistically upregulates N-Cadherin and
downregulates E-Cadherin at the transcriptional level. The HPA
database was utilized to search for an interaction network between
KMT5A and TWIST and discovered that there was indeed an interaction
between the two (Fig. 4E). Based on
the prediction of binding sites between FBP1 promoter and TWIST1,
dual-luciferase reporter gene experiment was used to verify whether
TWIST1 had an effect on promoter activation and regulation. Using
empty plasmids as controls, results revealed that overexpression of
TWIST1 can increase FBP1 promoter activity by ~5-fold, suggesting
that TWIST1 activates FBP1 promoter activity (Fig. 4F).
Discussion
Primary or acquired resistance to chemotherapy is a
major barrier to BRCA treatment. The main reasons for chemotherapy
failure include: i) Inadequate pharmacokinetic properties of the
drug (30); ii) tumor
cell-intrinsic factors, such as the expression of drug efflux
pumps, altered cellular homeostasis, and glucose metabolism; and
iii) the tumor microenvironment, characterized by hypoxia and
acidosis (31). The mechanism of
DTX resistance in BRCA is multifaceted. The present study focused
on drug resistance induced by changes in glucose metabolism, and
targeting glycolytic enzyme activities could be a useful strategy
for cancer therapy (32).
In the present study, it was revealed that KMT5A was
highly expressed in DTX-resistant BRCA cells. Analysis of protein
expression differences after KMT5A knockdown revealed significant
changes in signalling pathways related to glucose metabolism. FBP1,
a key gluconeogenic enzyme, was significantly upregulated.
Dual-luciferase reporter assays confirmed that KMT5A inhibited the
expression of FBP1, and glucose uptake and lactic acid production
experiments revealed that KMT5A inhibited gluconeogenesis and
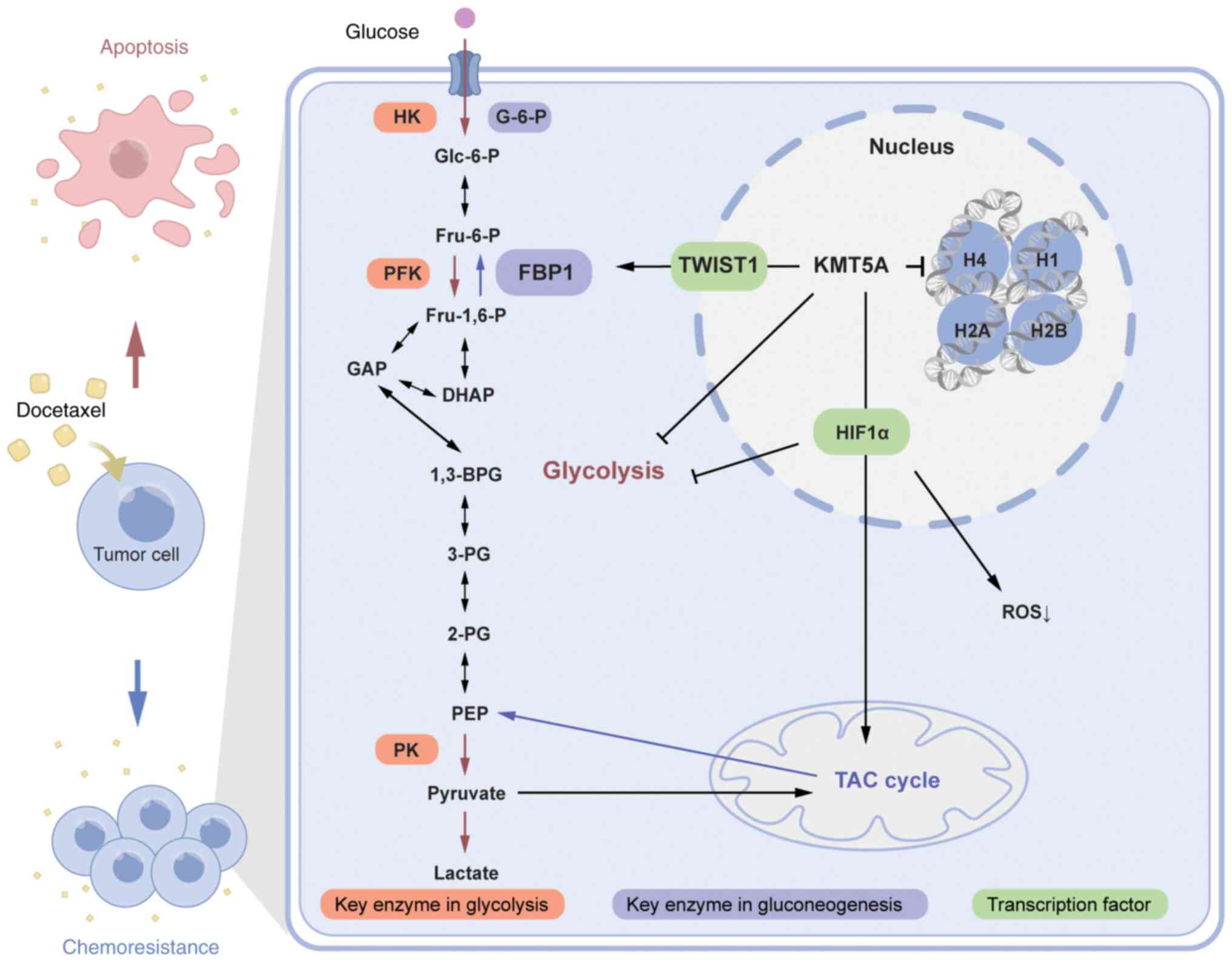
promoted anaerobic glycolysis of glucose. Because KMT5A inhibits
the transcription factor TWIST1, it was verified that TWIST1
promotes the expression of FBP1 through a dual-luciferase reporter
assay. Therefore, KMT5A reduces the ratio of G2/M phase cells and
reduces the sensitivity of cells to DTX by decreasing the ability
of TWIST1 to promote FBP1 expression. Conversely, after the
upregulation of FBP1 and the downregulation of KMT5A, the
sensitivity of MDA-MB-231 cells to DTX increased. Due to the lack
of experiments to directly verify the interaction between KMT5A and
TWIST1, the correlation between the two genes via several websites
were analysed and it was deduced that KMT5A indirectly affects
TWIST1. Moreover, Yang et al (29) revealed that KMT5A confers dual
transcriptional activity on TWIST1 in BRCA, which also confirms
this point.
A previous study by the authors demonstrated a
positive correlation between KMT5A and HIF1α and HIF1α target genes
and validated KMT5A as a novel metabolic reprogramming regulator
(20) (Fig. 5). Ectopic expression of miR-335 or
depletion of its target gene KMT5A can enhance the sensitivity of
paclitaxel-resistant BRCA cells to paclitaxel (32). Additionally, overexpression of KMT5A
induces DTX chemoresistance in BRCA. However, the possible
mechanism through which KMT5A promotes chemotherapy resistance in
BRCA through regulating glucose metabolism has not been reported,
and the present study research shed additional light on this
topic.
Some limitations should also be considered and
discussed when interpreting these results and presenting their
contributions. First, on account of cumulative evidence, It was
concluded that KMT5A plays an important role in BRCA development by
inducing cancer cell multiplication and chemotherapy resistance.
However, additional functional experiments are needed to further
explore the complex underlying mechanisms in vitro and in
vivo. Due to the small sample size of the present study's
trial, the expression level of KMT5A in different subtypes of BRCA
was revealed through the UALCAN database and the IHC of different
subtypes of BRCA was not discussed. While KMT5A may decrease the
sensitivity of BRCA to DTX, it may also promote the metastasis of
some types of BRCA. For example, Yu et al (33) discovered that KMT5A expression is
significantly associated with activated Hippo/YAP signaling and
promoted triple-negative BRCA metastasis. Moreover, due to the
relatively small sample size of the survival and KMT5A expression
analyses, replication studies in larger populations and more
ethnicities are needed to validate these results.
In conclusion, the findings of the present study
confirmed that KMT5A induces DTX resistance in BRCA by affecting
glucose metabolism and that the knockdown or inhibition of KMT5A
expression may reverse chemotherapy resistance in patients and
improve the prognosis of patients with BRCA. Future studies should
focus on investigating the detailed mechanisms by which KMT5A
regulates the activity of other metabolic pathways and its possible
regulatory role in other tumors.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Science
Foundation of China (grant no. 8177101282).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. The data generated in the
present study may be found in the iProX database under accession
number IPX0008291000 or at the following URL: https://www.iprox.cn//page/SCV017.html?query=IPX0008291000.
Authors' contributions
XP, LM, XC, FT and XZ made substantial contributions
to the conception. XZ made substantial contributions to the study
design. XP and LM participated in the laboratory work and data
analyses. XP and XZ confirm the authenticity of all the raw data.
All authors participated in drafting or revising the content for
important intellectual content, and read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
YS-2017-006) by the ethics committee of Shanghai Sixth Peoples
Hospital Affiliated to Shanghai Jiao Tong University School of
Medicine (Shanghai, China). Written informed consent was obtained
from all the participants before the enrolment and publication of
the present study. The present study was certifiably performed
following the 1964 declaration of HELSINKI and later
amendments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Use of artificial intelligence tools
During the preparation of this work, artificial
intelligence tools were used to improve the readability and
language of the manuscript, and subsequently, the authors revised
and edited the content produced by the artificial intelligence
tools as necessary, taking full responsibility for the ultimate
content of the present manuscript.
Glossary
Abbreviations
Abbreviations:
|
BRCA
|
breast cancer
|
|
DTX
|
docetaxel
|
|
FBP1
|
fructose-1,6-bisphosphatase
|
|
IC50
|
half maximal inhibitory
concentration
|
|
KMT5A
|
lysine methyltransferase 5A
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
References
|
1
|
Miller KD, Nogueira L, Devasia T, Mariotto
AB, Yabroff KR, Jemal A, Kramer J and Siegel RL: Cancer treatment
and survivorship statistics, 2022. CA Cancer J Clin. 72:409–436.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:E279–E289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vasan N, Baselga J and Hyman DM: A view on
drug resistance in cancer. Nature. 575:299–309. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen H, Zhang M and Deng Y: Long noncoding
RNAs in Taxane resistance of breast cancer. Int J Mol Sci.
24:122532023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sajid A, Rahman H and Ambudkar SV:
Advances in the structure, mechanism, and targeting of
chemoresistance-linked ABC transporters. Nat Rev Cancer.
23:762–779. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cao X, Hou J, An Q, Assaraf YG and Wang X:
Towards the overcoming of anticancer drug resistance mediated by
p53 mutations. Drug Resist Updat. 49:1006712020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Di Pietro A, Dayan G, Conseil G, Steinfels
E, Krell T, Trompier D, Baubichon-Cortay H and Jault J:
P-glycoprotein-mediated resistance to chemotherapy in cancer cells:
Using recombinant cytosolic domains to establish structure-function
relationships. Braz J Med Biol Res. 32:925–939. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bukowski K, Kciuk M and Kontek R:
Mechanisms of multidrug resistance in cancer chemotherapy. Int J
Mol Sci. 21:32332020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dominiak A, Chełstowska B, Olejarz W and
Nowicka G: Communication in the cancer microenvironment as a target
for therapeutic interventions. Cancers (Basel). 12:12322020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bishayee K, Lee SH and Park YS: The
Illustration of altered glucose dependency in drug-resistant cancer
cells. Int J Mol Sci. 24:139282023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu L, Zhang L, Sun J, Hu X, Kalvakolanu
DV, Ren H and Guo B: Roles for the methyltransferase SETD8 in DNA
damage repair. Clin Epigenetics. 14:342022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Al Temimi AHK, Martin M, Meng Q, Lenstra
DC, Qian P, Guo H, Weinhold E and Mecinović J: Lysine Ethylation by
histone lysine methyltransferases. Chembiochem. 21:392–400. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kukita A, Sone K, Kaneko S, Kawakami E,
Oki S, Kojima M, Wada M, Toyohara Y, Takahashi Y, Inoue F, et al:
The Histone Methyltransferase SETD8 regulates the expression of
tumor suppressor genes via H4K20 methylation and the p53 signalling
pathway in endometrial cancer cells. Cancers (Basel). 14:53672022.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wada M, Kukita A, Sone K, Hamamoto R,
Kaneko S, Komatsu M, Takahashi Y, Inoue F, Kojima M, Honjoh H, et
al: Epigenetic modifier SETD8 as a therapeutic target for
high-grade serous ovarian cancer. Biomolecules. 10:16862020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liao T, Wang YJ, Hu JQ, Wang Y, Han LT, Ma
B, Shi RL, Qu N, Wei WJ, Guan Q, et al: Histone methyltransferase
KMT5A gene modulates oncogenesis and lipid metabolism of papillary
thyroid cancer in vitro. Oncol Rep. 39:2185–2192.
2018.PubMed/NCBI
|
|
16
|
Herviou L, Ovejero S, Izard F,
Karmous-Gadacha O, Gourzones C, Bellanger C, De Smedt E, Ma A,
Vincent L, Cartron G, et al: Targeting the methyltransferase SETD8
impairs tumor cell survival and overcomes drug resistance
independently of p53 status in multiple myeloma. Clin Epigenetics.
13:1742021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li B, Qiu B, Lee DS, Walton ZE, Ochocki
JD, Mathew LK, Mancuso A, Gade TP, Keith B, Nissim I and Simon MC:
Fructose-1,6-bisphosphatase opposes renal carcinoma progression.
Nature. 513:251–255. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li H, Li M, Pang Y, Liu F, Sheng D and
Cheng X: Fructose-1,6-bisphosphatase-1 decrease may promote
carcinogenesis and chemoresistance in cervical cancer. Mol Med Rep.
16:8563–8571. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li H, Qi Z, Niu Y, Yang Y, Li M, Pang Y,
Liu M, Cheng X, Xu M and Wang Z: FBP1 regulates proliferation,
metastasis, and chemoresistance by participating in C-MYC/STAT3
signalling axis in ovarian cancer. Oncogene. 40:5938–5949. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang R, Yu Y, Zong X, Li X, Ma L and
Zheng Q: Monomethyltransferase SETD8 regulates breast cancer
metabolism via stabilizing hypoxia-inducible factor 1α. Cancer
Lett. 390:1–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Thul PJ and Lindskog C: The human protein
atlas: A spatial map of the human proteome. Protein Sci.
27:233–244. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ghandi M, Huang FW, Jané-Valbuena J,
Kryukov GV, Lo CC, McDonald ER III, Barretina J, Gelfand ET,
Bielski CM, Li H, et al: Next-generation characterization of the
cancer cell line encyclopedia. Nature. 569:503–508. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kenicer J, Spears M, Lyttle N, Taylor KJ,
Liao L, Cunningham CA, Lambros M, MacKay A, Yao C, Reis-Filho J and
Bartlett JM: Molecular characterisation of isogenic taxane
resistant cell lines identify novel drivers of drug resistance. BMC
Cancer. 14:7622014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jørgensen S, Elvers I, Trelle MB, Menzel
T, Eskildsen M, Jensen ON, Helleday T, Helin K and Sørensen CS: The
histone methyltransferase SET8 is required for S-phase progression.
J Cell Biol. 179:1337–1345. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sousa-Pimenta M, Estevinho LM, Szopa A,
Basit M, Khan K, Armaghan M, Ibrayeva M, Sönmez Gürer E, Calina D,
Hano C and Sharifi-Rad J: Chemotherapeutic properties and
side-effects associated with the clinical practice of terpene
alkaloids: Paclitaxel, docetaxel, and cabazitaxel. Front Pharmacol.
14:11573062023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma A, Yu W, Li F, Bleich RM, Herold JM,
Butler KV, Norris JL, Korboukh V, Tripathy A, Janzen WP, et al:
Discovery of a selective, substrate-competitive inhibitor of the
lysine methyltransferase SETD8. J Med Chem. 57:6822–6833. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang F, Sun L, Li Q, Han X, Lei L, Zhang H
and Shang Y: SET8 promotes epithelial-mesenchymal transition and
confers TWIST dual transcriptional activities. EMBO J. 31:110–123.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Cao C, Tan X, Liao X, Du X, Wang
X, Liu T, Gong D, Hu Z and Tian X: SETD8, a frequently mutated gene
in cervical cancer, enhances cisplatin sensitivity by impairing DNA
repair. Cell Biosci. 13:1072023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Phan LM, Yeung SC and Lee MH: Cancer
metabolic reprogramming: Importance, main features, and potentials
for precise targeted anticanceranti-cancer therapies. Cancer Biol
Med. 11:1–19. 2014.PubMed/NCBI
|
|
32
|
Wang Y, Wang H, Ding Y, Li Y, Chen S,
Zhang L, Wu H, Zhou J, Duan K, Wang W, et al: N-peptide of vMIP-II
reverses paclitaxel-resistance by regulating miRNA-335 in breast
cancer. Int J Oncol. 51:918–930. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu B, Su J, Shi Q, Liu Q, Ma J, Ru G,
Zhang L, Zhang J, Hu X and Tang J: KMT5A-methylated SNIP1 promotes
triple-negative breast cancer metastasis by activating YAP
signaling. Nature Commun. 13:21922022. View Article : Google Scholar : PubMed/NCBI
|