Introduction
Chronic myeloid leukemia (CML) is a life-threatening
hematological malignancy driven by intricate and multifactorial
pathogenic mechanisms that underlie its clinical heterogeneity and
therapeutic challenges. The Philadelphia chromosome (Ph), arising
from a reciprocal translocation t(9;22)(q34;q11.2), is detected in
up to 95% of adult CML cases (1).
This chromosomal aberration generates an abnormal BCR-ABL1
fusion transcript, with molecular detection of this transcript
serving as a key component for genetic confirmation of CML
diagnosis (2,3). Recent advancements in molecular
biology have highlighted the emerging role of atypical fusion genes
in CML pathogenesis and progression (4,5). In
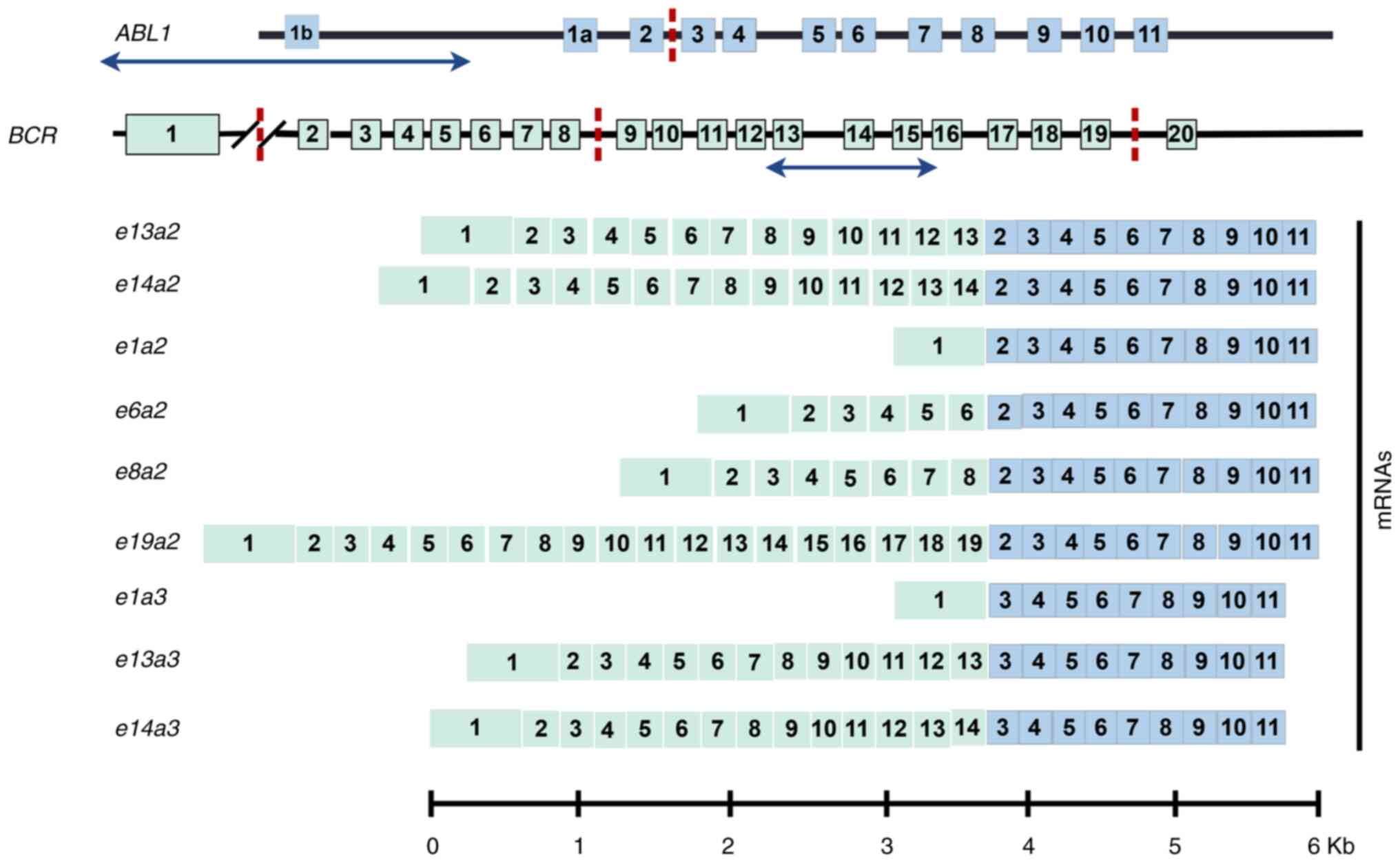
Ph-positive leukemia, the canonical BCR-ABL1 fusion gene
functions as a pivotal oncogenic driver. Variable chromosomal
breakpoints within BCR and ABL1 genes result in
distinct BCR-ABL1 transcript variants and corresponding
protein isoforms (6). The
e13a2 and e14a2 subtypes represent the most prevalent
BCR-ABL1 isoforms in patients with CML, both containing
intact sequences encoding the Src homology 3 domain(SH3), SH2 and
kinase domains of ABL1. Beyond these common variants, rare
fusion genotypes including e13a3, e14a3 and e1a3 have
been documented (7,8). The e13a3 and e14a3
subtypes, characterized by the absence of ABL1 exon 2,
collectively account for <1% of CML cases (9). These atypical fusion proteins exhibit
structural and functional alterations secondary to ABL1
truncation, potentially influencing leukemia biological behavior,
therapeutic responsiveness and clinical outcomes. Recent study has
revealed that these atypical variants may regulate cellular
signaling pathways and gene expression, influencing the progression
of leukemia (3,4). Simvastatin overcomes drug resistance
in chronic myeloid leukemia cells to imatinib by inhibiting the
PI3K/AKT survival signaling pathway and downregulating its
controlled anti-apoptotic proteins (10). Simultaneously, in combination with
imatinib, it interferes with Wnt/β-catenin signaling and increases
suppressive histone modification to decrease expression of the
oncogene. Through these multi-pathway effects, it ultimately
induces mitochondrial pathway apoptosis, thereby effectively
overcoming imatinib resistance (10). Furthermore, emerging targeted
therapies for these atypical variants are under investigation,
aiming to improve patient outcomes and overcome the limitations of
current treatment strategies (11).
Multicenter studies, including the European Treatment Outcome Study
(EUTOS) collaborative network, are advancing understanding of
atypical BCR-ABL1 fusion genes in CML (4,12).
These studies have developed protocols for monitoring these
variants using advanced techniques, such as reverse
transcription-quantitative (RT-q)PCR, as standard methods are not
applicable because its ‘standardized’ or ‘universal’ detection
tools (primers and probes) are designed for ‘typical’ or ‘common’
fusion variants. When atypical variants are encountered, these
tools cannot bind and recognize them effectively, leading to
detection failure (false-negative results). Efforts by EUTOS are
focused on refining treatment strategies and establishing
guidelines for managing these rare variants (13).
Guidelines from organizations, including the
National Comprehensive Cancer Network, emphasize the necessity of
detecting specific recurrent genetic abnormalities in bone marrow
nucleated cells or peripheral blood leukocytes for optimal risk
stratification and treatment planning (14). Recommended methodologies include
cytogenetic analysis (karyotyping), interphase fluorescence in
situ hybridization (FISH) and RT-PCR for fusion gene detection.
Previous investigations have implemented RT-qPCR for JAK2,
Calreticulin and myeloproliferative leukemia proto-oncogene gene
analysis (15), supplemented with
specialized primer sets targeting BCR exon (e)1, 12 and 3 to
identify BCR-ABL1 breakpoints, demonstrating comprehensive
coverage of previously reported uncommon breakpoints (11,16).
Bone marrow smear examination combined with FISH and karyotyping
provides preliminary evidence of BCR-ABL1 fusion (17). Next-generation sequencing (NGS)
enables genomic analysis through fragmentation of genomic DNA or
transcriptomic RNA, library preparation and high-throughput
sequencing via fluorescence signal detection during
polymerase/ligase-mediated nucleotide incorporation (18). For non-IS standardized transcripts,
quantitative calibration and reporting methods, such as relative
ABL1 copy number analysis and laboratory-built reference
curves, are recommended for improved quantification (19). Droplet digital (dd)PCR), with its
defined detection limit and quantification limit, is a key tool for
monitoring residual disease levels in these variants, with
variant-specific primer design and stringent quality control
procedures essential to ensure accuracy. Whole-genome sequencing
(WGS) using exon capture techniques facilitates detection of
BCR-ABL1 fusions through comprehensive genomic interrogation
(20). Additional methodologies,
such as nested PCR coupled with agarose gel electrophoresis, have
utility in detecting these transcripts, offering enhanced
sensitivity and specificity compared with conventional techniques
while enabling amplification of extended DNA fragments (21). Recent guidelines from European
LeukemiaNet 2023 and EUTOS suggest regular monitoring of measurable
residual disease using advanced techniques and more frequent
follow-up for patients with atypical transcripts to improve patient
management (6,22). Ongoing multicenter collaborations,
such as the EUTOS study, are key in providing robust data on the
clinical outcomes of these variants. These studies aim to validate
the prognostic value of atypical BCR-ABL1 fusion genes and refine
treatment strategies for these rare subtypes. Atypical
BCR-ABL1 testing should involve multiplex RT-PCR and NGS,
followed by ddPCR for minimal residual disease monitoring,
providing a structured approach to managing cases with atypical
BCR-ABL1 fusion genes (Fig.
1), with follow-up frequency and therapeutic adjustments based
on patient response.
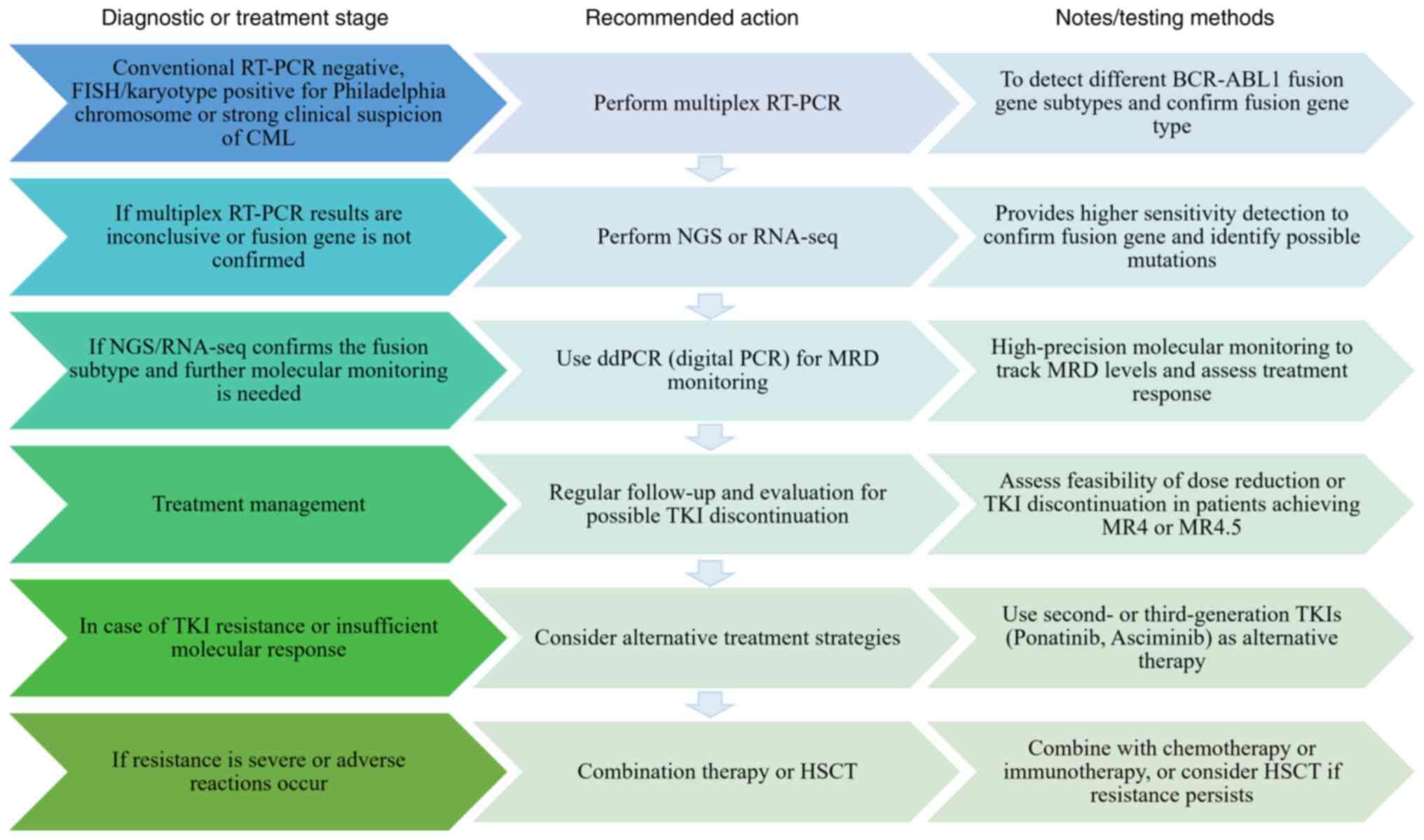 | Figure 1.Clinical roadmap for atypical
BCR-ABL1 testing. BCR, breakpoint cluster region; ABL1,
abelson leukemia1; RT-q, reverse transcription-quantitative; NGS,
next-generation sequencing; CML, chronic myeloid leukemia; TKI,
tyrosine kinase inhibitor; seq, sequencing; dd, digital droplet;
MRD, minimal residual disease; MR, molecular response; HSCT,
hematopoietic stem cell transplantation. |
Current research on atypical fusion genes in
leukemia remains exploratory, with knowledge gaps persisting. The
low incidence of these genetic variants in leukemia populations has
resulted in limited case reports (23,24),
posing diagnostic and therapeutic challenges for clinicians
managing patients with atypical fusion-positive CML. The present
study aimed to review the structural characteristics therapeutic
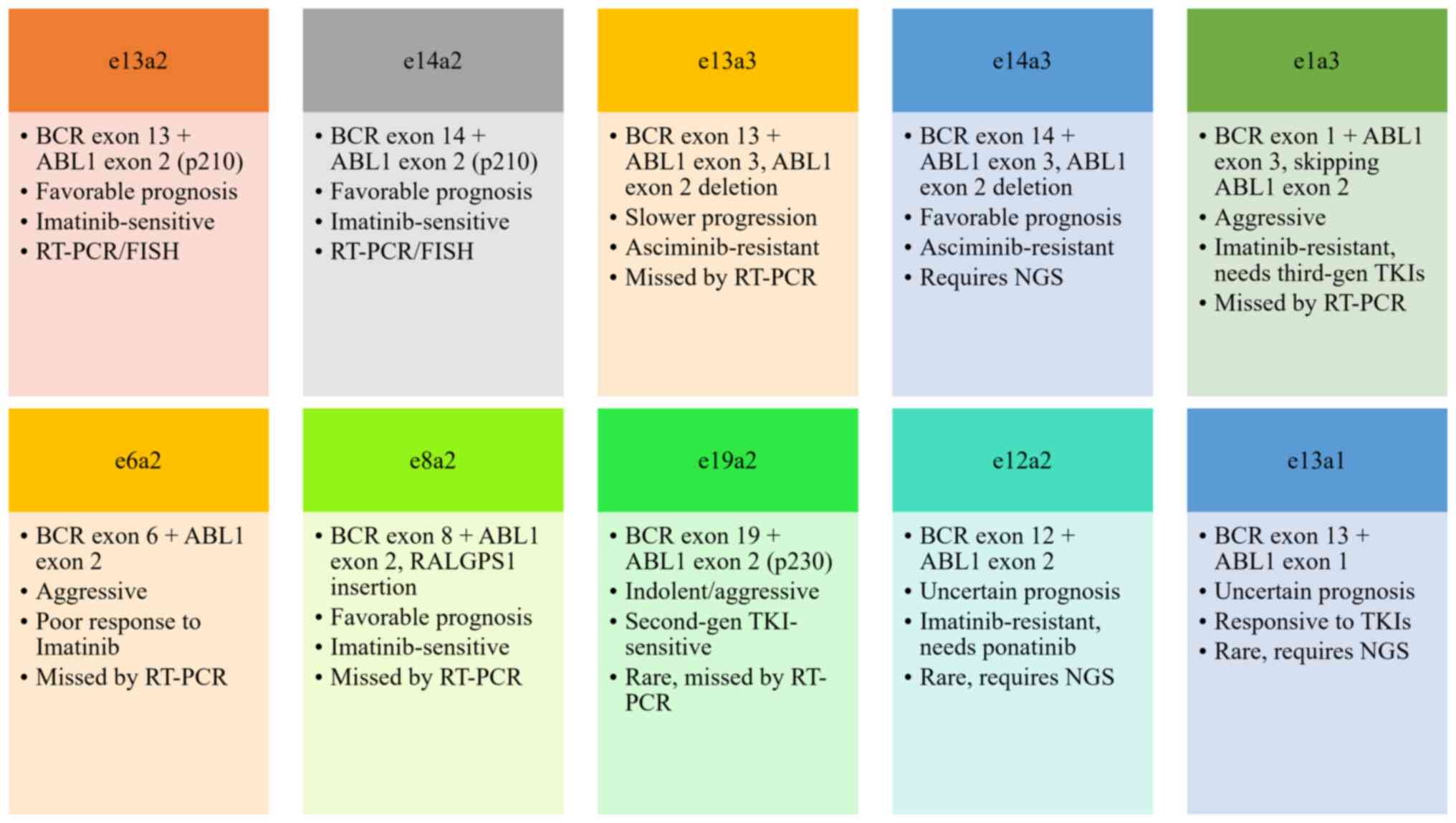
management, and prognostic implications of the e13a3, e14a3,
e1a3, e1a2, e6a2, e8a2, e19a2, e12a2 and e13a1 BCR-ABL1
fusion transcripts to delineate their clinical significance
(Figs. 2 and 3).
Materials and methods
The literature search was conducted using
PubMed(pubmed.ncbi.nlm.nih.gov/), Embase
(embase.com/landing?status=grey) and Web of
Science(webofscience.com/wos/) from January 2000 to July 2025 using
the following search strategy: ((BCR-ABL[Title/Abstract] OR
BCR::ABL1[Title/Abstract]) AND (atypical[Title/Abstract] OR
rare[Title/Abstract] OR e13a3 OR e14a3 OR e1a3 OR e1a2 OR e6a2 OR
e8a2 OR e19a2 OR e12a2 OR e18a2 OR e13a1) AND (CML[Title/Abstract]
OR ‘chronic myeloid leukemia’[MeSH Terms] OR Ph +
ALL[Title/Abstract]). A two-step ‘include-then-exclude’ process was
performed: All case reports, series or retrospective studies in
which atypical BCR-ABL1 transcripts were confirmed at the RNA or
DNA level, the diagnosis met World Health Organization(WHO)
criteria for CML or acute lymphoblastic leukemia(Ph+
ALL) and both treatment details and evaluable follow-up outcomes
were provided were eligible (25,26);
conversely, reviews, editorials, animal studies lacking primary
data and duplicate publications with overlapping cases were
excluded, retaining only the most complete dataset for each
patient. A total of two reviewers independently screened
titles/abstracts, extracted data. Discrepancies resolved by a third
reviewer. Because study designs varied widely, the present review
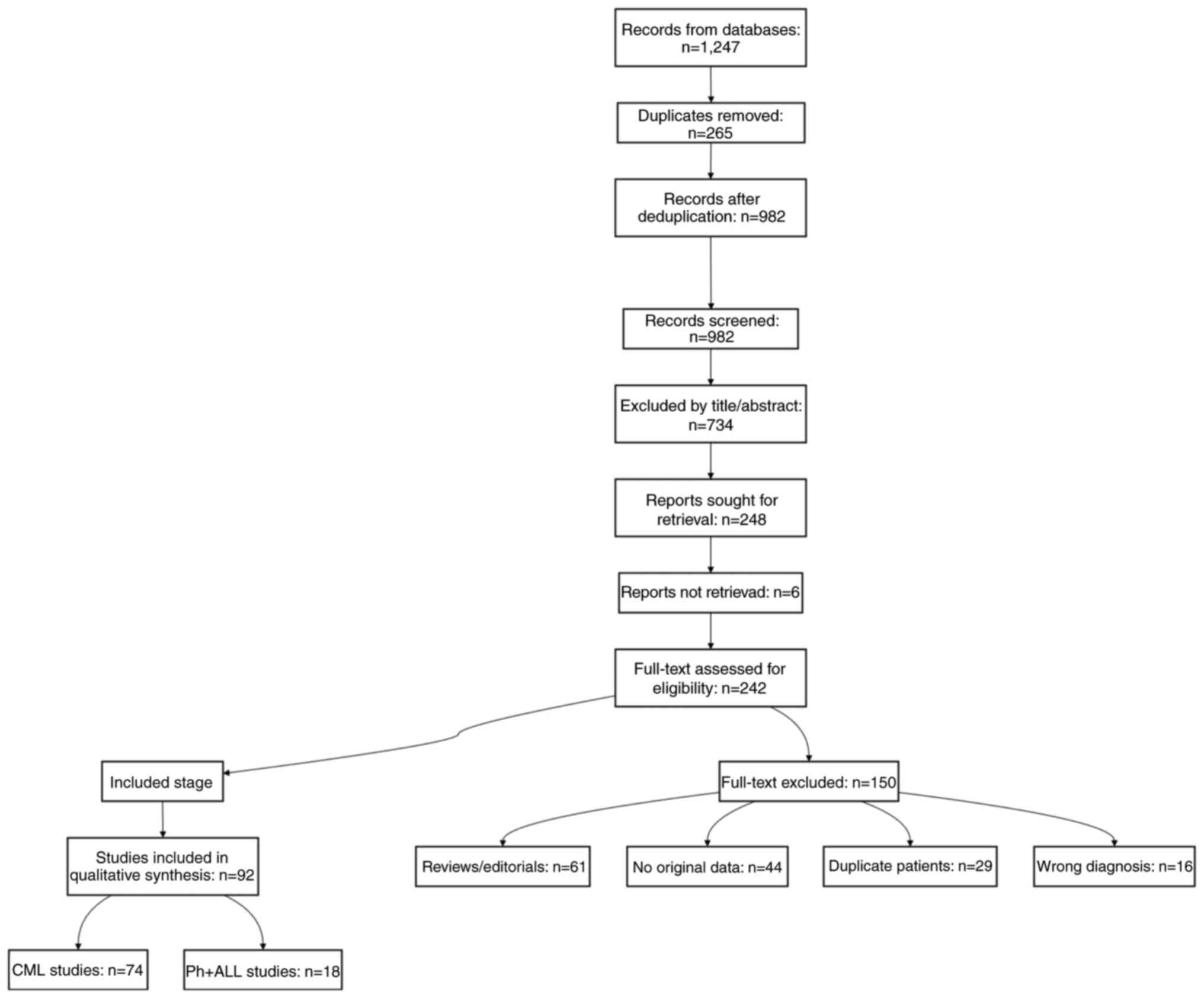
conducted a descriptive synthesis. The PRISMA flowchart (Fig. 4) documents the systematic selection
process. For each transcript subtype, the strength of evidence was
graded hierarchically: Grade A (robust), ≥10 clinically annotated
cases with a median follow-up ≥1 year; grade B (moderate), 3–9
cases or follow-up <1 year and grade C (limited), 1–2 cases or
in vitro data only (Table
I).
 | Table I.Evidence levels. |
Table I.
Evidence levels.
| Subtype | Total cases
(CML/Ph+ ALL) | Evidence type | Median follow-up,
months (range) | Outcome
definition | Evidence grade | (Refs.) |
|---|
| e13a3 | 38 (34/4) | Case series | 24.0
(6.0–120.0) | CCyR, MMR, OS | B | (17,27,30,34,37,40,41,43,44) |
| e14a3 | 25 (22/3) | Case reports | 18.0
(3.0–96.0) | CCyR, MMR | B | (11,21,53–57) |
| e1a3 | 12 (9/3) | Case reports | 15.0
(3.0–60.0) | CNS relapse,
OS | C | (60,64,69,72) |
| e1a2 | 41 (41/0) | Retrospective
cohort | 69.5
(12.0–120.0) | OS, blast
crisis | A | (81,91,92,95–97) |
| e6a2 | 18 (15/3) | Case reports | 12.0
(1.0–48.0) | ASCT outcomes | C | (63,65,66,100,102–104,140) |
| e8a2 | 5 (4/1) | Case reports | 6.0 (3.0–36.0) | CHR, CMR | C | (107,109,110,116,141) |
| e19a2 | 22 (20/2) | Case series | 30.0
(6.0–108.0) | MMR, TFR | B | (118,119,121,122,125,129,142–145) |
| e12a2 | 3 (3/0) | Case reports | 72.0
(60.0–84.0) | MR4 sustained | C | (130,146) |
e13a3 variant
Structural characteristics
The e13a3 (b2a3) BCR-ABL1 transcript is
generated through direct linkage of e13 of the BCR gene to
e3 (a3) of the ABL1 gene, resulting in deletion of
ABL1 e2 (a2). This fusion produces a truncated
protein that retains constitutively activated TK activity (27). The lack of the Src homology 3 (SH3)
domain in this variant is linked to its unique structural
properties, contributing to the formation of an SH3-deficient
isoform. SH3 deficiency can impact downstream signaling, enhancing
kinase activity and potentially promoting leukemogenesis (28). SH3-deficient variants such as
e13a3 are associated with altered protein interactions and
subcellular localization, potentially affecting cell signaling
pathways and contributing to disease progression. In murine models,
e13a3, as an SH3-deficient variant, shows slower disease
progression compared with canonical isoforms, though it retains
leukemogenic potential, capable of inducing CML (29). This slower progression may be
influenced by changes in cell adhesion, which alter the interaction
between leukemic cells and the microenvironment, affecting disease
dynamics. The SH3 domain normally serves as a negative regulator of
ABL1 TK activity, and its deletion in the e13a3 variant
enhances kinase activity (7). The
e13a3 fusion breakpoint resides within the major breakpoint
cluster region, resulting in the production of a 210 kDa (p210)
fusion protein. Notably, the absence of the SH3 domain in the
truncated ABL1 moiety induces structural alterations in the
chimeric protein. This aberrant protein retains constitutively
activated TK activity, which drives leukemic cell proliferation and
inhibits differentiation (27).
Compared with canonical fusion subtypes such as e14a2 or
e13a2, this variant exhibits a unique genomic architecture
(30). The e13a3 transcript
is predominantly observed in patients with chronic phase CML, with
rare case reports in Ph chromosome-positive ALL (31–33).
Notably, a Chinese study initially failed to detect the
e13a3 fusion using RT-qPCR, underscoring the risk of missing
rare fusion subtypes when employing primer sets targeting
conventional breakpoints, even in cases with confirmed
t(9;22) translocation (17).
Conversely, another study (34)
documented a CML case with a normal karyotype and negative RT-PCR
findings, where subsequent FISH analysis revealed BCR-ABL1
fusion. This highlights the importance of multimodal diagnostic
approaches, particularly when conventional methods yield equivocal
results.
Therapeutic management
Imatinib, a first-generation TKI is widely utilized
in e13a3 variant CML. Most Ph-positive CML patients
receiving 400 mg/day imatinib achieve complete cytogenetic
remission (CCyR) within 6–12 months and maintain durable responses
(35,36). McCarron et al (37) reported a 66-year-old male patient
with Ph-positive CML who attained progressively deepening
cytogenetic responses and declining BCR-ABL1/ABL1 ratios
following sustained 400 mg/day imatinib therapy. In Ph-negative CML
cohorts (representing 5–10% of cases, characterized by
CR-ABL1 rearrangements undetectable by conventional
cytogenetics) (38,39), studies (34,40)
have evaluated second-generation TKIs including nilotinib. These
agents demonstrate efficacy in achieving CCyR and major molecular
response (MMR) in Ph-positive populations (17,27),
which is consistent with the report by Zhou et al (41). Dasatinib, another second-generation
TKI, has also been employed in this context. Mechanistic and in
vitro studies indicate that the e13a3 variant may
exhibit resistance to asciminib, with clinical evidence remaining
limited (7). Resistance observed in
e13a3 variant CML is largely based on laboratory-based
research (7,42), and there is insufficient clinical
data to support these findings.
Combination strategies integrating TKIs with
chemotherapy have been explored. One notable example is the use of
the ponatinib-fludarabine + Low-dose Cytarabine (Ara-C) +
Granulocyte Colony-Stimulating Factor(G-CSF) + idarubicin regimen
followed by allogeneic stem cell transplantation (ASCT), which
resulted in molecular negativity and full donor chimerism at 19
months post-transplant in one case (43). Massimino et al (44) implemented a tailored approach in an
89-year-old male patient with mild renal impairment, including
initial hydroxyurea (2,000 mg/day) for leukocytosis management,
transitioning to dasatinib 100 mg/day, which achieved a deep
molecular response (MR), characterized by a further reduction in
transcript levels to undetectable. This strategy has also been used
in other studies (17,30).
Prognostic outcomes
Most studies suggest that e13a3-positive
patients exhibit a lower risk of progression to accelerated phase
or blast crisis, with superior long-term event-free survival
compared with rare variants such as e1a2 or e19a2
(12,45). Most patients attain deep, sustained
responses following TKI monotherapy or combination regimens. A
patient achieved complete hematological remission (CHR) at 2 months
and MMR with RT-PCR negativity by 8 months, maintaining remission
for 2 years (44). Another case
demonstrated FISH-confirmed CCyR (0% BCR-ABL1 fusion) at 6
months, sustained beyond 24 months (34). While certain TKI-treated cases
exhibit persistent low-level e13a3 transcripts despite CCyR
(37), combination therapies have
shown favorable results. In addition to treatment efficacy, it is
key to evaluate how the treatment regimen affects quality of life.
Long-term use of TKIs can lead to side effects such as chronic
fatigue, nausea and musculoskeletal pain, which may limit the
ability to perform daily tasks and participate in social activities
(46,47). Balancing treatment effectiveness
with the impact on physical and emotional wellbeing is essential
for optimal clinical decision-making. Resistance to treatment can
develop due to ABL kinase mutations such as T315I or activation of
compensatory signaling pathways such as PI3K/AKT and SRC, which
allow the leukemic cells to survive despite the presence of TKIs
(48). These mechanisms of
resistance contribute to treatment failure and disease progression.
In these cases, next-generation TKIs such as ponatinib and
asciminib, which target resistant mutations, can be effective,
although they may be associated with more severe side effects
(49–51). Combining TKIs with other therapeutic
modalities, including chemotherapy or stem cell transplantation,
may be necessary for patients with resistant disease to achieve
long-term disease control (52).
Evidence on e13a3 variant outcomes is summarized in Table II. Further multi-center studies are
needed to validate these prognostic outcomes in CML.
 | Table II.Summary of existing reports on cases
associated with the e13a3 variant. |
Table II.
Summary of existing reports on cases
associated with the e13a3 variant.
| Type | Country | Age, years | Sex | Method | Treatment | Outcome | Follow-up
duration | Diagnosis
stage | Mutations | Adjustment |
Transplantation | Adverse
reactions | Cause of death | (Refs.) |
|---|
| CML Ph(−) | Japan | 37 | F | FISH, RT-PCR | TKI: Imatinib, 400
mg/day | CHR, MMR | 12 months | Chronic phase | None | No adjustment | No
transplantation | Fatigue | None | (40) |
| CML Ph(−) | China | 24 | F | FISH, RT-PCR, gene
sequencing | TKI: Imatinib 400
mg/day | CCyR | 18 months | Chronic phase | None | No adjustment | No
transplantation | Nausea | None | (34) |
| CML Ph(+) | China | 39 | M | RT-PCR, Sanger
sequencing | TKI: Nilotinib, 300
mg/day | MMR | 12 months | Chronic phase | None | No adjustment | No
transplantation | Fatigue | None | (41) |
| CML Ph(+) | UK | 30 | M | RT-PCR | TKI: Ponatinib, 45
mg/day. Non-TKI: FLAG-IDA, allo-ASCT | MMR | 24 months | Chronic phase | None | No adjustment | Allo-ASCT | Gastrointestinal
disturbances | None | (43) |
| CML Ph(+) | Japan | 49 | M | RT-PCR, FISH | TKI: Imatinib, 400
mg/day. Non-TKI: Interferon, hydroxyurea | MMR | 12 months | Chronic phase | None | No adjustment | No
transplantation | Nausea | None | (30) |
| CML Ph(+) | Korea | 57 | M | RT-PCR, Sanger
sequencing, multiplex RT-PCR | TKI: Nilotinib, 300
mg/day. Non-TKI: Interferon, Hydroxyurea | CCyR, MMR | 18 months | Chronic phase | None | No adjustment | No
transplantation | Fatigue | None | (27) |
| CML Ph(+) | China | 32 | M | RT-qPCR, FISH,
karyotype analysis | TKI: 300 mg/day
nilotinib. Non-TKI: Hydroxyurea | CCyR | 18 months | Chronic phase | None | No adjustment | No
transplantation | Gastrointestinal
disturbances | None | (17) |
| CML Ph(+) | Italy | 89 | M | G-banding,
multiplex RT-PCR, Sanger sequencing | TKI: Dasatinib, 50
mg/day. Non-TKI: Hydroxyurea | CCyR, but e13a3
BCR-ABL1 transcript remains | 24 months | Chronic phase | None | No adjustment | No
transplantation | Nausea | None | (44) |
| CML Ph(+) | Ireland | 66 | M | Genetic analysis,
qPCR | TKI: Imatinib, 400
mg/day | BCR-ABL1
transcripts are decreased | 12 months | Chronic phase | None | No adjustment | No
transplantation | Nausea | None | (37) |
e14a3 variant
Structural characteristics
In this fusion transcript, the ABL1
breakpoint resides within intron 2, generating a chimeric mRNA
linking BCR e14 to ABL1 e3. This rearrangement
induces structural and functional alterations in the fusion
protein, dysregulating intracellular signaling pathways to promote
leukemic cell proliferation, survival and immune evasion. A
previous study employed customized RT-PCR coupled with Sanger
sequencing to confirm this fusion mRNA (53), concurrently identifying
non-synonymous mutations in TP53, FMS-like tyrosine kinase
3, KIT Proto-Oncogene, Receptor Tyrosine Kinase(KIT) and
paired box 5, underscoring the molecular heterogeneity. Another
case report documented methylenetetrahydrofolate reductase mutation
in a patient with CML harboring this BCR-ABL1 fusion,
providing insights for future investigations (54). A study identified a BCR-ABL1
fusion in a rare CML case, where the breakpoints occurred at
BCR intron 14 and ABL1 intron 2, using NGS (53). This unique fusion led to a
compromised SH3 domain, which was associated with altered drug
response and distinct clinical manifestations (53). These findings emphasize the critical
role of SH3 domain loss in modulating therapeutic outcomes and the
molecular heterogeneity underlying CML.
Therapeutic management
TKI monotherapy with imatinib or nilotinib has been
used in e14a3-positive cases. A Chinese study (11) reported a 67-year-old Ph-positive
female with coexisting e13a3 and e14a3 variants, the
first documented instance of dual rare BCR-ABL1 fusions in
China, who achieved therapeutic response with imatinib. Vaniawala
et al (55) reported
e14a3 BCR-ABL1 fusion in a 30-year-old male managed solely
with imatinib. Nilotinib was similarly employed to treat a
52-year-old male by Massimino et al (56), with both cases achieving a treatment
response.
Personalized combination regimens have also been
explored. In a study by Lyu et al (53), hydroxyurea was initially
administered for rapid leukocytosis control prior to TKI
initiation, with subsequent imatinib dose reduction (from 400 to
300 mg/day) due to intolerance, emphasizing individualized dosing.
A previous study (57) reported
sequential intolerance to imatinib and dasatinib, ultimately
transitioning to hydroxyurea monotherapy.
Prognostic outcomes
Most patients with e14a3 variant CML exhibit
favorable prognoses, achieving sustained hematological, cytogenetic
and molecular remissions. In a Chinese cohort (11), imatinib monotherapy induced rapid
MR, with CCyR attainment within 3 months and notable
BCR-ABL1-e14a3 transcript reduction. Nilotinib-treated cases
similarly demonstrated favorable outcomes (56). Combination regimens have shown
variable efficacy: One study reported TKI monotherapy achieving CHR
at 2 months, MMR at 3 months and sustained transcript negativity
for 9 years without kinase domain mutations (21). Adjuvant agents such as interferon,
hydroxyurea, and aspirin were incorporated, though immunomodulatory
effects of interferon yielded inconsistent results compared with
prior reports (57,58). Conversely, a patient requiring
hydroxyurea-nilotinib combination therapy achieved hematological
and molecular remission after dose adjustment (54). Key e14a3 variant case reports
and outcomes are summarized in Table
III.
 | Table III.Summary of existing reports on cases
associated with the e14a3 variant. |
Table III.
Summary of existing reports on cases
associated with the e14a3 variant.
| Type | Country | Age, years | Sex | Method | Treatment | Outcome | Follow-up
duration | Diagnosis
stage | Mutations | Adjustment |
Transplantation | Adverse
reactions | Cause of death | (Refs.) |
|---|
| CML Ph(+) | China | 24 | M | RT-PCR, Sanger
sequencing, NGS | TKI: Imatinib, 400
mg/day. Non- TKI: Hydroxyurea | Imatinib is
tolerated after lowering the dosage | 18 months | Chronic phase | None | Dose was decreased
to 300 mg/day | No
transplantation | Fatigue | None | (53) |
| CML Ph(+) | USA | 81 | M | FISH, Cytogenetic
analysis, RT-qPCR, DNA Sequencing | TKI: Dasatinib, 50
mg/day; imatinib, 400 mg/day. Non-TKI: Hydroxyurea | PCyR | 12 months | Chronic phase | None | No adjustment | No
transplantation | Fatigue,
gastrointestinal disturbances | None | (57) |
| CML Ph(+) | USA | 54 | F | RT-qPCR, FISH | TKI: Nilotinib, 300
mg/day Non-TKI: Hydroxyurea | CHR, MMR | 18 months | Chronic phase | None | No adjustment | No
transplantation | Nausea,
fatigue | None | (54) |
| CML Ph(+) | China | 67 | F | RT-qPCR | TKI: Imatinib, 400
mg/day | CCyR | 24 months | Chronic phase | None | No adjustment | No
transplantation | Fatigue | None | (11) |
| CML Ph(+) | China | 41 | F | FISH, nested PCR,
qPCR | TKI: Imatinib, 400
mg/day; dasatinib, 50 mg/day; nilotinib, 300 mg/day. Non-TKI:
Hydroxyurea, interferon | MMR | 24 months | Chronic phase | None | No adjustment | No
transplantation | Gastrointestinal
disturbances | None | (21) |
| CML Ph(+) | Italy | 52 | M | Cytogenetic
analysis, multiplex, RT-PCR | TKI: Imatinib, 400
mg/day; dasatinib, 50 mg/day; nilotinib 300 mg/day | CHR, CCyR | 18 months | Chronic phase | None | No adjustment | No
transplantation | Nausea,
fatigue | None | (56) |
| CML Ph(+) | India | 30 | M | Cytogenetic
analysis, FISH, RT-PCR | TKI: Imatinib, 400
mg/day. Non-TKI: Allo-ASCT | CHR | 24 months | Chronic phase | None | No adjustment | Allo-ASCT | None | None | (55) |
e1a3 variant
Structural characteristics
The e1a3 transcript arises from direct fusion
of e1 of the BCR gene to e3 (a3) of the
ABL1 gene, skipping ABL1 e2 (a2). This structural
alteration results in a truncated fusion protein lacking
approximately two-thirds of the sequence encoding the SH3 domain
within the ABL1 moiety (8).
Distinct from common variants, its unique fusion junction may
perturb subcellular localization, substrate specificity and
signaling pathways, thereby disrupting cellular homeostasis. This
transcript is relatively rare, with a single case (4.8%) identified
among patients with CML in a Syrian study (59). Additionally, its occurrence has been
documented in Ph+ ALL and AML (31). Some researchers have posited that a
subset of e1a3 BCR-ABL1-positive ALL cases may represent
undiagnosed CML in lymphoid blast crisis, requiring exclusion
through comprehensive clinical history review (60). A previous study (61) identified multiple atypical
BCR-ABL1 transcripts in CML, challenging prior assumptions
of singular fusion dominance. Conventional RT-PCR frequently fails
to detect these transcripts, often yielding false-negative results
(62), while RNA sequencing
(RNA-seq) uncovers their presence, highlighting the necessity for
advanced molecular diagnostics in clinical practice. The
e1a3 and e6a2 BCR-ABL1 transcripts are characterized
by unique fusion breakpoints within the ABL1 gene (63,64).
These isoforms are less common and associated with more aggressive
disease progression, including early blast crisis and resistance to
standard TKIs (65,66). These isoforms demonstrate TKI
resistance and often require multimodal therapy, including the use
of third-generation TKIs or stem cell transplantation (67,68).
Therapeutic strategies
Unlike e13a3 and e14a3 variants, dasatinib serves as
the primary TKI for e1a3-positive CML. The majority of reported
cases demonstrate an indolent clinical course (62,69). A
previous study (64) reported a
patient achieving CCyR following immediate dasatinib initiation
(140 mg/day). An 80-year-old Ph-positive male treated with 400
mg/day imatinib attained rapid CCyR and hematological normalization
but subsequently developed lymphoblastic crisis at 5 months,
suggesting a risk of ALL transformation (8). Combination therapies, including
dasatinib with nilotinib or ponatinib, have shown variable efficacy
(68,70); the T315I mutation frequently serves
as the primary resistance mechanism, necessitating the switch to
third-generation TKIs (71).
Innovative approaches, such as third-generation TKIs
combined with ASCT, have been employed in a previous study
(72). A 56-year-old female patient
maintained disease-free status post-ASCT with continued
olverembatinib therapy, underscoring the potential of
next-generation TKIs and ASCT in managing this rare subtype
(72).
Prognostic outcomes
Prognoses for e1a3 variant CML patients
exhibit marked heterogeneity. A Japanese male (64) achieved CCyR by 6 months with
dasatinib, despite presenting with extramedullary leukemia lacking
leukocytosis, which is rare in CML. A previous case (72) demonstrated sustained remission
post-ASCT and olverembatinib maintenance, yet developed isolated
central nervous system (CNS) infiltration without
hematological/cytogenetic relapse, implicating the CNS as a
potential sanctuary site. Due to the blood-brain barrier and
relatively immune-privileged status, conventional systemically
administered chemotherapeutic and targeted therapeutic agents often
fail to achieve effective concentrations within the CNS. This
allows cancer cells to evade treatment, survive, and cause a
relapse in this sanctuary site, while the rest of the body may
still be in a state of remission. A patient harboring the
e1a3 fusion, typically associated with aggressive disease,
maintained stable, untreated CML, challenging the association
between BCR-ABL1 variants and clinical severity (64). Key e1a3 variant case reports
and outcomes are summarized in Table
IV.
 | Table IV.Cases associated with the e1a3
variant. |
Table IV.
Cases associated with the e1a3
variant.
| Type | Country | Age, years | Gender | Method | Treatment | Outcome | Follow-up
duration | Diagnosis
stage | Mutations | Adjustment |
Transplantation | Adverse
reactions | Cause of death | (Refs.) |
|---|
| CML Ph(+) | Japan | 49 | M | FISH, RT-PCR,
Sanger Sequencing | TKI: Dasatinib, 50
mg/day | CCyR | 12 months | Chronic phase | None | No adjustment | No
transplantation | Fatigue, mild
nausea | None | (64) |
| CML Ph(+) | Spain | 80 | M | FISH, RT-PCR,
Cytomolecular assays | TKI: Imatinib, 400
mg/day | Lymphoblastic
crisis | 6 months | Blast crisis | None | No adjustment | No
transplantation | Severe fatigue,
gastrointestinal disturbances | Lymphoblastic
transformation | (60) |
| CML Ph(+) | Spain | 75 | F | RT-PCR | TKI: Imatinib, 400
mg/day | In good
condition | 24 months | Chronic phase | None | No TKI | No treatment | No
transplantation | None | (69) |
| CML Ph(+) | China | 56 | F | FISH, RT-PCR,
FC | TKI: Flumatinib, 50
mg/day; orebatinib, 50 mg/day. Non-TKI: Chemotherapy,
allo-ASCT | Disease-free | 24 months | Chronic phase | None | No adjustment | Yes, Allo-ASCT | None | None | (71) |
e1a2 variant
Structural characteristics
The e1a2 variant arises from fusion between
e1 of the BCR gene and e2 of the ABL1 gene,
generating a chimeric protein with distinct structural and
functional properties. By contrast with the canonical p210 isoform,
the p190 variant lacks central e13 and 14 of the BCR gene.
Despite this truncation, the p190 fusion protein retains notably
enhanced TK activity, which remains sufficient to drive
leukemogenesis (73,74). The e1a2 transcript is rare in
CML, accounting for ~1.8% of cases in a cohort of 2,322 patients
treated with TKIs, including 1,326 male and 996 female patients,
with a median age of 48 years (range 18–88) (14,75),
In the aforementioned study, 41 patients (1.8%) exhibited the
e1a2 fusion, confirmed by RT-PCR. This variant is associated
with a distinct phenotype marked by monocytosis, absence of
basophilia and blast crisis presentation at initial diagnosis in
61% of cases, significantly higher than in patients with canonical
transcripts (76). In a study by
Gong et al (52), 16 of 41
patients with the e1a2 transcript presented with blast
crisis at initial diagnosis, two had accelerated phase and 23 were
in chronic phase (76) The
frequency of monocytosis at initial diagnosis was confirmed in 10
patients with available blood counts, showing a median of 11.5%
(range, 5–36%), with seven patients exhibiting monocytosis >10%
(76). In Ph-positive adult ALL,
the e1a2 variant accounts for 61.2% of cases, as reported in
a national cohort of 67 patients with Ph+ ALL, and is
typically associated with elevated leukocyte count and lymphoid
lineage differentiation (77).
RT-PCR for BCR-ABL1 detection, and Sanger sequencing are
employed to confirm atypical fusion transcripts.
Therapeutic strategies
Imatinib remains the primary therapeutic agent for
e1a2-positive leukemia. A standard initial dose of 400
mg/day is administered in patients with ALL and CML, with dose
escalation or TKI switching considered for suboptimal responders.
However, it is important to consider the impact of side effects on
daily life (78). Common adverse
reactions such as fatigue, gastrointestinal disturbance and skin
rashes can significantly disrupt daily activities and affect the
overall quality of life (79).
These side effects should be weighed when selecting the most
appropriate treatment regimen, and supportive care may be necessary
to improve patient comfort during therapy (80). A previous study (81) documented a patient with Ph-negative
CML achieving molecular remission (undetectable e1a2
BCR-ABL1 by RT-PCR) after 2 months of imatinib monotherapy,
alongside rare cyclical leukocyte fluctuations and spontaneous
normalization without intervention. However, resistance to TKIs is
a major challenge, particularly in patients with mutations in the
ABL kinase domain, such as the T315I mutation, which notably
impairs the binding of TKIs to the BCR-ABL1 fusion protein
(82,83). These mutations lead to decreased
efficacy of first- and second-generation TKIs. Additionally,
compensatory signaling pathways, including the PI3K/AKT and SRC
kinase pathways, may be activated, allowing leukemic cells to
bypass the inhibition of BCR-ABL1, contributing to treatment
resistance (84). To overcome these
mechanisms of resistance, third-generation TKIs such as ponatinib
and asciminib, which are designed to target BCR-ABL1 with
T315I mutations and other resistant forms, show promising results
(50,51,85).
However, these agents also cause more severe side effects,
including cardiovascular complications, which should be managed.
Combining TKIs with other therapeutic strategies, such as
chemotherapy or immunotherapy, may provide an alternative approach
to overcome resistance, but this requires consideration of the
risk-to-benefit ratio for each patient (86–89).
Combination approaches are frequently employed,
often incorporating consolidation chemotherapy post-remission
(68,90). Japanese protocols (91) combine hydroxyurea with TKIs. For
relapsed/refractory cases, TKI substitution or chemotherapy
intensification may be pursued. A previous study (92) reported initial imatinib-induced
symptom resolution and 2.5-log BCR-ABL1 reduction, followed
by leukemic transformation at 6 months necessitating high-dose
chemotherapy and nilotinib. Transcript isoform switching may be a
potential molecular mechanism underlying disease recurrence
(93).
Prognostic outcomes
Prognostic heterogeneity characterizes e1a2
variant CML. While some patients achieve durable remission with
imatinib-based regimens (81,92,94,95),
clonal dynamics complicate outcomes. Multivariable analyses have
confirmed that e1a2 BCR-ABL1 serves as an independent
adverse prognostic factor, with a median OS of 69.5 months. Given
its clinical behavior resembling Ph-positive ALL, certain
researchers advocate classifying e1a2 as a distinct
high-risk subtype of CML (76). One
case (96) harbored dual
e13a3 and e1a2 clones, developing imatinib resistance
linked to e1a2 persistence despite achieving CHR, ultimately
progressing to blast crisis and death. Notably, resistance occurred
without ABL1 kinase domain mutations, suggesting alternative
mechanisms. A separate study (97)
described extramedullary blast crisis at TKI initiation, mirroring
e1a3 cases, yet subsequent multimodal therapy (TKIs, ASCT)
achieved sustained complete molecular remission (CMR) for >48
months. These findings underscore the necessity for comprehensive
molecular monitoring and adaptive therapeutic strategies. Key
e1a2 variant case reports and outcomes are summarized in
Table V.
 | Table V.Cases associated with the e1a2
variant. |
Table V.
Cases associated with the e1a2
variant.
| Type | Country | Age | Gender | Method | Treatment | Outcome | Follow-up
duration | Diagnosis
stage | Mutations | Adjustment |
Transplantation | Adverse
reactions | Cause of death | (Refs.) |
|---|
| CML Ph(+) | USA | 23 | F | G-banding, FISH,
PCR | TKI: Imatinib, 400
mg/day; dasatinib, 50 mg/day. Non-TKI: Chemotherapy, ASCT | MMR | 12 months | Blast crisis | None | No adjustment | Yes, Allo-ASCT | Fatigue,
nausea | None | (97) |
| CML Ph(−) | Serbia | 32 | M | FISH, RT-PCR,
Southern blotting | TKI: Imatinib, 400
mg/day | CHR, MMR | 18 months | Blast crisis | None | No adjustment | No
transplantation | Mild nausea,
fatigue | None | (81) |
| CML Ph(+) | Japan | 77 | M | RT-PCR | Non-TKI:
Hydroxyurea | Death | 6 months | Blast crisis | None | No treatment | No
transplantation | Severe fatigue, GI
disturbances | DIC and respiratory
failure | (91) |
| CML Ph(+) | UK | 53 | M | Sanger sequencing,
cytogenetic analysis | TKI: Imatinib, 400
mg/day; nilotinib, 300 mg/day Non-TKI: FLAG-Ida, ASCT | MMR | 18 months | Blast crisis | None | No adjustment | Allo-ASCT | Mild fatigue | None | (92) |
| CML Ph(+) | Ireland | 61 | F | RT-PCR, | TKI: Imatinib, 400
mg/day. Non-TKI: Hydroxyurea | CHR, CCyR | 24 months | Blast crisis | None | No adjustment | No
transplantation | Mild fatigue | None | (95) |
| CML Ph(+) | Spain | 79 | F | Cytogenetic
analysis, FISH, RT-PCR, Southern blotting | TKI: Imatinib 400
mg/day | Death | 12 months | Blast crisis | None | No adjustment | No
transplantation | Severe fatigue, GI
disturbances | Death | (96) |
e6a2 variant
Structural characteristics
e6a2 variant results from fusion between e6
of the BCR gene and e2 of the ABL1 gene. This
rearrangement alters the fusion protein structure, conferring
distinct TK activity and protein interaction profiles compared with
common variants, thereby dysregulating multiple intracellular
signaling pathways to drive leukemogenesis (98). This transcript accounts for
0.02–2.30% of all BCR-ABL1-positive CML cases. Although most
patients present in chronic phase, up to 40% of cases are diagnosed
in accelerated phase or blast crisis, with this variant frequently
demonstrating an aggressive clinical course (9,65).
This transcript was also detected in a case of ABL (99). Notably, conventional RT-qPCR may
fail to detect the e6a2 BCR-ABL1 transcript, necessitating
specialized RT-PCR strategies for rare fusion detection (66). Zagaria et al (100) employed ddPCR for e6a2
transcript quantification, leveraging its high sensitivity,
absolute quantification without standard curves and multiplexing
capabilities. Concurrent additional sex combs-like 1(ASXL1)
mutations, identified via NGS in e6a2-positive cases, may
synergize with the fusion to promote acute transformation (65). Domains retained or missing in each
subtype fusion protein are presented in Table VI.
 | Table VI.Comparison of BCR-ABL1 fusion
protein domains. |
Table VI.
Comparison of BCR-ABL1 fusion
protein domains.
| Fusion protein | Retained
domains | Missing
domains | Biological
implications |
|---|
| e6a2 | BCR coiled-coil,
DBL/PH, TK | SH3 | Loss of SH3 domain
may contribute to dysregulated signaling, associated with
resistance to TKI therapy |
| e13a3 | BCR coiled-coil,
DBL/PH, TK | SH3 | Loss of SH3
enhances kinase activity, promoting unregulated cell proliferation
and leukemia development |
| e14a3 | BCR coiled-coil,
DBL/PH, TK | SH3 | Alterations in SH3
may affect cellular signaling, potentially influencing TKI
response |
| e1a3 | BCR coiled-coil,
TK | DBL/PH, SH3 | Missing SH3 and
DBL/PH domains disrupt signaling pathways, leading to aggressive
disease progression. |
Therapeutic strategies
TKIs including imatinib (101), nilotinib (63) and dasatinib (102) are used for e6a2 variant
management, with imatinib remaining the cornerstone. However,
certain scholars advocate upfront use of second-generation TKIs or
ASCT to circumvent suboptimal responses to imatinib (103). In one CML case (102), initial hydroxyurea therapy for
thrombocytosis was discontinued due to neutropenia, followed by
successful imatinib 400 mg/day administration. In vitro
sensitivity assay measuring Crk-like protein and phosphorylated-Src
family kinase (Tyr416) inhibition confirmed imatinib
responsiveness, guiding therapeutic decisions (103).
Combination regimens integrating chemotherapy and
TKIs are employed in refractory cases. Crampe et al
(65) reported a patient achieving
hematological and morphological remission (BCR-ABL1/ABL1:
0.06%) with imatinib dose escalation (from 400 to 600 mg/day),
though subsequent sepsis necessitated allogeneic ASCT. Furthermore,
targeted therapies may confer prognostic benefits in patients
harboring co-occurring ASXL1 mutations.
Prognostic outcomes
Prognoses for e6a2 variant CML exhibit
marked variability, with frequent fatal outcomes. While some
achieve sustained remission post-TKI monotherapy (63,102)
or ASCT (65), others experience
rapid progression. Prognostic indices indicate that despite a
subset of patients exhibiting low-risk Sokal scores (63), OS rates remain inferior to those
observed in patients with common transcript subtypes. A patient
with Ph-positive CML maintained CCyR for 6 months on dasatinib
despite notable eosinophilic hyperplasia with atypical precursors
(a morphology potentially linked to the e6a2 transcript)
(100). Conversely, Beel et
al (66) documented rapid blast
crisis within 3 months of TKI initiation, culminating in fatal
multidrug-resistant bacteremia post-ASCT. Rohon et al
(103) advocated early ASCT or
clinical trial enrollment for e6a2-positive cases following
short-term TKI or dual Src/ABL inhibitor therapy.
Aggressive presentations include iliac sarcoma at
diagnosis (104) and novel
BCR-ABL1 kinase domain mutations (K245E, L284S)
emerging during imatinib therapy, culminating in blast crisis CML
and death (105). These findings
underscore the need for personalized strategies addressing
variant-specific biology. Key e6a2 variant case reports and
outcomes are summarized in Table
VII.
 | Table VII.Cases associated with the e6a2
variant. |
Table VII.
Cases associated with the e6a2
variant.
| Type | Country | Age, years | Gender | Method | Treatment | Outcome | Follow-up
duration | Diagnosis
stage | Mutations | Adjustment |
Transplantation | Adverse
reactions | Cause of death | (Refs.) |
|---|
| CML Ph(+) | Italy | 49 | M | Cytogenetic
testing, FISH, RT-PCR, ddPCR | TKI: Dasatinib, 100
mg/day | CCR | 12 months | Chronic phase | None | No adjustment | No
transplantation | Mild fatigue | None | (100) |
| CML Ph(+) | Ireland | 48 | F | Sanger sequencing,
RT-qPCR, NGS | TKI: Imatinib, 400
mg/day. Non-TKI: Induction therapy, allo-ASCT | CHR, MMR | 18 months | Chronic phase | None | Dose reduction | Allo-ASCT | Mild nausea,
fatigue | None | (65) |
| CML Ph(+) | Belgium | 57 | M | TaqMan RQ-PCR,
FISH, RT-qPCR | TKI: Imatinib, 400
mg/day. Non-TKI: Leukapheresis and chemotherapy, Allo-ASCT | Death | 6 months | Accelerated
phase | None | No adjustment | Allo-ASCT | Severe nausea, GI
distress | Sepsis | (66) |
| CML Ph(+) | Czech Republic | 51 | M | RT-PCR,
sequencing | TKI: Imatinib, 400
mg/day. Non-TKI: Hydroxyurea, G-CSF, allo-ASCT | MMR | 24 months | Chronic phase | None | No adjustment | Allo-ASCT | Mild fatigue | None | (103) |
| CML Ph(+) | Italy | 43 | M | FISH, RT-PCR | TKI: Imatinib, 400
mg/day. Non-TKI: Hydroxyurea | CCyR | 18 months | Chronic phase | None | No adjustment | No
transplantation | Mild nausea | None | (102) |
| CML Ph(+) | Italy | 46 | F | RT-PCR, Sanger
sequencing | TKI: Nilotinib 300
mg/day | CHR, CCyR | 12 months | Chronic phase | None | No adjustment | No
transplantation | Mild nausea,
fatigue | None | (63) |
| CML Ph(+) | Portugal | 18 | F | RT-PCR | TKI: Imatinib, 400
mg/day. Non-TKI: Radiotherapy, chemotherapy | Death | 3 months | Acute lymphoblastic
leukemia | None | No adjustment | No
transplantation | Severe nausea,
fatigue | Leukemic
transformation | (104) |
| CML Ph(+) | Germany | 48 | M | FISH, multiple
RT-PCR | TKI: Imatinib, 400
mg/day; dasatinib 50 mg/day | Death | 8 months | Chronic phase | None | No adjustment | No
transplantation | Severe fatigue, GI
distress | Sepsis | (140) |
e8a2 variant
Structural characteristics
The e8a2 variant is rare in CML cases. It is
characterized by fusion between e8 of the BCR gene and exon
a2 of ABL1, with insertion of a 127-bp sequence from e8 of
Ral GEF with PH domain and SH3 binding motif 1 (106). Studies demonstrate that the
generation of this transcript requires at least three chromosomal
breaks (106,107): The first occurs within ABL1
intron 1b, causing inversion and insertion of this region
downstream of BCR e8; the second occurs at BCR intron
8, facilitating fusion of BCR e8 with ABL1 a2; the
third may involve additional chromosomes, forming a four-way
translocation (107). While the
BCR-ABL1 e8a2 transcript is predominantly observed in CML,
its occurrence in ALL remains rare (106,108). A Uruguayan study (109) documented a rare four-way
translocation t(1;17;9;22)(p35;q24;q44;q11) in a 51-year-old
female patient with e8a2-positive CML, highlighting the
complexity of chromosomal rearrangements in leukemogenesis.
Researchers (110) have identified
somatic mutations in tumor protein p53 binding protein 2 and
cadherin-10 via whole-exome sequencing, absent in typical CML or
healthy controls, suggesting potential BCR-ABL1-driven
mutagenesis. Burmeister et al (107) proposed a mechanistic model for
cryptic exon activation, generating transcripts containing 55-bp
ABL1 intron 1b sequences. While the 55-bp insertion has been
suggested as a potential prerequisite for sustaining kinase
activity (111), documented cases
demonstrate that insertion-free e8a2 retains
oncoprotein-coding capacity, suggesting molecular heterogeneity
(112,113). The e8a2 and e19a2
variants are rare and associated with complex chromosomal
rearrangements. These variants can be considered distinct from
typical BCR-ABL transcripts. due to their distinct
structural features, including the involvement of additional
chromosomal breaks that contribute to unique functional properties
of the fusion proteins. These isoforms are less commonly associated
with early TKI resistance but are often found in patients with
advanced disease or when conventional diagnostic techniques
fail.
Therapeutic strategies
TKI regimens (imatinib, dasatinib, nilotinib)
constitute the primary therapeutic approach for
e8a2-positive CML, often supplemented by individualized
protocols. Dasatinib is prioritized as frontline therapy (110), with adjunctive measures such as
thromboprophylaxis and allopurinol administration tailored to
patient-specific factors, including age, comorbidity and disease
phase. Close clinical monitoring ensures timely regimen
optimization.
Prognostic outcomes
Most patients with e8a2 variant CML show a tendency
towards favorable outcomes under TKI therapy (114); while initial studies associated
the e8a2 transcript with thrombocytosis and suggested a poorer
prognosis, more recent findings indicate that prognosis may vary,
and the evidence remains inconclusive (107,115). Imatinib-treated cases typically
demonstrate robust responses, while interferon-intolerant patients
achieve CHR and CMR (BCR-ABL1 <0.001%) within 6 weeks, sustained
beyond 6 months (110). Tchirkov
et al (116) validated
real-time RT-PCR for precise molecular monitoring, enabling
therapeutic efficacy assessment and relapse prediction. Key e8a2
variant case reports and outcomes are summarized in Table VIII.
 | Table VIII.Cases associated with the e8a2
variant. |
Table VIII.
Cases associated with the e8a2
variant.
| Type | Country | Age | Gender | Method | Treatment | Outcome | Follow-up
duration | Diagnosis
stage | Mutations | Adjustment |
Transplantation | Adverse
reactions | Cause of death | (Refs.) |
|---|
| CML Ph(+) | Uruguay | 51 | F | GTG-banding, FISH,
RT-PCR | TKI: Imatinib, 400
mg/day. Non-TKI: Hydroxyurea | CCyR, MMR, CHR | 12 months | Chronic phase | None | No adjustment | No
transplantation | Mild fatigue,
nausea | None | (109) |
| CML Ph(+) | Korea | 46 | M | FISH, RT-PCR | TKI: Imatinib, 400
mg/day | CHR | 10 months | Chronic phase | None | No adjustment | No
transplantation | Mild nausea | None | (141) |
| CML Ph(+) | France | 43 | M | RT-PCR, cytogenetic
testing, FISH | TKI: Imatinib, 400
mg/day | MMR, CCyR | 15 months | Chronic phase | None | No adjustment | No
transplantation | Mild fatigue, GI
upset | None | (116) |
| CML Ph(+) | China | 49 | M | FISH, RT-PCR,
Sanger sequencing, RQ-PCR | TKI: Dasatinib, 50
mg/day. Non-TKI: Hydroxyurea, interferon | MMR, CHR | 18 months | Chronic phase | None | No adjustment | No
transplantation | Mild nausea | None | (110) |
| CML Ph(+) | Germany | 74 | M | RT-PCR, multiple
PCR, RT-qPCR | TKI: Nilotinib, 300
mg/day. Non-TKI: Hydroxyurea | BCR-ABL1 levels are
significantly decreased | 24 months | Chronic phase | None | No adjustment | No
transplantation | Mild fatigue | None | (108) |
e19a2 variant
Structural characteristics
The e19a2 (µ-BCR-ABL1) transcript arises from
aberrant fusion of BCR intron 19 to ABL1 exon a2, encoding a
230-kDa fusion protein (117). Its
formation involves submicroscopic insertion events, resulting in
FISH-negative/RT-PCR-positive detection. While p230 retains BCR
oligomerization domains and ABL1 TK activity, structural divergence
from p210 may compromise kinase-dependent signaling efficiency.
Sequencing of cDNA microproducts (118) has identified mutations in ABL1
e4-9, while WGS uncovered a 122-kb ABL1 insertion into the BCR
locus (117).
Therapeutic strategies
Management of e19a2-positive leukemia
involves sequential or combinatorial TKI regimens. Patients
frequently achieve MMR through sequential use of nilotinib,
dasatinib or ponatinib. Imatinib, though initially employed, is
often substituted due to resistance, thrombocytopenia, fluid
retention or drug interactions (119,120). Dose escalation may partially
restore hematological/cytogenetic responses in resistant cases. For
imatinib-resistant patients harboring the E355G mutation,
second-generation TKIs such as nilotinib induce major cytogenetic
responses, offering alternative therapeutic avenues (121). Allogeneic ASCT is utilized in
select cases (122), primarily
because it remains the only potentially curative treatment for CML.
It becomes a critical salvage treatment option when patients
develop resistance to or intolerable severe side effects from
multiple TKIs, or when the disease progresses from the chronic
phase to the prognostically unfavorable accelerated or blast
phase.
Prognostic outcomes
The prognosis of patients with e19a2 variant
CML is influenced by genetic architecture, therapeutic regimen and
individual comorbidities. Evidence indicates a trend towards
favorable outcomes, but individual responses differ (119,122). While studies have linked the
e19a2 transcript to an indolent phenotype (115,123), accumulating cases demonstrate
clinical courses indistinguishable from classic CML (117), with potential heightened
aggressiveness (124).
Second-generation TKIs such as nilotinib and dasatinib demonstrate
robust efficacy, exemplified by a 72-year-old patient with
chronic-phase CML who achieved CCyR at 6 months and MMR at 12
months with dasatinib combined with hydroxyurea and interferon
adjuncts, underscoring its utility as frontline therapy (120). Disease progression may occur in
certain cases, manifesting as leukocytosis or marrow dysplasia.
Notably, a patient managed with nilotinib required dose
interruptions due to grade 2 hepatotoxicity yet maintained
sustained CCyR and deep MR during long-term follow-up, aligning
with findings by Crampe et al (121) and Ernst et al (122), which confirmed nilotinib durable
efficacy following treatment interruptions (125). Hydroxyurea monotherapy has also
stabilized leukocyte counts without complications in select cases
(126–128).
Resistance mechanisms pose challenges. An Italian
study (118) reported a
dasatinib-resistant T315I mutation, typically associated with TKI
refractoriness, where dose escalation partially restored
hematological and cytogenetic responses, suggesting salvage
potential in mutation-positive patients. Clonal evolution,
including double Ph chromosomes and tetraploidy detected via FISH
and cytogenetics in an imatinib-treated patient (129), culminated in fatal blast crisis
within 2 years, emphasizing the need for personalized strategies in
e19a2 BCR-ABL1-positive CML. e19a2 variant case
reports and outcomes are summarized in Table IX.
 | Table IX.Cases associated with the
e19a2 variant. |
Table IX.
Cases associated with the
e19a2 variant.
| Type | Country | Age, years | Gender | Method | Treatment | Outcome | Follow-up
duration | Diagnosis
stage | Mutations | Adjustment |
Transplantation | Adverse
reactions | Cause of death | (Refs.) |
|---|
| CML Ph(+) | Italy | 78 | F | FISH, RT-PCR | TKI: Imatinib, 400
mg; day; nilotinib, 300 mg/day; dasatinib. 50 mg/day. Non-TKI:
Interferon, cytarabine | Drug
resistance | 18 months | Chronic phase | None | No adjustment | No
transplantation | Mild fatigue | None | (118) |
| CML Ph(+) | Japan | 85 | F | FISH, RT-PCR | TKI: Imatinib, 400
mg/day Non-TKI: Hydroxyurea | Death | 12 months | Chronic phase | None | No adjustment | No
transplantation | Mild nausea | Death | (129) |
| CML Ph(+) | Germany | 89 | F | FISH, RT-PCR,
direct sequencing | TKI: Imatinib, 400
mg/day; dasatinib, 50 mg/day; nilotinib. 300 mg/day. Non-TKI:
Hydroxyurea | No complications
observed, but WBC count remained high | 24 months | Chronic phase | None | No adjustment | No
transplantation | Mild fatigue | None | (142) |
| CML Ph(+) | Tunisia | 34 | F | RT-PCR, FISH | TKI: Imatinib, 400
mg/day nilotinib, 300 mg/day. Non-TKI: Hydroxyurea | Initial MCyR, but
TKI resistance develops later | 15 months | Chronic phase | None | No adjustment | No
transplantation | Mild nausea | None | (143) |
| CML Ph(+) | Japan | 72 | F | RT-qPCR, FISH | TKI: Nilotinib 300
mg/day | CCyR, MMR | 18 months | Chronic phase | None | No adjustment | No
transplantation | Mild fatigue | None | (129) |
| CML Ph(+) | Ireland | 26 | M | RT-PCR, Direct
sequencing | TKI: Imatinib, 400
mg/day Non-TKI: Interferon, hydroxyurea therapy | e19a2 BCR-ABL1
remains | 24 months | Chronic phase | None | No adjustment | No
transplantation | Mild fatigue | None | (144) |
| CML Ph(+) | Japan | 77 | F | RT-qPCR, | TKI: Imatinib, 400
mg/day; nilotinib, 300 mg/day | MMR | 20 months | Chronic phase | None | No adjustment | No
transplantation | Mild fatigue | None | (125) |
| CML Ph(+) | Ireland | 53 | F | Bone marrow
morphology, cytogenetics, molecular analysis | TKI: Imatinib, 400
mg/day; nilotinib, 300 mg/day Non-TKI: G-CSF | MMR | 24 months | Chronic phase | None | No adjustment | No
transplantation | Mild fatigue | None | (121) |
| CML Ph(+) | Germany | 33 | M | Multiple PCR,
Sanger sequencing | TKI: Ponatin, 45
mg/day; baxitinib, 5 mg/day. Non-TKI: Interferon | MMR | 18 months | Chronic phase | None | No adjustment | No
transplantation | Mild nausea | None | (122) |
| CML Ph(+) | France | 72 | F | FISH, PCR,
sequencing | TKI: Imatinib, 400
mg/day; dasatinib 50 mg/day | MMR | 24 months | Chronic phase | None | No adjustment | No
transplantation | Mild fatigue | None | (119) |
| CML Ph(-) | UK | 43 | F | G-banded chromosome
analysis, RT-qPCR | TKI: Imatinib, 400
mg/day dasatinib 50 mg/day. Non-TKI: Hydroxyurea | MMR | 18 months | Chronic phase | None | No adjustment | No
transplantation | Mild fatigue | None | (145) |
e12a2 variant
The e12a2 variant, a rare subtype, arises
from fusion between e12 of the BCR gene and exon a2 of
ABL1. Investigators employed primer sets (BCR-10 and ABL1-4)
in RT-PCR assays to detect uncommon e12a2 BCR-ABL1 fusion
transcripts, identifying an 18-bp insertion derived from
ABL1 intron 1b at the junctional site (130). Notably, this isoform may co-occur
with common transcripts, suggesting clonal heterogeneity or
molecular evolution during disease progression (130).
Therapeutic approaches involve sequential TKIs
(imatinib, dasatinib, nilotinib, bosutinib, ponatinib) (130). A 59-year-old male patient with CML
who developed resistance to imatinib (130) achieved CCyR and MR3 within 6
months of nilotinib escalation (800 mg/day). Due to cardiovascular
adverse events, therapy was subsequently transitioned to ponatinib
(15 mg/day), maintaining a MR4 for 6 years. However, management
complexity arises from frequent requirement for multiple drugs,
dose-limiting toxicity and treatment-associated burdens,
necessitating rigorous monitoring. The paucity of reported e12a2
BCR-ABL1 cases underscores the need for expanded cohort studies
to elucidate its impact on disease progression and prognosis.
e18a2 variant
The e18a2 transcript is a rare
BCR-ABL1 fusion variant arising from t(9;22)(q34;q11)
chromosomal translocation, which juxtaposes e18 of the BCR
gene with exon 2 (a2) of ABL1, encoding a 225 kDa
fusion protein (p225) (131,132). The breakpoint within the µ-BCR
region retains nearly complete BCR sequences, including
calcium-binding and GTPase-Activating Protein domains specific for
the Rac GTPase and coiled-coil oligomerization motifs, while
preserving the intact TK domain of ABL1 (133). The e18a2 transcript is rare
in CML, with an estimated incidence <1%. A recent study
documented a 49-year-old patient with CML initially misclassified
as e19a2-positive; relapse evaluation failed due to negative
conventional RT-qPCR targeting common isoforms, highlighting
diagnostic challenges (134).
Coexistence of e18a2 with e19a2 transcripts further
complicates detection (135).
Current therapeutic evidence for e18a2
remains sparse (4,134), though insights may be extrapolated
from other rare variants. In a 16-year-old female patient with CML
harboring e18a2 (136),
initial RQ-PCR failed to detect the transcript, yielding
false-negative results. Treatment commenced with hydroxyurea and
imatinib 600 mg/day, later decreased to 400 mg/day due to
thrombocytopenia. CHR was achieved by day 56, followed by major
cytogenetic response by day 106. Customized RQ-PCR monitoring
revealed a decline in tumor burden to 1×10−3 by month
15. Imatinib was safely re-escalated to 600 mg/day without relapse,
demonstrating favorable tolerability. The prognostic value of
e18a2 remains contentious due to limited sample sizes and
undefined molecular kinetics.
e13a1 variant
The e13a1 transcript is characterized by the
replacement of the terminal 38 bp of BCR e13 with a 37-bp
sequence derived from ABL1 intron 1–2/e1, resulting in
bidirectional disruption of exon junction architecture. Notably, a
G>A point mutation within the inserted sequence substitutes
glutamine with lysine at position 27 (137), potentially altering local charge
distribution and impacting drug-binding efficiency, though direct
experimental evidence remains lacking. A previous study documented
a 69-year-old patient with CML initially yielding negative results
with TaqMan RT-q and multiplex PCR assays; subsequent Sanger
sequencing of single-step PCR products confirmed the e13a1
transcript. The patient achieved sustained MR (BCR-ABL1/ABL1
levels ranging from MR4.5 to MMR) following imatinib therapy,
underscoring therapeutic efficacy while necessitating long-term
surveillance.
Other variants
Certain BCR-ABL1 transcripts reported in the
literature are rare (4,61), with their clinical significance
poorly defined. For example, e1a4 and e1a5 variants
have been described exclusively in Ph+ ALL (61), although large-scale epidemiological
data validating their prevalence or clinical relevance are lacking.
The e8a4 variant was detected in a patient with Sézary
syndrome (138); to the best of
our knowledge, however, there have been no subsequent studies
investigating this variant, and its direct association with
BCR-ABL1-driven oncogenesis requires further exploration.
The existence of these rare transcripts suggests certain variants
may emerge selectively within specific disease subtypes or
individuals. Nevertheless, due to limitations in detection
technologies and the paucity of reported cases, numerous potential
variants may remain undetected or systematically
uncharacterized.
Discussion
The growing recognition of atypical BCR-ABL1
fusion transcripts in CML underscores the need for nuanced
diagnostic and therapeutic strategies (4,26).
While canonical isoforms dominate clinical practice, atypical
variants such as e13a3, e14a3, e1a3, e1a2, e6a2 and
e8a2 exhibit distinct molecular architectures that notably
influence disease biology, therapeutic responsiveness and clinical
outcomes (139).
Advances in understanding atypical fusions have
revealed distinct structural configurations that alter fusion
protein function, dysregulating intracellular signaling,
proliferation, differentiation and apoptosis. Therapeutic
strategies combining TKIs, chemotherapy and ASCT demonstrate
variable efficacy across subtypes. While certain patients achieve
durable remission, others experience refractory disease or rapid
progression, highlighting pronounced inter-variant prognostic
heterogeneity. For example, e13a3 and e8a2 variants
are frequently associated with indolent disease course and
favorable responses to TKIs, as evidenced by sustained cytogenetic
and molecular remissions in multiple case series (7,44,110,116). Conversely, patients with
e1a2 and e6a2 isoforms experience more rapid disease
progression, including accelerated phase or blast crisis. In
refractory cases, 5-year OS rates are 40–70%. Notably, patients
with e1a2-positive CML often present with lymphoid blast
crisis-like features, including monocytosis and absence of
basophilia, mirroring Ph-positive ALL. Similarly, e6a2 cases
show elevated rates of clonal evolution and resistance mutations,
necessitating early escalation to second-generation TKIs or ASCT.
These findings emphasize that atypical transcripts are not
uniformly benign and require vigilant monitoring. TKI
responsiveness varies significantly across atypical variants. While
imatinib remains effective for e13a3 and e8a2, e1a2
and e6a2 subtypes often require early transition to second-
or third-generation TKIs due to intrinsic resistance. Dose
adjustments or combination regimens may salvage responses in
resistant cases, as demonstrated in patients with e6a2
achieving molecular remission post-ASCT. However, therapeutic
decisions must balance efficacy against toxicity, particularly in
elderly or comorbid populations. For example, dose reduction
mitigates hepatotoxicity while maintaining remission in
e14a3 cases.
Conventional diagnostic methods, such as standard
RT-PCR or FISH, may fail to detect rare fusion isoforms due to
primer mismatches or cryptic chromosomal rearrangements. For
example, e1a3 and e6a2 transcripts are frequently
missed by routine assays, leading to delayed diagnosis and
inappropriate therapeutic choices. Complementary techniques,
including multiplex RT-PCR, nested PCR and RNA-seq, are essential
for identifying atypical breakpoints and coexisting mutations that
may drive disease progression. ddPCR further enhances sensitivity
for minimal residual disease monitoring, particularly for
low-abundance transcripts such as e6a2.
Data on atypical transcripts derive predominantly
from case reports and small cohorts, limiting statistical power and
generalizability. To address these challenges, multicenter
collaborative studies are required to expand case accrual and
establish robust genomic databases. Mechanistic investigations
should delineate molecular pathways and crosstalk between atypical
fusions and ancillary signaling networks, informing precision
therapeutics. Diagnostic innovation should prioritize
high-sensitivity/specificity assays for rare fusion detection,
enabling early intervention. Therapeutic development requires
variant-tailored approaches integrating genomic profiling, disease
stage and patient comorbidities, alongside intensified research
into resistance mechanisms and salvage strategies. Longitudinal
studies assessing long-term outcomes and survivorship are key to
optimize holistic care, refine prognostic stratification and
ultimately improve survival for patients with atypical
BCR-ABL1-positive leukemia.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Taishan Youth Scholar
Foundation of Shandong Province (grant no. tsqn201812140).
Availability of data and materials
Not applicable.
Authors' contributions
XZ wrote the manuscript and constructed figures and
tables. AL, DK and PZ revised the manuscript. YS and NS designed
the methodology. Data authentication is not applicable. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Foroni L, Wilson G, Gerrard G, Mason J,
Grimwade D, White HE, de Castro DG, Austin S, Awan A, Burt E, et
al: Guidelines for the measurement of BCR-ABL1 transcripts in
chronic myeloid leukaemia. Br J Haematol. 153:179–1790. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stella S, Gottardi EM, Favout V, Barragan
Gonzalez E, Errichiello S, Vitale SR, Fava C, Luciano L, Stagno F,
Grimaldi F, et al: The Q-LAMP method represents a valid and rapid
alternative for the detection of the BCR-ABL1 rearrangement in
Philadelphia-positive leukemias. Int J Mol Sci. 20:61062019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jajosky AN and Lichtman MA: Uncommon
phenotypes of BCR::ABL1-positive chronic myelogenous leukemia.
Haematologica. 110:1912–1920. 2025.PubMed/NCBI
|
|
4
|
Breccia M: Atypical CML: Diagnosis and
treatment. Hematology Am Soc Hematol Educ Program. 2023:476–482.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan Z, Shi L, Li W, Liu W, Galderisi C,
Spittle C, Spittle C and Li J: A novel Next-generation sequencing
assay for the identification of BCR::ABL1 Transcript type and
accurate and sensitive detection of TKI-resistant mutations. J Appl
Lab Med. 9:886–900. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cross NCP, Ernst T, Branford S, Cayuela
JM, Deininger M, Fabarius A, Kim DDH, Machova Polakova K, Radich
JP, Hehlmann R, et al: European LeukemiaNet laboratory
recommendations for the diagnosis and management of chronic myeloid
leukemia. Leukemia. 37:2150–2167. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Leske IB and Hantschel O: The e13a3 (b2a3)
and e14a3 (b3a3) BCR::ABL1 isoforms are resistant to asciminib.
Leukemia. 38:2041–2045. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Z: The e1a3 BCR-ABL1 fusion
transcript in Philadelphia chromosome-positive acute lymphoblastic
leukaemia: A case report. Hematology. 28:21860402023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Baccarani M, Castagnetti F, Gugliotta G,
Rosti G, Soverini S, Albeer A and Pfirrmann M; International
BCR-ABL Study Group, : The proportion of different BCR-ABL1
transcript types in chronic myeloid leukemia. An international
overview. Leukemia. 33:1173–1183. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ding L, Chen Q, Chen K, Jiang Y, Li G,
Chen Q, Bai D, Gao D, Deng M, Zhang H and Xu B: Simvastatin
potentiates the cell-killing activity of imatinib in
imatinib-resistant chronic myeloid leukemia cells mainly through
PI3K/AKT pathway attenuation and Myc downregulation. Eur J
Pharmacol. 913:1746332021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Zhang Y, Meng X, Chen S, Wang T,
Zhang L and Ma X: Chronic myeloid leukemia with two rare fusion
gene transcripts of atypical BCR::ABL: A case report and literature
review. Medicine (Baltimore). 103:e367282024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schäfer V, White HE, Gerrard G, Möbius S,
Saussele S, Franke GN, Mahon FX, Talmaci R, Colomer D, Soverini S,
et al: Assessment of individual molecular response in chronic
myeloid leukemia patients with atypical BCR-ABL1 fusion
transcripts: Recommendations by the EUTOS cooperative network. J
Cancer Res Clin Oncol. 147:3081–3089. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hoffmann VS, Baccarani M, Hasford J,
Castagnetti F, Di Raimondo F, Casado LF, Turkina A, Zackova D,
Ossenkoppele G, Zaritskey A, et al: Treatment and outcome of 2904
CML patients from the EUTOS population-based registry. Leukemia.
31:593–60. 20171 View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hughes T, Deininger M, Hochhaus A,
Branford S, Radich J, Kaeda J, Baccarani M, Cortes J, Cross NC,
Druker BJ, et al: Monitoring CML patients responding to treatment
with tyrosine kinase inhibitors: Review and recommendations for
harmonizing current methodology for detecting BCR-ABL transcripts
and kinase domain mutations and for expressing results. Blood.
108:28–37. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin X, Huang H and Chen P: Retrospective
analysis of the clinical features of 172 patients with
BCR-ABL1-negative chronic myeloproliferative neoplasms. Mol
Cytogenet. 13:82020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang Z, Wang W, Toruner GA, Hu S, Fang H,
Xu J, You MJ, Medeiros LJ, Khoury JD and Tang G: Optical genome
mapping for detection of BCR::ABL1-another tool in our toolbox.
Genes (Basel). 15:13572024. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu B, Zhang W and Ma H: Complete
cytogenetic response to Nilotinib in a chronic myeloid leukemia
case with a rare e13a3(b2a3) BCR-ABL fusion transcript: A case
report. Mol Med Rep. 13:2635–2638. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Avila M and Meric-Bernstam F:
Next-generation sequencing for the general cancer patient. Clin Adv
Hematol Oncol. 17:447–454. 2019.PubMed/NCBI
|
|
19
|
Sorokin M, Rabushko E, Rozenberg JM,
Mohammad T, Seryakov A, Sekacheva M and Buzdin A: Clinically
relevant fusion oncogenes: Detection and practical implications.
Ther Adv Med Oncol. 14:175883592211441082022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee H, Seo J, Shin S, Lee ST and Choi JR:
Development and validation of sensitive BCR::ABL1 fusion gene
quantitation using next-generation sequencing. Cancer Cell Int.
23:1062023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang X, Sun H, Su Y and Yi H: Long-term
molecular remission after treatment with imatinib in a chronic
myeloid leukemia patient with extreme thrombocytosis harboring rare
e14a3 (b3a3) BCR::ABL1 transcript: A case report. Curr Oncol.
29:8171–8179. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Blachly JS, Walter RB and Hourigan CS: The
present and future of measurable residual disease testing in acute
myeloid leukemia. Haematologica. 107:2810–2822. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Denk D, Bradtke J, König M and Strehl S:
PAX5 fusion genes in t(7;9)(q11.2;p13) leukemia: A case report and
review of the literature. Mol Cytogenet. 7:132014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kosik P, Skorvaga M and Belyaev I:
Incidence of preleukemic fusion genes in healthy subjects.
Neoplasma. 63:659–672. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Elgehama A, Chen W, Pang J, Mi S, Li J,
Guo W, Wang X, Gao J, Yu B, Shen Y and Xu Q: Blockade of the
interaction between Bcr-Abl and PTB1B by small molecule SBF-1 to
overcome Imatinib-resistance of chronic myeloid leukemia cells.
Cancer Lett. 372:82–88. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Szuber N, Orazi A and Tefferi A: Chronic
neutrophilic leukemia and atypical chronic myeloid leukemia: 2024
update on diagnosis, genetics, risk stratification, and management.
Am J Hematol. 99:1360–1387. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ha J, Cheong JW, Shin S, Lee ST and Choi
JR: Chronic myeloid leukemia with rare variant b2a3 (e13a3)
BCR-ABL1 fusion. Ann Lab Med. 36:287–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hai A, Kizilbash NA, Zaidi SHH, Alruwaili
J and Shahzad K: Differences in structural elements of Bcr-Abl
oncoprotein isoforms in Chronic Myelogenous Leukemia.
Bioinformation. 10:108–114. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gross AW, Zhang X and Ren R: Bcr-Abl with
an SH3 deletion retains the ability to induce a myeloproliferative
disease in mice, yet c-Abl activated by an SH3 deletion induces
only lymphoid malignancy. Mol Cell Biol. 19:6918–6928. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu LG, Tanaka H, Ito K, Kyo T, Ito T and
Kimura A: Chronic myelogenous leukemia with e13a3 (b2a3) type of
BCR-ABL transcript having a DNA breakpoint between ABL exons a2 and
a3. Am J Hematol. 74:268–272. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Burmeister T, Schwartz S, Taubald A, Jost
E, Lipp T, Schneller F, Diedrich H, Thomssen H, Mey UJ, Eucker J,
et al: Atypical BCR-ABL mRNA transcripts in adult acute
lymphoblastic leukemia. Haematologica. 92:1699–1702. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Crampe M, Kearney L, O'Brien D, Bacon CL,
O'Shea D and Langabeer SE: Molecular monitoring in adult
philadelphia Chromosome-positive acute lymphoblastic leukemia with
the variant e13a3 BCR-ABL1 fusion. Case Rep Hematol.
2019:96350702019.PubMed/NCBI
|
|
33
|
Phan CL, Tan SN, Tan SM, Kadir SSSA, Ramli
NLM, Lim TO and Ng CC: A variant e13a3 BCR-ABL1 fusion transcript
in refractory adult B-cell acute lymphoblastic leukemia achieving
complete remission with CAR-Tcell therapy. Cancer Genet.
250-251:20–24. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Duan MH, Li H and Cai H: A rare e13a3
(b2a3) BCR-ABL1 fusion transcript with normal karyotype in chronic
myeloid leukemia: The challenges in diagnosis and monitoring
minimal residual disease (MRD). Leuk Res. 59:8–11. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Waclaw J, Sacha T and Stoklosa T: Imatinib
in the treatment of chronic myeloid leukemia: Current perspectives
on optimal dose. Blood and Lymphatic Cancer: Targets Ther.
5:101–108. 2015.
|
|
36
|
Fava C, Rege-Cambrin G and Saglio G:
Imatinib: The First-Line CML Therapy. Chronic Myeloid Leukemia
[Internet]. Hehlmann R: Cham: Springer International Publishing;
pp. 49–59. 2021, https://doi.org/10.1007/978-3-030-71913-5_4
View Article : Google Scholar
|
|
37
|
McCarron SL, Langabeer SE, Bolger K,
Haslam K, Crampe M, Kelly J and Morrell R: Molecular response to
imatinib in chronic myeloid leukaemia with a variant e13a3 BCR-ABL1
fusion. Med Oncol. 32:4522015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Marzocchi G, Castagnetti F, Luatti S,
Baldazzi C, Stacchini M, Gugliotta G, Amabile M, Specchia G,
Sessarego M, Giussani U, et al: Variant Philadelphia
translocations: Molecular-cytogenetic characterization and
prognostic influence on frontline imatinib therapy, a GIMEMA
Working Party on CML analysis. Blood. 117:6793–6800. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lim TH, Tien SL, Lim P and Lim AST: The
incidence and patterns of BCR/ABL rearrangements in chronic myeloid
leukaemia (CML) using fluorescence in situ hybridisation (FISH).
Ann Acad Med Singap. 34:533–538. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Masuko M, Furukawa T, Abe T, Wada R,
Maruyama S, Kitajima T, Shibasaki Y, Toba K, Okada M and Aizawa Y:
A chronic myeloid leukemia patient with atypical karyotype and
BCR-ABL e13a3 transcript caused by complex chromosome
rearrangement. Int J Hematol. 90:230–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou X, Li MR and Shan NN: Chronic myeloid
leukemia with the e13a3 atypical fusion gene: A case report. Oncol
Lett. 29:3192025. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Soverini S: Resistance mutations in CML
and how we approach them. Hematology Am Soc Hematol Educ Program.
2023:469–475. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu D, Claudiani S, Naresh K, Mucklow S,
Neelakantan P, Yebra E, Apperley JF, Khorashad J and Milojkovic D:
Blast crisis of chronic myeloid leukemia with plasmacytoid
dendritic cell phenotype associated with a rare fusion transcript,
e13a3 BCR-ABL1. Leuk Lymphoma. 60:3090–3091. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Massimino M, Stella S, Tirrò E, Consoli
ML, Pennisi MS, Puma A, Vitale SR, Romano C, Zammit V, Stagno F, et
al: Efficacy of dasatinib in a very elderly CML patient expressing
a rare E13a3 Bcr-Abl1 fusion transcript: A case report. Anticancer
Res. 39:3949–3954. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang YL, Bagg A, Pear W, Nowell PC and
Hess JL: Chronic myelogenous leukemia: Laboratory diagnosis and
monitoring. Genes Chromosomes Cancer. 32:97–11. 20011 View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Caldemeyer L, Dugan M, Edwards J and Akard
L: Long-term side effects of tyrosine kinase inhibitors in chronic
myeloid leukemia. Curr Hematol Malig Rep. 11:71–79. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Narra RK, Flynn KE and Atallah E: Chronic
myeloid leukemia-the promise of tyrosine kinase inhibitor
discontinuation. Curr Hematol Malig Rep. 12:415–4123. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Santos FPS and Ravandi F: Advances in
treatment of chronic myelogenous Leukemia-new treatment options
with tyrosine kinase inhibitors. Leuk Lymphoma. 50 (Suppl
2):S16–S26. 2009. View Article : Google Scholar
|
|
49
|
Hughes TP, Mauro MJ, Cortes JE, Minami H,
Rea D, DeAngelo DJ, Breccia M, Goh YT, Talpaz M, Hochhaus A, et al:
Asciminib in chronic myeloid leukemia after ABL kinase inhibitor
failure. N Engl J Med. 381:2315–2326. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Haddad FG, Issa GC, Jabbour E and Yilmaz
M: Ponatinib for the treatment of adult patients with resistant or
intolerant Chronic-phase chronic myeloid leukemia. Expert Opin
Pharmacother. 23:751–758. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cortes JE, Sasaki K, Kim DW, Hughes TP,
Etienne G, Mauro MJ, Hochhaus A, Lang F, Heinrich MC, Breccia M, et
al: Asciminib monotherapy in patients with chronic-phase chronic
myeloid leukemia with the T315I mutation after ≥1 prior tyrosine
kinase inhibitor: 2-year follow-up results. Leukemia. 38:1522–1533.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Singh VK and Coumar MS: Chronic myeloid
leukemia: Existing therapeutic options and strategies to overcome
drug resistance. Mini Rev Med Chem. 19:333–345. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lyu X, Yang J, Wang X, Hu J, Liu B, Zhao
Y, Guo Z, Liu B, Fan R and Song Y: A novel BCR-ABL1 fusion gene
identified by next-generation sequencing in chronic myeloid
leukemia. Mol Cytogenet. 9:472016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chisti MM and Sanders DS: Chronic myeloid
leukemia with b3a3 (e14a3) fusion: A rare BCR/ABL rearrangement
presenting with thrombocytosis-Does MTHFR polymorphism matter. Case
Rep Oncol. 11:485–492. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vaniawala S, Acharya A, Parekh H and
Mukhopadhyaya PN: Rare e14a3 (b3a3) BCR-ABL fusion in chronic
myeloid leukemia in India: The threats and challenges in monitoring
minimal residual disease (MRD). Anal Cell Pathol (Amst). 36:85–92.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Massimino M, Stella S, Tirrò E, Consoli
ML, Pennisi MS, Puma A, Vitale SR, Romano C, Zammit V, Stagno F, et
al: Rapid decline of Philadelphia-positive metaphases after
nilotinib treatment in a CML patient expressing a rare e14a3
BCR-ABL1 fusion transcript: A case report. Oncol Lett.
18:2648–2653. 2019.PubMed/NCBI
|
|
57
|
Jinawath N, Norris-Kirby A, Smith BD,
Gocke CD, Batista DA, Griffin CA and Murphy KM: A rare e14a3 (b3a3)
BCR-ABL fusion transcript in chronic myeloid leukemia: Diagnostic
challenges in clinical laboratory practice. J Mol Diagn.
11:359–363. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
O'Brien SG, Guilhot F, Larson RA, Gathmann
I, Baccarani M, Cervantes F, Cornelissen JJ, Fischer T, Hochhaus A,
Hughes T, et al: Imatinib Compared with Interferon and Low-dose
cytarabine for newly diagnosed chronic-phase chronic myeloid
leukemia. N Engl J Med. 348:994–1004. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Farhat-Maghribi S, Habbal W and Monem F:
Frequency of BCR-ABL transcript types in Syrian CML patients. J
Oncol. 2016:84208532016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Martinez-Serra J, Del Campo R, Gutierrez
A, Antich JL, Ginard M, Durán MA, Bento L, Ros T, Amat JC, Vidal C,
et al: Chronic myeloid leukemia with an e1a3 BCR-ABL fusion
protein: Transformation to lymphoid blast crisis. Biomark Res.
2:142014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sun H, Yan Z and Zhang S: Three atypical
BCR/ABL transcripts detected simultaneously in a
Philadelphia-positive acute lymphoblastic leukemia patient showing
resistance to tyrosine kinase inhibitors. Int J Hematol.
117:134–136. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Shin SY, Cho JH, Kim HJ, Jang JH, Lee ST
and Kim SH: Two cases of acute lymphoblastic leukemia with an e1a3
BCR-ABL1 fusion transcript. Ann Lab Med. 35:159–161. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Manzella L, Tirrò E, Vitale SR, Puma A,
Consoli ML, Tambè L, Pennisi MS, DI Gregorio S, Romano C, Tomarchio
C, et al: Optimal response in a patient with CML expressing
BCR-ABL1 E6A2 fusion transcript with nilotinib therapy: A case
report. In Vivo. 34:1481–1486. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Miyashita N, Onozawa M, Suto K, Fujisawa
S, Okazaki N, Hidaka D, Ohigashi H, Yasumoto A, Sugita J, Hashimoto
D, et al: Aleukemic extramedullary blast crisis as an initial
presentation of chronic myeloid leukemia with E1A3 BCR-ABL1 fusion
transcript. Intern Med. 61:1049–1054. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Crampe M, Haslam K, Groarke E, Kelleher E,
O'Shea D, Conneally E and Langabeer SE: Chronic myeloid leukemia
with an e6a2 BCR-ABL1 fusion transcript: Cooperating mutations at
blast crisis and molecular monitoring. Case Rep Hematol.
2017:90717022017.PubMed/NCBI
|
|
66
|
Beel KA, Lemmens J, Vranckx H, Maertens J
and Vandenberghe P: CML with e6a2 BCR-ABL1 transcript: An
aggressive entity? Ann Hematol. 90:1241–1243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hochhaus A, Breccia M, Saglio G,
García-Gutiérrez V, Réa D, Janssen J and Apperley J: Expert
Opinion-management of chronic myeloid leukemia after resistance to
second-generation tyrosine kinase inhibitors. Leukemia.
34:1495–1502. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hochhaus A, Ernst T, Eigendorff E and La
Rosée P: Causes of resistance and treatment choices of second- and
third-line treatment in chronic myelogenous leukemia patients. Ann
Hematol. 94 (Suppl 2):S133–S140. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Roman J, Jimenez A, Barrios M, Castillejo
JA, Maldonado J and Torres A: E1A3 as a unique, naturally occurring
BCR-ABL transcript in an indolent case of chronic myeloid
leukaemia. Br J Haematol. 114:635–637. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chen R and Chen B: The role of dasatinib
in the management of chronic myeloid leukemia. Drug Des Devel Ther.
9:773–779. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sheets JW, Eulitt P, He R, Olteanu H,
Coombs CC, Foster MC, Montgomery ND and Zeidner JF: Philadelphia
chromosome-positive acute myeloid leukemia with e1a3 BCR-ABL1
fusion transcript. Hemasphere. 4:e4842020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Qiang X, Wen Q, Li J, Chen S, Tao T, Zhang
H, Wang P, Peng X, Feng Y and Zhang X: Isolated central nervous
system infiltrated and progressed to acute myeloid leukemia from
chronic myeloid leukemia with e1a3 BCR-ABL1 transcript: A rare case
report and literature review. Cancer Manag Res. 17:35–43. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Soverini S, Albano F, Bassan R, Fabbiano
F, Ferrara F, Foà R, Olivieri A, Rambaldi A, Rossi G, Sica S, et
al: Next-generation sequencing for BCR-ABL1 kinase domain mutations
in adult patients with Philadelphia chromosome-positive acute
lymphoblastic leukemia: A position paper. Cancer Med. 9:2960–2970.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kearney L, Crampe M and Langabeer SE:
Frequency and spectrum of atypical BCR-ABL1 transcripts in chronic
myeloid leukemia. Exp Oncol. 42:78–79. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Arana-Trejo RM, Ruíz Sánchez E,
Ignacio-Ibarra G, Báez de la Fuente E, Garces O, Gómez Morales E,
Castro Granados M, Ovilla Martínez R, Rubio-Borja ME, Solís Anaya
L, et al: BCR/ABL p210, p190 and p230 fusion genes in 250 Mexican
patients with chronic myeloid leukaemia (CML). Clin Lab Haematol.
24:145–150. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Gong Z, Medeiros LJ, Cortes JE, Zheng L,
Khoury JD, Wang W, Tang G, Loghavi S, Luthra R, Yang W, et al:
Clinical and prognostic significance of e1a2 BCR-ABL1 transcript
subtype in chronic myeloid leukemia. Blood Cancer J. 7:e5832017.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Bhreathnach Ú, Kearney L and Langabeer SE:
Prevalence of atypical BCR-ABL1 transcript types in adult
Philadelphia chromosome-positive acute lymphoblastic leukemia:
Implications for measurable residual disease. Hematol Transfus Cell
Ther. 44:130–131. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wasif K, Wasif N and Saif MW:
Imatinib-induced ototoxicity in a patient with gastrointestinal
stromal tumor (GIST). Cureus. 8:e8482016.PubMed/NCBI
|
|
79
|
Shin H, Choi SY, Kee KM, Kim SH, Yang SY,
Jung SY, Noh H, Zang DY, Kim DW and Lee JI: Comprehensive analyses
of safety and efficacy toward individualizing imatinib dosage in
patients with chronic myeloid leukemia. Int J Hematol. 111:417–426.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Joensuu H, Trent JC and Reichardt P:
Practical management of tyrosine kinase inhibitor-associated side
effects in GIST. Cancer Treat Rev. 37:75–88. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Radojkovic M, Ristic S, Pavlovic S and
Colovic M: Molecular response to imatinib in patient with Ph
negative p190 BCR-ABL transcript positive chronic myeloid leukemia
with cyclic leukocytosis. Leuk Res. 33:e10–e12. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Sun J, Hu R, Han M, Tan Y, Xie M, Gao S
and Hu JF: Mechanisms underlying therapeutic resistance of tyrosine
kinase inhibitors in chronic myeloid leukemia. Int J Biol Sci.
20:175–181. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Moore FR, Yang F and Press RD: Detection
of BCR-ABL1 kinase domain mutations causing imatinib resistance in
chronic myelogenous leukemia. Methods Mol Biol. 999:25–39. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ben-Batalla I, Erdmann R, Jørgensen H,
Mitchell R, Ernst T, von Amsberg G, Schafhausen P, Velthaus JL,
Rankin S and Clark RE: Axl blockade by BGB324 inhibits BCR-ABL
tyrosine kinase Inhibitor-sensitive and -resistant chronic myeloid
leukemia. Clin Cancer Res. 23:2289–2300. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yeung DT, Shanmuganathan N and Hughes TP:
Asciminib: A new therapeutic option in chronic-phase CML with
treatment failure. Blood. 139:3474–3479. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Cioccio J and Claxton D: Therapy of acute
myeloid leukemia: Therapeutic targeting of tyrosine kinases. Expert
Opin Investig Drugs. 28:337–349. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Pan Q, Lu Y, Xie L, Wu D, Liu R, Gao W,
Luo K, He B and Pu Y: Recent advances in boosting EGFR tyrosine
kinase inhibitors-based cancer therapy. Mol Pharm. 20:829–852.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Illmer T and Ehninger G: FLT3 kinase
inhibitors in the management of acute myeloid leukemia. Clin
Lymphoma Myeloma. 8 (Suppl 1):S24–S34. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Cui Q, Liang P, Dai H, Cui W, Cai M, Ding
Z, Ma Q, Yin J, Li Z, Liu S, et al: Case report: CD38-directed
CAR-T cell therapy: A novel immunotherapy targeting CD38-positive
blasts overcomes TKI and chemotherapy resistance of myeloid chronic
myeloid leukemia in blastic phase. Front Immunol. 13:10129812022.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Khan AM, Munir A, Asrani R and Najjar S:
Acute myeloid leukemia with Philadelphia chromosome,
near-tetraploidy, and 5q deletion. Cureus. 11:e56062019.PubMed/NCBI
|
|
91
|
Ohsaka A, Shiina S, Kobayashi M, Kudo H
and Kawaguchi R: Philadelphia Chromosome-positive chronic myeloid
leukemia expressing p190(BCR-ABL). Intern Med. 41:1183–1187. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Tucker D, Hamilton MS, Kerr JP, Wickham C
and Hunter H: Lytic bone disease as the presenting feature of
Philadelphia-positive monosomy 7 myelodysplasia progressing to
acute myeloid leukaemia. Gene. 501:219–221. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Stella S, Massimino M, Tirrò E, Vitale SR,
Scalise L, Leotta S, Pennisi MS, Puma A, Romano C, Stagno F, et al:
B-ALL relapses after autologous stem cell transplantation
associated with a shift from e1a2 to e14a2 BCR-ABL transcripts: A
case report. Anticancer Res. 39:431–435. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Balatzenko G, Guenova M, Kalinova I,
Belcheva M, Hristozova H and Kaleva V: Simultaneous occurrence of
ETV6-RUNX1 and BCR-ABL1 (e1a2) transcripts in a child with B-cell
acute lymphoblastic leukemia. Cancer Genet Mar. 206:97–101. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Langabeer SE, Crampe M, Haslam K, Kelly J
and Cahill MR: Sustained clinical remission despite suboptimal
molecular response to imatinib in e1a2 BCR-ABL chronic myeloid
leukemia. Leuk Res. 34:e176–e177. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Agirre X, Román-Gómez J, Vázquez I,
Jiménez-Velasco A, Larráyoz MJ, Lahortiga I, Andreu EJ, Márquez J,
Beltrán de Heredia JM, Odero MD, et al: Coexistence of different
clonal populations harboring the b3a2 (p210) and e1a2 (p190)
BCR-ABL1 fusion transcripts in chronic myelogenous leukemia
resistant to imatinib. Cancer Genet Cytogenet. 60:22–26. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Ai DI, Liu W, Lu G, Patel KP and Chen ZI:
Extramedullary blast crisis as initial presentation in chronic
myeloid leukemia with the e1a2 BCR-ABL1 transcript: A case report.
Mol Clin Oncol. 3:1319–1322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Dupont M, Jourdan E and Chiesa J:
Identification of E6A2 BCR-ABL fusion in a Philadelphia-positive
CML. Leukemia. 14:2011–2012. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Staal-Viliare A, Latger-Cannard V, Rault
JP, Didion J, Grégoire MJ, Bologna S, Witz B, Jonveaux P, Lecompte
T and Rio Y: A case of de novo acute basophilic leukaemia:
Diagnostic criteria and review of the literature. Ann Biol Clin
(Paris). 64:361–365. 2006.(In French). PubMed/NCBI
|
|
100
|
Zagaria A, Anelli L, Coccaro N, Tota G,
Casieri P, Cellamare A, Impera L, Brunetti C, Minervini A,
Minervini CF, et al: BCR-ABL1 e6a2 transcript in chronic myeloid
leukemia: Biological features and molecular monitoring by droplet
digital PCR. Virchows Arch. 467:357–363. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Popovici C, Cailleres S, David M,
Lafage-Pochitaloff M, Sainty D and Mozziconacci MJ: E6a2 BCR-ABL
fusion with BCR exon 5-deleted transcript in a Philadelphia
positive CML responsive to Imatinib. Leuk Lymphoma. 46:1375–1377.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Breccia M, Cannella L, Diverio D, Streponi
P, Nanni M, Stefanizzi C, Natalino F, Mecarocci S and Alimena G:
Isolated thrombocytosis as first sign of chronic myeloid leukemia
with e6a2 BCR/ABL fusion transcript, JAK2 negativity and complete
response to imatinib. Leuk Res. 32:177–180. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Rohon P, Divoka M, Calabkova L, Mojzikova
R, Katrincsakova B, Rusinakova Z, Lapcikova A, Raida L, Faber E,
Jarosova M, et al: Identification of e6a2 BCR-ABL fusion in a
Philadelphia-positive CML with marked basophilia: Implications for
treatment strategy. Biomed Pap Med Fac Univ Palacky Olomouc Czech
Repub. 155:187–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Torres F, Ivanova-Dragoeva A, Pereira M,
Veiga J, Rodrigues AS, Sousa AB, Tavares P and Fernandes AR: An
e6a2 BCR-ABL fusion transcript in a CML patient having an iliac
chloroma at initial presentation. Leuk Lymphoma. 48:1034–1037.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Schnittger S, Bacher U, Kern W, Haferlach
T, Hertenstein B and Haferlach C: A new case with rare e6a2 BCR-ABL
fusion transcript developing two new resistance mutations during
imatinib mesylate, which were replaced by T315I after subsequent
dasatinib treatment. Leukemia. 22:856–858. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
McCarron SL, Kelly J, Coen N, McCabe S,
Fay M, O'Dwyer M, Hayden PJ and Langabeer SE: A novel e8a2 BCR-ABL1
fusion with insertion of RALGPS1 exon 8 in a patient with relapsed
Philadelphia chromosome-positive acute lymphoblastic leukemia. Leuk
Lymphoma. 52:919–921. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Burmeister T, Bullinger L and le Coutre P:
The recurrent atypical e8a2 BCR::ABL1 transcript with insertion of
an inverted 55 base pair ABL1 Intron 1b sequence: A detailed
molecular analysis. Acta Haematol. 146:413–418. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Mroczkowska A, Jaźwiec B, Urbańska-Rakus
J, Szymanowska S, Tessmann A, Pająk S, Machnik K, Haus O and Wróbel
T: A case report of pediatric acute lymphoblastic leukemia with
e8a2 BCR/ABL1 fusion transcript. BMC Med Genomics. 15:202022.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Riva E, Manrique Arechavaleta G, De
Almeida C, Costa V, Fernandez Del Campo M, Ifran González S and
Uriarte R: A novel e8a2 BCR-ABL1 fusion with insertion of MAST2
exon 2 in a four-way translocation t (1;17;9;22) (p35;q24;q44;q11)
in a patient with chronic myeloid leukemia. Leuk Lymphoma.
57:203–205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhang Y, Cheng Z, Yan WZ, Liu SF, Hu CH
and Zhang GS: Molecular characterization and therapeutic reaction
to dasatinib in a CML patient harboring a novel e8a2 BCR-ABL1
transcript with a somatic mutation in TP53BP2 and cadherin-10
genes. Leuk Lymphoma. 59:233–236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Branford S and Apperley JF: Measurable
residual disease in chronic myeloid leukemia. Haematologica.
107:2794–809. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Cundell MJ, Hutter LH, Nunes Bastos R,
Poser E, Holder J, Mohammed S, Novak B and Barr FA: A PP2A-B55
recognition signal controls substrate dephosphorylation kinetics
during mitotic exit. J Cell Biol. 214:539–554. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Gelens L, Qian J, Bollen M and Saurin AT:
The importance of Kinase-phosphatase integration: Lessons from
mitosis. Trends Cell Biol. 28:6–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Jin C, Zhu X, Xiao M, Liu S, Liu X, Liu J,
Xu X, Yi S and Meng L: A Novel e8a2BCR-ABL1 fusion transcript
without insertion sequence in a patient with chronic myeloid
leukemia. Ann Lab Med. 38:169–171. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Langabeer SE, McCarron SL, Kelly J,
Krawczyk J, McPherson S, Perera K and Murphy PT: Chronic myeloid
leukemia with e19a2 BCR-ABL1 transcripts and marked thrombocytosis:
The role of molecular monitoring. Case Rep Hematol.
2012:4587162012.PubMed/NCBI
|
|
116
|
Tchirkov A, Couderc JL, Périssel B, Goumy
C, Regnier A, Uhrhammer N, Verrelle P and Berger M: Major molecular
response to imatinib in a patient with chronic myeloid leukemia
expressing a novel form of e8a2 BCR-ABL transcript. Leukemia.
20:167–168. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
May PC, Reid AG, Robinson ME, Khorashad
JS, Milojkovic D, Claudiani S; Genomics England Research
Consortium, ; Willis F, Apperley JF and Innes AJ: FISH-negative
BCR::ABL1-positive e19a2 chronic myeloid leukaemia: The most
cryptic of insertions. BMC Med Genomics. 16:1722023. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Cea M, Cirmena G, Garuti A, Rocco I,
Palermo C, Cagnetta A, Moran E, Colombo N, Grasso R, Fugazza G, et
al: A T315I mutation in e19a2 BCR/ABL1 chronic myeloid leukemia
responding to dasatinib. Leuk Res. 34:e240–e242. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Popovici C, Charbonnier A, Gisserot O,
Aguilon P, Rémy V, Olschwang S and Mozziconacci MJ: Y253H mutation
appearing in a micro-BCR-ABL (e19a2) CML. Leuk Res. 32:361–362.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Ikeda K, Harada-Shirado K, Matsumoto H,
Noji H, Ogawa K and Takeishi Y: Molecular response of e19a2
BCR-ABL1 chronic myeloid leukemia with double Philadelphia
chromosome to dasatinib. J Clin Oncol. 34:e130–e133. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Crampe M, Garry J, Langabeer SE and Murphy
PT: Sustained molecular response with nilotinib in
Imatinib-intolerant chronic myeloid leukaemia with an e19a2
BCR-ABL1 fusion. Hematol Oncol Stem Cell Ther. 9:168–169. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ernst P, Rinke J, Franke GN, Dicker F,
Haferlach T, Ernst T and Hochhaus A: Treatment-free remission after
third-line therapy with asciminib in chronic myeloid leukemia with
an atypical e19a2 BCR::ABL1 transcript and T315I mutation.
Leukemia. 38:2037–2040. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Mondal BC, Majumdar S, Dasgupta UB,
Chaudhuri U, Chakrabarti P and Bhattacharyya S: e19a2 BCR-ABL
fusion transcript in typical chronic myeloid leukaemia: A report of
two cases. J Clin Pathol. 59:1102–1103. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
McCarron SL, Maher K, Kelly J, Ryan MF and
Langabeer SE: Rapid evolution to blast crisis associated with a
Q252H ABL1 kinase domain mutation in e19a2 BCR-ABL1 chronic myeloid
leukaemia. Case Rep Hematol. 2013:4907402013.PubMed/NCBI
|
|
125
|
Kajiguchi T, Okuno S, Ohno T and Abe A:
Molecular response to nilotinib in a patient with
imatinib-intolerant e19a2-positive chronic myeloid leukemia. Intern
Med. 53:2801–2804. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Santos FK de S and Maia CN: Patients with
sickle cell disease taking hydroxyurea in the hemocentro regional
de Montes Claros. Rev Bras Hematol Hemoter. 33:105–109. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Krakoff IH: Clinical and Pharmacologic
Effects of Hydroxyurea. Antineoplastic and Immunosuppressive
Agents: Part II [Internet]. Sartorelli AC and Johns DG: Springer;
Berlin, Heidelberg: pp. 789–792. 1975, https://doi.org/10.1007/978-3-642-65806-8_43
View Article : Google Scholar
|
|
128
|
Kamidani R, Chiba N, Kuroda A, Uchida A
and Okada H: Successful therapeutic leukapheresis for chronic
myeloid leukemia identified by persistent erection: A case report.
Cureus. 16:e613512024.PubMed/NCBI
|
|
129
|
Oshikawa G, Kurosu T, Arai A, Murakami N
and Miura O: Clonal evolution with double Ph followed by
tetraploidy in imatinib-treated chronic myeloid leukemia with e19a2
transcript in transformation. Cancer Genet Cytogenet. 199:56–61.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Stella S, Massimino M, Tirrò E, Vitale SR,
Accurso V, Puma A, Pennisi MS, DI Gregorio S, Romano C, DI Raimondo
F, et al: Detection and clinical implications of a novel BCR-ABL1
E12A2 Insertion/deletion in a CML patient expressing the E13A2
isoform. Anticancer Res. 39:6965–6971. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Alikian M, Gale RP, Apperley JF and Foroni
L: Molecular techniques for the personalised management of patients
with chronic myeloid leukaemia. Biomol Detect Quantif. 11:4–20.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Suttorp M, Millot F, Sembill S, Deutsch H
and Metzler M: Definition, epidemiology, pathophysiology, and
essential criteria for diagnosis of pediatric chronic myeloid
leukemia. Cancers (Basel). 13:7982021. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Wu CC, Beird HC, Zhang J and Futreal PA:
FusionPathway: Prediction of pathways and therapeutic targets
associated with gene fusions in cancer. PLoS Comput Biol.
14:e10062662018. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Pretzsch T, Progscha S and Burmeister T:
Diagnostic ambiguity caused by an atypical e18a2 BCR::ABL1
transcript in a chronic myeloid leukemia patient. Case Rep Hematol.
2024:94391342024.PubMed/NCBI
|
|
135
|
Sheng HX, Zhou LN, Chen JL, Hu GL, Wang
QH, Gao DG, Liao L, Yang Y, Sun T, Chen H and Zhang B: Concurrence
of e18a2 and e19a2 in Ph (+) chronic myelogenous leukemia: A case
report and literature review. Zhonghua Xue Ye Xue Za Zhi.
38:799–802. 2017.(In Chinese). PubMed/NCBI
|
|
136
|
van der Velden VHJ, Beverloo HB, Hoogeveen
PG and Zwaan CM: A novel BCR-ABL fusion transcript (e18a2) in a
child with chronic myeloid leukemia. Leukemia. 21:833–835. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Naumann N, Bross-Bach U, Seifarth W,
Fabarius A, Hofmann WK, Saußele S and Spiess B: A new aberrantly
spliced BCR-ABL1 transcript variant (e13a1) identified in routine
monitoring using different quantitative reverse transcription
polymerase chain reaction techniques in a patient with chronic
myeloid leukemia. EJHaem. 3:1339–1342. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Callet-Bauchu E, Salles G, Gazzo S, Dalle
S, Berger F and Hayette S: Identification of a novel e8/a4 BCR/ABL
fusion transcript in a case of a transformed Sézary syndrome.
Haematologica. 92:1277–1278. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Wang H, Han C, Gong B, Liu Y, Liu K, Gu R,
Wang Y, Wei H, Mi Y, Liu B and Wang J: Clinical features and
treatment response to TKIs in chronic myeloid leukemia patients
with atypical BCR::ABL1 transcripts. Leuk Res. 155:1077332025.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Zangrando A, Intini F, te Kronnie G and
Basso G: Validation of NG2 antigen in identifying BP-ALL patients
with MLL rearrangements using qualitative and quantitative flow
cytometry: A prospective study. Leukemia. 22:858–861. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Park IJ, Lim YA, Lee WG, Park JS, Kim HC,
Lee HJ and Cho SR: A case of chronic myelogenous leukemia with e8a2
fusion transcript. Cancer Genet Cytogenet. 185:106–108. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Lee J, Kim DS, Lee HS, Choi SI and Cho YG:
Concurrence of e1a2 and e19a2 BCR-ABL1 fusion transcripts in a
typical case of chronic myeloid leukemia. Ann Lab Med. 37:74–76.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Bennour A, Beaufils N, Sennana H, Meddeb
B, Saad A and Gabert J: E355G mutation appearing in a patient with
e19a2 chronic myeloid leukaemia resistant to imatinib. J Clin
Pathol. 63:737–740. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Langabeer SE, McCarron SL, Carroll P,
Kelly J, O'Dwyer M and Conneally E: Molecular response to first
line nilotinib in a patient with e19a2 BCR-ABL1 chronic myeloid
leukemia. Leuk Res. 35:e169–e170. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Jeon EK, Lim J, Kim M, Yahng SA, Lee SE,
Cho BS, Kim YJ, Kim HJ, Min CK, Cho SG, et al: The first case of
acute lymphoblastic leukemia with the e19a2 BCR-ABL1 transcript:
Imatinib therapy followed by unrelated donor transplantation
induces a durable molecular response. Leukemia. 25:366–367. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Qin YZ, Jiang Q, Jiang H, Lai YY, Shi HX,
Chen WM, Yu L and Huang XJ: Prevalence and outcomes of uncommon
BCR-ABL1 fusion transcripts in patients with chronic myeloid
leukaemia: Data from a single centre. Br J Haematol. 182:693–700.
2018. View Article : Google Scholar : PubMed/NCBI
|