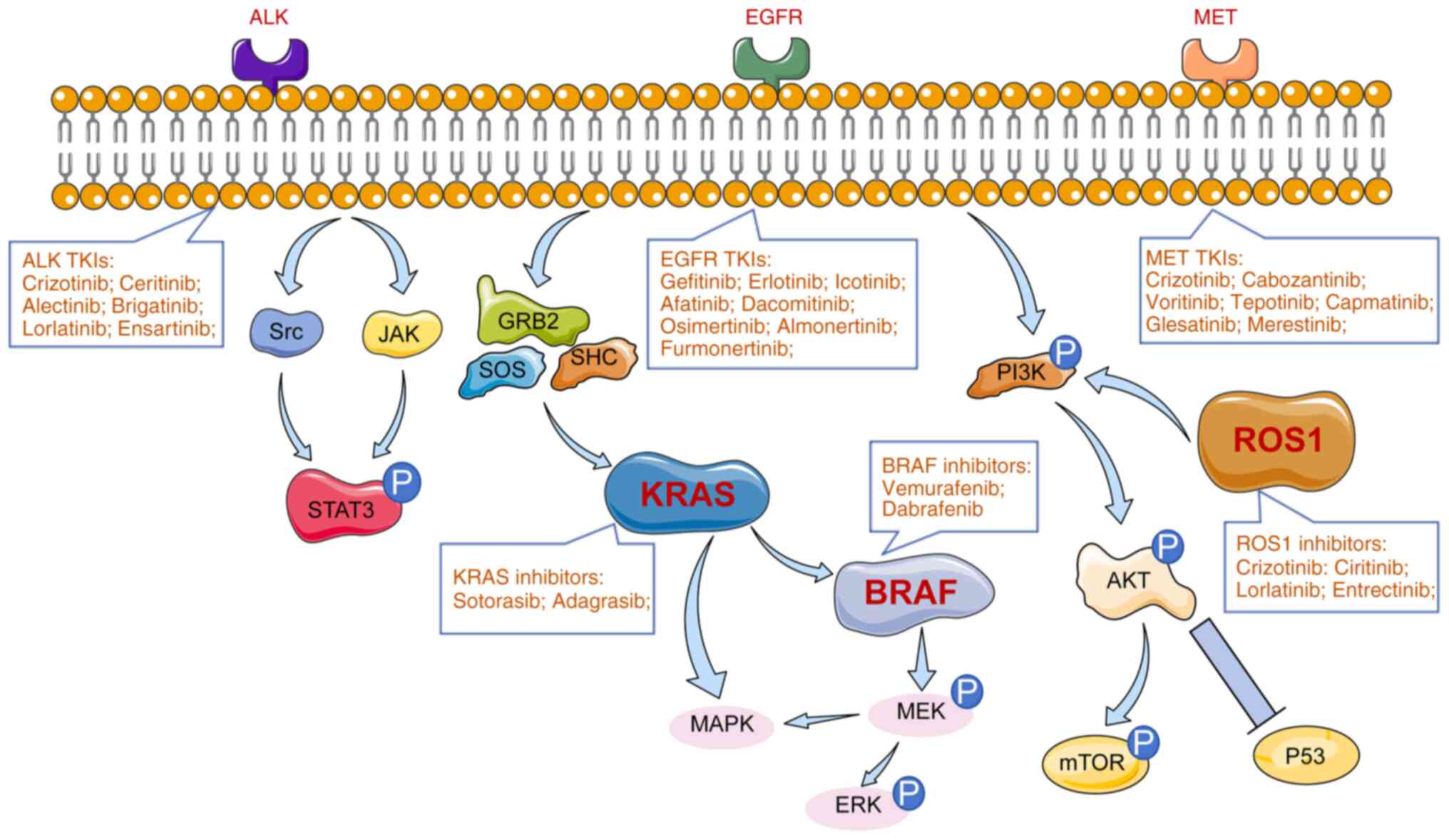
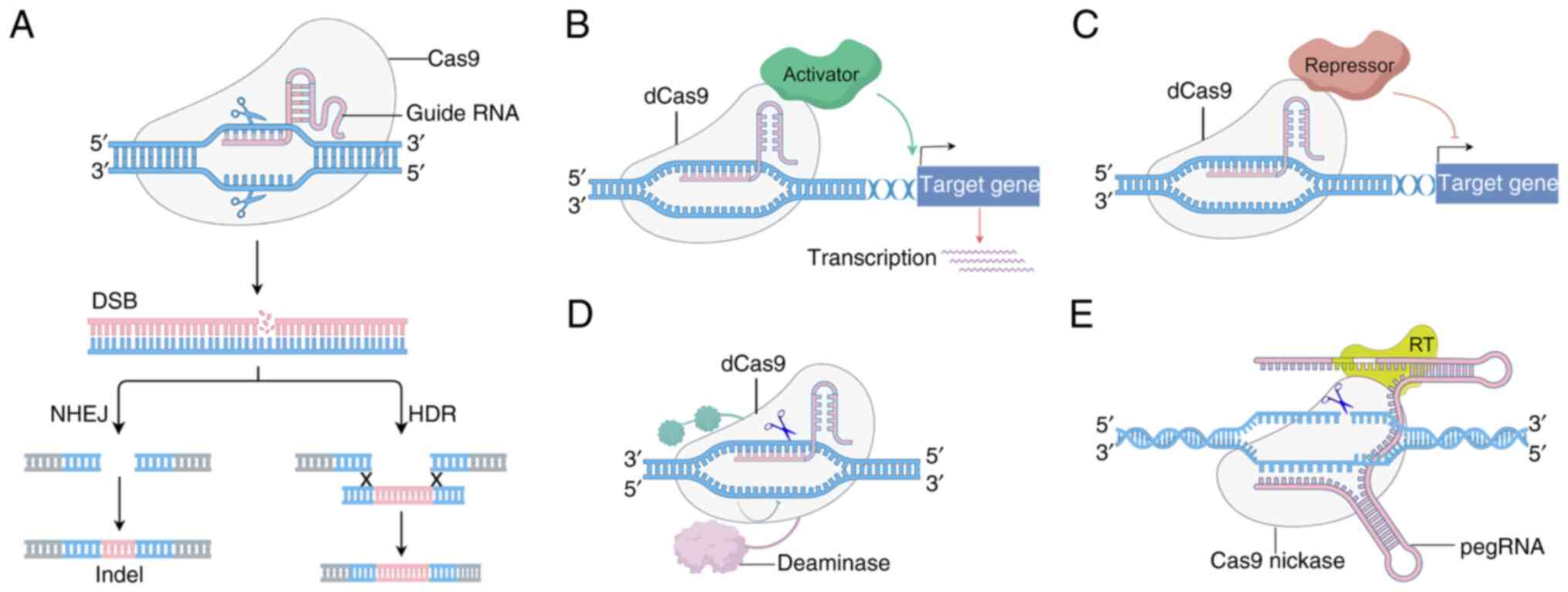
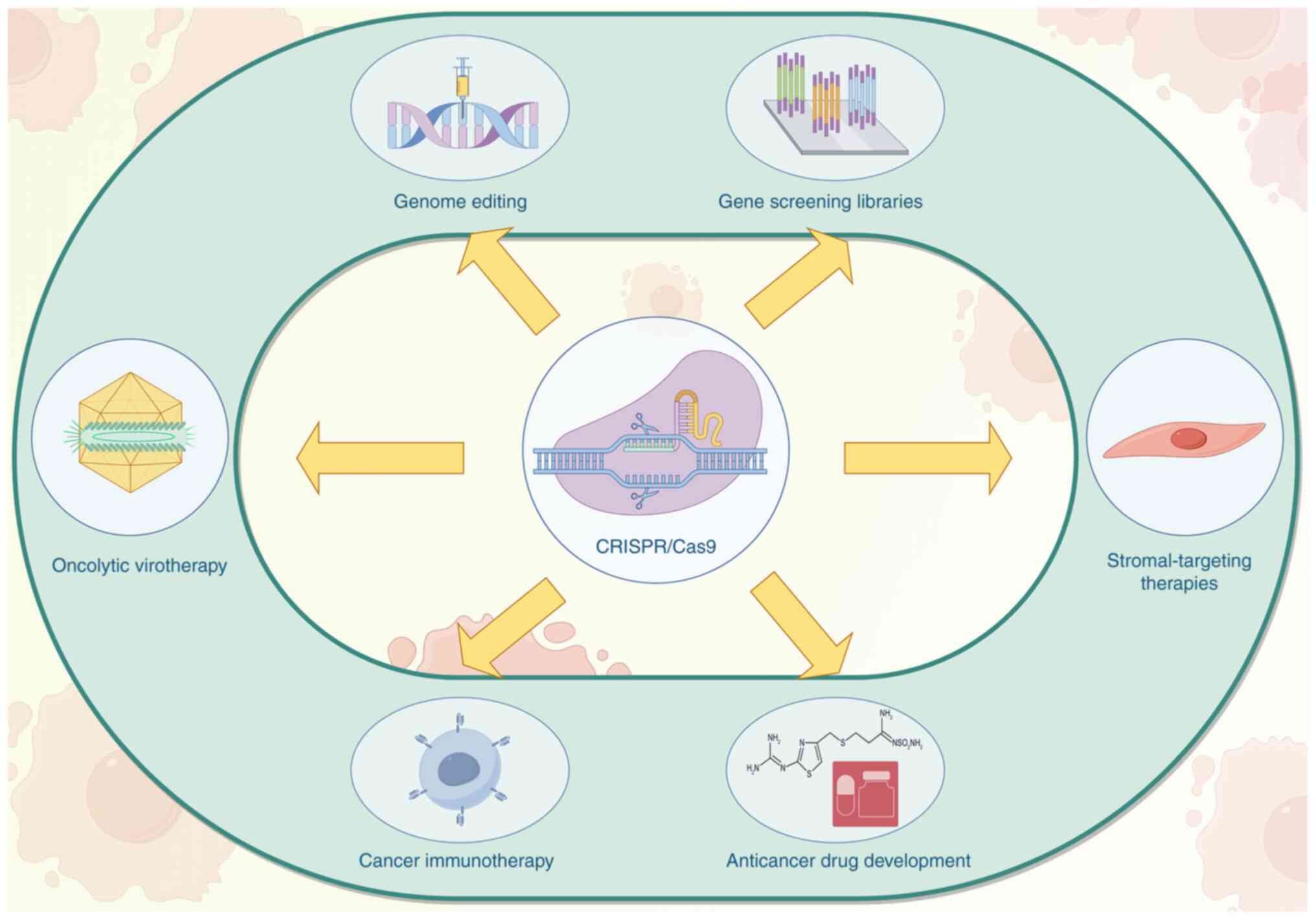
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cooper AJ, Kobayashi Y, Kim D, Clifford
SE, Kravets S, Dahlberg SE, Chambers ES, Li J, Rangachari D, Nguyen
T, et al: Identification of a RAS-activating TMEM87A-RASGRF1 fusion
in an exceptional responder to sunitinib with non-small cell lung
cancer. Clin Cancer Res. 26:4072–4079. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsuji T, Ozasa H, Aoki W, Aburaya S,
Yamamoto Funazo T, Furugaki K, Yoshimura Y, Yamazoe M, Ajimizu H,
Yasuda Y, et al: YAP1 mediates survival of ALK-rearranged lung
cancer cells treated with alectinib via pro-apoptotic protein
regulation. Nat Commun. 11:742020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jamroskovic J, Doimo M, Chand K, Obi I,
Kumar R, Brännström K, Hedenström M, Nath Das R, Akhunzianov A,
Deiana M, et al: Quinazoline ligands induce cancer cell death
through selective STAT3 inhibition and G-quadruplex stabilization.
J Am Chem Soc. 142:2876–2888. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cho SW, Kim S, Kim JM and Kim JS: Targeted
genome engineering in human cells with the Cas9 RNA-guided
endonuclease. Nat Biotechnol. 31:230–232. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jinek M, East A, Cheng A, Lin S, Ma E and
Doudna J: RNA-programmed genome editing in human cells. Elife.
2:e004712013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jinek M, Chylinski K, Fonfara I, Hauer M,
Doudna JA and Charpentier E: A programmable dual-RNA-guided DNA
endonuclease in adaptive bacterial immunity. Science. 337:816–821.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sharma G, Sharma AR, Bhattacharya M, Lee
SS and Chakraborty C: CRISPR-Cas9: A preclinical and clinical
perspective for the treatment of human diseases. Mol Ther.
29:571–586. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu X, Ma W, Mei C, Chen X, Yao Y, Liu Y,
Qin X and Yuan Y: Description of CRISPR/Cas9 development and its
prospect in hepatocellular carcinoma treatment. J Exp Clin Cancer
Res. 39:972020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meng H, Nan M, Li Y, Ding Y, Yin Y and
Zhang M: Application of CRISPR-Cas9 gene editing technology in
basic research, diagnosis and treatment of colon cancer. Front
Endocrinol (Lausanne). 14:11484122023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang F, Wang H, Fan S, Qiu H, Li X, Shi G,
Li Z, Luan X and Wu H: Advances in synthetic lethality in potential
oncology therapeutic approaches. Curr Top Med Chem. Jan
30–2025.(Epub ahead of print). View Article : Google Scholar
|
|
13
|
Dimitri A, Herbst F and Fraietta JA:
Engineering the next-generation of CAR T-cells with CRISPR-Cas9
gene editing. Mol Cancer. 21:782022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Akram F, Haq IU, Ahmed Z, Khan H and Ali
MS: CRISPR-Cas9, a promising therapeutic tool for cancer therapy: A
review. Protein Pept Lett. 27:931–944. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lou K, Steri V, Ge AY, Hwang YC,
Yogodzinski CH, Shkedi AR, Choi ALM, Mitchell DC, Swaney DL, Hann
B, et al: KRASG12C inhibition produces a driver-limited
state revealing collateral dependencies. Sci Signal.
12:eaaw94502019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Midha A, Dearden S and McCormack R: EGFR
mutation incidence in non-small-cell lung cancer of adenocarcinoma
histology: A systematic review and global map by ethnicity
(mutMapII). Am J Cancer Res. 5:2892–2911. 2015.PubMed/NCBI
|
|
17
|
Thai AA, Solomon BJ, Sequist LV, Gainor JF
and Heist RS: Lung cancer. Lancet. 398:535–554. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Demirci Y, Zhang B and Unver T:
CRISPR/Cas9: An RNA-guided highly precise synthetic tool for plant
genome editing. J Cell Physiol. 233:1844–1859. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lattanzi A, Meneghini V, Pavani G, Amor F,
Ramadier S, Felix T, Antoniani C, Masson C, Alibeu O, Lee C, et al:
Optimization of CRISPR/Cas9 delivery to human hematopoietic stem
and progenitor cells for therapeutic genomic rearrangements. Mol
Ther. 27:137–150. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Ma S, Sun L, Zhang T, Chang J, Lu W,
Chen X, Liu Y, Wang X, Shi R, et al: Programmable single and
multiplex base-editing in bombyx mori using RNA-guided cytidine
deaminases. G3 (Bethesda). 8:1701–1709. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Donohoue PD, Pacesa M, Lau E, Vidal B,
Irby MJ, Nyer DB, Rotstein T, Banh L, Toh MS, Gibson J, et al:
Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides
mitigates off-target activity in T cells. Mol Cell.
81:3637–3649.e5. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cai W and Wang M: Engineering nucleic acid
chemistry for precise and controllable CRISPR/Cas9 genome editing.
Sci Bull (Beijing). 64:1841–1849. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tian X, Gu T, Patel S, Bode AM, Lee MH and
Dong Z: CRISPR/Cas9-an evolving biological tool kit for cancer
biology and oncology. NPJ Precis Oncol. 3:82019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rotow J and Bivona TG: Understanding and
targeting resistance mechanisms in NSCLC. Nat Rev Cancer.
17:637–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dilly J, Hoffman MT, Abbassi L, Li Z,
Paradiso F, Parent BD, Hennessey CJ, Jordan AC, Morgado M, Dasgupta
S, et al: Mechanisms of resistance to oncogenic KRAS inhibition in
pancreatic cancer. Cancer Discov. 14:2135–2161. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Drosten M and Barbacid M: Targeting the
MAPK pathway in KRAS-driven tumors. Cancer Cell. 37:543–550. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Isermann T, Sers C, Der CJ and Papke B:
KRAS inhibitors: Resistance drivers and combinatorial strategies.
Trends Cancer. 11:91–116. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ou SHI, Jänne PA, Leal TA, Rybkin II,
Sabari JK, Barve MA, Bazhenova L, Johnson ML, Velastegui KL,
Cilliers C, et al: First-in-human phase I/IB dose-finding study of
adagrasib (MRTX849) in patients with advanced KRASG12C
solid tumors (KRYSTAL-1). J Clin Oncol. 40:2530–2538. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Salgia R, Pharaon R, Mambetsariev I, Nam A
and Sattler M: The improbable targeted therapy: KRAS as an emerging
target in non-small cell lung cancer (NSCLC). Cell Rep Med.
2:1001862021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Skoulidis F, Li BT, Dy GK, Price TJ,
Falchook GS, Wolf J, Italiano A, Schuler M, Borghaei H, Barlesi F,
et al: Sotorasib for lung cancers with KRAS p.G12C mutation. N Engl
J Med. 384:2371–2381. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao Q, Ouyang W, Kang B, Han X, Xiong Y,
Ding R, Li Y, Wang F, Huang L, Chen L, et al: Selective targeting
of the oncogenic KRAS G12S mutant allele by CRISPR/Cas9 induces
efficient tumor regression. Theranostics. 10:5137–5153. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dompe N, Klijn C, Watson SA, Leng K, Port
J, Cuellar T, Watanabe C, Haley B, Neve R, Evangelista M and Stokoe
D: A CRISPR screen identifies MAPK7 as a target for combination
with MEK inhibition in KRAS mutant NSCLC. PLoS One.
13:e01992642018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li K, Yang M, Liang N and Li S:
Determining EGFR-TKI sensitivity of G719X and other uncommon EGFR
mutations in non-small cell lung cancer: Perplexity and solution
(review): Perplexity and solution. Oncol Rep. 37:1347–1358. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fukuoka M, Yano S, Giaccone G, Tamura T,
Nakagawa K, Douillard JY, Nishiwaki Y, Vansteenkiste J, Kudoh S,
Rischin D, et al: Multi-institutional randomized phase II trial of
gefitinib for previously treated patients with advanced
non-small-cell lung cancer (The IDEAL 1 Trial) [corrected]. J Clin
Oncol. 21:2237–2246. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tsao MS, Sakurada A, Cutz JC, Zhu CQ,
Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, et
al: Erlotinib in lung cancer-molecular and clinical predictors of
outcome. N Engl J Med. 353:133–144. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shi Y, Zhang L, Liu X, Zhou C, Zhang L,
Zhang S, Wang D, Li Q, Qin S, Hu C, et al: Icotinib versus
gefitinib in previously treated advanced non-small-cell lung cancer
(ICOGEN): A randomised, double-blind phase 3 non-inferiority trial.
Lancet Oncol. 14:953–961. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang JCH, Wu YL, Schuler M, Sebastian M,
Popat S, Yamamoto N, Zhou C, Hu CP, O'Byrne K, Feng J, et al:
Afatinib versus cisplatin-based chemotherapy for EGFR
mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6):
Analysis of overall survival data from two randomised, phase 3
trials. Lancet Oncol. 16:141–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu YL, Cheng Y, Zhou X, Lee KH, Nakagawa
K, Niho S, Tsuji F, Linke R, Rosell R, Corral J, et al: Dacomitinib
versus gefitinib as first-line treatment for patients with
EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): A
randomised, open-label, phase 3 trial. Lancet Oncol. 18:1454–1466.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Soria JC, Ohe Y, Vansteenkiste J,
Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura
F, Nogami N, Kurata T, et al: Osimertinib in untreated EGFR-mutated
advanced non-small-cell lung cancer. N Engl J Med. 378:113–125.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang JCH, Camidge DR, Yang CT, Zhou J, Guo
R, Chiu CH, Chang GC, Shiah HS, Chen Y, Wang CC, et al: Safety,
efficacy, and pharmacokinetics of almonertinib (HS-10296) in
pretreated patients with EGFR-mutated advanced NSCLC: A
multicenter, open-label, phase 1 trial. J Thorac Oncol.
15:1907–1918. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shi Y, Hu X, Zhang S, Lv D, Wu L, Yu Q,
Zhang Y, Liu L, Wang X, Cheng Y, et al: Efficacy, safety, and
genetic analysis of furmonertinib (AST2818) in patients with EGFR
T790M mutated non-small-cell lung cancer: A phase 2b, multicentre,
single-arm, open-label study. Lancet Respir Med. 9:829–839. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chmielecki J, Gray JE, Cheng Y, Ohe Y,
Imamura F, Cho BC, Lin MC, Majem M, Shah R, Rukazenkov Y, et al:
Candidate mechanisms of acquired resistance to first-line
osimertinib in EGFR-mutated advanced non-small cell lung cancer.
Nat Commun. 14:10702023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang ZF, Ren SX, Li W and Gao GH:
Frequency of the acquired resistant mutation T790 M in non-small
cell lung cancer patients with active exon 19Del and exon 21 L858R:
A systematic review and meta-analysis. BMC Cancer. 18:1482018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Arcila ME, Oxnard GR, Nafa K, Riely GJ,
Solomon SB, Zakowski MF, Kris MG, Pao W, Miller VA and Ladanyi M:
Rebiopsy of lung cancer patients with acquired resistance to EGFR
inhibitors and enhanced detection of the T790M mutation using a
locked nucleic acid-based assay. Clin Cancer Res. 17:1169–1180.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kuiper JL, Heideman DAM, Thunnissen E,
Paul MA, van Wijk AW, Postmus PE and Smit EF: Incidence of T790M
mutation in (sequential) rebiopsies in EGFR-mutated NSCLC-patients.
Lung Cancer. 85:19–24. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yu HA, Arcila ME, Rekhtman N, Sima CS,
Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M and Riely GJ:
Analysis of tumor specimens at the time of acquired resistance to
EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers.
Clin Cancer Res. 19:2240–2247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cross DAE, Ashton SE, Ghiorghiu S,
Eberlein C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MRV, Ward RA,
Mellor MJ, et al: AZD9291, an irreversible EGFR TKI, overcomes
T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer
Discov. 4:1046–1061. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rosell R, Moran T, Queralt C, Porta R,
Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M,
et al: Screening for epidermal growth factor receptor mutations in
lung cancer. N Engl J Med. 361:958–967. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ichihara E and Lovly CM: Shades of T790M:
Intratumor heterogeneity in EGFR-mutant lung cancer. Cancer Discov.
5:694–696. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Oxnard GR, Hu Y, Mileham KF, Husain H,
Costa DB, Tracy P, Feeney N, Sholl LM, Dahlberg SE, Redig AJ, et
al: Assessment of resistance mechanisms and clinical implications
in patients with EGFR T790M-positive lung cancer and acquired
resistance to osimertinib. JAMA Oncol. 4:1527–1534. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Park HR, Kim TM, Lee Y, Kim S, Park S, Ju
YS, Kim M, Keam B, Jeon YK, Kim DW and Heo DS: Acquired resistance
to third-generation EGFR tyrosine kinase inhibitors in patients
with de novo EGFRT790M-mutant NSCLC. J Thorac Oncol.
16:1859–1871. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Vad-Nielsen J, Staunstrup NH, Kjeldsen ML,
Dybdal N, Flandin G, De Stradis C, Daugaard TF, Vilsbøll-Larsen T,
Maansson CT, Doktor TK, et al: Genome-wide epigenetic and
mRNA-expression profiling followed by CRISPR/Cas9-mediated
gene-disruptions corroborate the MIR141/MIR200C-ZEB1/ZEB2-FGFR1
axis in acquired EMT-associated EGFR TKI-resistance in NSCLC cells.
Transl Lung Cancer Res. 12:42–65. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zeng H, Castillo-Cabrera J, Manser M, Lu
B, Yang Z, Strande V, Begue D, Zamponi R, Qiu S, Sigoillot F, et
al: Genome-wide CRISPR screening reveals genetic modifiers of
mutant EGFR dependence in human NSCLC. Elife. 8:e502232019.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang TH, Wu CC, Huang KY, Leu YL, Yang SC,
Chen CL and Chen CY: Integrated omics analysis of non-small-cell
lung cancer cells harboring the EGFR C797S mutation reveals the
potential of AXL as a novel therapeutic target in TKI-resistant
lung cancer. Cancers (Basel). 13:1112020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Guernet A, Mungamuri SK, Cartier D,
Sachidanandam R, Jayaprakash A, Adriouch S, Vezain M, Charbonnier
F, Rohkin G, Coutant S, et al: CRISPR-barcoding for intratumor
genetic heterogeneity modeling and functional analysis of oncogenic
driver mutations. Mol Cell. 63:526–538. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Devarakonda S, Morgensztern D and Govindan
R: Genomic alterations in lung adenocarcinoma. Lancet Oncol.
16:e342–e351. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang SS, Nagasaka M, Zhu VW and Ou SHI:
Going beneath the tip of the iceberg. Identifying and understanding
EML4-ALK variants and TP53 mutations to optimize treatment of ALK
fusion positive (ALK+) NSCLC. Lung Cancer. 158:126–136. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Soda M, Choi YL, Enomoto M, Takada S,
Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K,
Hatanaka H, et al: Identification of the transforming EML4-ALK
fusion gene in non-small-cell lung cancer. Nature. 448:561–566.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sasaki T, Rodig SJ, Chirieac LR and Jänne
PA: The biology and treatment of EML4-ALK non-small cell lung
cancer. Eur J Cancer. 46:1773–1780. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa
K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, et al:
First-line crizotinib versus chemotherapy in ALK-positive lung
cancer. N Engl J Med. 371:2167–2177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Camidge DR, Bang YJ, Kwak EL, Iafrate AJ,
Varella-Garcia M, Fox SB, Riely GJ, Solomon B, Ou SH, Kim DW, et
al: Activity and safety of crizotinib in patients with ALK-positive
non-small-cell lung cancer: Updated results from a phase 1 study.
Lancet Oncol. 13:1011–1019. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Riely GJ, Wood DE, Ettinger DS, Aisner DL,
Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, et
al: Non-small cell lung cancer, version 4.2024, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw.
22:249–274. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Gadgeel SM, Shaw AT, Govindan R, Gandhi L,
Socinski MA, Camidge DR, De Petris L, Kim DW, Chiappori A,
Moro-Sibilot DL, et al: Pooled analysis of CNS response to
alectinib in two studies of pretreated patients with ALK-positive
non-small-cell lung cancer. J Clin Oncol. 34:4079–4085. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Soria JC, Tan DSW, Chiari R, Wu YL,
Paz-Ares L, Wolf J, Geater SL, Orlov S, Cortinovis D, Yu CJ, et al:
First-line ceritinib versus platinum-based chemotherapy in advanced
ALK-rearranged non-small-cell lung cancer (ASCEND-4): A randomised,
open-label, phase 3 study. Lancet. 389:917–929. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Peters S, Camidge DR, Shaw AT, Gadgeel S,
Ahn JS, Kim DW, Ou SI, Pérol M, Dziadziuszko R, Rosell R, et al:
Alectinib versus crizotinib in untreated ALK-positive
non-small-cell lung cancer. N Engl J Med. 377:829–838. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Camidge DR, Kim HR, Ahn MJ, Yang JC, Han
JY, Lee JS, Hochmair MJ, Li JY, Chang GC, Lee KH, et al: Brigatinib
versus crizotinib in ALK-positive non-small-cell lung cancer. N
Engl J Med. 379:2027–2039. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shaw AT, Bauer TM, de Marinis F, Felip E,
Goto Y, Liu G, Mazieres J, Kim DW, Mok T, Polli A, et al:
First-line lorlatinib or crizotinib in advanced ALK-positive lung
cancer. N Engl J Med. 383:2018–2029. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sullivan I and Planchard D: ALK inhibitors
in non-small cell lung cancer: The latest evidence and
developments. Ther Adv Med Oncol. 8:32–47. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ma D, Zhang Y, Xing P, Hao X, Wang M, Wang
Y, Shan L, Xin T, Liang H, Du Y, et al: Clinical features and
outcomes of ALK rearranged non-small cell lung cancer with primary
resistance to crizotinib. Thorac Cancer. 10:1213–1219. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Matikas A, Kentepozidis N, Georgoulias V
and Kotsakis A: Management of resistance to crizotinib in
anaplastic lymphoma kinase-positive non-small-cell lung cancer.
Clin Lung Cancer. 17:474–482. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Spaans JN and Goss GD: Trials to overcome
drug resistance to EGFR and ALK targeted therapies-past, present,
and future. Front Oncol. 4:2332014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kong X, Pan P, Sun H, Xia H, Wang X, Li Y
and Hou T: Drug discovery targeting anaplastic lymphoma kinase
(ALK). J Med Chem. 62:10927–10954. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Maddalo D, Manchado E, Concepcion CP,
Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N,
de Stanchina E, et al: In vivo engineering of oncogenic chromosomal
rearrangements with the CRISPR/Cas9 system. Nature. 516:423–427.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Drilon A, Somwar R, Wagner JP, Vellore NA,
Eide CA, Zabriskie MS, Arcila ME, Hechtman JF, Wang L, Smith RS, et
al: A novel crizotinib-resistant solvent-front mutation responsive
to cabozantinib therapy in a patient with ROS1-rearranged lung
cancer. Clin Cancer Res. 22:2351–2358. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Awad MM, Katayama R, McTigue M, Liu W,
Deng YL, Brooun A, Friboulet L, Huang D, Falk MD, Timofeevski S, et
al: Acquired resistance to crizotinib from a mutation in CD74-ROS1.
N Engl J Med. 368:2395–2401. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Shaw AT, Solomon BJ, Chiari R, Riely GJ,
Besse B, Soo RA, Kao S, Lin CC, Bauer TM, Clancy JS, et al:
Lorlatinib in advanced ROS1-positive non-small-cell lung cancer: A
multicentre, open-label, single-arm, phase 1–2 trial. Lancet Oncol.
20:1691–1701. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Lim SM, Kim HR, Lee JS, Lee KH, Lee YG,
Min YJ, Cho EK, Lee SS, Kim BS, Choi MY, et al: Open-label,
multicenter, phase II study of ceritinib in patients with
non-small-cell lung cancer harboring ROS1 rearrangement. J Clin
Oncol. 35:2613–2618. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Dziadziuszko R, Krebs MG, De Braud F,
Siena S, Drilon A, Doebele RC, Patel MR, Cho BC, Liu SV, Ahn MJ, et
al: Updated integrated analysis of the efficacy and safety of
entrectinib in locally advanced or metastatic ROS1 fusion-positive
non-small-cell lung cancer. J Clin Oncol. 39:1253–1263. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Drilon A, Siena S, Dziadziuszko R, Barlesi
F, Krebs MG, Shaw AT, de Braud F, Rolfo C, Ahn MJ, Wolf J, et al:
Entrectinib in ROS1 fusion-positive non-small-cell lung cancer:
Integrated analysis of three phase 1–2 trials. Lancet Oncol.
21:261–270. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Shaw AT, Ou SHI, Bang YJ, Camidge DR,
Solomon BJ, Salgia R, Riely GJ, Varella-Garcia M, Shapiro GI, Costa
DB, et al: Crizotinib in ROS1-rearranged non-small-cell lung
cancer. N Engl J Med. 371:1963–1971. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Huber KVM, Salah E, Radic B, Gridling M,
Elkins JM, Stukalov A, Jemth AS, Göktürk C, Sanjiv K, Strömberg K,
et al: Stereospecific targeting of MTH1 by (S)-crizotinib as an
anticancer strategy. Nature. 508:222–227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Choi PS and Meyerson M: Targeted genomic
rearrangements using CRISPR/Cas technology. Nat Commun. 5:37282014.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Sato H, Schoenfeld AJ, Siau E, Lu YC, Tai
H, Suzawa K, Kubota D, Lui AJW, Qeriqi B, Mattar M, et al: MAPK
pathway alterations correlate with poor survival and drive
resistance to therapy in patients with lung cancers driven by ROS1
fusions. Clin Cancer Res. 26:2932–2945. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Skead G and Govender D: Gene of the month:
MET. J Clin Pathol. 68:405–409. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Drilon A, Cappuzzo F, Ou SHI and Camidge
DR: Targeting MET in lung cancer: Will expectations finally be MET?
J Thorac Oncol. 12:15–26. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Pasquini G and Giaccone G: C-MET
inhibitors for advanced non-small cell lung cancer. Expert Opin
Investig Drugs. 27:363–375. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Bubendorf L, Dafni U, Schöbel M, Finn SP,
Tischler V, Sejda A, Marchetti A, Thunnissen E, Verbeken EK, Warth
A, et al: Prevalence and clinical association of MET gene
overexpression and amplification in patients with NSCLC: Results
from the European thoracic oncology platform (ETOP) lungscape
project. Lung Cancer. 111:143–149. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Fujino T, Suda K and Mitsudomi T: Emerging
MET tyrosine kinase inhibitors for the treatment of non-small cell
lung cancer. Expert Opin Emerg Drugs. 25:229–249. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Mathieu LN, Larkins E, Akinboro O, Roy P,
Amatya AK, Fiero MH, Mishra-Kalyani PS, Helms WS, Myers CE, Skinner
AM, et al: FDA approval summary: Capmatinib and tepotinib for the
treatment of metastatic NSCLC harboring MET exon 14 skipping
mutations or alterations. Clin Cancer Res. 28:249–254. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Togashi Y, Mizuuchi H, Tomida S, Terashima
M, Hayashi H, Nishio K and Mitsudomi T: MET gene exon 14 deletion
created using the CRISPR/Cas9 system enhances cellular growth and
sensitivity to a MET inhibitor. Lung Cancer. 90:590–597. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Fernandes M, Hoggard B, Jamme P, Paget S,
Truong MJ, Grégoire V, Vinchent A, Descarpentries C, Morabito A,
Stanislovas J, et al: MET exon 14 skipping mutation is a hepatocyte
growth factor (HGF)-dependent oncogenic driver in vitro and in
humanised HGF knock-in mice. Mol Oncol. 17:2257–2274. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Negrao MV, Raymond VM, Lanman RB,
Robichaux JP, He J, Nilsson MB, Ng PKS, Amador BE, Roarty EB, Nagy
RJ, et al: Molecular landscape of BRAF-mutant NSCLC reveals an
association between clonality and driver mutations and identifies
targetable non-V600 driver mutations. J Thorac Oncol. 15:1611–1623.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Marchetti A, Felicioni L, Malatesta S,
Grazia Sciarrotta M, Guetti L, Chella A, Viola P, Pullara C,
Mucilli F and Buttitta F: Clinical features and outcome of patients
with non-small-cell lung cancer harboring BRAF mutations. J Clin
Oncol. 29:3574–3579. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yao Z, Torres NM, Tao A, Gao Y, Luo L, Li
Q, de Stanchina E, Abdel-Wahab O, Solit DB, Poulikakos PI and Rosen
N: BRAF mutants evade ERK-dependent feedback by different
mechanisms that determine their sensitivity to pharmacologic
inhibition. Cancer Cell. 28:370–383. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Degirmenci U, Wang M and Hu J: Targeting
aberrant RAS/RAF/MEK/ERK signaling for cancer therapy. Cells.
9:1982020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Planchard D, Kim TM, Mazieres J, Quoix E,
Riely G, Barlesi F, Souquet PJ, Smit EF, Groen HJ, Kelly RJ, et al:
Dabrafenib in patients with BRAF(V600E)-positive advanced
non-small-cell lung cancer: A single-arm, multicentre, open-label,
phase 2 trial. Lancet Oncol. 17:642–650. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Hyman DM, Puzanov I, Subbiah V, Faris JE,
Chau I, Blay JY, Wolf J, Raje NS, Diamond EL, Hollebecque A, et al:
Vemurafenib in multiple nonmelanoma cancers with BRAF V600
mutations. N Engl J Med. 373:726–736. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Lin L, Asthana S, Chan E, Bandyopadhyay S,
Martins MM, Olivas V, Yan JJ, Pham L, Wang MM, Bollag G, et al:
Mapping the molecular determinants of BRAF oncogene dependence in
human lung cancer. Proc Natl Acad Sci USA. 111:E748–E757. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Vaishnavi A, Juan J, Jacob M, Stehn C,
Gardner EE, Scherzer MT, Schuman S, Van Veen JE, Murphy B, Hackett
CS, et al: Transposon mutagenesis reveals RBMS3 silencing as a
promoter of malignant progression of BRAFV600E-driven lung
tumorigenesis. Cancer Res. 82:4261–4273. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Nishinarita N, Igawa S, Kasajima M,
Kusuhara S, Harada S, Okuma Y, Sugita K, Ozawa T, Fukui T,
Mitsufuji H, et al: Smoking history as a predictor of epidermal
growth factor receptor tyrosine kinase inhibitors in patients with
non-small cell lung cancer harboring EGFR mutations. Oncology.
95:109–115. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Fu S, Liu C, Huang Q, Fan S, Tang H, Fu X,
Ai B, Liao Y and Chu Q: Estrogen receptor β1 activation accelerates
resistance to epidermal growth factor receptor-tyrosine kinase
inhibitors in non-small cell lung cancer. Oncol Rep. 39:1313–1321.
2018.PubMed/NCBI
|
|
102
|
Girard N: Optimizing outcomes in EGFR
mutation-positive NSCLC: Which tyrosine kinase inhibitor and when?
Future Oncol. 14:1117–1132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Lategahn J, Keul M and Rauh D: Lessons to
be learned: The molecular basis of kinase-targeted therapies and
drug resistance in non-small cell lung cancer. Angew Chem Int Ed
Engl. 57:2307–2313. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Hsu KH, Huang YH, Tseng JS, Chen KC, Ku
WH, Su KY, Chen JJW, Chen HW, Yu SL, Yang TY and Chang GC: High
PD-L1 expression correlates with primary resistance to EGFR-TKIs in
treatment naïve advanced EGFR-mutant lung adenocarcinoma patients.
Lung Cancer. 127:37–43. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Terai H, Kitajima S, Potter DS, Matsui Y,
Quiceno LG, Chen T, Kim TJ, Rusan M, Thai TC, Piccioni F, et al: ER
stress signaling promotes the survival of cancer ‘persister cells’
tolerant to EGFR tyrosine kinase inhibitors. Cancer Res.
78:1044–1057. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Krall EB, Wang B, Munoz DM, Ilic N,
Raghavan S, Niederst MJ, Yu K, Ruddy DA, Aguirre AJ, Kim JW, et al:
KEAP1 loss modulates sensitivity to kinase targeted therapy in lung
cancer. Elife. 6:e189702017. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Gannon HS, Kaplan N, Tsherniak A, Vazquez
F, Weir BA, Hahn WC and Meyerson M: Identification of an
‘exceptional responder’ cell line to MEK1 inhibition: Clinical
implications for MEK-targeted therapy. Mol Cancer Res. 14:207–215.
2016. View Article : Google Scholar : PubMed/NCBI
|