Introduction
Circulating tumor cells (CTCs) are shed from primary
tumors and circulate in the peripheral blood. CTCs are considered
the origin of metastases, and analyzing CTCs holds the potential
for early metastasis detection and monitoring of therapeutic
effects in malignant tumors (1,2). Among
the available CTC detection devices, CellSearch (Veridex, LLC) is a
semi-automatic system that utilizes antibodies against epithelial
cell adhesion molecule (EpCAM), and is the only Food and Drug
Administration (FDA)-approved device for clinical CTC detection
(3). CellSearch consistently yields
reproducible results and demonstrates its clinical relevance in
epithelial cancers (4,5). However, epithelial tumor cells undergo
epithelial-mesenchymal transition (EMT) to enhance their migration
and invasiveness, resulting in the downregulation of epithelial
markers such as EpCAM (6).
Consequently, EpCAM-dependent systems, including CellSearch, may be
limited in that they cannot capture CTCs undergoing EMT (EMT-CTCs),
which is a pivotal subtype implicated in metastasis (7,8). To
address this limitation, a novel polymeric microfluidic device
‘CTC-chip’ (9) was developed based
on the conventional CTC-chip designed by Nagrath et al
(10). The CTC-chip can capture
diverse types of CTCs expressing targeted tumor cell antigens by
easily attaching various antibodies to numerous microposts on the
chip surface (11–13), and its clinical utility was
previously reported by our group (14,15).
In a previous study by our group, CTCs were undetectable in 40% of
patients with lung cancer and metastases, indicating the need to
develop an EpCAM-independent capture system using other antibodies
(15). Recently, Satelli et
al (16) identified
cell-surface vimentin (CSV) as an EMT-CTC marker. The CSV
monoclonal antibody clone 84-1 primarily associates with tumor
cells and demonstrates clinical utility (17–19).
In the present study, CSV expression was validated in lung and
colon cancer cell lines, and the capture efficiency of this
polymeric CTC-chip coated with an anti-CSV antibody was assessed as
a novel marker for mesenchymal CTCs, with the aim of establishing
an EMT-CTC capture system.
Materials and methods
Cell lines
The human lung cancer cell lines PC9 (Riken
BioResource Research Center) and H441 [American Type Culture
Collection (ATCC)], and the human colorectal cancer cell line DLD1
(ATCC) were cultured in RPMI-1640 medium (Wako Pure Chemical
Industries) supplemented with 10% fetal bovine serum (FBS; Thermo
Fisher Scientific, Inc.). The human lung cancer cell line A549
(ATCC) was cultured in Eagle's minimal essential medium (Wako Pure
Chemical Industries) supplemented with 10% FBS and 1% non-essential
amino acids (Wako Pure Chemical Industries). All cells were
cultured in a humidified atmosphere containing 5% CO2 at
37°C.
Flow cytometry
Adhered cells were harvested using 1 mM EDTA (Wako
Pure Chemical Industries) in PBS at 37°C to maintain cell surface
protein integrity, as it was found that CSV is degraded by trypsin
treatment (20). Subsequently,
cells were blocked with Protein Block (Dako) for 15 min and then
sequentially incubated with primary antibodies, including an
anti-EpCAM antibody (clone: HEA125; cat. no. sc-59906; Santa Cruz
Biotechnology, Inc.), an anti-CSV antibody (clone: 84-1; cat. no.
H00007431-M08; Abnova), an anti-E-cadherin antibody (clone: 36;
cat. no. 610182; BD Biosciences), or an anti-Vimentin antibody
(clone: RV202; cat. no. 562337; BD Biosciences), each diluted to
1:100, for 60 min at room temperature. Following primary antibody
incubation, the cells were incubated with a goat anti-mouse IgG
antibody conjugated with FITC (cat. no. 349031; BD Biosciences),
diluted to 1:20, for 30 min at room temperature. Flow cytometry was
performed using an EC800 cell analyzer (Sony Biotechnology) and
FlowJo software version 10 (Becton-Dickinson and Co.) was used for
data analysis. The mean fluorescence intensity was determined by
comparison with the negative control.
Preparation of CTC-chip
The polymeric CTC-chip system was utilized following
a two-step coating process with antibodies to capture CTCs, as
previously described (11).
Initially, the chip (cat. no. G4B442; Cytona, Inc.) was incubated
overnight at 4°C with a goat anti-mouse IgG antibody (cat. no.
1031-01; SouthernBiotech) in PBS at a concentration of 200 µg/ml.
Subsequently, the chip was incubated for 60 min at room temperature
with either an anti-EpCAM antibody (20 µg/ml; clone HEA125) or an
anti-CSV antibody (10 and 100 µg/ml; clone 84-1) to capture the
tumor cells. The chip coated with the respective antibody was
denoted as the ‘EpCAM-chip’ and ‘CSV-chip’, respectively. After
incubation, the chip surface was washed with PBS and kept moist.
The chip is typically used following a 60-min antibody
reaction.
Sample preparation and evaluation of
cell-capture efficacy
Cells were labeled with the CellTrace™
carboxyfluorescein succinimidyl ester (CFSE) Cell Proliferation Kit
(Thermo Fisher Scientific, Inc.) and suspended in 1 ml PBS
containing 5% bovine serum albumin (Nacalai Tesque, Inc.) or blood
sampled from a healthy volunteer (100 cells/ml). The cell
suspension sample (1 ml) was applied to the CTC-chip and sent to
the chip using a syringe pump at a constant flow rate of 1.0 ml/h.
Images and videos of the cells on the chip were captured using a
fluorescence microscope CKX41 (Olympus Corp.) and a digital video
camera (Sony Biotechnology, Inc.), respectively. The actual number
of cells (N-total) that were sent into the chip was determined by
counting the number of cells that passed through the chip inlet and
the number of captured cells (N-captured) was determined by
counting the CFSE-labelled cells remaining on the chip. Cell
capture efficiency was calculated as N-captured/N-total. The
healthy volunteer (author MK) donated blood and provided formal
written informed consent for its use in this study for experimental
purposes. Data collection from healthy subjects was included in the
Ethics Committee approval (approval no. H26-15).
Statistical analysis
The average and standard error of the capture
efficiency were calculated from 3 independent experimental repeats.
All analyses were performed using SPSS (version 27.0; IBM
Corp.).
Results
Classification of cell lines based on
E-cadherin and vimentin expression
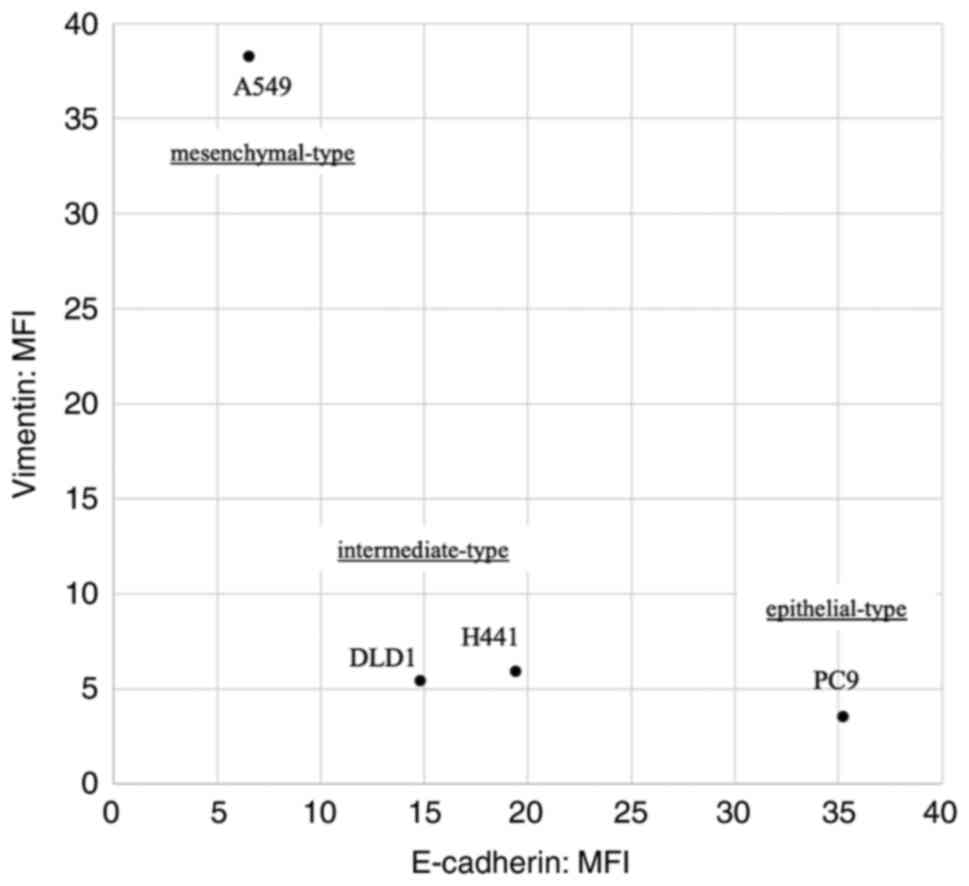
The classification of the cell lines based on flow
cytometric analysis of E-cadherin and vimentin expression is
summarized in Fig. 1. According to
these results, the cell lines were classified into three types:
Epithelial (PC9; high E-cadherin expression and low vimentin
expression), mesenchymal (A549; low E-cadherin expression and high
vimentin expression) and intermediate (H441 and DLD1).
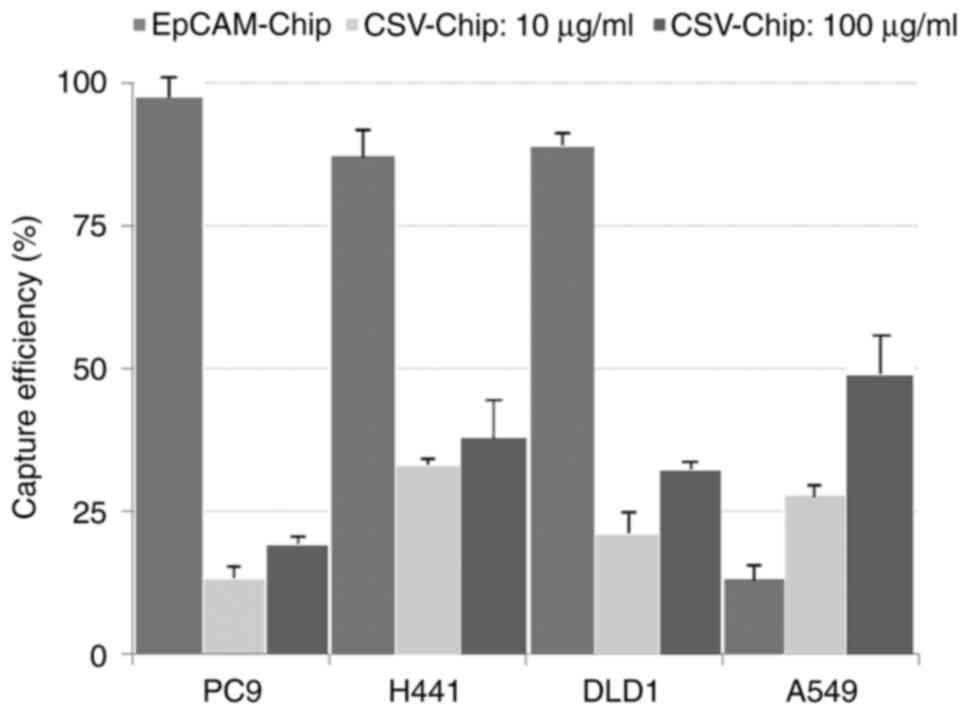
EpCAM expression and capture
efficiency
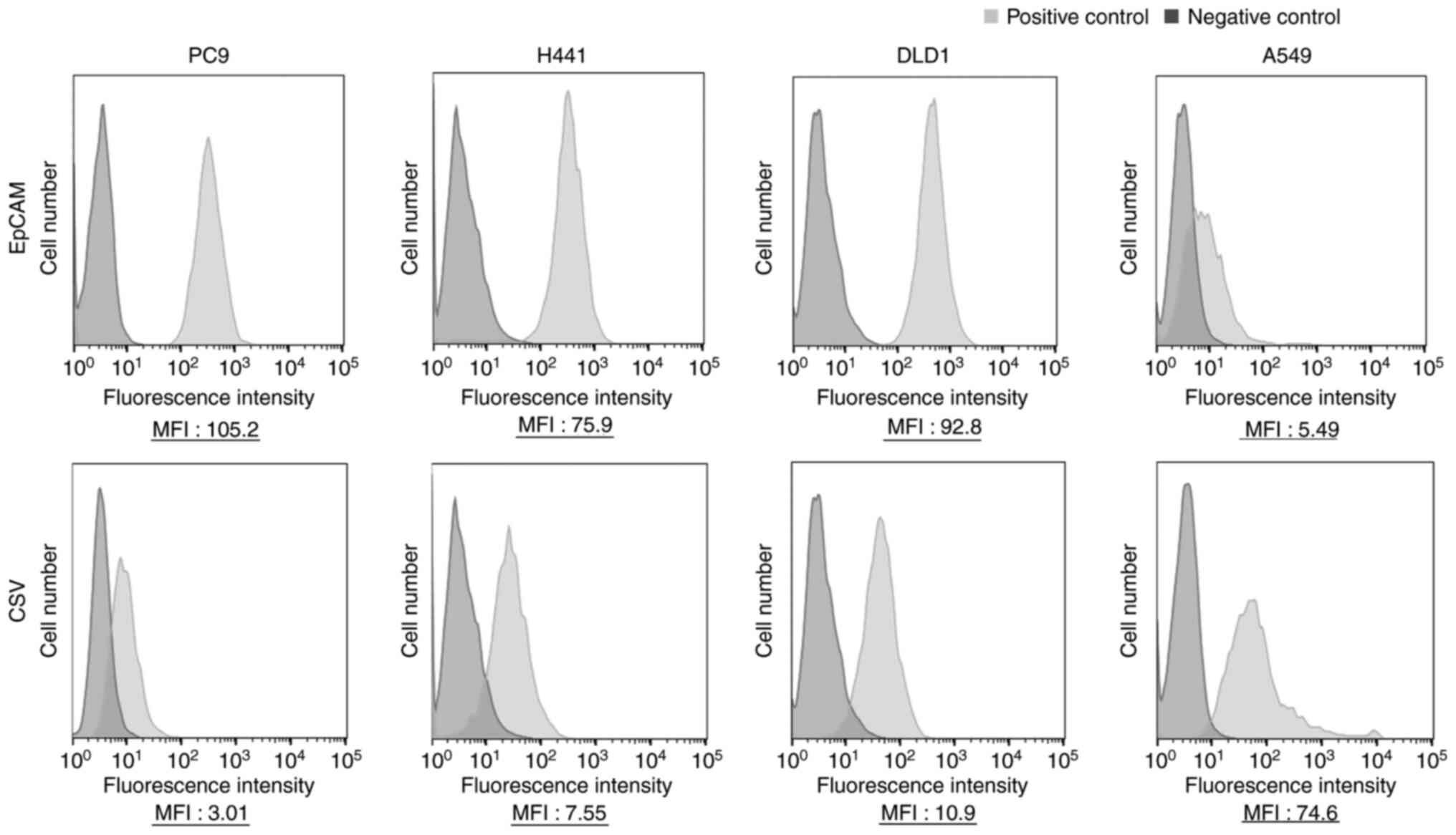
EpCAM was strongly expressed in the epithelial and
intermediate types but weakly expressed in the mesenchymal type
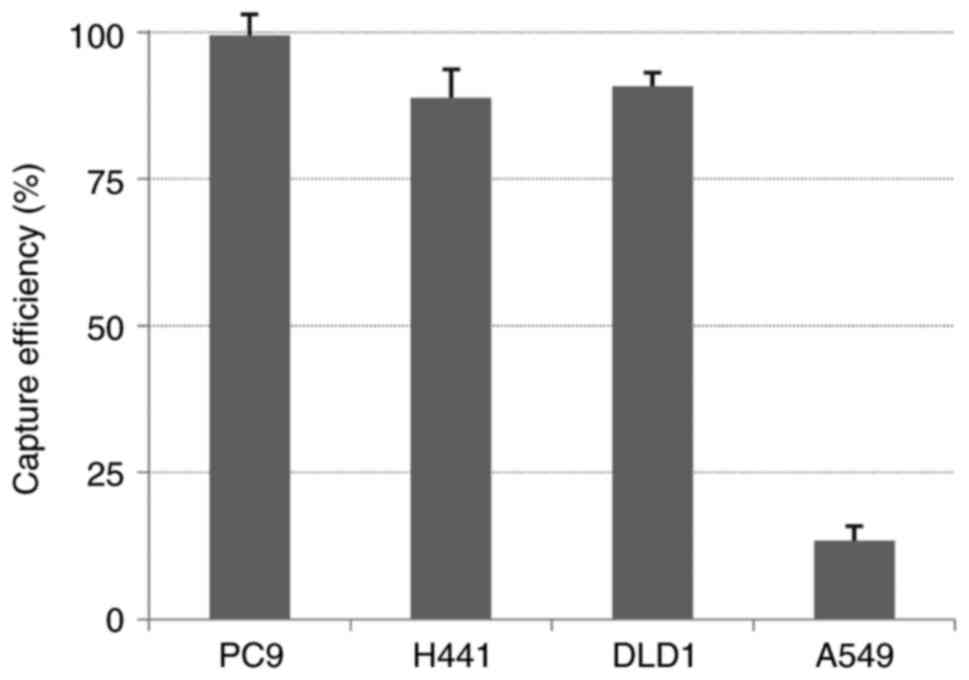
(Fig. 2). The capture efficiency of
the EpCAM-chip was in accordance with the expression intensity of
EpCAM. High capture rates were observed in the epithelial and
intermediate types (average capture efficiencies: PC9, 99.4%; H441,
88.8%; and DLD1, 90.8%), and a low capture rate was observed in the
mesenchymal type (average capture efficiency: A549, 13.4%)
(Fig. 3).
CSV expression and capture
efficiency
The expression of CSV was inversely in accordance
with that of EpCAM and increased in the following order:
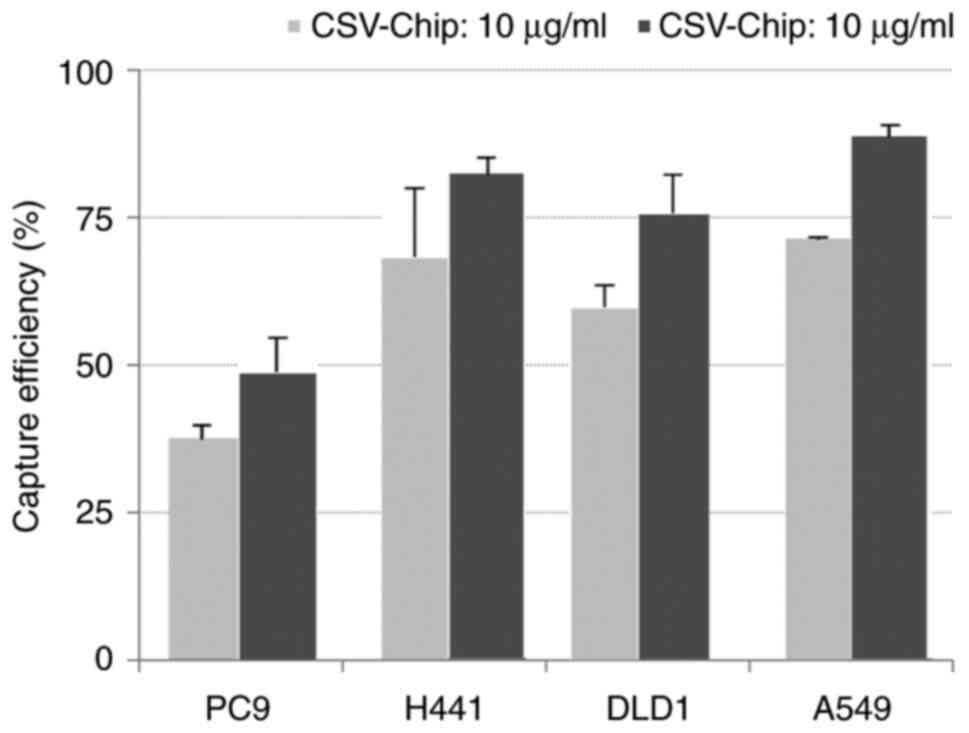
Mesenchymal, intermediate and epithelial (Fig. 3). The results obtained by examining
the capture efficiency of the CSV-chip when tumor cells were spiked
in PBS or blood at two concentrations (anti-CSV antibody: 10 and
100 µg/ml) are provided in Figs. 4
and 5. When spiked in PBS, the
average capture efficiencies of PC9, H441, DLD1 and A549 were 37.1,
68.2, 60.4 and 72.4%, at a concentration of 10 µg/ml, and 48.7,
82.1, 75.5 and 88.4%, at a concentration of 100 µg/ml, respectively
(Fig. 4). When spiked in blood, the
average capture efficiencies were 13.7, 33.0, 21.2 and 27.7% at a
concentration of 10 µg/ml, and 19.5, 38.2, 33.0 and 46.8% at a
concentration of 100 µg/ml (Fig.
5), respectively. The capture efficiency of the CSV-chip was
also in accordance with the expression intensity of CSV.
Discussion
The present study focused on CSV, a novel marker for
EMT-CTCs, as an alternative to traditional EpCAM-based capture
methods. The findings demonstrated that the CSV-chip effectively
captured mesenchymal-type cells, which are often overlooked by
EpCAM-dependent systems. This advancement represents a significant
improvement in cancer diagnostics and monitoring, offering a more
comprehensive tool for detecting metastatic cancer and studying
EMT-CTCs, which are pivotal subtypes involved in invasive growth
and metastatic spread.
Histological evaluation remains a cornerstone of
oncology diagnostics, providing essential insight into prognosis
and treatment planning for various cancer types. Traditionally,
this method relies on invasive tissue biopsies, which pose several
challenges, including patient discomfort, potential complications
and the risk of sampling errors due to tumor heterogeneity. By
contrast, liquid biopsies offer a less invasive alternative by
enabling real-time monitoring, comprehensive profiling and early
detection of relapse (21,22). Although CTCs are promising
biomarkers for liquid biopsies, their clinical utility has been
limited by low detection rates. The CellSearch system, which is the
only FDA-approved device for clinical CTC detection, relies on an
EpCAM-based capture method (3) that
can overlook tumor cells that have undergone EMT. This limitation
underscores the need for alternative detection methods and further
refinements (7,8).
EMT is a critical process in tumor invasion and
metastasis (6), which highlights
the necessity for EpCAM-independent capture systems. Vimentin, a
key marker of EMT, has been associated with tumor metastasis
(23–25). Traditionally, the expression of
vimentin in blood cells has previously restricted its use in CTC
capture (26). However, recent
research has identified a subset of aggressive CTCs that express
vimentin on their surface (CSV) and exhibit an EMT phenotype. This
finding suggests a higher likelihood of tumor recurrence and a
poorer prognosis for patients with CSV-positive CTCs (19). The development and validation of
monoclonal antibody clone 84-1, specific to CSV, represents a
significant advancement in targeting and capturing these cells
(17–19).
In the present study, by conjugating anti-CSV
antibodies to a universal CTC-chip designed to accommodate various
antibodies, a system for capturing EMT-CTCs was developed. This
system allowed for a more nuanced understanding of the role of
CSV-positive CTCs in cancer progression and metastasis. The
comprehensive classification of lung and colon cancer cell lines
based on EMT markers revealed that EpCAM was strongly expressed in
epithelial-type cells but was significantly reduced in
mesenchymal-type cells. Conversely, CSV expression was more
prevalent in cells with mesenchymal traits, highlighting its
potential as a specific marker for mesenchymal tumors.
Despite these encouraging results, the CSV-chip
demonstrated only moderate capture efficiency when cells were
spiked into blood samples, likely due to the inhibitory effects of
blood components on antigen-antibody interactions (12). This suggests that, while the
CSV-chip shows considerable promise, optimizing its conditions and
making additional refinements are essential for enhancing its
clinical performance. The present study identified a CSV
concentration of 100 µg/ml as optimal based on blood-based
concentration studies; however, further research is required to
validate this concentration for clinical applications. In addition,
pretreatment processes such as hemolysis, CD45 depletion or
peripheral blood mononuclear cell fractionation should be further
explored to enhance the capture efficiency and ensure more accurate
detection.
Recent studies have highlighted the increased counts
of CTCs and the higher proportions of CSV-positive CTCs in patients
with lymph nodes or distant metastases (27). Notably, patients with CSV-positive
CTCs experienced significantly shorter disease-free survival
compared to those with EpCAM-targeted CTCs, whose prognosis
remained unaffected (18,28,29).
These findings underscore the critical role of CSV-positive CTCs in
patient prognosis and support ongoing investigations into antibody
therapeutics targeting CSV to disrupt tumor-forming cells (30,31).
Our previous reports examining the prognostic significance of
EpCAM-CTCs have demonstrated the clinical impact of CTC counts
(15), and the study of CSV-CTCs
provides an opportunity to gain deeper insights into the
complexities of CTCs and their role in cancer progression.
Despite these promising results, one limitation of
the present study was the absence of clinical sample data. To
address this limitation, work is in progress to condition and
collect additional clinical samples for analysis to validate the
efficacy and clinical relevance of the CSV-chip in real-world
settings. This validation is essential for the broader application
and acceptance of the CSV-chip in clinical practice. The ongoing
investigation into CSV-CTCs is expected to refine our understanding
of CTCs in clinical contexts and potentially enhance the prognostic
and therapeutic strategies in cancer care.
In conclusion, the development of the CSV-chip
represents a significant advancement in CTC detection technology.
By enabling the capture of a broader range of CTCs, including those
undergoing EMT, the CSV-chip enhances our ability to monitor and
understand metastasis more effectively. Future research should
focus on validating these findings in patient samples and
integrating this technology with downstream molecular analyses.
This approach will provide deeper insights into the biology of
metastasis and guide personalized cancer treatment strategies,
ultimately contributing to more effective management of metastatic
cancer.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
MK, KY and FT conceived and designed the study. TK,
TM, RO, HM, MT and KK assisted with the study setup and provided
critical insights. MK, TK and MM performed cell experiments. TO
provided the CTC-chip and technical support. MK drafted the
manuscript. FT was involved in the project management and
supervised the study. MK and FT confirm the authenticity of all the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was reviewed and approved by the
Institutional Review Board of the University of Occupational and
Environmental Health, Japan (Approval No: 10-127). The healthy
volunteer (author MK) donated blood and provided formal written
informed consent for its use in this study for experimental
purposes. Data collection from healthy subjects was included in the
Ethics Committee approval (approval no. H26-15).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EpCAM
|
epithelial cell adhesion molecule
|
|
CTCs
|
circulating tumor cells
|
|
EMT
|
epithelial-mesenchymal transition
|
|
CSV
|
cell surface vimentin
|
References
|
1
|
Lozar T, Gersak K, Cemazar M, Kuhar CG and
Jesenko T: The biology and clinical potential of circulating tumor
cells. Radiol Oncol. 53:131–147. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Deng Z, Wu S, Wang Y and Shi D:
Circulating tumor cell isolation for cancer diagnosis and
prognosis. EBioMedicine. 83:1042372022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Allard WJ, Matera J, Miller MC, Repollet
M, Connelly MC, Rao C, Tibbe AG, Uhr JW and Terstappen LW: Tumor
cells circulate in the peripheral blood of all major carcinomas but
not in healthy subjects or patients with nonmalignant diseases.
Clin Cancer Res. 10:6897–6904. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cohen SJ, Punt CJ, Iannotti N, Saidman BH,
Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, et
al: Relationship of circulating tumor cells to tumor response,
progression-free survival, and overall survival in patients with
metastatic colorectal cancer. J Clin Oncol. 26:3213–3221. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tanaka F, Yoneda K, Kondo N, Hashimoto M,
Takuwa T, Matsumoto S, Okumura Y, Rahman S, Tsubota N, Tsujimura T,
et al: Circulating tumor cell as a diagnostic marker in primary
lung cancer. Clin Cancer Res. 15:6980–6986. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brabletz S, Schuhwerk H, Brabletz T and
Stemmler MP: Dynamic EMT: A multi-tool for tumor progression. EMBO
J. 40:e1086472021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu G, Lu Z, Li C, Huang X and Luo Q:
Prognostic and therapeutic significance of circulating tumor cell
phenotype detection based on epithelial-mesenchymal transition
markers in early and midstage colorectal cancer first-line
chemotherapy. Comput Math Methods Med. 2021:22945622021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao Y, Fan WH, Song Z, Lou H and Kang X:
Comparison of circulating tumor cell (CTC) detection rates with
epithelial cell adhesion molecule (EpCAM) and cell surface vimentin
(CSV) antibodies in different solid tumors: A retrospective study.
PeerJ. 9:e107772021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ohnaga T, Shimada Y, Moriyama M, Kishi H,
Obata T, Takata K, Okumura T, Nagata T, Muraguchi A and Tsukada K:
Polymeric microfluidic devices exhibiting sufficient capture of
cancer cell line for isolation of circulating tumor cells. Biomed
Microdevices. 15:611–616. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nagrath S, Sequist LV, Maheswaran S, Bell
DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky
A, et al: Isolation of rare circulating tumour cells in cancer
patients by microchip technology. Nature. 450:1235–1239. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chikaishi Y, Yoneda K, Ohnaga T and Tanaka
F: EpCAM-independent capture of circulating tumor cells with a
‘universal CTC-chip’. Oncol Rep. 37:77–82. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuwata T, Yoneda K, Mori M, Kanayama M,
Kuroda K, Kaneko MK, Kato Y and Tanaka F: Detection of circulating
tumor cells (CTCs) in malignant pleural mesothelioma (MPM) with the
‘universal’ CTC-chip and an anti-podoplanin antibody NZ-1.2. Cells.
9:8882020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanayama M, Oyama R, Mori M, Taira A,
Shinohara S, Kuwata T, Takenaka M, Yoneda K, Kuroda K, Ohnaga T, et
al: Novel circulating tumor cell-detection chip combining
conventional podoplanin and EGFR antibodies for all histological
malignant pleural mesothelioma. Oncol Lett. 22:5222021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoneda K, Kuwata T, Chikaishi Y, Mori M,
Kanayama M, Takenaka M, Oka S, Hirai A, Imanishi N, Kuroda K, et
al: Detection of circulating tumor cells with a novel microfluidic
system in malignant pleural mesothelioma. Cancer Sci. 110:726–733.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanayama M, Kuwata T, Mori M, Nemoto Y,
Nishizawa N, Oyama R, Matsumiya H, Taira A, Shinohara S, Takenaka
M, et al: Prognostic impact of circulating tumor cells detected
with the microfluidic ‘universal CTC-chip’ for primary lung cancer.
Cancer Sci. 113:1028–1037. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Satelli A, Mitra A, Brownlee Z, Xia X,
Bellister S, Overman MJ, Kopetz S, Ellis LM, Meng QH and Li S:
Epithelial-mesenchymal transitioned circulating tumor cells capture
for detecting tumor progression. Clin Cancer Res. 21:899–906. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Satelli A, Batth I, Brownlee Z, Mitra A,
Zhou S, Noh H, Rojas CR, Li H, Meng QH and Li S: EMT circulating
tumor cells detected by cell-surface vimentin are associated with
prostate cancer progression. Oncotarget. 8:49329–49337. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei T, Zhang X, Zhang Q, Yang J, Chen Q,
Wang J, Li X, Chen J, Ma T, Li G, et al: Vimentin-positive
circulating tumor cells as a biomarker for diagnosis and treatment
monitoring in patients with pancreatic cancer. Cancer Lett.
452:237–243. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li H, Zhu YZ, Xu L, Han T, Luan J, Li X,
Liu Y, Wang Z, Liu Q, Kong X, et al: Exploring new frontiers: Cell
surface vimentin as an emerging marker for circulating tumor cells
and a promising therapeutic target in advanced gastric cancer. J
Exp Clin Cancer Res. 43:1292024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Riegger J and Brenner RE: Increase of cell
surface vimentin is associated with vimentin network disruption and
subsequent stress-induced premature senescence in human
chondrocytes. Elife. 12:e914532023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Markou A, Tzanikou E and Lianidou E: The
potential of liquid biopsy in the management of cancer patients.
Semin Cancer Biol. 84:69–79. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Alix-Panabières C, Marchetti D and Lang
JE: Liquid biopsy: From concept to clinical application. Sci Rep.
13:216852023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Strouhalova K, Přechová M, Gandalovičová
A, Brábek J, Gregor M and Rosel D: Vimentin intermediate filaments
as potential target for cancer treatment. Cancers (Basel).
12:1842020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Usman S, Waseem NH, Nguyen TKN, Mohsin S,
Jamal A, Teh MT and Waseem A: Vimentin is at the heart of
epithelial mesenchymal transition (EMT) mediated metastasis.
Cancers (Basel). 13:49852021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Paulin D, Lilienbaum A, Kardjian S,
Agbulut O and Li Z: Vimentin: Regulation and pathogenesis.
Biochimie. 197:96–112. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ramos I, Stamatakis K, Oeste CL and
Pérez-Sala D: Vimentin as a multifaceted player and potential
therapeutic target in viral infections. Int J Mol Sci. 21:46752020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xie X, Wang L, Wang X, Fan WH, Qin Y, Lin
X, Xie Z, Liu M, Ouyang M, Li S and Zhou C: Evaluation of cell
surface vimentin positive circulating tumor cells as a diagnostic
biomarker for lung cancer. Front Oncol. 11:6726872021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu J, Yang M, Peng T, Liu Y and Cao Y:
Evaluation of cell surface vimentin positive circulating tumor
cells as a prognostic biomarker for stage III/IV colorectal cancer.
Sci Rep. 13:187912023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Batth IS, Dao L, Satelli A, Mitra A, Yi S,
Noh H, Li H, Brownlee Z, Zhou S, Bond J, et al: Cell surface
vimentin-positive circulating tumor cell-based relapse prediction
in a long-term longitudinal study of postremission neuroblastoma
patients. Int J Cancer. 147:3550–3559. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Noh H, Zhao Q, Yan J, Kong LY,
Gabrusiewicz K, Hong S, Xia X, Heimberger AB and Li S: Cell surface
vimentin-targeted monoclonal antibody 86C increases sensitivity to
temozolomide in glioma stem cells. Cancer Lett. 433:176–185. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Batth IS and Li S: Discovery of
cell-surface vimentin (CSV) as a sarcoma target and development of
CSV-targeted IL12 immune therapy. Adv Exp Med Biol. 1257:169–178.
2020. View Article : Google Scholar : PubMed/NCBI
|