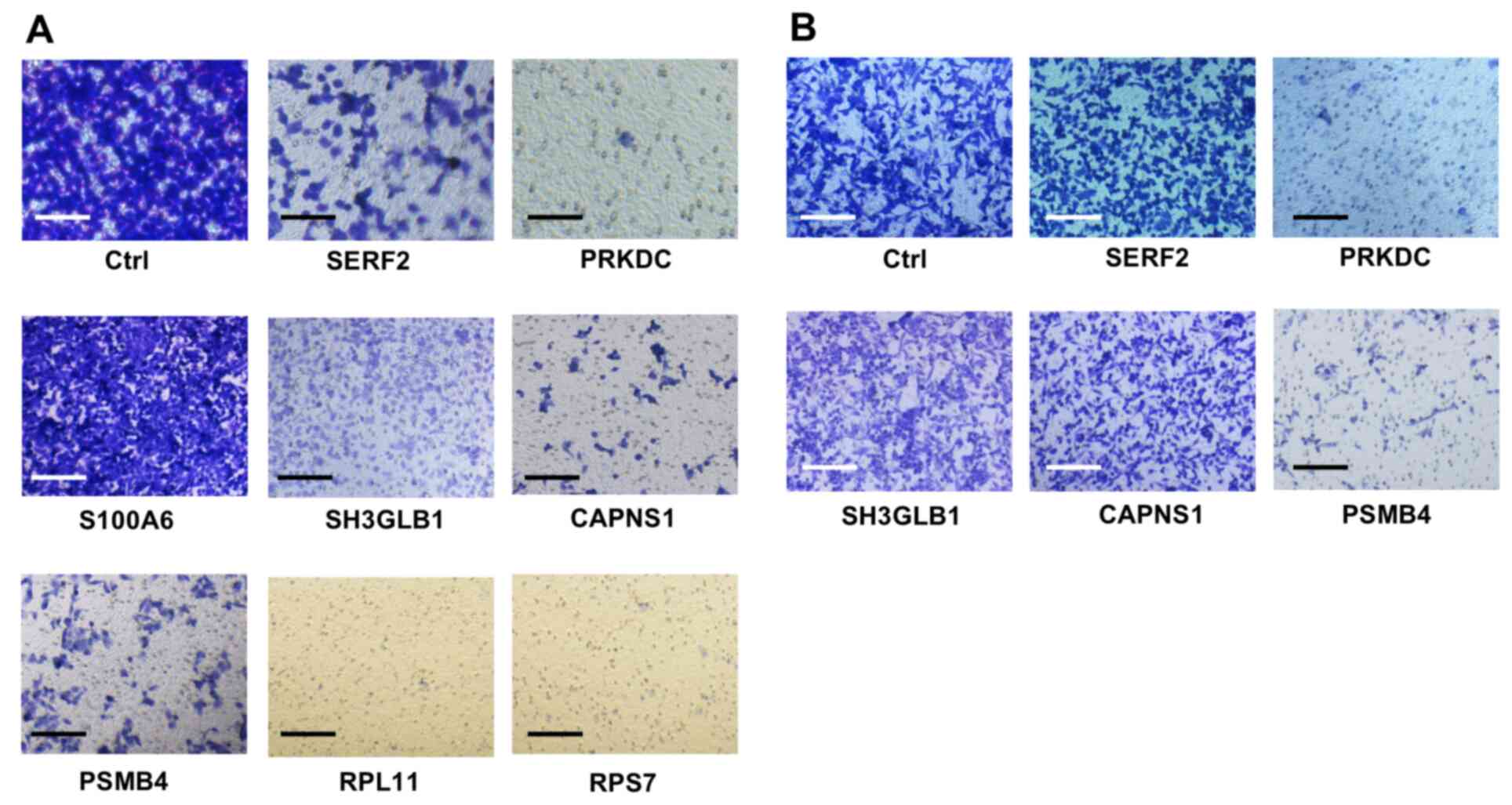
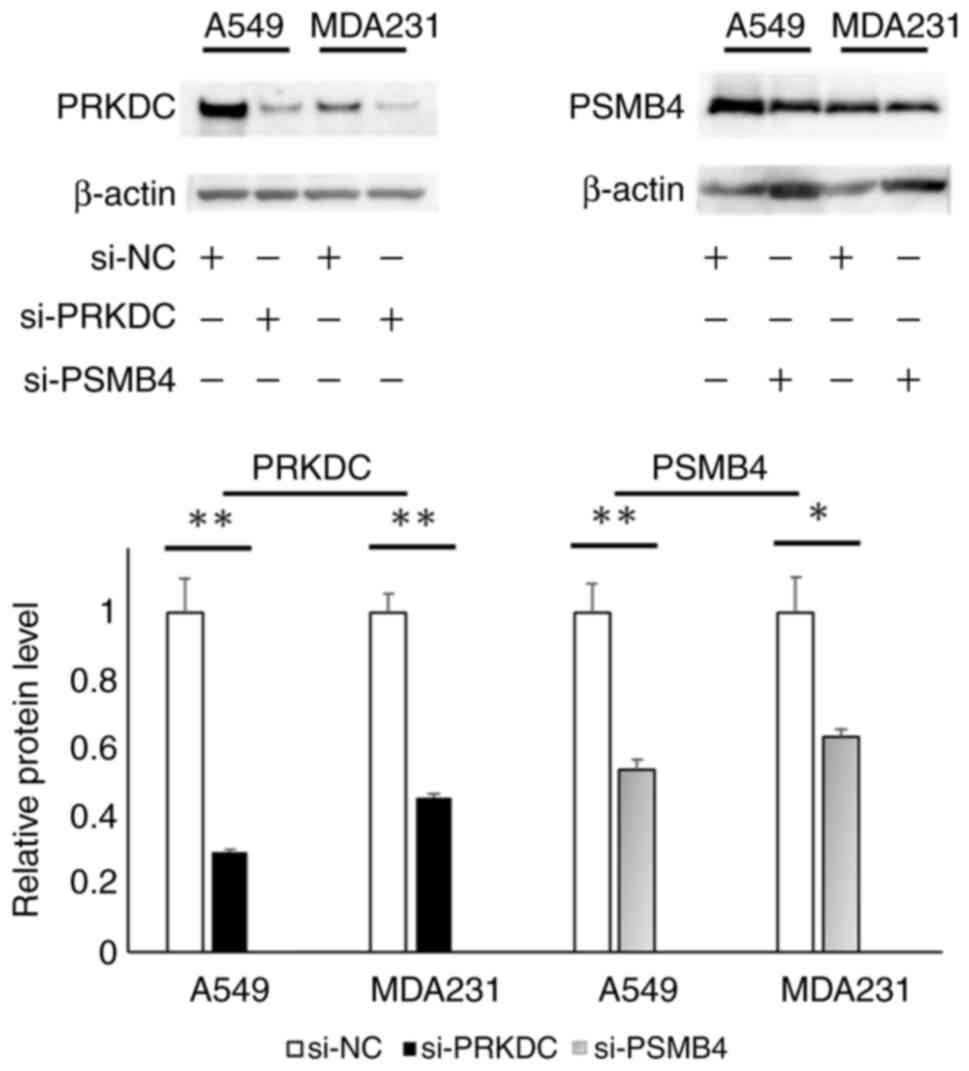
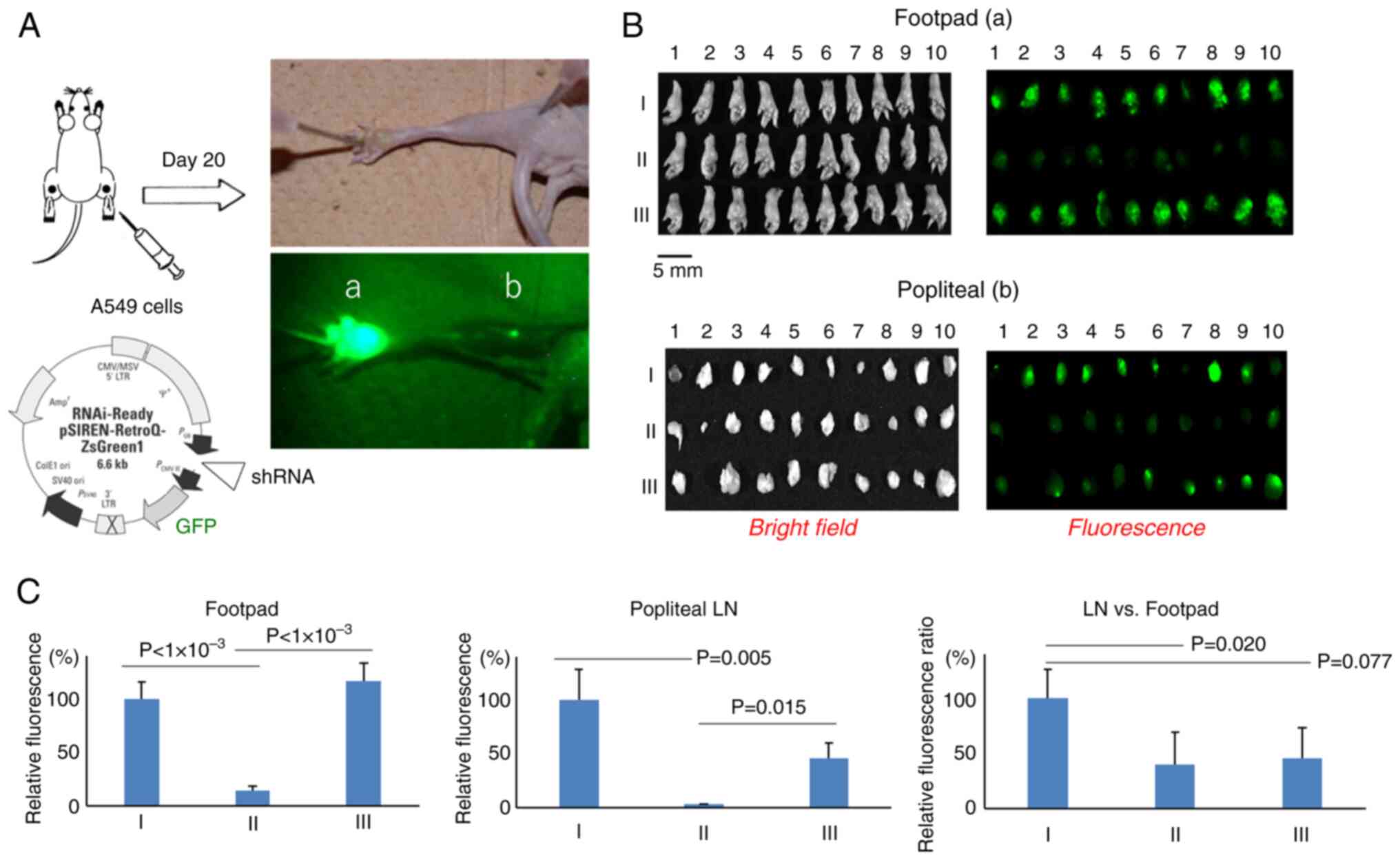
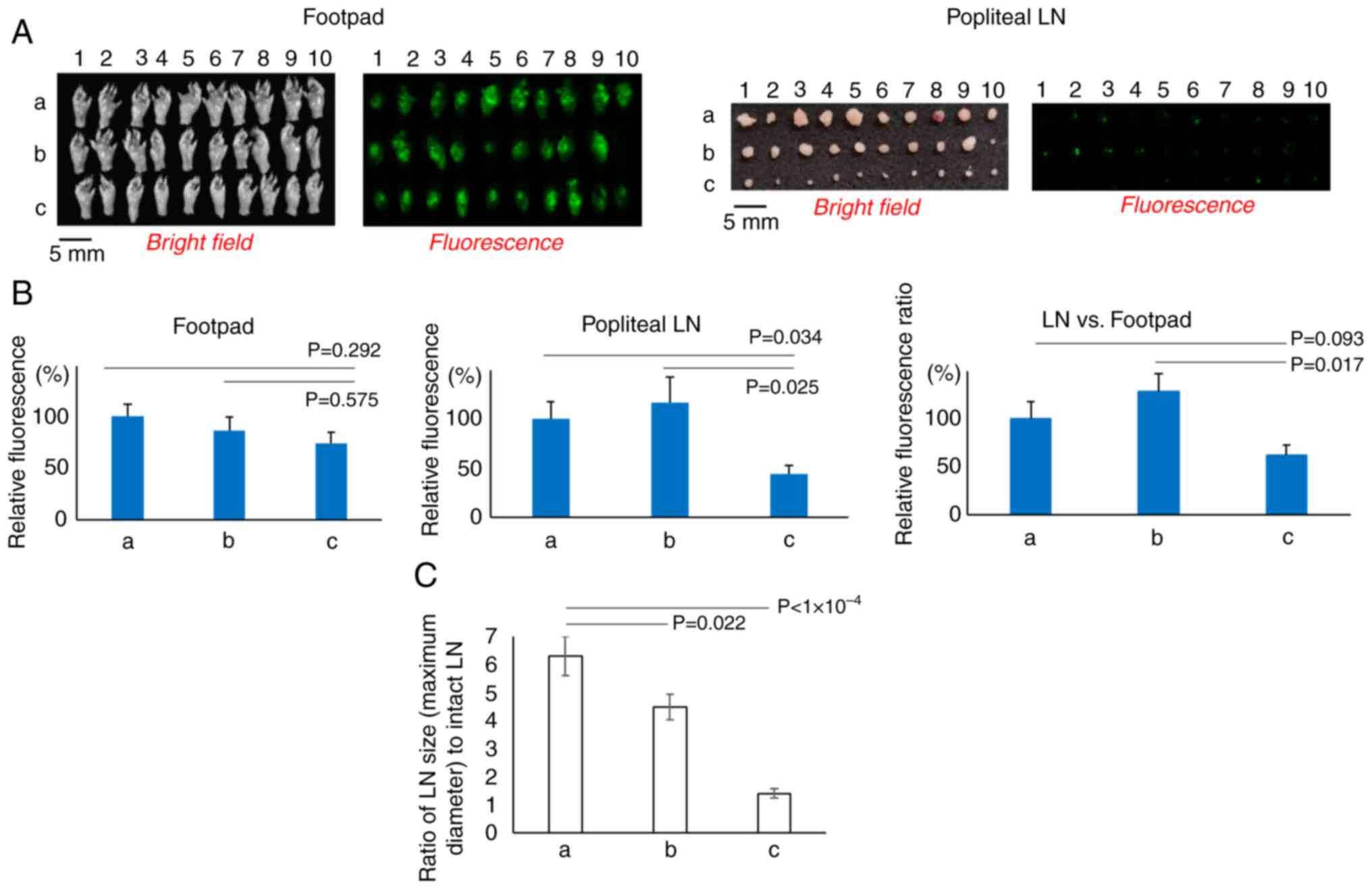
|
1
|
Pavlidis N and Pentheroudakis G: Cancer of
unknown primary site. Lancet. 379:1428–1435. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rassy E, Parent P, Lefort F, Boussios S,
Baciarello G and Pavlidis N: New rising entities in cancer of
unknown primary: Is there a real therapeutic benefit? Crit Rev
Oncol Hematol. 147:1028822020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Greco FA and Pavlidis N: Treatment for
patients with unknown primary carcinoma and unfavorable prognostic
factors. Semin Oncol. 36:65–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pavlidis N, Khaled H and Gaafar R: A mini
review on cancer of unknown primary site: A clinical puzzle for the
oncologists. J Adv Res. 6:375–382. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fizazi K, Greco FA, Pavlidis N, Daugaard
G, Oien K and Pentheroudakis G; ESMO Guidelines Committee, :
Cancers of unknown primary site: ESMO clinical practice guidelines
for diagnosis, treatment and follow-up. Ann Oncol. 26 (Suppl
5):v133–v138. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hayashi H, Kurata T, Takiguchi Y, Arai M,
Takeda K, Akiyoshi K, Matsumoto K, Onoe T, Mukai H, Matsubara N, et
al: Randomized phase II trial comparing site-specific treatment
based on gene expression profiling with carboplatin and paclitaxel
for patients with cancer of unknown primary site. J Clin Oncol.
37:570–579. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rassy E, Labaki C, Chebel R, Boussios S,
Smith-Gagen J, Greco FA and Pavlidis N: Systematic review of the
CUP trials characteristics and perspectives for next-generation
studies. Cancer Treat Rev. 107:1024072022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kurahashi I, Fujita Y, Arao T, Kurata T,
Koh Y, Sakai K, Matsumoto K, Tanioka M, Takeda K, Takiguchi Y, et
al: A microarray-based gene expression analysis to identify
diagnostic biomarkers for unknown primary cancer. PLoS One.
8:e632492013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arao T, Fukumoto H, Takeda M, Tamura T,
Saijo N and Nishio K: Small in-frame deletion in the epidermal
growth factor receptor as a target for ZD6474. Cancer Res.
64:9101–9104. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tanaka K, Arao T, Maegawa M, Matsumoto K,
Kaneda H, Kudo K, Fujita Y, Yokote H, Yanagihara K, Yamada Y, et
al: SRPX2 is overexpressed in gastric cancer and promotes cellular
migration and adhesion. Int J Cancer. 124:1072–1080. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Clough E and Barrett T: The gene
expression omnibus database. Methods Mol Biol. 1418:93–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lenburg ME, Liou LS, Gerry NP, Frampton
GM, Cohen HT and Christman MF: Previously unidentified changes in
renal cell carcinoma gene expression identified by parametric
analysis of microarray data. BMC Cancer. 3:312003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pawitan Y, Bjöhle J, Amler L, Borg AL,
Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, et al: Gene
expression profiling spares early breast cancer patients from
adjuvant therapy: Derived and validated in two population-based
cohorts. Breast Cancer Res. 7:R953–R964. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
International Genetics Consortium, .
Expression project for oncology (expO). Gene Expression Omnibus,
GSE2109. 2005.Available from:. http://www.intgen.org/
|
|
16
|
Luesch H, Chanda SK, Raya RM, DeJesus PD,
Orth AP, Walker JR, Belmonte JCI and Schultz PG: A functional
genomics approach to the mode of action of apratoxin A. Nat Chem
Biol. 2:158–167. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bild AH, Yao G, Chang JT, Wang Q, Potti A,
Chasse D, Joshi NR, Harpole D, Lancaster JM, Berchuck A, et al:
Oncogenic pathway signatures in human cancers as a guide to
targeted therapies. Nature. 439:353–357. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dyrskjøt L, Kruhøffer M, Thykjaer T,
Marcussen N, Jensen JL, Møller K and Ørntoft TF: Gene expression in
the urinary bladder: A common carcinoma in situ gene expression
signature exists disregarding histopathological classification.
Cancer Res. 64:4040–4048. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gemma A, Li C, Sugiyama Y, Matsuda K,
Seike Y, Kosaihira S, Minegishi Y, Noro R, Nara M, Seike M, et al:
Anticancer drug clustering in lung cancer based on gene expression
profiles and sensitivity database. BMC Cancer. 6:1742006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rinaldi A, Kwee I, Taborelli M, Largo C,
Uccella S, Martin V, Poretti G, Gaidano G, Calabrese G, Martinelli
G, et al: Genomic and expression profiling identifies the B-cell
associated tyrosine kinase Syk as a possible therapeutic target in
mantle cell lymphoma. Br J Haematol. 132:303–316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bachtiary B, Boutros PC, Pintilie M, Shi
W, Bastianutto C, Li JH, Schwock J, Zhang W, Penn LZ, Jurisica I,
et al: Gene expression profiling in cervical cancer: An exploration
of intratumor heterogeneity. Clin Cancer Res. 12:5632–5640. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pyeon D, Newton MA, Lambert PF, den Boon
JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH,
Smith EM, et al: Fundamental differences in cell cycle deregulation
in human papillomavirus-positive and human papillomavirus-negative
head/neck and cervical cancers. Cancer Res. 67:4605–4619. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Xia XQ, Jia Z, Sawyers A, Yao H,
Wang-Rodriquez J, Mercola D and McClelland M: In silico estimates
of tissue components in surgical samples based on expression
profiling data. Cancer Res. 70:6448–6455. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kaneda H, Arao T, Tanaka K, Tamura D,
Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y,
et al: FOXQ1 is overexpressed in colorectal cancer and enhances
tumorigenicity and tumor growth. Cancer Res. 70:2053–2063. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Picot J, Guerin CL, Le Van Kim C and
Boulanger CM: Flow cytometry: Retrospective, fundamentals and
recent instrumentation. Cytotechnology. 64:109–130. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen Z, Zhuo W, Wang Y, Ao X and An J:
Down-regulation of layilin, a novel hyaluronan receptor, via RNA
interference, inhibits invasion and lymphatic metastasis of human
lung A549 cells. Biotechnol Appl Biochem. 50:89–96. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kura Y, De Velasco MA, Sakai K, Uemura H,
Fujita K and Nishio K: Exploring the relationship between
ulcerative colitis, colorectal cancer, and prostate cancer. Hum
Cell. 37:1706–1718. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao Y, Thomas HD, Batey A, Cowell IG,
Richardson CJ, Griffin RJ, Calvert AH, Newell DR, Smith GCM and
Curtin NJ: Preclinical evaluation of a potent novel DNA-dependent
protein kinase inhibitor NU7441. Cancer Res. 66:5354–5362. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen D, Frezza M, Schmitt S, Kanwar J and
Dou QP: Bortezomib as the first proteasome inhibitor anticancer
drug: Current status and future perspectives. Curr Cancer Drug
Targets. 11:239–253. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lengrand J, Pastushenko I, Vanuytven S,
Song Y, Venet D, Sarate RM, Bellina M, Moers V, Boinet A, Sifrim A,
et al: Pharmacological targeting netrin-1 inhibits EMT in cancer.
Nature. 620:402–408. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu K, Newbury PA, Glicksberg BS, Zeng
WZD, Paithankar S, Andrechek ER and Chen B: Evaluating cell lines
as models for metastatic breast cancer through integrative analysis
of genomic data. Nat Commun. 10:21382019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dylgjeri E and Knudsen KE: DNA-PKcs: A
targetable protumorigenic protein kinase. Cancer Res. 82:523–533.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xiang Z, Hou G, Zheng S, Lu M, Li T, Lin
Q, Liu H, Wang X, Guan T, Wei Y, et al: ER-associated degradation
ligase HRD1 links ER stress to DNA damage repair by modulating the
activity of DNA-PKcs. Proc Natl Acad Sci USA. 121:e24030381212024.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Caron P, Pankotai T, Wiegant WW,
Tollenaere MAX, Furst A, Bonhomme C, Helfricht A, de Groot A,
Pastink A, Vertegaal ACO, et al: WWP2 ubiquitylates RNA polymerase
II for DNA-PK-dependent transcription arrest and repair at DNA
breaks. Genes Dev. 33:684–704. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kotula E, Berthault N, Agrario C, Lienafa
MC, Simon A, Dingli F, Loew D, Sibut V, Saule S and Dutreix M:
DNA-PKcs plays role in cancer metastasis through regulation of
secreted proteins involved in migration and invasion. Cell Cycle.
14:1961–1972. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou X, Xu R, Wu Y, Zhou L and Xiang T:
The role of proteasomes in tumorigenesis. Genes Dis. 11:1010702023.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Narayanan S, Cai CY, Assaraf YG, Guo HQ,
Cui Q, Wei L, Huang JJ, Ashby CR Jr and Chen ZS: Targeting the
ubiquitin-proteasome pathway to overcome anti-cancer drug
resistance. Drug Resist Updat. 48:1006632020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chhabra S: Novel proteasome inhibitors and
histone deacetylase inhibitors: Progress in myeloma therapeutics.
Pharmaceuticals (Basel). 10:402017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gandolfi S, Laubach JP, Hideshima T,
Chauhan D, Anderson KC and Richardson PG: The proteasome and
proteasome inhibitors in multiple myeloma. Cancer Metastasis Rev.
36:561–584. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hideshima T and Anderson KC: Biologic
impact of proteasome inhibition in multiple myeloma cells-from the
aspects of preclinical studies. Semin Hematol. 49:223–227. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu YH, Hong CW, Wang YC, Huang WJ, Yeh YL,
Wang BJ, Wang YJ and Chiu HW: A novel histone deacetylase inhibitor
TMU-35435 enhances etoposide cytotoxicity through the proteasomal
degradation of DNA-PKcs in triple-negative breast cancer. Cancer
Lett. 400:79–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Byers LA, Diao L, Wang J, Saintigny P,
Girard L, Peyton M, Shen L, Fan Y, Giri U, Tumula PK, et al: An
epithelial-mesenchymal transition gene signature predicts
resistance to EGFR and PI3K inhibitors and identifies Axl as a
therapeutic target for overcoming EGFR inhibitor resistance. Clin
Cancer Res. 19:279–290. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rassy E, Boussios S and Pavlidis N:
Genomic correlates of response and resistance to immune checkpoint
inhibitors in carcinomas of unknown primary. Eur J Clin Invest.
51:e135832021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tanizaki J, Yonemori K, Akiyoshi K, Minami
H, Ueda H, Takiguchi Y, Miura Y, Segawa Y, Takahashi S, Iwamoto Y,
et al: Open-label phase II study of the efficacy of nivolumab for
cancer of unknown primary. Ann Oncol. 33:216–226. 2022. View Article : Google Scholar : PubMed/NCBI
|