Introduction
Cancer of unknown primary (CUP) is defined as
histologically confirmed metastatic cancer in which the primary
site cannot be found even after extensive standard investigations
(1). At present, the favorable risk
subgroup for CUP includes patients with neuroendocrine carcinomas
of unknown primary, peritoneal adenocarcinomatosis of a serous
papillary subtype, isolated axillary nodal metastases in female
patients, squamous cell carcinoma involving non-supraclavicular
cervical lymph nodes (LNs), single metastatic deposit from unknown
primary, and men with blastic bone metastases and prostate-specific
antigen expression (2). Novel
favorable subsets of CUP have emerged, including colorectal, lung
and renal CUP, which underlines the importance of cancer-specific
treatments (2). Patients with CUP
who are categorized into the unfavorable subset (~80% of patients)
receive empiric chemotherapy with a platinum-taxane regimen.
However, the prognosis remains poor, with a median survival period
of 6–12 months (3,4). The poor prognosis of CUP is
attributable to its clinical heterogeneity originating from various
types of cancers, which makes a single empiric regimen inefficient
(5).
Our previous randomized controlled study (6) assessed whether site-specific therapy
based on prediction of the primary site may improve the outcomes in
untreated patients with CUP. Genome-wide gene expression profiling
of CUP was performed in the study using a microarray to predict the
primary site in each patient with CUP (6). However, substantial shortcomings have
been identified in the existing research comparing site-specific
treatment and empiric chemotherapy. These deficiencies include
patient accrual problems (oversampling treatment-resistant tumor
types and long recruitment), study design limitations
(observational and problematic trials), heterogeneity among the CUP
classifiers (epigenetic vs. transcriptomic profiling) and
incomparable therapies (7).
Treatment based on the prediction of the primary site did not lead
to any improvement of the overall survival compared with empiric
chemotherapy in our previous study (6); however, gene profiling analyses of
patients with CUP and comparison of the gene expression patterns in
these patients with those in tumors from 24 known primary sites
revealed several genes that were uniquely upregulated in CUP
(8). Of the ~22,000 genes mounted
on the microarray, 44 genes that were upregulated in CUPs were
identified in our previous study (8).
The early metastasis in the natural history of CUP
remains poorly understood. The present study identified genes that
could serve a role in the metastasis in CUP. Small interfering RNA
(siRNA) knockdown screens were used to assess the genes upregulated
in the present CUP cohort to determine their impact on the
migration of cancer cells and further characterize candidate genes
using an in vivo metastasis model.
Materials and methods
Cell culture and reagents
The A549 human lung cancer cell line (cat. no.
CCL-185) and the MDA231 breast cancer cell line (cat. no. HTB-26)
were procured from American Type Culture Collection, and cultured
in DMEM (Thermo Fisher Scientific, Inc.) with 10% FBS
(MilliporeSigma) in accordance with the instructions provided by
the suppliers. The cells were maintained in a 5% CO2
humidified atmosphere at 37°C.
Cell viability assay
The proliferation rate of cultured cells was
analyzed using MTT (MilliporeSigma) as previously described
(9). The assay was performed in
triplicate where applicable.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA (1 µg) from cells, extracted using ISOGEN
(Nippon Gene Co., Ltd.), was converted into cDNA using a GeneAmp
RNA-PCR kit (Thermo Fisher Scientific, Inc.). Reverse transcription
was performed at 37°C for 2 h. qPCR was performed using TB Green
Premix Ex Taq II (Takara Bio, Inc.), including TB Green as an
intercalator that emits fluorescence when bound to double-stranded
DNA. The thermocycling conditions were as follows: 95°C for 1 min
and 50 cycles of 95°C for 5 sec and 60°C for 30 sec. Quantification
based on the cycle threshold (Ct) values (2−ΔΔCq method)
(10) was performed using an
Applied Biosystems 7900 HT Fast Real-time PCR System (Thermo Fisher
Scientific, Inc.) as previously described (11). The primers used to amplify the
target genes are listed in Table
SI. GAPDH was used to normalize the expression levels in
quantitative analyses. For the siRNAs listed in Table I, the analyses were conducted once,
but in triplicate in case the migration rate was <50%. For a few
siRNAs that impaired the viability of the cells, it was not
possible to perform the analyses (see below; Table I).
 | Table I.Effect of siRNA-induced knockdown of
genes on cell migration. |
Table I.
Effect of siRNA-induced knockdown of
genes on cell migration.
|
|
| A549 cells | MDA231 |
|---|
|
|
|
|
|
|---|
| siRNA | siRNA
(EHU-)a | Viability,
%b | Fold change of the
expression of the gene targeted by the siRNAc | Migration rate,
%d | Viability,
%b | Fold change of the
expression of the gene targeted by the siRNAc | Migration rate,
%d |
|---|
|
Controle | SIC001 | 100.0 | 1.000 | 100.0 | 100.0 | 1.000 | 100.0 |
| S100A4 | 125681 | >90.0 | 0.069 | >90.0 |
|
|
|
| HSPA8 | 115141 | >90.0 | 0.199 | >90.0 |
|
|
|
| TIMP1 | 156431 | >90.0 | 0.069 | >90.0 |
|
|
|
| LGALS1 | 034721 | >90.0 | 0.035 | >90.0 |
|
|
|
| SERF2 | 096201 | >90.0 | 0.559±0.033 | 44.3±8.1 | >90.0 | 0.652 | >90.0 |
| PRKDC | 123791 | 51.0±3.1 | 0.087±0.020 | <1.0 | 75.2±2.9 | 0.107±0.018 | 1.9±1.5 |
| NEDD8 | 112521 | >90.0 | 0.008 | >90.0 |
|
|
|
| APOC | 125971 | >90.0 | 0.314 | >90.0 |
|
|
|
| YWHAZ | 076001 | >90.0 | 0.363 | >90.0 |
|
|
|
| SUB1 | 031131 | >90.0 | 0.037 | >90.0 |
|
|
|
| NOLA3 | 104671 | >90.0 | 0.044 | >90.0 |
|
|
|
| SNRPD2 | 146751 | 50.0±4.6 | 0.019 | 67.2±5.4 |
|
|
|
| ATP5H | 128441 | >90.0 | 0.110 | >90.0 |
|
|
|
| TCEB2 | 150621 | >90.0 | 0.034 | 71.7±6.9 |
|
|
|
| OAZ1 | 034471 | >90.0 | 0.044 | >90.0 |
|
|
|
| NPIPL3 | 106191 | 79.3±4.0 | 0.953 | >90.0 |
|
|
|
| SRGN | 143531 | >90.0 | 0.017 | >90.0 |
|
|
|
| S100A6 | 120391 | >90.0 | 0.019 | >90.0 |
|
|
|
| PFN1 | 106911 | >90.0 | 0.035 | >90.0 |
|
|
|
| EIF5A | 147491 | >90.0 | 0.015 | >90.0 |
|
|
|
| S100A11 | 147921 | 46.3±6.4 | 0.017 | 55.6±3.9 |
|
|
|
| NUTF2 | 159321 | 64.7±7.1 | 0.021 | 50.7±2.5 |
|
|
|
| STK17A | 018811 | >90.0 | 0.167 | >90.0 |
|
|
|
| YWHAH | 004271 | >90.0 | 0.096 | >90.0 |
|
|
|
| SELT | 117491 | >90.0 | 0.017 | >90.0 |
|
|
|
| CST3 | 019031 | >90.0 | 0.046 | >90.0 |
|
|
|
| GDI2 | 109311 | >90.0 | 0.095 | >90.0 |
|
|
|
| VDAC3 | 108731 | >90.0 | 0.048 | >90.0 |
|
|
|
| TYROBP | 033221 | >90.0 | 0.346 | >90.0 |
|
|
|
| VIM | 151861 | >90.0 | 0.057 | >90.0 |
|
|
|
| LSM7 | 057161 | >90.0 | 0.042 | >90.0 |
|
|
|
| GSTP1 | 009201 | >90.0 | 0.051 | >90.0 |
|
|
|
| NDUFS8 | 053841 | >90.0 | 0.047 | >90.0 |
|
|
|
| POLR2J | 117061 | >90.0 | 0.853 | >90.0 |
|
|
|
| RPS7 | 110521 | 25.8±2.1 | 0.013±0.005 | <1.0 | <1.0 | n.p. | n.p. |
| RPL11 | 108131 | 18.8±3.2 | 0.002±0.001 | <1.0 | <1.0 | n.p. | n.p. |
| RPLP2 | 055751 | <1.0 | n.p. | n.p. |
|
|
|
| RPL18A | 147231 | <1.0 | n.p. | n.p. |
|
|
|
| RPL36 | 056091 | <1.0 | n.p. | n.p. |
|
|
|
| RPS10 | 110371 | <1.0 | n.p. | n.p. |
|
|
|
| IMP3f | 085841 | 58.2±7.4 | 0.257 | >90.0 |
|
|
|
| CALM3f | 143521 | >90.0 | 0.023 | >90.0 |
|
|
|
| AP2S1f | 135191 | >90.0 | 0.009 | >90.0 |
|
|
|
|
ARL6IP4f | 087871 | >90.0 | 0.135 | >90.0 |
|
|
|
|
ATP6VOBf | 142591 | >90.0 | 0.038 | >90.0 |
|
|
|
|
SH3GLB1f | 098381 | >90.0 | 0.123±0.011 | 43.9±6.0 | >90.0 | 0.058 | >90.0 |
| CAPNS1f | 047191 | >90.0 | 0.031±0.009 | 15.9±2.3 | >90.0 | 0.022 | >90.0 |
| PGK1f | 105941 | >90.0 | 0.232 | >90.0 |
|
|
|
| PSMB4f | 025191 | >90.0 | 0.051±0.004 | 21.8±2.5 | >90.0 | 0.033±0.008 | 17.3±4.3 |
| PDCD6f | 094521 | >90.0 | 0.030 | >90.0 |
|
|
|
Western blot analysis
Total proteins were extracted from cells using RIPA
buffer (FUJIFILM Wako Pure Chemical Corporation) containing
protease inhibitor mix Complete™ (Roche Diagnostics).
The protein concentrations were determined using a BCA Protein
Assay Kit (Pierce; Thermo Fisher Scientific, Inc.). The proteins
were boiled at 100°C, loaded (20 mg per lane), separated by
SDS-PAGE (5–20%) and transferred to PVDF membranes. After 1 h of
blocking with 3% (w/v) BSA (Merck KGaA) at room temperature, the
membranes were incubated overnight with primary antibodies at 4°C.
After being rinsed twice with TBS buffer (pH 8.0) containing 0.1%
Tween-20, the membranes were incubated with HRP-labeled secondary
antibodies against mouse IgG (100-fold dilution; cat. no. 7076;
Cell Signaling Technology, Inc.) at room temperature for 1 h. An
enhanced chemiluminescence solution (GE Healthcare) was used for
color development. β-actin was used as the internal standard. The
experiment was performed in triplicate. Antibodies against protein
kinase DNA-activated catalytic subunit (PRKDC)/DNA-dependent
protein kinase catalytic subunit (DNA-PK) (1,000-fold dilution;
cat. no. 12311; Cell Signaling Technology, Inc.), proteasome
subunit β type-4 (PSMB4) (200-fold dilution; cat. no. sc-390878;
Santa Cruz Biotechnology, Inc.) and β-actin (200-fold dilution;
cat. no. sc-47778; Santa Cruz Biotechnology, Inc.) were used.
ImageJ software ver. 1.54 g (National Institutes of Health) was
used for densitometry.
Migration assay
The migration assay was performed using the Boyden
chamber method using polycarbonate membranes with a pore size of 8
µm (Chemotaxicell; Kurabo Bio-Medical Department; Kurabo
Industries, Ltd.) as previously described (11). The membranes were coated with
fibronectin (50 µg/ml) on the outer side at room temperature for 1
h and dried for 2 h at room temperature. The cells
(2×104 cells/well) were then seeded into the upper
chambers containing 500 µl migrating medium (DMEM containing 0.5%
FBS), and the upper chambers were placed into the lower chambers
containing 700 µl DMEM with 10% FBS. After incubation at 37°C for
24 h, the media in the upper chambers were aspirated, and the
non-migrated cells on the inner sides of the membranes were removed
using a cotton swab. The cells that had migrated to the outer side
of the membranes were fixed with 100% ethanol at room temperature
for 5 min, stained with 0.1% crystal violet solution at room
temperature for 15 min and observed under a light microscope. The
number of migrated cells in five fields per chamber was averaged.
The migration rate was verified in triplicate for each gene where
applicable (Table I).
Selection of genes for the migration
assay
The selection of genes was based on our previous
expression microarray analysis for CUP (GSE42392) (8), which was compared with microarray
datasets for several cancer types obtained from the Gene Expression
Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) (12). The datasets used were GSE781
(13), GSE1456 (14), GSE2109 (15), GSE2742 (16), GSE3149 (17), GSE3167 (18), GSE4127 (19), GSE4176 (20), GSE5787 (21), GSE6791 (22) and GSE8218 (23).
Transfection of siRNA
Predesigned siRNAs (MISSION® siRNA)
targeting 50 genes and a nonspecific target (MISSION®
siRNA Universal Negative Control; Table
I) were purchased from MilliporeSigma. Cells were transfected
with each siRNA (10 nM) using RNAiMAX (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions as
previously described (24),
incubated at 37°C for 48 h, and then trypsinized and seeded
immediately for the migration assay.
Plasmid construction, virus production
and stable transfection
shRNAs targeting PRKDC or PSMB4 were constructed
using oligonucleotides encoding siRNA directed against the
respective genes (5′-CCTGAAGTCTTTACAACATATCTC-3′ and
5′-CCGCAACATCTCTCGCATTATCTC-3′ for PRKDC and PSMB4 shRNA,
respectively). The oligonucleotides were cloned into RNAi-Ready
pSIREN RetroQZsGreen (Takara Bio, Inc.), which is a
self-inactivating retroviral expression vector. This dual
expression vector encodes green fluorescent protein (GFP) under the
control of the cytomegalovirus promotor and shRNA under the control
of the U6 promoter. The viral vectors constructed were designated
as pSIREN-shPRKDC, pSIREN-shPSMB4 and a control vector
(pSIREN-RetroQZsGreen vector without oligonucleotide inserted).
Each of the pSIREN-RetroQZsGreen constructs (45 µg) was
co-transfected with a pVSV-G vector (Takara Bio, Inc.) constituting
the viral envelope (molar ratio of 3:1 for pSIREN-RetroQZsGreen
constructs:pVSV-G vector) into GP2-293 packaging cells
(2×107 cells; Takara Bio, Inc.) using the FuGENE6
transfection reagent (Roche Diagnostics). After 48 h at 37°C, the
culture medium was collected, and the viral particles were
concentrated by centrifugation at 15,000 × g for 3 h at 4°C. The
viral pellet was then resuspended in fresh DMEM. The viral vector
titer was calculated using a Retrovirus Titer Set (Takara Bio,
Inc.) according to the manufacturer's instructions. The minimal MOI
was then determined to be 4 for all the constructs by counting the
GFP-positive A549 cells infected by serial dilutions of the
virus-containing media. At 3 days after the infection with the
virus in A549 cells (1×105 cells), the cells were
trypsinized, propagated for 1 week and resuspended in DMEM
(5×106 cells/ml). Subsequently, 1 ml of the resuspended
solution was processed for cell sorting using a FACSCalibur Flow
Cytometer (BD Biosciences). This device consists of a flow
cytometer and a fluorescence activated cell sorter (FACS) in which
GFP-positive cells excited by the 488 nm laser line emit green
fluorescence at 509 nm, which is optimally detected by the FL-1
detector, and the GFP-positive cells were captured by a catcher
tube for sorting (25). The
software used for analysis was CellQuest software Ver. 3.3 (BD
Biosciences). The subsequent experiment (LN metastasis assay) was
performed immediately after the sorted cells reached the number
required for inoculation into mice (1×107 cells for each
construct) during the culture for ~1 week.
Animal models and LN metastasis
assay
A total of 60 nude mice (BALB/c nu/nu; 5-week-old;
female; mean weight, 17.8 g; CLEA Japan, Inc.) were used for the
in vivo experiments. Experimemts were approved by the
Institutional Animal Care Committee of the Kindai University
Faculty of Medicine (approval no. KDMS-25-019; Osaka-Sayama,
Japan). The mice were housed at the Animal Laboratory, Kindai
University Faculty of Medicine (Osaka-Sayama, Japan), and
maintained in a specific pathogen-free vivarium at 18–23°C with 50%
humidity under a 12/12 h dark/light cycle. The animals had free
access to drinking water and were fed a pelleted basal diet ad
libitum.
The in vivo study consisted of two parts that
estimated two effects on the potential of the cells to metastasize
from footpad to popliteal LN: Experiment 1, the effect of
downregulation of PSMB4 and PRKDC; and experiment 2, the effect of
inhibitors of PSMB4 and PRKDC. For each of them, a single
experiment was performed and 30 mice were used. For experiment 1,
the cells (A549 cells bearing a control vector, pSIREN-shPRKDC, or
pSIREN-shPSMB4) were resuspended in Hanks' Balanced Salt Solution
(MilliporeSigma) and injected subcutaneously (1×106
cells) into the hind footpads of mice (10 mice per group) on day 0
in accordance with the method described for a previous study
(26). On the final day (day 20),
mice were euthanized using CO2 flowing from a compressed
cylinder into a chamber (under ambient conditions) containing the
mice so that 100% CO2 gradually filled the chamber
(30%/min) (27). Next, the primary
tumors and popliteal LNs were visualized by fluorescence imaging
and images were captured using a macro-imaging station consisting
of a SBIG cooled CCD camera model ST-7XME (Santa Barbara Imaging)
mounted onto a dark box. The integrated density of the fluorescence
spots was quantified using ImageJ software ver. 1.54 g (National
Institutes of Health). For experiment 2, the DNA-PK inhibitor
NU7441 (10 mg/kg; FUJIFILM Wako Pure Chemical Corporation)
(28) and the proteasome inhibitor
bortezomib (1 mg/kg; Takeda Pharmaceutical Company, Ltd.) (29), and vehicle (DMSO) were administered
intravenously alone (9–10 mice/group) 1 week after inoculation of
A549 cells bearing a control vector, and this administration was
repeated twice a week for 3 weeks (2nd to 4th weeks). The LNs were
resected (on day 35), and their maximum diameters were measured.
Fluorescence images were also captured for the primary tumors and
popliteal LNs. The animal details and protocol were otherwise as
aforementioned.
Statistical analysis
Data are presented as the mean ± SD, unless
otherwise noted. An unpaired t-test was used for comparisons
between two groups. Comparisons among multiple groups were
conducted using one-way ANOVA, followed by the Student-Newman-Keuls
test. These analyses were performed as a single experiment.
Statistical analyses were performed using SigmaPlot v13.0 (Grafiti
LLC). P<0.05 was considered to indicate a statistically
significant difference.
Results
Selection of candidate genes in
CUP
In our previous microarray study, 44 genes that were
upregulated in CUPs compared with non-CUPs as a whole were
identified (8). Of the 44 genes, 40
were selected as candidates for the next screening using the
migration assay (Table I). Three
pseudogene-like genes (LOC392501, LOC442171 and LOC646417) with
unknown functions and one gene without an available siRNA (LAPTM5)
were excluded. As additional candidates for the screening, 10 genes
that were screened out by the comparison of gene expression
profiles between CUP and individual cancer types of known primary
site were selected (8). These 10
genes were most highly expressed in CUP of all cancer types
examined, and each of the genes showed ≥1.5-fold enriched
expression in CUP compared with the second most enriched cancer
type for each gene (Table SII). A
total of 50 genes were selected (Table
I) for the migration assay.
Screening for genes associated with
the metastatic potential in CUP
For all 50 aforementioned genes, specific siRNAs
were obtained from the same commercial source (MISSION®
siRNA; Table I). For screening,
A549 cells were transfected with siRNAs, and genes were evaluated
for suppressed migration after siRNA knockdown. Images for cell
migration assays are shown in Figs.
1 and S1. The migration assay
showed that A549 cells transfected with some siRNAs exhibited poor
migration, meaning the number of A549 cells migrating to the outer
side of the porous membrane in the Boyden chamber was decreased
compared with that of the cells transfected with control siRNA
(Fig. 1A). For each of the 50
siRNAs transfected into A549 cells, the present study aimed to
determine the cell viability, the fold-change in the expression of
the targeted gene and the cell migration rate (Table I). The results revealed that
transfection with siRNAs targeting SERF2, PRKDC, SH3GLB1, CAPNS1,
PSMB4, RPS7 and RPL11 reduced the migration of A549 cells by
>50% (Fig. 1A; Table I).
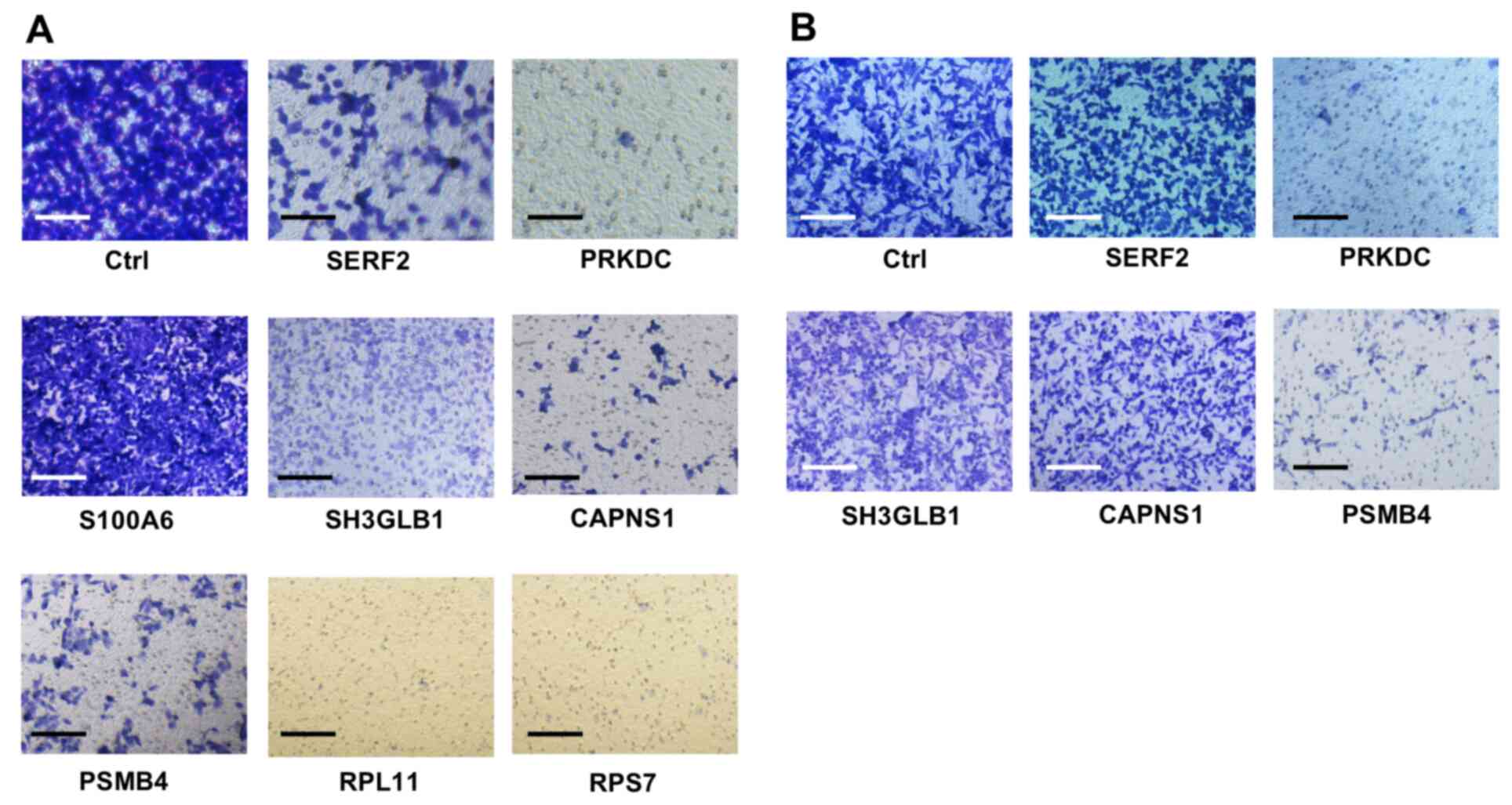 | Figure 1.Migration assays were performed using
the Boyden chamber method. After transfection of each siRNA into
the (A) A549 and (B) MDA231 cells, the cells were incubated for 48
h before the migration assay. The cells that had migrated to the
outer side of the membranes within 24 h were fixed and stained. A
representative image of a triplicate analysis of each siRNA is
shown. Scale bar, 20 µm. siRNA, small interfering RNA; Ctrl,
control siRNA; SERF2, small EDRK-rich factor 2; PRKDC,
DNA-dependent protein kinase catalytic subunit; S100A6, S100
calcium-binding protein A6; SH3GLB1, SH3-domain GRB2-like
endophilin B1; CAPNS1, calpain, small subunit 1; PSMB4, proteasome
subunit β type-4; RPL11, ribosomal protein L11; RPS7, ribosomal
protein S7. |
Our previous study identified six genes encoding
ribosomal proteins (RPLP2, RPL18A, RPL36, RPS10, RPS7 and RPL11) as
CUP-associated (−upregulated) genes (8). siRNAs against four of these ribosomal
protein genes (RPLP2, RPL18A, RPL36 and RPS10) almost completely
impaired the viability of A549 cells, and it was not possible to
perform the expression assay and migration assay using siRNAs of
these genes (Table I). The other
two siRNAs (targeting RPS7 and RPL11) abolished the viability of
MDA231 cells (Table I), indicating
that reduced expression of any single ribosomal protein molecule
could be critical for cell survival.
The present study subsequently focused on the siRNAs
against the five genes (SERF2, PRKDC, SH3GLB1, CAPNS1 and PSMB4)
based on their ability to reduce migration to less than half of
that of the control without affecting the cell viability (>50%
of the control; Table I). The
present study further analyzed their influence on the migration of
another cell line (MDA231). Only siRNAs specific for PRKDC and
PSMB4 markedly reduced the migration of the cells (Fig. 1B; Table
I). These siRNAs did not affect the viability of MDA231 cells,
resulting in a cell viability rate of 75.2% for PRKDC-specific
siRNA and >90% for PSMB4-specific siRNA. Furthermore, a marked
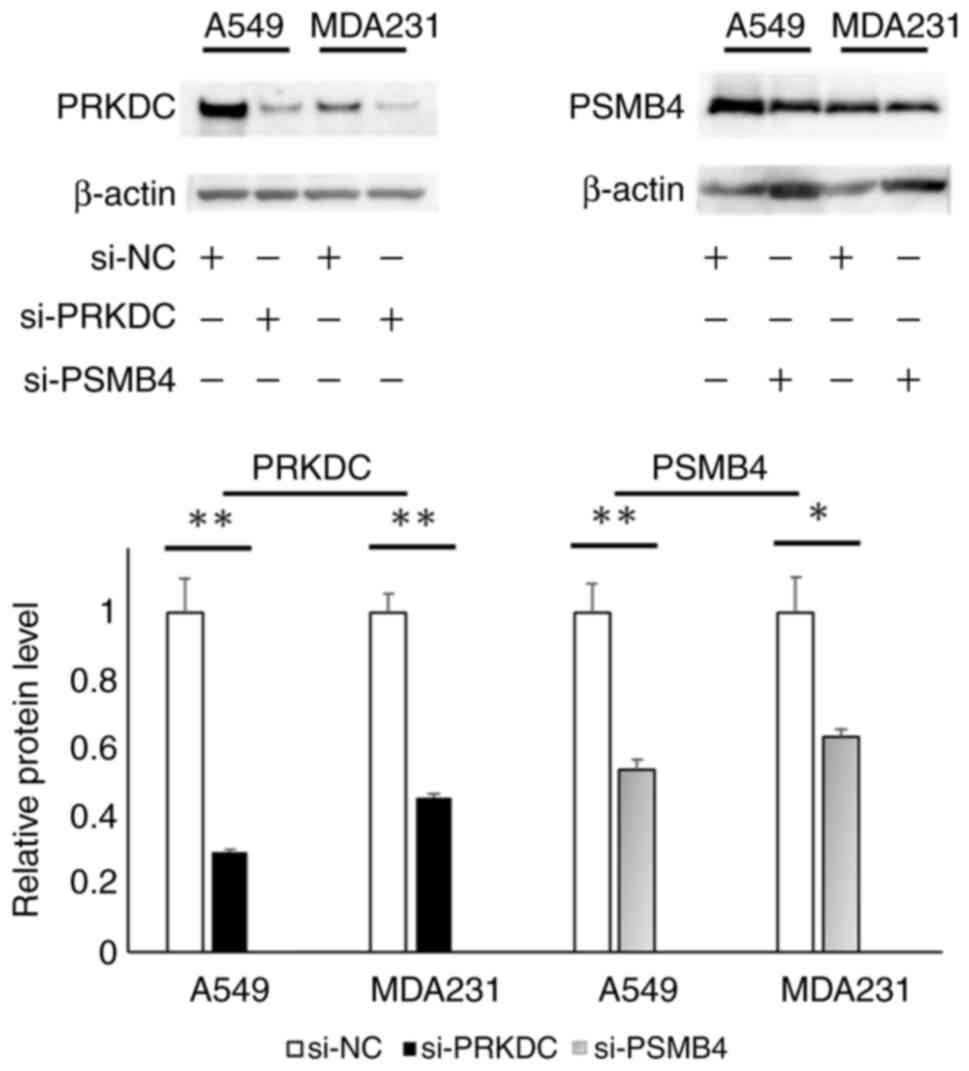
reduction in the expression of the respective genes was observed.
The siRNAs targeting PRKDC and PSMB4 reduced the expression levels
of the corresponding genes (0.107- and 0.033-fold changes,
respectively), which was similar to the trends observed for A549
cells (Table I). Western blotting
confirmed that the expression of PRKDC and PSMB4 was reduced by the
siRNA-induced mRNA knockdown (Fig.
2).
Effect of downregulation of the
candidate genes on LN metastasis
To determine whether downregulation of the two
genes, PRKDC and PSMB4, might affect the potential of the cells to
metastasize to the LNs in vivo, cell lines that stably
expressed both GFP and shRNA targeting the respective genes were
engineered. A control cell line transfected with a control vector
that expressed GFP alone was also established. The stable cell
lines for these constructs were isolated by FACS using GFP as a
reporter (Fig. S2A-C) and they
showed stable shRNA-based knockdown of PRKDC and PSMB4 during the
passage culture for ≥3 months. Western blotting confirmed that the
PRKDC and PSMB4 proteins expressed in A549 cells were reduced for
the respective constructs compared with the control vector
(Fig. S3A). The growth rate
remained unaltered in the A549 cells transfected with shRNA
targeting PSMB4 (>90% of the rate in the control cells), but the
cells transfected with shRNA targeting PRKDC exhibited an ~25%
reduction in the growth rate relative to that in the control cells
at 72 h (Fig. S3B). To evaluate
the metastasis-promoting potential of these two genes, cells
(1×106) containing the respective shRNA constructs were
subcutaneously injected into the footpads of BALB/c nude mice. On
day 20, the fluorescence intensities of GFP expressed in the
footpad and popliteal LNs were measured (Fig. 3A). The fluorescence intensities were
lower after injection of cells expressing PRKDC-shRNA than after
injection of cells transfected with a control vector (~14.8%),
whereas fluorescence intensities following injection of the cells
expressing PSMB4-shRNA were comparable to those following injection
of cells containing the control vector (Fig. 3B and C). A proportionate reduction
in fluorescence intensities was also observed in the popliteal LNs
in both the PRKDC-shRNA and PSMB4-shRNA groups, with popliteal
LN/footpad fluorescence intensity ratios of 39.4 and 45.5%,
respectively, compared with the control group (Fig. 3C). It was attempted to perform a
similar assay using MDA231-shRNA constructs; however, this cell
line failed to become engrafted into the footpads of the mice (data
not shown).
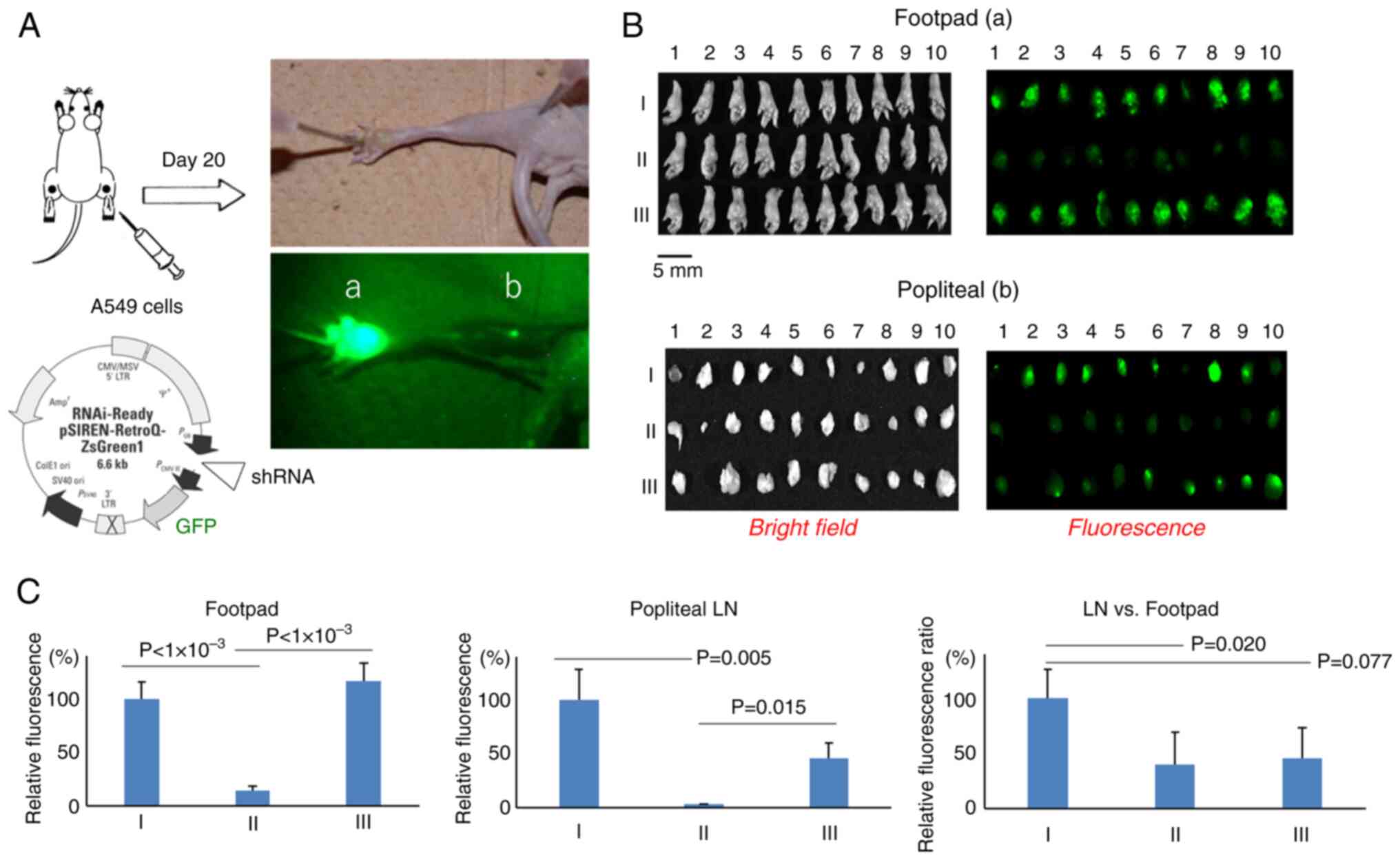 | Figure 3.Quantification of cells migrating
from the footpad to the popliteal LNs. (A) On day 20 after
inoculation of the cells into the footpad, the mice were sacrificed
and fixed, and the fluorescence in the (a) footpad and (b) LNs was
visualized and quantified. (B) Bright-field images (left) and
fluorescence images (right) of the resected footpad (top) and LNs
(bottom) are shown for: I, A549 cells harboring a control vector;
II, A549 cells expressing PRKDC shRNA; and III, A549 cells
expressing PSMB4 shRNA. Scale bar, 5 mm. (C) Relative fluorescence
levels of control vector (I), PRKDC shRNA (II) and PSMB4 shRNA
(III) in the footpad (left) and LN (middle), and the LN/footpad
fluorescence ratio (right). Data are presented as the mean ±
standard error of the mean. LN, lymph node; PRKDC, DNA-dependent
protein kinase catalytic subunit; PSMB4, proteasome subunit β
type-4; shRNA, short hairpin RNA; GFP, green fluorescent
protein. |
The present study subsequently examined whether
known inhibitors of PRKDC (DNA-PK) or PSMB4 (proteasome) could
alter the migration of cells. The inhibitors were injected
intravenously into the tails of mice that had been inoculated with
A549 cells harboring a control vector expressing GFP. NU7441, a
DNA-PK inhibitor, did not significantly change the fluorescence
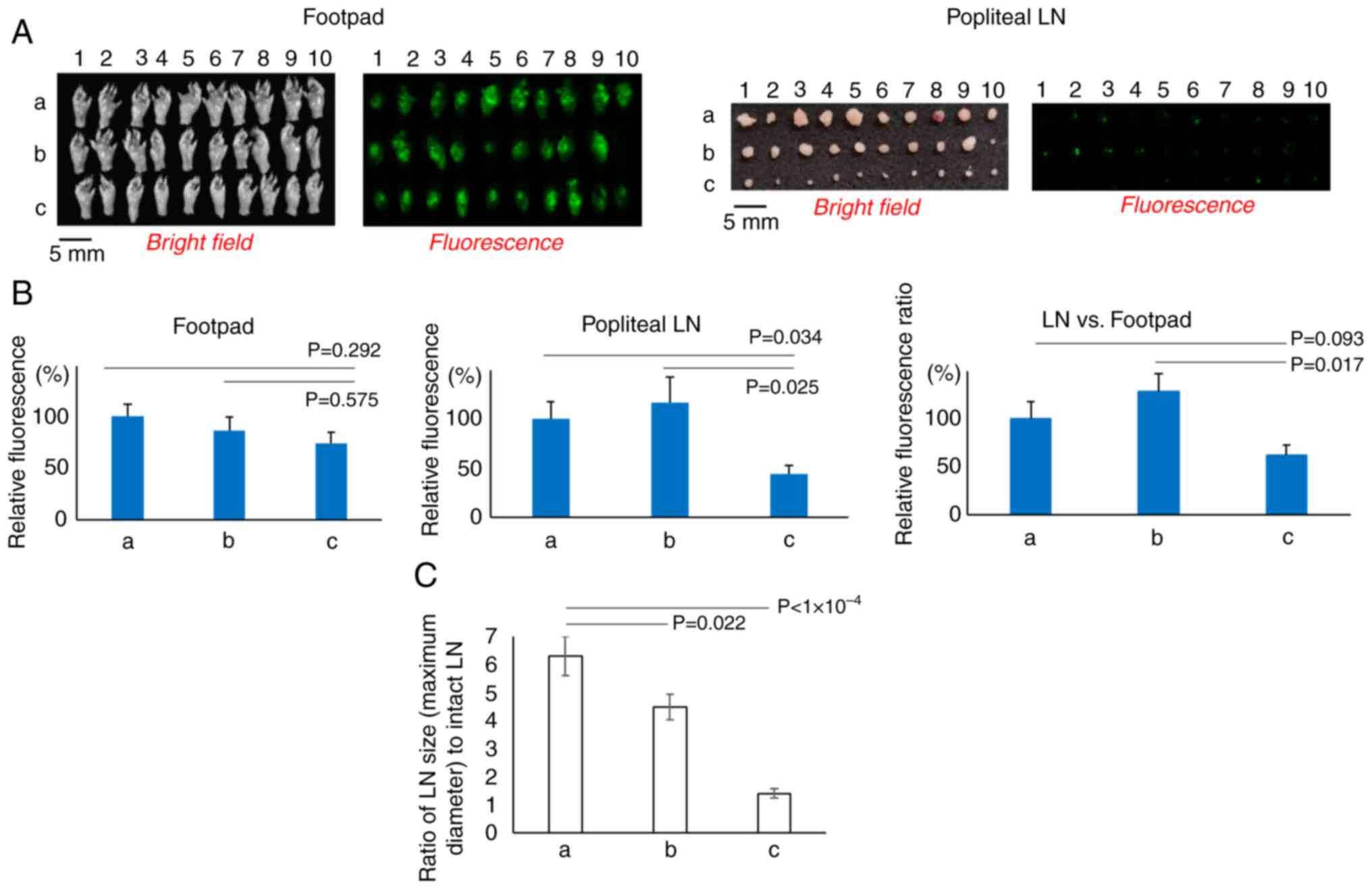
intensity in either the footpads or popliteal LNs (Fig. 4A and B). By contrast, bortezomib, a
proteasome inhibitor, markedly retarded the migration of the cells
to the popliteal LNs, although the cell growth rate in the footpads
was comparable to that in the control animals, resulting in a lower
popliteal/footpad fluorescence intensity ratio compared with that
in the control group (P=0.093) and in the animals injected with
NU7441 (P=0.017) (Fig. 4B).
A marked difference in the size of the metastatic
LNs was observed between the groups that were and were not treated
with bortezomib (Fig. 4C). Compared
with the control group (vehicle), the bortezomib-treated group
showed significantly smaller (undeveloped) tumors at the metastatic
sites (LNs; P<10−4). The mean LN maximum diameters in
the group treated with vehicle and in the group treated with NU7447
(DNA-PK inhibitor) were 6.3- and 4.5-fold greater than the diameter
of intact LNs, whereas they were only 1.4-fold greater in the group
treated with bortezomib.
Discussion
Our previous study analyzed tumor mRNA samples from
60 patients with CUP using microarray analysis and constructed a
normalized gene expression profile specific to CUPs, which
identified a number of genes that were upregulated in the CUP
(8).
In the present study, to further narrow down the
genes closely related to the development of CUP among these
candidate genes, cell-based siRNA screening was performed in
vitro. A549 and MDA231 cells were selected for the screening
because they are among the most widely used cell lines as models
for research on the metastasis of lung cancer and breast cancer,
respectively (30,31). Furthermore, our previous study
demonstrated that the gene expression profile of CUP closely
resembled that of lung adenocarcinoma (8).
Individual knockdown of several candidate genes in
A549 or MDA231 cells using specific siRNAs resulted in restricted
migration of the cells. siRNAs against PRKDC and PSMB4 restricted
the migration in both cell lines, suggesting that these genes might
be involved in the metastatic ability of CUP. Furthermore, shRNAs
for PRKDC and PSMB4 also suppressed the migration of A549 cells
in vivo, with significance observed for shRNA for PRKDC,
whereas knockdown of PRKDC tended to slow the growth of the cells
as well. The PRKDC gene encodes DNA-PKcs, a large subunit (~469
KDa) of DNA-PK, which is an abundantly expressed kinase in higher
eukaryotes (32). As a DNA damage
repair protein, it drives several pathways that promote metastasis
and tumor growth (33,34). In melanoma cells, DNA-PKcs may
control the secretion of numerous proteins involved in metastasis,
such as matrix metalloproteinases, thereby regulating the tumor
microenvironment (35). A similar
regulation in favor of metastasis could occur in CUPs exhibiting
upregulation of DNA-PKcs (8).
PSMB4 encodes the β7 subunit of proteasome 20S. The
20S proteasome is the catalytic core of the proteasome complex with
a concentric circular structure, including two α rings and two β
rings, each ring consisting of 7 subunits, α1-7 and β1-7,
respectively (36). Proteasomes
affect tumor development by regulating tumor signaling pathways,
including NF-κB signaling. They recognize and degrade ubiquitinated
IκB, an inhibitor of NF-κB, thereby activating NF-κB signaling
(37). Proteasomes are also
required to maintain homeostasis in proliferating tumor cells by
eliminating the accumulation of misfolded proteins (38). In multiple myeloma (MM), plasma
cells secrete several immunoglobulins, which are macromolecules
that are synthesized and folded in the endoplasmic reticulum (ER)
(36). Therefore, proteasome
inhibitors that are widely used in the treatment of MM can block
the degradation of IκB and trigger the accumulation of misfolded
proteins, inhibiting NF-κB activity and inducing ER stress,
respectively, ultimately leading to cell death of the MM cells
(39,40). Furthermore, DNA-PKcs is an
additional target that is cleaved and inactivated by proteasome
inhibitors in MM cells (40,41).
In the present study, bortezomib, the first
proteasome inhibitor developed as a chemotherapy drug (29), markedly restricted the metastatic
ability of A549 cells in vivo. The target of this drug has
been identified as the β5 subunit of proteasome 20S (39). Subunits other than the β7 subunit
(encoded by PSMB4) include the β5 subunit, which was also
upregulated in both CUPs (GSE42392) (8) and A549 cells compared with small
airway epithelial cells (GSE4824) (42) (Table
SIII). The trend was also noted in other tumors located in the
liver or head and neck regions (Table
SIII), with each of these tumors identified as a potential
treatment target for bortezomib (36,37).
The development of immune checkpoint inhibitors
(ICIs) has markedly changed the treatment paradigms for numerous
cancer types. Among patients with CUP, 28% exhibit one or more
predictive biomarkers for ICIs. For example, programmed
death-ligand 1 is expressed on ≥5% tumor cells in 22.5% or on
lymphocytes in 58.7% of such patients. Microsatellite
instability-high is observed in 1.8% and tumor mutational burden
(TMB) ≥17 mutations per megabase of the tumor genome is present in
11.8% of these patients (43).
Although these biomarkers have not yet been
validated in patients with CUP, those with CUP with TMB >10
mutations per megabase generally experience improved outcomes when
treated with ICIs (43), and our
previous study recently demonstrated the clinical benefits of
nivolumab in patients with CUP (44). The present study demonstrated a
putative role for the proteasome in the progression of CUP that
could be prevented by proteasome-targeted therapy. Proteasome
inhibitors combined with ICIs may be an additional therapeutic
option for CUP, for which there are limited treatment options.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Mrs. Tomoko Kitayama
(Department of Genome Biology, Kindai University Faculty of
Medicine, Osaka-Sayama, Japan) and Mr. Kentaro Egawa (Center for
Animal Experiment, Kindai University Faculty of Medicine,
Osaka-Sayama, Japan) for help with the animal experiments.
Funding
The present study was supported by the Grant-in-Aid for
Scientific Research (C) (grant no. 17K07204) of Ministry of
Education, Culture, Sports, Science and Technology of Japan, and by
the Japan Agency for Medical Research and Development (grant no.
201438137A).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YF designed the study and drafted the manuscript. YF
and MADV performed the experiments and collected data. YF, MADV,
HH, KNa and KNi analyzed and interpreted the data, and revised the
manuscript. YF and MADV confirmed the authenticity of all the raw
data. All authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Animal Care Committee of the Kindai University Faculty of Medicine
(approval no. KDMS-25-019; Osaka-Sayama, Japan) and carried out in
compliance with the standards for the use of laboratory
animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CUP
|
cancer of unknown primary
|
|
PRKDC
|
protein kinase DNA-activated catalytic
subunit
|
|
PSMB4
|
proteasome subunit β type-4
|
References
|
1
|
Pavlidis N and Pentheroudakis G: Cancer of
unknown primary site. Lancet. 379:1428–1435. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rassy E, Parent P, Lefort F, Boussios S,
Baciarello G and Pavlidis N: New rising entities in cancer of
unknown primary: Is there a real therapeutic benefit? Crit Rev
Oncol Hematol. 147:1028822020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Greco FA and Pavlidis N: Treatment for
patients with unknown primary carcinoma and unfavorable prognostic
factors. Semin Oncol. 36:65–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pavlidis N, Khaled H and Gaafar R: A mini
review on cancer of unknown primary site: A clinical puzzle for the
oncologists. J Adv Res. 6:375–382. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fizazi K, Greco FA, Pavlidis N, Daugaard
G, Oien K and Pentheroudakis G; ESMO Guidelines Committee, :
Cancers of unknown primary site: ESMO clinical practice guidelines
for diagnosis, treatment and follow-up. Ann Oncol. 26 (Suppl
5):v133–v138. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hayashi H, Kurata T, Takiguchi Y, Arai M,
Takeda K, Akiyoshi K, Matsumoto K, Onoe T, Mukai H, Matsubara N, et
al: Randomized phase II trial comparing site-specific treatment
based on gene expression profiling with carboplatin and paclitaxel
for patients with cancer of unknown primary site. J Clin Oncol.
37:570–579. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rassy E, Labaki C, Chebel R, Boussios S,
Smith-Gagen J, Greco FA and Pavlidis N: Systematic review of the
CUP trials characteristics and perspectives for next-generation
studies. Cancer Treat Rev. 107:1024072022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kurahashi I, Fujita Y, Arao T, Kurata T,
Koh Y, Sakai K, Matsumoto K, Tanioka M, Takeda K, Takiguchi Y, et
al: A microarray-based gene expression analysis to identify
diagnostic biomarkers for unknown primary cancer. PLoS One.
8:e632492013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arao T, Fukumoto H, Takeda M, Tamura T,
Saijo N and Nishio K: Small in-frame deletion in the epidermal
growth factor receptor as a target for ZD6474. Cancer Res.
64:9101–9104. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tanaka K, Arao T, Maegawa M, Matsumoto K,
Kaneda H, Kudo K, Fujita Y, Yokote H, Yanagihara K, Yamada Y, et
al: SRPX2 is overexpressed in gastric cancer and promotes cellular
migration and adhesion. Int J Cancer. 124:1072–1080. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Clough E and Barrett T: The gene
expression omnibus database. Methods Mol Biol. 1418:93–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lenburg ME, Liou LS, Gerry NP, Frampton
GM, Cohen HT and Christman MF: Previously unidentified changes in
renal cell carcinoma gene expression identified by parametric
analysis of microarray data. BMC Cancer. 3:312003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pawitan Y, Bjöhle J, Amler L, Borg AL,
Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, et al: Gene
expression profiling spares early breast cancer patients from
adjuvant therapy: Derived and validated in two population-based
cohorts. Breast Cancer Res. 7:R953–R964. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
International Genetics Consortium, .
Expression project for oncology (expO). Gene Expression Omnibus,
GSE2109. 2005.Available from:. http://www.intgen.org/
|
|
16
|
Luesch H, Chanda SK, Raya RM, DeJesus PD,
Orth AP, Walker JR, Belmonte JCI and Schultz PG: A functional
genomics approach to the mode of action of apratoxin A. Nat Chem
Biol. 2:158–167. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bild AH, Yao G, Chang JT, Wang Q, Potti A,
Chasse D, Joshi NR, Harpole D, Lancaster JM, Berchuck A, et al:
Oncogenic pathway signatures in human cancers as a guide to
targeted therapies. Nature. 439:353–357. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dyrskjøt L, Kruhøffer M, Thykjaer T,
Marcussen N, Jensen JL, Møller K and Ørntoft TF: Gene expression in
the urinary bladder: A common carcinoma in situ gene expression
signature exists disregarding histopathological classification.
Cancer Res. 64:4040–4048. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gemma A, Li C, Sugiyama Y, Matsuda K,
Seike Y, Kosaihira S, Minegishi Y, Noro R, Nara M, Seike M, et al:
Anticancer drug clustering in lung cancer based on gene expression
profiles and sensitivity database. BMC Cancer. 6:1742006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rinaldi A, Kwee I, Taborelli M, Largo C,
Uccella S, Martin V, Poretti G, Gaidano G, Calabrese G, Martinelli
G, et al: Genomic and expression profiling identifies the B-cell
associated tyrosine kinase Syk as a possible therapeutic target in
mantle cell lymphoma. Br J Haematol. 132:303–316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bachtiary B, Boutros PC, Pintilie M, Shi
W, Bastianutto C, Li JH, Schwock J, Zhang W, Penn LZ, Jurisica I,
et al: Gene expression profiling in cervical cancer: An exploration
of intratumor heterogeneity. Clin Cancer Res. 12:5632–5640. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pyeon D, Newton MA, Lambert PF, den Boon
JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH,
Smith EM, et al: Fundamental differences in cell cycle deregulation
in human papillomavirus-positive and human papillomavirus-negative
head/neck and cervical cancers. Cancer Res. 67:4605–4619. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Xia XQ, Jia Z, Sawyers A, Yao H,
Wang-Rodriquez J, Mercola D and McClelland M: In silico estimates
of tissue components in surgical samples based on expression
profiling data. Cancer Res. 70:6448–6455. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kaneda H, Arao T, Tanaka K, Tamura D,
Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y,
et al: FOXQ1 is overexpressed in colorectal cancer and enhances
tumorigenicity and tumor growth. Cancer Res. 70:2053–2063. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Picot J, Guerin CL, Le Van Kim C and
Boulanger CM: Flow cytometry: Retrospective, fundamentals and
recent instrumentation. Cytotechnology. 64:109–130. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen Z, Zhuo W, Wang Y, Ao X and An J:
Down-regulation of layilin, a novel hyaluronan receptor, via RNA
interference, inhibits invasion and lymphatic metastasis of human
lung A549 cells. Biotechnol Appl Biochem. 50:89–96. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kura Y, De Velasco MA, Sakai K, Uemura H,
Fujita K and Nishio K: Exploring the relationship between
ulcerative colitis, colorectal cancer, and prostate cancer. Hum
Cell. 37:1706–1718. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao Y, Thomas HD, Batey A, Cowell IG,
Richardson CJ, Griffin RJ, Calvert AH, Newell DR, Smith GCM and
Curtin NJ: Preclinical evaluation of a potent novel DNA-dependent
protein kinase inhibitor NU7441. Cancer Res. 66:5354–5362. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen D, Frezza M, Schmitt S, Kanwar J and
Dou QP: Bortezomib as the first proteasome inhibitor anticancer
drug: Current status and future perspectives. Curr Cancer Drug
Targets. 11:239–253. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lengrand J, Pastushenko I, Vanuytven S,
Song Y, Venet D, Sarate RM, Bellina M, Moers V, Boinet A, Sifrim A,
et al: Pharmacological targeting netrin-1 inhibits EMT in cancer.
Nature. 620:402–408. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu K, Newbury PA, Glicksberg BS, Zeng
WZD, Paithankar S, Andrechek ER and Chen B: Evaluating cell lines
as models for metastatic breast cancer through integrative analysis
of genomic data. Nat Commun. 10:21382019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dylgjeri E and Knudsen KE: DNA-PKcs: A
targetable protumorigenic protein kinase. Cancer Res. 82:523–533.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xiang Z, Hou G, Zheng S, Lu M, Li T, Lin
Q, Liu H, Wang X, Guan T, Wei Y, et al: ER-associated degradation
ligase HRD1 links ER stress to DNA damage repair by modulating the
activity of DNA-PKcs. Proc Natl Acad Sci USA. 121:e24030381212024.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Caron P, Pankotai T, Wiegant WW,
Tollenaere MAX, Furst A, Bonhomme C, Helfricht A, de Groot A,
Pastink A, Vertegaal ACO, et al: WWP2 ubiquitylates RNA polymerase
II for DNA-PK-dependent transcription arrest and repair at DNA
breaks. Genes Dev. 33:684–704. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kotula E, Berthault N, Agrario C, Lienafa
MC, Simon A, Dingli F, Loew D, Sibut V, Saule S and Dutreix M:
DNA-PKcs plays role in cancer metastasis through regulation of
secreted proteins involved in migration and invasion. Cell Cycle.
14:1961–1972. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou X, Xu R, Wu Y, Zhou L and Xiang T:
The role of proteasomes in tumorigenesis. Genes Dis. 11:1010702023.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Narayanan S, Cai CY, Assaraf YG, Guo HQ,
Cui Q, Wei L, Huang JJ, Ashby CR Jr and Chen ZS: Targeting the
ubiquitin-proteasome pathway to overcome anti-cancer drug
resistance. Drug Resist Updat. 48:1006632020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chhabra S: Novel proteasome inhibitors and
histone deacetylase inhibitors: Progress in myeloma therapeutics.
Pharmaceuticals (Basel). 10:402017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gandolfi S, Laubach JP, Hideshima T,
Chauhan D, Anderson KC and Richardson PG: The proteasome and
proteasome inhibitors in multiple myeloma. Cancer Metastasis Rev.
36:561–584. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hideshima T and Anderson KC: Biologic
impact of proteasome inhibition in multiple myeloma cells-from the
aspects of preclinical studies. Semin Hematol. 49:223–227. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu YH, Hong CW, Wang YC, Huang WJ, Yeh YL,
Wang BJ, Wang YJ and Chiu HW: A novel histone deacetylase inhibitor
TMU-35435 enhances etoposide cytotoxicity through the proteasomal
degradation of DNA-PKcs in triple-negative breast cancer. Cancer
Lett. 400:79–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Byers LA, Diao L, Wang J, Saintigny P,
Girard L, Peyton M, Shen L, Fan Y, Giri U, Tumula PK, et al: An
epithelial-mesenchymal transition gene signature predicts
resistance to EGFR and PI3K inhibitors and identifies Axl as a
therapeutic target for overcoming EGFR inhibitor resistance. Clin
Cancer Res. 19:279–290. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rassy E, Boussios S and Pavlidis N:
Genomic correlates of response and resistance to immune checkpoint
inhibitors in carcinomas of unknown primary. Eur J Clin Invest.
51:e135832021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tanizaki J, Yonemori K, Akiyoshi K, Minami
H, Ueda H, Takiguchi Y, Miura Y, Segawa Y, Takahashi S, Iwamoto Y,
et al: Open-label phase II study of the efficacy of nivolumab for
cancer of unknown primary. Ann Oncol. 33:216–226. 2022. View Article : Google Scholar : PubMed/NCBI
|