|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rajurkar S, Mambetsariev I, Pharaon R,
Leach B, Tan T, Kulkarni P and Salgia R: Non-small cell lung cancer
from genomics to therapeutics: A framework for community practice
integration to arrive at personalized therapy strategies. J Clin
Med. 9:18702020. View Article : Google Scholar : PubMed/NCBI
|
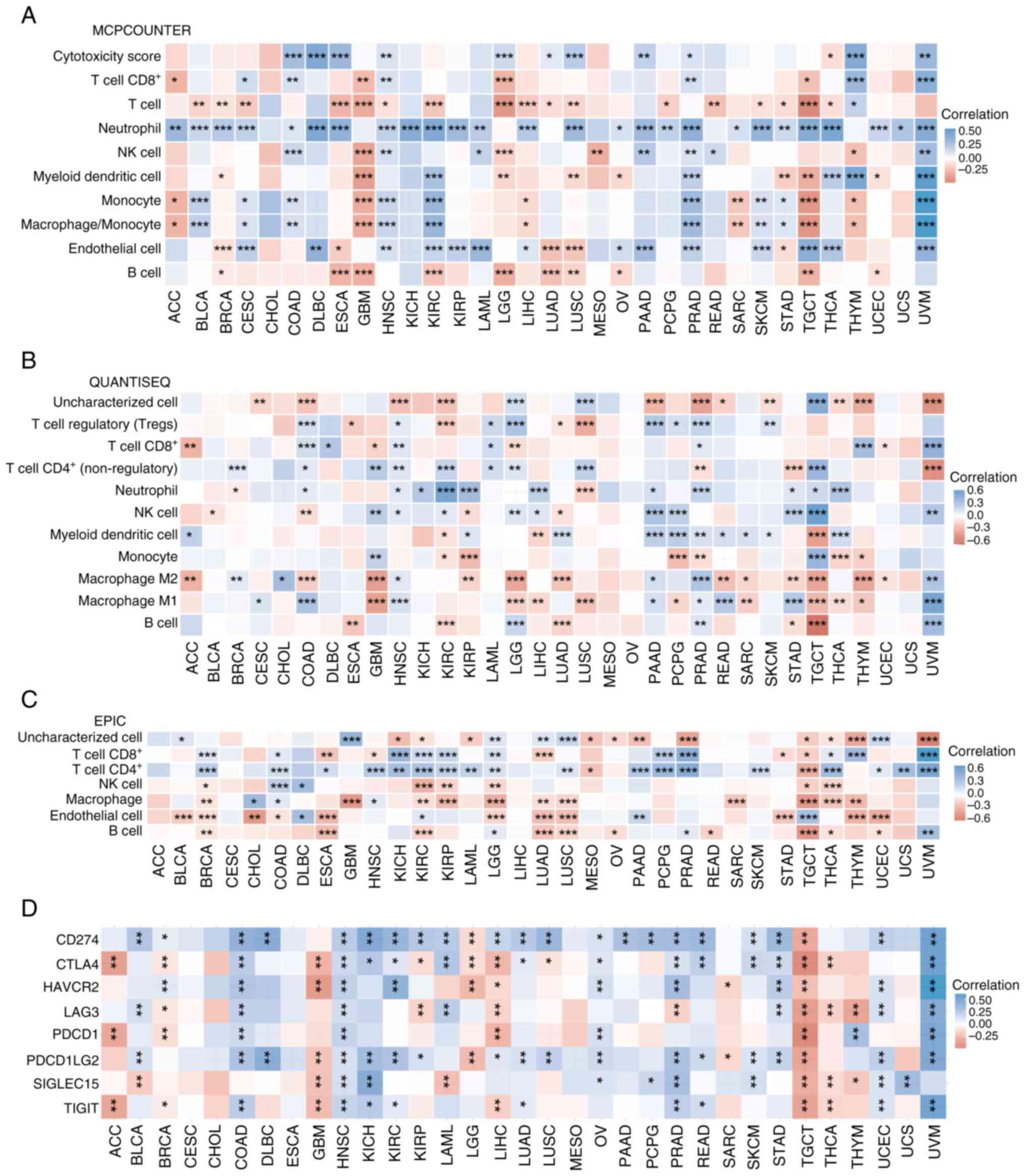
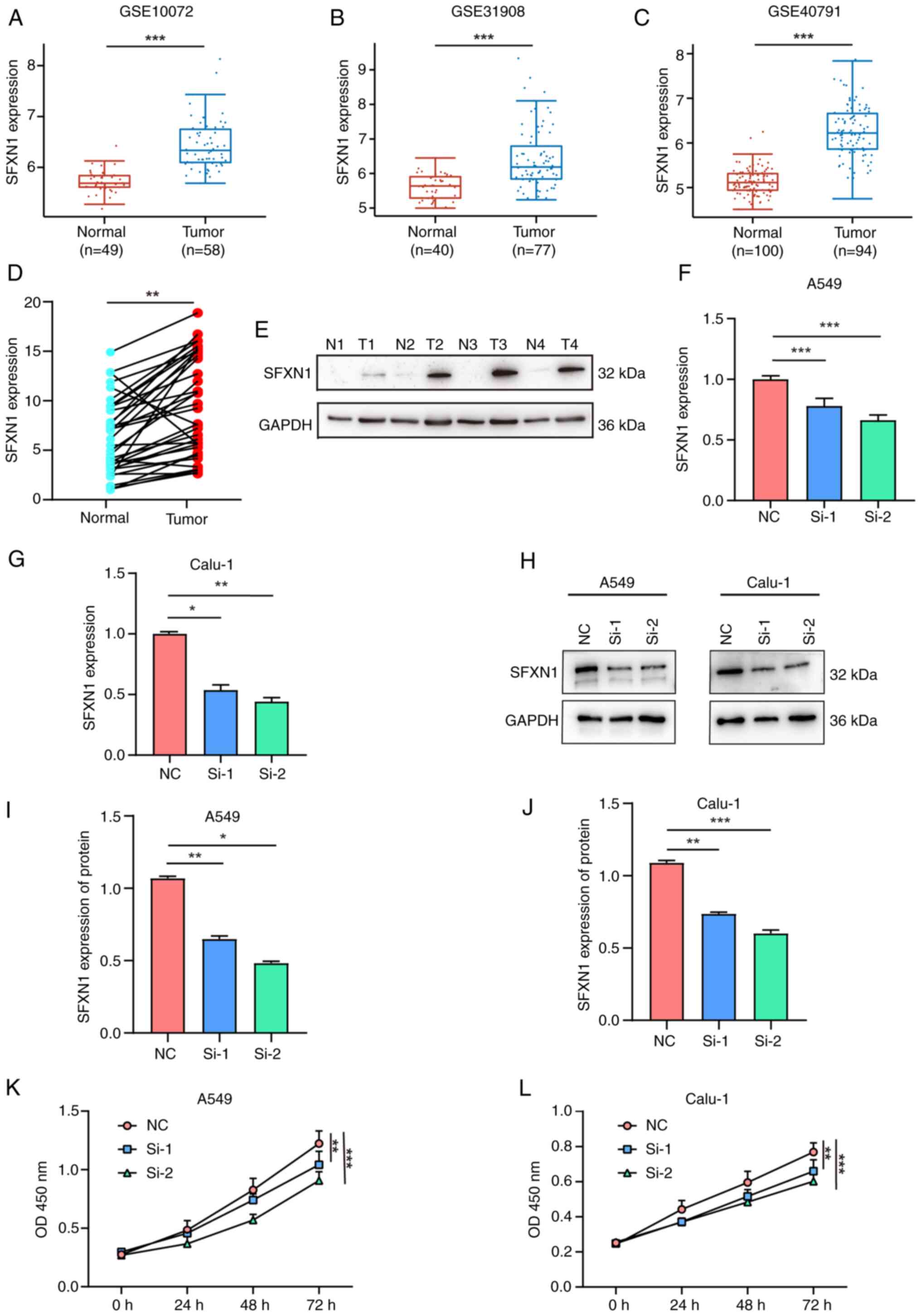
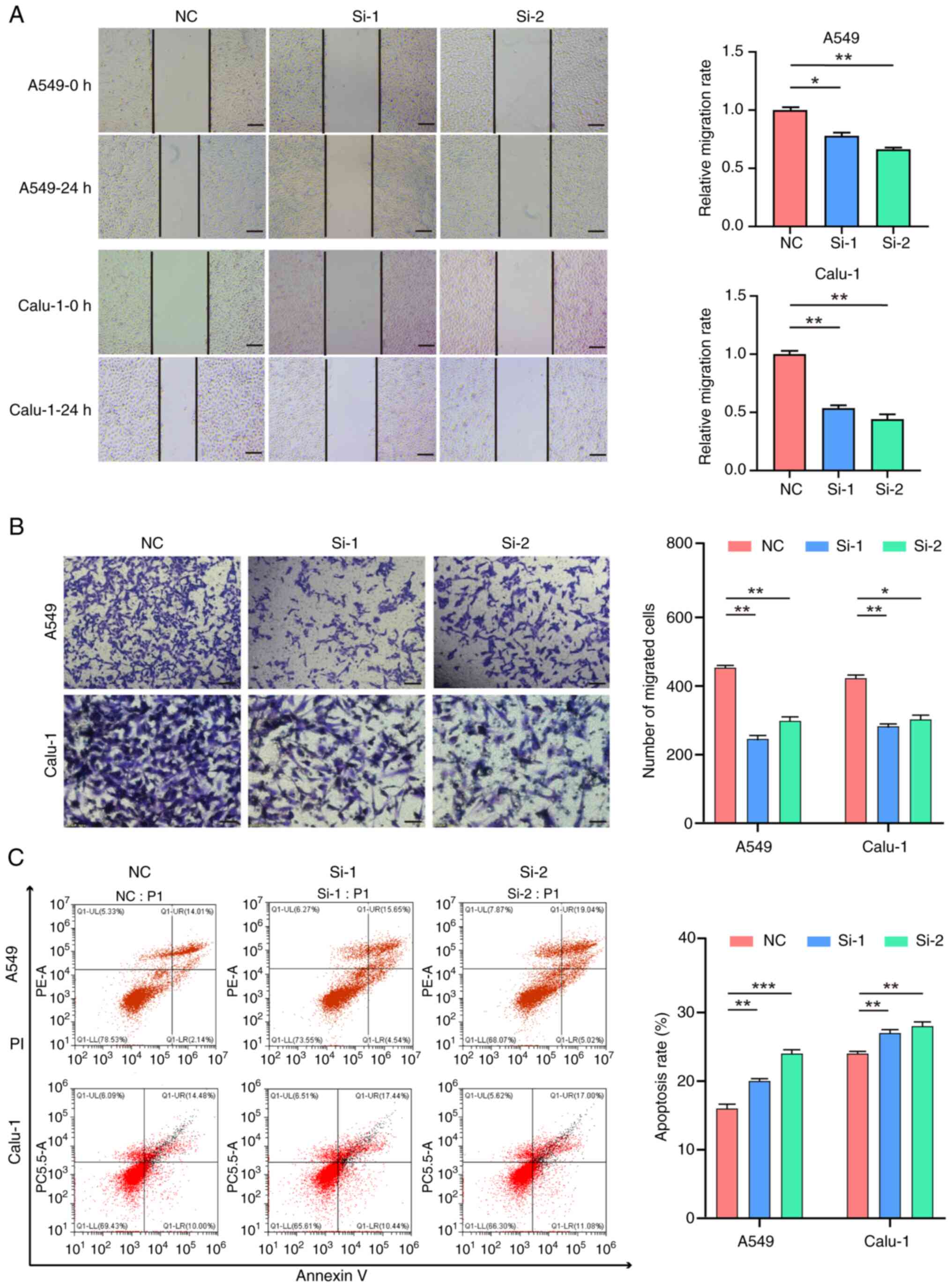
|
3
|
Tifoun N, De Las Heras JM, Guillaume A,
Bouleau S, Mignotte B and Le Floch N: Insights into the roles of
the sideroflexins/SLC56 family in iron homeostasis and iron-sulfur
biogenesis. Biomedicines. 9:1032021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Miotto G, Tessaro S, Rotta GA and Bonatto
D: In silico analyses of Fsf1 sequences, a new group of fungal
proteins orthologous to the metazoan sideroblastic anemia-related
sideroflexin family. Fungal Genet Biol. 44:740–753. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burgos-Barragan G, Wit N, Meiser J,
Dingler FA, Pietzke M, Mulderrig L, Pontel LB, Rosado IV, Brewer
TF, Cordell RL, et al: Mammals divert endogenous genotoxic
formaldehyde into one-carbon metabolism. Nature. 548:549–554. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ducker GS and Rabinowitz JD: One-carbon
metabolism in health and disease. Cell Metab. 25:27–42. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fu TF, Rife JP and Schirch V: The role of
serine hydroxymethyltransferase isozymes in one-carbon metabolism
in MCF-7 cells as determined by (13)C NMR. Arch Biochem Biophys.
393:42–50. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rivell A, Petralia RS, Wang YX, Mattson MP
and Yao PJ: Sideroflexin 3 is a mitochondrial protein enriched in
neurons. Neuromolecular Med. 21:314–321. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mon EE, Wei FY, Ahmad RNR, Yamamoto T,
Moroishi T and Tomizawa K: Regulation of mitochondrial iron
homeostasis by sideroflexin 2. J Physiol Sci. 69:359–373. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fleming MD, Campagna DR, Haslett JN,
Trenor CC III and Andrews NC: A mutation in a mitochondrial
transmembrane protein is responsible for the pleiotropic
hematological and skeletal phenotype of flexed-tail (f/f) mice.
Genes Dev. 15:652–657. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kory N, Wyant GA, Prakash G, Uit de Bos J,
Bottanelli F, Pacold ME, Chan SH, Lewis CA, Wang T, Keys HR, et al:
SFXN1 is a mitochondrial serine transporter required for one-carbon
metabolism. Science. 362:eaat95282018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weston C, Klobusicky J, Weston J, Connor
J, Toms SA and Marko NF: Aberrations in the iron regulatory gene
signature are associated with decreased survival in diffuse
infiltrating gliomas. PLoS One. 11:e01665932016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen Q, Wang R, Zhang J and Zhou L:
Sideroflexin1 as a novel tumor marker independently predicts
survival in lung adenocarcinoma. Transl Cancer Res. 8:1170–1178.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yagi K, Shimada S, Akiyama Y, Hatano M,
Asano D, Ishikawa Y, Ueda H, Watanabe S, Akahoshi K, Ono H, et al:
Loss of SFXN1 mitigates lipotoxicity and predicts poor outcome in
non-viral hepatocellular carcinoma. Sci Rep. 13:94492023.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li N, Wang W, Zhou H, Wu Q, Duan M, Liu C,
Wu H, Deng W, Shen D and Tang Q: Ferritinophagy-mediated
ferroptosis is involved in sepsis-induced cardiac injury. Free
Radic Biol Med. 160:303–318. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Navani S: Manual evaluation of tissue
microarrays in a high-throughput research project: The contribution
of Indian surgical pathology to the human protein atlas (HPA)
project. Proteomics. 16:1266–1270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
18
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu RZ, Graham K, Glubrecht DD, Germain
DR, Mackey JR and Godbout R: Association of FABP5 expression with
poor survival in triple-negative breast cancer: Implication for
retinoic acid therapy. Am J Pathol. 178:997–1008. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ryan BM, Zanetti KA, Robles AI, Schetter
AJ, Goodman J, Hayes RB, Huang WY, Gunter MJ, Yeager M, Burdette L,
et al: Germline variation in NCF4, an innate immunity gene, is
associated with an increased risk of colorectal cancer. Int J
Cancer. 134:1399–1407. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nicolau-Neto P, Da Costa NM, de Souza
Santos PT, Gonzaga IM, Ferreira MA, Guaraldi S, Moreira MA, Seuánez
HN, Brewer L, Bergmann A, et al: Esophageal squamous cell carcinoma
transcriptome reveals the effect of FOXM1 on patient outcome
through novel PIK3R3 mediated activation of PI3K signaling pathway.
Oncotarget. 9:16634–16647. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Verduci L, Ferraiuolo M, Sacconi A, Ganci
F, Vitale J, Colombo T, Paci P, Strano S, Macino G, Rajewsky N and
Blandino G: The oncogenic role of circPVT1 in head and neck
squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD
transcription-competent complex. Genome Biol. 18:2372017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nazarov PV, Muller A, Kaoma T, Nicot N,
Maximo C, Birembaut P, Tran NL, Dittmar G and Vallar L: RNA
sequencing and transcriptome arrays analyses show opposing results
for alternative splicing in patient derived samples. BMC Genomics.
18:4432017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Selamat SA, Chung BS, Girard L, Zhang W,
Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al:
Genome-scale analysis of DNA methylation in lung adenocarcinoma and
integration with mRNA expression. Genome Res. 22:1197–1211. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pei H, Li L, Fridley BL, Jenkins GD,
Kalari KR, Lingle W, Petersen G, Lou Z and Wang L: FKBP51 affects
cancer cell response to chemotherapy by negatively regulating Akt.
Cancer Cell. 16:259–266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cui J, Chen Y, Chou WC, Sun L, Chen L, Suo
J, Ni Z, Zhang M, Kong X, Hoffman LL, et al: An integrated
transcriptomic and computational analysis for biomarker
identification in gastric cancer. Nucleic Acids Res. 39:1197–1207.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lánczky A and Győrffy B: Web-based
survival analysis tool tailored for medical research (KMplot):
Development and implementation. J Med Internet Res. 23:e276332021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao J, Yang X, Chen S, Wang J, Fan X, Fu S
and Yang L: The predictive efficacy of tumor mutation burden in
immunotherapy across multiple cancer types: A meta-analysis and
bioinformatics analysis. Transl Oncol. 20:1013752022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fumet JD, Truntzer C, Yarchoan M and
Ghiringhelli F: Tumour mutational burden as a biomarker for
immunotherapy: Current data and emerging concepts. Eur J Cancer.
131:40–50. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ansari A, Ray SK, Sharma M, Rawal R and
Singh P: Tumor mutational burden as a biomarker of immunotherapy
response: An immunogram approach in onco-immunology. Curr Mol Med.
24:1461–1469. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Strickler JH, Hanks BA and Khasraw M:
Tumor mutational burden as a predictor of immunotherapy response:
Is more always better? Clin Cancer Res. 27:1236–1241. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Steuer CE and Ramalingam SS: Tumor
mutation burden: Leading immunotherapy to the era of precision
medicine? J Clin Oncol. 36:631–632. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang G, Zheng RY and Jin ZS: Correlations
between microsatellite instability and the biological behaviour of
tumours. J Cancer Res Clin Oncol. 145:2891–2899. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yuan H, Yan M, Zhang G, Liu W, Deng C,
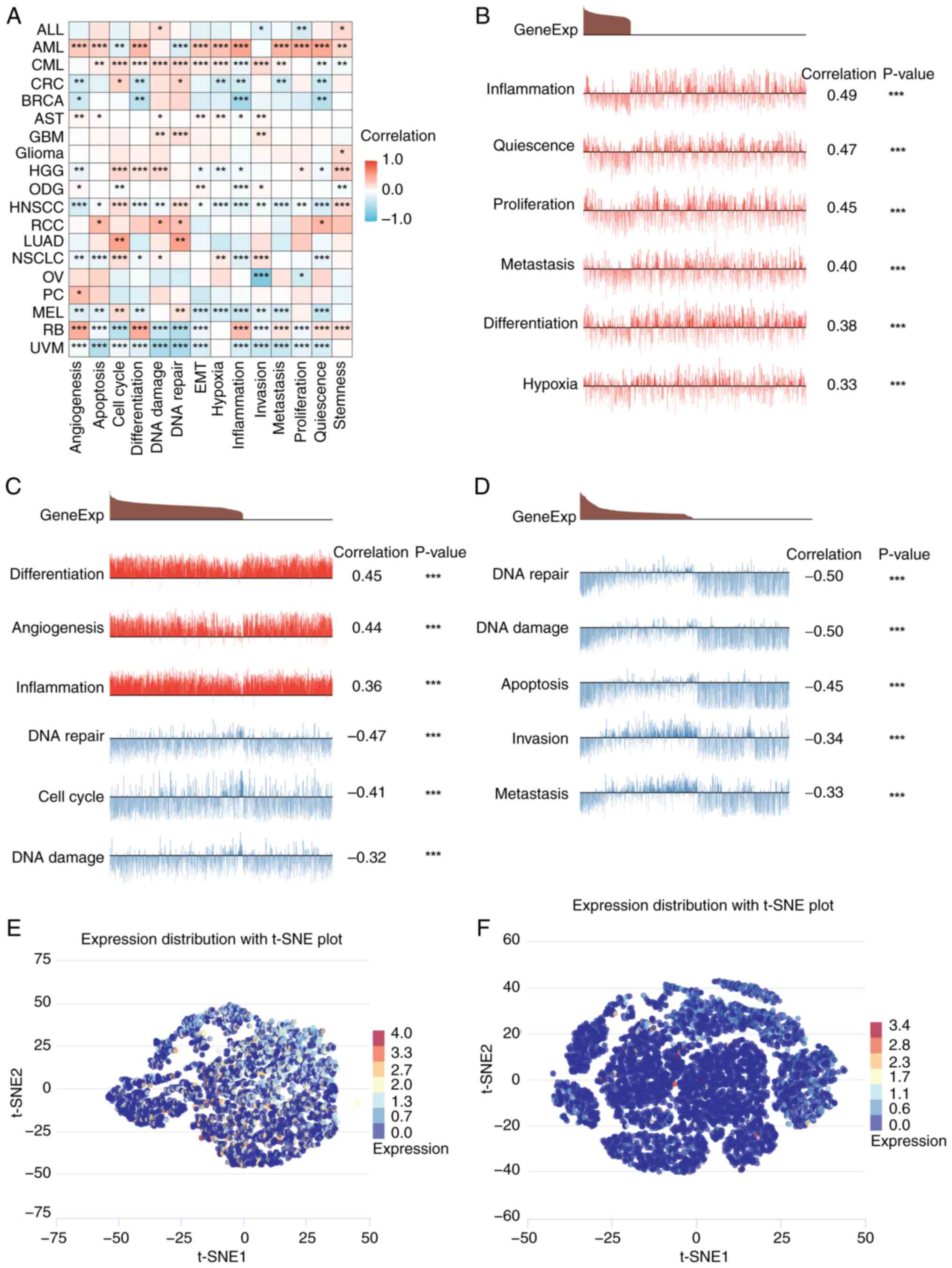
Liao G, Xu L, Luo T, Yan H, Long Z, et al: CancerSEA: A cancer
single-cell state atlas. Nucleic Acids Res. 47(D1): D900–D908.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Schag CC, Heinrich RL and Ganz PA:
Karnofsky performance status revisited: Reliability, validity, and
guidelines. J Clin Oncol. 2:187–193. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Landi MT, Dracheva T, Rotunno M, Figueroa
JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et
al: Gene expression signature of cigarette smoking and its role in
lung adenocarcinoma development and survival. PLoS One.
3:e16512008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu J, Zhou L, Huang L, Gu J, Li S, Liu B,
Feng J and Zhou Y: Nomogram integrating gene expression signatures
with clinicopathological features to predict survival in operable
NSCLC: A pooled analysis of 2164 patients. J Exp Clin Cancer Res.
36:42017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang Y, Foreman O, Wigle DA, Kosari F,
Vasmatzis G, Salisbury JL, van Deursen J and Galardy PJ: USP44
regulates centrosome positioning to prevent aneuploidy and suppress
tumorigenesis. J Clin Invest. 122:4362–4374. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Acoba MG, Alpergin ESS, Renuse S,
Fernández-Del-Río L, Lu YW, Khalimonchuk O, Clarke CF, Pandey A,
Wolfgang MJ and Claypool SM: The mitochondrial carrier SFXN1 is
critical for complex III integrity and cellular metabolism. Cell
Rep. 34:1088692021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ye X, Xu J, Cheng C, Yin G, Zeng L, Ji C,
Gu S, Xie Y and Mao Y: Isolation and characterization of a novel
human putative anemia-related gene homologous to mouse
sideroflexin. Biochem Genet. 41:119–125. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tang M, Huang Z, Luo X, Liu M, Wang L, Qi
Z, Huang S, Zhong J, Chen JX, Li L, et al: Ferritinophagy
activation and sideroflexin1-dependent mitochondria iron overload
is involved in apelin-13-induced cardiomyocytes hypertrophy. Free
Radic Biol Med. 134:445–457. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huang H, Zheng J, Shen N, Wang G, Zhou G,
Fang Y, Lin J and Zhao J: Identification of pathways and genes
associated with synovitis in osteoarthritis using bioinformatics
analyses. Sci Rep. 8:100502018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: an iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Andriani L, Ling YX, Yang SY, Zhao Q, Ma
XY, Huang MY, Zhang YL, Zhang FL, Li DQ and Shao ZM: Sideroflexin-1
promotes progression and sensitivity to lapatinib in
triple-negative breast cancer by inhibiting TOLLIP-mediated
autophagic degradation of CIP2A. Cancer Lett. 597:2170082024.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li Y, Yang W, Liu C, Zhou S, Liu X, Zhang
T, Wu L, Li X, Zhang J and Chang E: SFXN1-mediated immune cell
infiltration and tumorigenesis in lung adenocarcinoma: A potential
therapeutic target. Int Immunopharmacol. 132:1119182024. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sadeghi Rad H, Monkman J, Warkiani ME,
Ladwa R, O'Byrne K, Rezaei N and Kulasinghe A: Understanding the
tumor microenvironment for effective immunotherapy. Med Res Rev.
41:1474–1498. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Filipovic A, Miller G and Bolen J:
Progress toward identifying exact proxies for predicting response
to immunotherapies. Front Cell Dev Biol. 8:1552020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Reck M, Schenker M, Lee KH, Provencio M,
Nishio M, Lesniewski-Kmak K, Sangha R, Ahmed S, Raimbourg J, Feeney
K, et al: Nivolumab plus ipilimumab versus chemotherapy as
first-line treatment in advanced non-small-cell lung cancer with
high tumour mutational burden: Patient-reported outcomes results
from the randomised, open-label, phase III CheckMate 227 trial. Eur
J Cancer. 116:137–147. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pfeifer GP: Defining driver DNA
methylation changes in human cancer. Int J Mol Sci. 19:11662018.
View Article : Google Scholar : PubMed/NCBI
|