Introduction
Esophageal cancer is one of the most common solid
tumors and the seventh leading cause of cancer-related death
worldwide (1). Despite advancements
in treatment approaches, the prognosis for patients with advanced
esophageal cancer remains poor (2).
The most common histologic subtypes of esophageal cancer are
esophageal squamous cell carcinoma (ESCC) and esophageal
adenocarcinoma. In Asia, ESCC accounts for a particularly high
proportion of cases.
Regarding ESCC, differentiation grades are
classified into three categories: Well-differentiated, moderately
differentiated, and poorly differentiated, according to the Union
for International Cancer Control (UICC) Staging Manual, 8th
edition. Well-differentiated tumors are characterized by prominent
keratinization, including pearl formation, along with a minor
component of non-keratinizing basal-like cells. Moderately
differentiated tumors exhibit a range of histological features,
from parakeratotic changes to poorly keratinizing lesions. Poorly
differentiated tumors mainly consist of basal-like cells forming
both large and small nests, often accompanied by central necrosis.
Furthermore, these nests can form sheets or pavement-like
arrangements of tumor cells and may occasionally include small
numbers of parakeratotic or keratinizing cells (3). Poorly differentiated ESCC typically
has a greater propensity for early lymphatic metastasis and skip
metastasis to distant lymph node stations, leading to a poorer
prognosis (4,5).
Meanwhile, microRNAs (miRNAs or miRs) have gained
attention for their oncogenic or tumor-suppressive functions. These
endogenous, small, non-coding, single-stranded RNAs, measuring
20–25 nucleotides, regulate target gene expression at the
post-transcriptional level by binding to complementary sequences
(6,7). Therefore, it was hypothesized that
specific miRNAs and their target mRNAs play crucial roles in
ESCC.
In the present study, it was aimed to identify
miRNAs associated with tumor characteristics in ESCC by comparing
miRNA expression between poorly differentiated (por) and well- to
moderately differentiated (non-por) ESCC using a comprehensive
miRNA array-based approach with clinical tissue samples and GEO
datasets from the National Library of Medicine. It was also aimed
to identify mRNA targeted by specific miRNAs to better understand
the mechanisms of tumor progression and potential therapeutic
targets in ESCC.
Materials and methods
Microarray and bioinformatics
To identify potential biomarkers, microarray
analyses of ESCC tissue samples were first performed using 3D-Gene
miRNA oligo chips (Toray Industries, Inc.), each containing 2,565
genes. miRNA expression in tumor tissues was collected from
patients with ESCC who underwent curative resection at our
institution, comparing miRNA expression between non-por and por
ESCC within the same specimens. RNAs were labeled using the 3D-Gene
miRNA labeling kit (Toray Industries, Inc.). Fluorescent signals
were scanned with a 3D-Gene scanner 3000 (Toray Industries, Inc.)
and analyzed using the 3D-Gene Extraction software (Toray
Industries, Inc.). Next, our analysis was supplemented with a GEO
dataset (GSE43732) (8), a publicly
accessible repository that includes miRNA expression profiles from
primary lesions of 119 patients with ESCC. miRNA expression was
compared between patients with por and non-por ESCC and
additionally performed overall survival (OS) analysis according to
tumor differentiation status.
Patients and samples
The study population comprised a retrospective
cohort of patients with consecutive ESCC who underwent
esophagectomy at the Department of Digestive Surgery, University of
Yamanashi (Chuo, Japan) between February 2008 and January 2023. To
eliminate any confounding effects of chemotherapy or radiotherapy,
169 patients who received neoadjuvant chemotherapy (n=144) or
neoadjuvant chemoradiotherapy (n=25) were excluded. An additional
11 cases were excluded due to non-curative resection. Furthermore,
30 cases unsuitable for RNA extraction were removed because the
tissue samples were too shallow for adequate RNA retrieval. As a
result, a final cohort of 61 patients with ESCC were eligible for
the present study (Fig. S1).
Clinicopathological characteristics of patients (including sex and
age distribution) are listed in Table
I.
 | Table I.Clinicopathological characteristics
of patients. |
Table I.
Clinicopathological characteristics
of patients.
|
| Expression level of
miR-100-5p | Expression level of
miR-203a-3p |
|---|
|
|
|
|
|---|
|
Characteristics | Low (n=13) | High (n=48) | P-value | Low (n=36) | High (n=25) | P-value |
|---|
| Age
(years)a | 72 (43–83) | 78 (60–83) | 0.12 | 74 (43–83) | 74 (57–83) | 0.65 |
| Sexb |
|
|
|
|
|
|
|
Male | 12 (92.3) | 36 (75.0) | 0.26 | 29 (80.5) | 19 (76.0) | 0.75 |
|
Female | 1 (7.7) | 12 (25.0) |
| 7 (19.5) | 6 (24.0) |
|
| Tumor
differentiationb |
|
|
|
|
|
|
|
Well | 0 (0) | 18 (37.5) | <0.001 | 14 (38.9) | 4 (16.0) | 0.01 |
|
Moderately | 2 (15.4) | 21 (43.8) |
| 8 (22.2) | 15 (60.0) |
|
|
Poorly | 11 (84.6) | 9 (18.7) |
| 14 (38.9) | 6 (24.0) |
|
| Pathological T
stageb |
|
|
|
|
|
|
| T1 | 0 (0) | 17 (35.4) | 0.012 | 5 (13.9) | 12 (48.0) | 0.008 |
| T2, T3,
T4 | 13 (100) | 31 (64.6) |
| 31 (86.1) | 13 (52.0) |
|
| Pathological N
stageb |
|
|
|
|
|
|
| N0 | 3 (23.1) | 23 (48.0) | 0.13 | 11 (30.6) | 15 (60.0) | 0.03 |
| N1, N2,
N3 | 10 (76.9) | 25 (52.0) |
| 25 (69.4) | 10 (40.0) |
|
| Lymphatic
invasionb |
|
|
|
|
|
|
|
ly0 | 4 (30.8) | 23 (47.9) | 0.35 | 13 (36.1) | 14 (56.0) | 0.19 |
| ly1,
ly2, ly3 | 9 (69.2) | 25 (52.1) |
| 23 (63.9) | 11 (44.0) |
|
| Vascular
invasionb |
|
|
|
|
|
|
| v0 | 3 (23.1) | 14 (29.2) | 1.00 | 8 (22.2) | 9 (36.0) | 0.26 |
| v1, ν2,
ν3 | 10 (76.9) | 34 (70.8) |
| 28 (77.8) | 16 (64.0) |
|
Tumor specimens and resected lymph nodes obtained at
the time of surgery were immediately fixed in 10% neutral-buffered
formalin at room temperature for 24 h and embedded in paraffin
after fixation (FFPE). The clinical and pathological tumor stages
of esophageal cancer were classified according to the UICC TNM
staging, 8th Edition (3).
Patients were followed up with physical
examinations, blood tests, and computed tomography every 4 months
for at least 1 year, and every 6 months thereafter. The present
study was approved (approval no. 2723) by the Ethics Committee of
Yamanashi University (Chuo, Japan) and conducted in accordance with
the ethical standards of the Declaration of Helsinki and its
amendments.
Histopathological evaluation and
immunohistochemical (IHC) staining analyses
Serial sections were obtained from FFPE tissue
blocks for IHC staining and RNA extraction. FFPE sections (5 µm)
were used for IHC. After deparaffinization with xylene, dehydration
using a graded ethanol series, and antigen retrieval at 120°C for
15 min endogenous peroxidase activity was inhibited
(Peroxidase-Blocking Solution; cat. no. S2023; Dako; Agilent
Technologies, Inc.), and non-specific binding was blocked (Protein
Block Serum-Free; cat. no. X0909; Dako; Agilent Technologies, Inc).
The sections were then incubated with a polyclonal FKBP5
antibody (1:50; cat. no. 14155-1; Proteintech Group, Inc.) at room
temperature for 1.5 h. Subsequently, slides were treated with an
anti-rabbit secondary antibody (EnVision+ HRP; cat. no.
K4003; Dako; Agilent Technologies, Inc.) at room temperature for 60
min. DAB working solution was used to stain positive cells. The
results were observed and recorded using optical microscopy
(BZ-X810; Keyence Corporation).
Each slide was scored according to the H-score,
which is calculated by multiplying two parameters: the proportion
of positive cells (1–100%) and the intensity of staining (1=weak;
2=moderate; 3=strong; range, 0–300).
Western blot analysis
Total proteins were obtained from the harvested
KYSE70 cells, and protein concentrations were determined using the
bicinchoninic acid (BCA) protein assay kit (Thermo Fisher
Scientific, Inc.). Cell lysate proteins (20 µg/lane) were separated
by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) and then transferred onto polyvinylidene fluoride
membranes. After blocking with StartingBlock Blocking Buffer
(Thermo Fisher Scientific, Inc.) at room temperature for 1 h, the
membranes were incubated overnight at 4°C with the following
antibodies: E-cadherin (1:1,000; cat. no. 20874-1-AP; Proteintech
Group, Inc.), Vimentin (1:2,000; cat. no. 10366-1-AP; Proteintech
Group, Inc.), phosphorylated (p-)PI3K (1:1,000; cat. no. 17366;
Cell Signaling Technology, Inc.), PI3K (1:1,000; cat. no. 11889;
Cell Signaling Technology, Inc.), p-AKT (1:1,000; cat. no. 4060;
Cell Signaling Technology, Inc.), AKT (1:1,000; cat. no. 4691; Cell
Signaling Technology, Inc.), GAPDH (1:50,000; cat. no. 60004-1-lg;
Proteintech Group, Inc.). Then, the membranes were incubated with
goat anti-rabbit or anti-mouse IgG conjugated to horseradish
peroxidase (1:10,000; cat. nos. ab6721 and ab6789; Abcam) buffer
for 1 h at room temperature. The membranes then were probed with
the indicated antibodies, and proteins were detected by a western
blot HRP Substrate (Takara Bio, Inc.). Densitometric analysis of
the protein bands was performed using FIJI (ImageJ; version 1.54q,
National Institutes of Health).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
FFPE tissue samples were cut into 10-µm-thick
sections, and total RNA was extracted using the miRNeasy FFPE kit
(Qiagen GmbH) and NucleoSpin miRNA (Takara Bio, Inc.), following
the manufacturer's protocols. A NanoDrop 2000 spectrophotometer
(Thermo Fisher Scientific, Inc.) was used to measure the total RNA
concentration.
Reverse transcription was performed using the
High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher
Scientific, Inc.) and the TaqMan MicroRNA Reverse Transcription Kit
(Thermo Fisher Scientific, Inc.). Total RNA and miRNA levels were
quantified by RT-qPCR following standard procedures.
RT-qPCR conditions were as follows: Initial
denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for
15 sec and 60°C for 60 sec. Total RNA levels were normalized to the
endogenous control gene glyceraldehyde-3-phosphate dehydrogenase
(GAPDH), while miRNA levels were normalized to RNU6B.
The following primers were used for the TaqMan assay
(Thermo Fisher Scientific, Inc.): human hsa-miR-100-5p (cat. no.
78224_mir; 5′-AACCCGUAGAUCCGAACUUGUG-3′), hsa-miR-203a-3p (cat. no.
478316_mir; 5′-GUGAAAUGUUUAGGACCACUAG-3′), FKBP5 (cat. no.
Hs01561006_m1), GAPDH (cat. no. Hs02786624_g1) and RNU6B (cat. no.
001093; 5′-CGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTTT-3′). ΔCq
values for all RNAs were calculated relative to the control genes
GAPDH and RNU6B. Relative mRNA and miRNA expression levels were
evaluated using the 2−ΔΔCq method (9).
Cell culture
The poorly differentiated ESCC cell lines [KYSE70
(JCRB0190), KYSE110 (JCRB1064), and TE5 (RCB1949)] and the well- to
moderately differentiated ESCC cell lines [TE4 (RCB2097), TE8
(RCB2098), and TE11 (RCB2100)] were used in the present study.
KYSE70 and KYSE110 were obtained from the Japanese Collection of
Research Bioresources (JCRB) Cell Bank, while TE series cell lines
were obtained from the RIKEN BRC Cell Bank in 2024. All cell lines
were confirmed to be free of mycoplasma contamination, as
previously described (10–12).
KYSE70, TE4, TE8 and TE11 were cultured in Roswell
Park Memorial Institute (RPMI)-1640 medium (Nacalai Tesque, Inc.)
supplemented with 10% fetal bovine serum (FBS) and 1%
penicillin-streptomycin (Sigma-Aldrich; Merck KGaA). The other ESCC
cell lines were cultured in Ham's F-12 medium (FUJIFILM Wako Pure
Chemical Corporation) supplemented with 2% FBS and
penicillin-streptomycin (Sigma-Aldrich, Merck KGaA). All cells were
maintained in a humidified incubator at 37°C with 5% carbon
dioxide.
Transfection of KYSE70 with miRNA
mimics
The miRNA mimics for miR-100-5p (mirVana miRNA
mimic; cat. no. MC10188; 5′-AACCCGUAGAUCCGAACUUGUG-3′), miR-203a-3p
(mirVana miRNA mimic; cat. no. MC10152;
5′-GUGAAAUGUUUAGGACCACUAG-3′, and the miRNA mimic negative control
(mirVana miRNA Mimic Negative Control #1; cat. no. 4464058) were
purchased from Thermo Fisher Scientific, Inc. Transfections were
performed using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol.
The final concentration for miR-100-5p and
miR-203a-3p mimics, as well as the negative control, was 10
nM. For double transfection, both mimics were used at 10 nM each.
Total RNA and protein were collected 48 h post-transfection.
Transfection efficiency was assessed by quantifying miRNAs
expression levels using RT-qPCR.
Transfection of KYSE70 with siRNA
The siRNA for FKBP5 (Silencer Select siRNA;
cat. no. s5215) (sense sequence: GAGAAAGGCUUGUAUAGGAtt) and the
siRNA negative control (Silencer Select Negative Control #1; cat.
no. 4390843) were purchased from Thermo Fisher Scientific, Inc. The
transfection protocol and concentration were the same as those used
for the mimics. Knockdown efficiency of FKBP5 was confirmed
by western blotting.
Migration and invasion assays
The cell migration and invasion assays for KYSE70
cells were conducted using Transwell chambers. For invasion assays,
the chambers were pre-coated with Matrigel (Corning, Inc.), while
migration assays were performed without it. Cells
(1×105) suspended in serum-free medium were placed in
the upper chamber, while 10% FBS was added to the lower chamber.
After a 24-h incubation, cells were stained using the Differential
Quik III Stain Kit (Polysciences, Inc.). Following washing and
drying, images were captured and examined under the microscope.
Functional rescue experiments
To determine whether the effects of the
double-transfection of miR-100-5p and miR-203a-3p mimics on cell
migration and invasion were mediated through FKBP5
suppression, functional rescue experiments were performed by
co-transfecting the double-transfection condition with FKBP5
siRNA. KYSE70 cells were transfected with the double-transfection
of miR-100-5p and miR-203a-3p mimics (10 nM each), with or without
FKBP5 siRNA (10 nM). After 48 h, migration and invasion
assays were conducted as aforementioned. The number of migrated and
invaded cells in the double-transfection + FKBP5 siRNA group
was compared with that in the double-transfection group to assess
whether additional FKBP5 knockdown further modified the
miRNA-induced phenotype.
Cytotoxicity assay
To assess cell proliferation, KYSE70 cells
(1×104/well) were passaged in 96-well plates in
RPMI-1640 culture medium containing penicillin-streptomycin,
supplemented with 10% FBS. Cell proliferation was measured by the
OD values using Cell Counting Kit-8 (CCK-8; (Dojindo Laboratories,
Inc.). A total of 100 µl of CCK-8 solution was added to each well,
and the plates were incubated for 2 h at 37°C in a CO2
incubator. The absorbance was measured at 450 nm on a microplate
reader (Spectra Max ABS Plus, Molecular Devices, LLC).
Apoptosis assays
Apoptosis was assessed in KYSE70 cells following
FKBP5 knockdown. The siRNA transfection procedure and
concentrations were identical to those aformentioned. A total of 48
h after transfection, apoptotic activity was evaluated by both flow
cytometry and western blotting.
For flow cytometric analysis, cells were collected,
washed twice with cold PBS, and resuspended in binding buffer.
Annexin V-FITC and propidium iodide (PI) (Nacalai Tesque, Inc.)
were added according to the manufacturer's instructions, and
samples were analyzed using a flow cytometer (BD FACSCelesta; BD
Biosciences) with BD FACSDiva software (v8.0.1.1; BD Biosciences).
The proportions of viable, early apoptotic, and late apoptotic
cells were quantified.
For western blotting, protein expression of
apoptosis-related markers was examined using antibodies against
Caspase-3 (1:1,000; cat. no. 14220; Cell Signalling Technology,
Inc.) and Bcl-2 (1:1,000; cat. no. ab692; Abcam).
Bioinformatics analyses
RNA-seq transcriptome data of ESCC was obtained from
The Cancer Genome Atlas database (https://portal.gdc.cancer.gov/). Potential miRNA
target genes were predicted using TargetScan Human 8.0 (https://www.targetscan.org/vert_80/). To
investigate the biological functions of FKBP5, Pearson's
correlation coefficients were first calculated between FKBP5
expression and all other genes across ESCC samples. Genes with
|Pearson r|>0.3 and P<0.001 were considered significantly
correlated with FKBP5 and subjected to enrichment analysis.
Gene Ontology (GO) Biological Process and Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway enrichment were performed using
the R package clusterProfiler (version 4.8; http://bioconductor.org/packages/clusterProfiler),
with multiple-testing correction by Benjamini-Hochberg and
significance thresholds of adjusted P<0.05 and q-value
<0.05.
To assess pathway-level associations, single-sample
Gene Set Enrichment Analysis (GSEA) was applied using the GSVA
package (version 1.42; http://bioconductor.org/packages/GSVA). Each tumor
received a score for the following two gene sets from MSigDB:
HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION,
KEGG_PI3K_AKT_SIGNALING_PATHWAY (hsa04151).
Finally, pre-ranked GSEA was conducted with cluster
Profiler: GSEA by ranking all genes in descending order of
Pearson's correlation with FKBP5. Only the EMT (Hallmark)
and PI3K-Akt (KEGG) gene sets were evaluated for enrichment, using
an FDR cutoff of 0.25. All analyses were performed in R (version
4.4.0; R Foundation for Statistical Computing).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism version 10 (GraphPad Software, Inc.; Dotmatics). Unless
otherwise stated, quantitative values were expressed as the mean ±
standard deviation (SD). Statistical significance was assessed
using the Mann-Whitney U test, Fisher's exact test, and one-way
analysis of variance (ANOVA) for each time point, followed by
Tukey's post hoc test. OS was calculated from the date of surgery
to either death or the last follow-up. Relapse-free survival (RFS)
was measured from the date of surgery to recurrence, death, or the
last follow-up. Survival curves were generated using the
Kaplan-Meier method and compared using the log-rank test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Identification of miRNA
candidates
First, miRNA candidates were identified using a
miRNA array-based approach in combination with the GEO datasets.
The expression levels of each miRNA were compared between por and
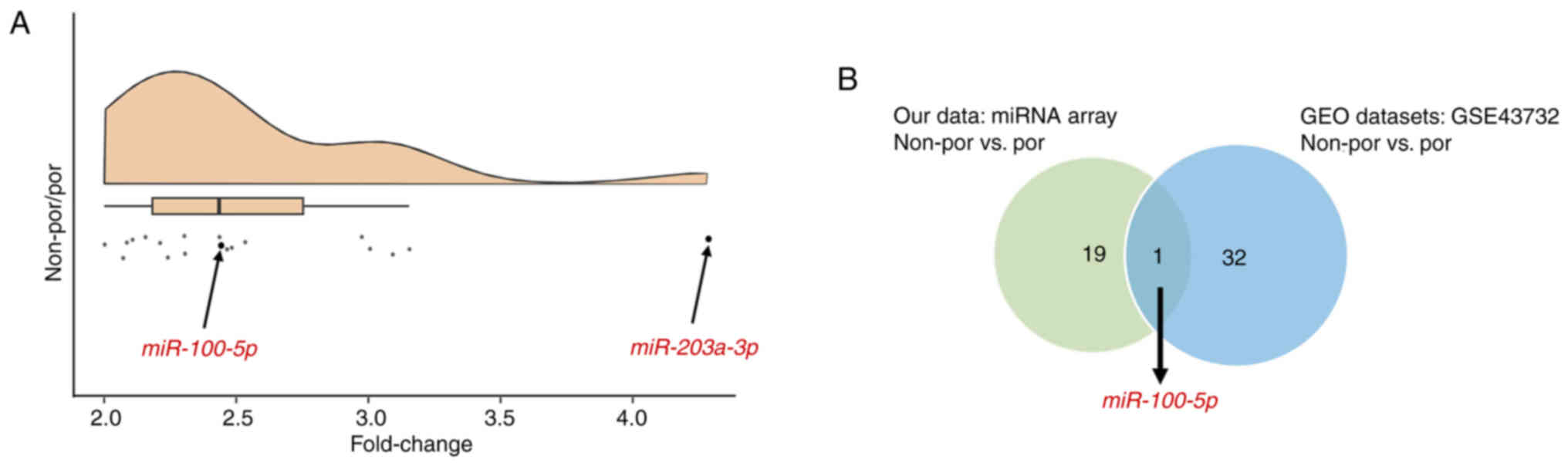
non-por ESCC. miR-203a-3p was selected, which exhibited a four-fold
difference in expression during microarray analysis, ensuring that
the signal intensity of each miRNA was adequate (Fig. 1A). Additionally, miR-100-5p was
selected, which exhibited a two-fold difference in expression
during microarray analysis, supported by the GEO datasets (Fig. 1A and B).
Furthermore, analysis of a publicly available GEO
dataset revealed a consistent trend toward poorer prognosis in
patients with por ESCC (Fig. S2).
Although this difference was not statistically significant, por
ESCC is generally considered more aggressive, and this trend may
have biological relevance. Consequently, miR-100-5p and miR-203a-3p
were chosen for further analyses in the present study.
Clinical significance of miR-100-5p
and 203a-3p in patients with ESCC
Next, the clinical significance of miR-100-5p and
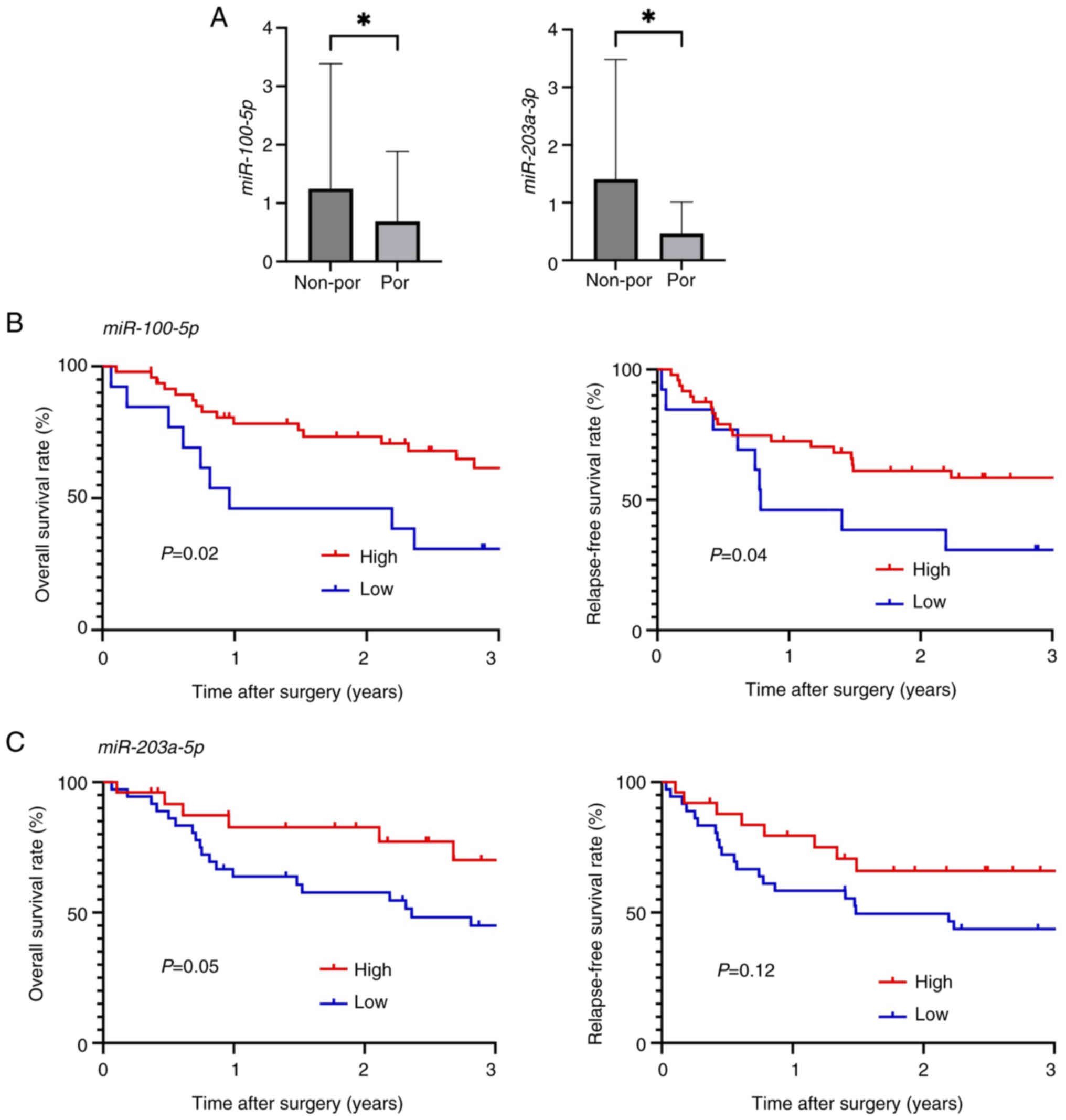
miR-203a-3p was examined in patients with ESCC. The expression
levels of miR-100-5p and miR-203a-3p were significantly lower in
patients with por ESCC than in patients with non-por ESCC (P=0.02
and P=0.04, respectively) (Fig.
2A). Based on these results, a receiver operating
characteristic curve was constructed according to histological
differentiation and divided the expression levels of miR-100-5p and
miR-203a-3p into high-expression and low-expression groups for
further analysis.
In the low miR-100-5p group, tumor
differentiation was poorer, and the depth of invasion was greater
compared with the high miR-100-5p group (P<0.001 and P=0.012,
respectively). Similarly, in the low miR-203a-3p group, tumor
differentiation was poorer, the depth of invasion was greater, and
lymph node metastasis occurred more frequently compared with the
high miR-203a-3p group (P=0.01, P=0.008, and P=0.03, respectively)
(Table I).
As demonstrated in Fig.
2B, OS and RFS rates were significantly worse in the low
miR-100-5p group compared with the high miR-100-5p group (P=0.02
and P=0.04, respectively). Additionally, the OS rate was
significantly worse in the low miR-203a-3p group than in the high
miR-203a-3p group, while the RFS rate was not statistically
significant (P=0.05 and P=0.12, respectively; Fig. 2C).
Expression and function of miR-100-5p
and miR-203a-3p in ESCC cell lines
Furthermore, the function of miR-100-5p and
miR-203a-3p was examined in ESCC cell lines. First, their
expression levels were measured in ESCC cell lines and it was found
that miR-100-5p and miR-203a-3p were significantly lower in poorly
differentiated ESCC cell lines compared with well- and moderately
differentiated cell lines (Fig.
3A). This expression pattern was consistent with our clinical
findings that patients with high expression of these miRNAs had
higher degree of differentiation and more favorable prognosis.
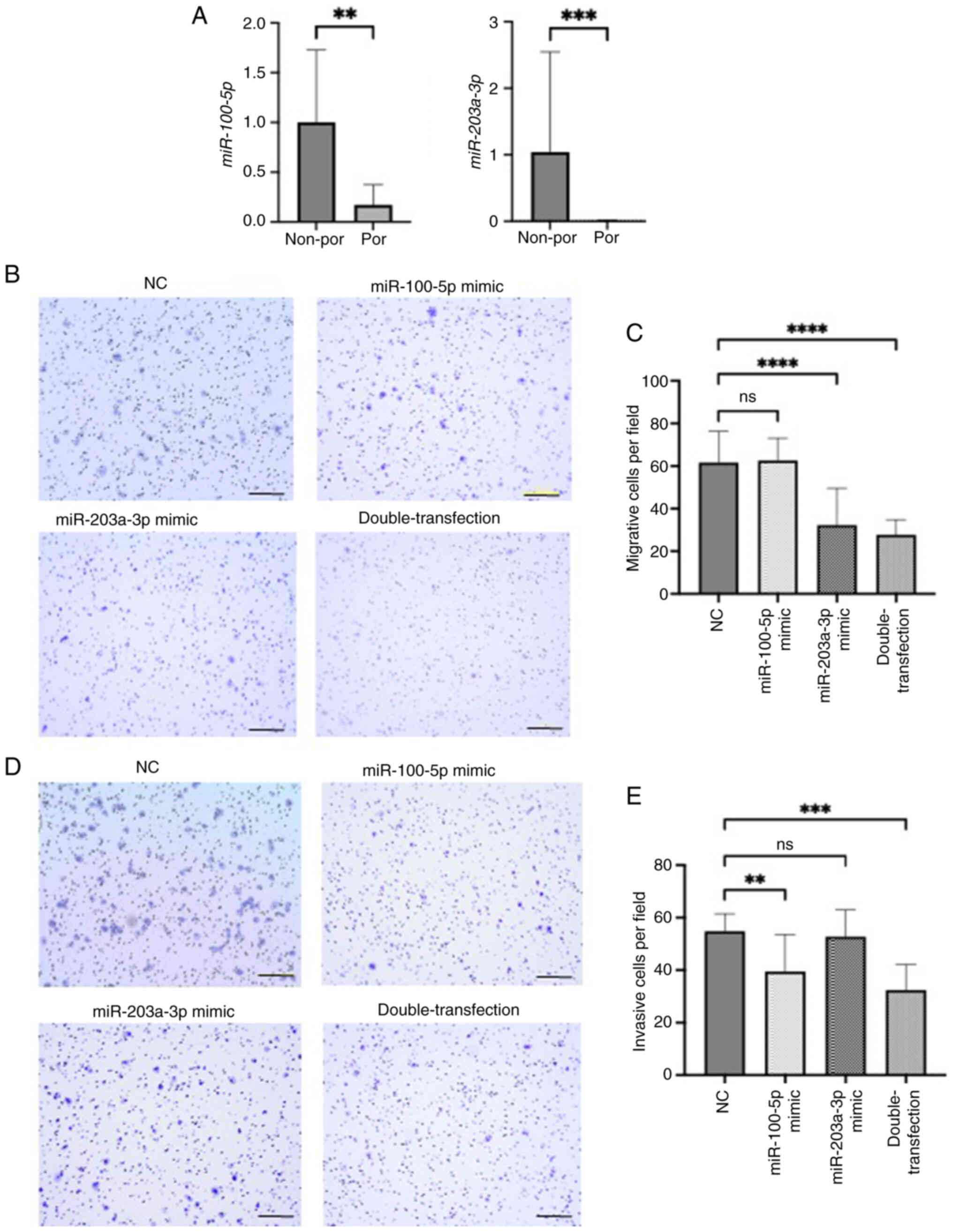 | Figure 3.(A) Relationship between tumor
differentiation and miR-100-5p and miR-203a-3p levels in non-por
and por cell lines. Representative images and quantitative
measurement of (B and C) migration and (D and E) invasion assays
induced by miR-100-5p and miR-203a-3p mimics in KYSE70 cells. For
double-transfection experiments, both miR-100-5p and miR-203a-3p
mimics were co-transfected at 10 nM each. Scale bar, 200 µm. All
the quantitative measurements were performed in triplicate, and
data for relative quantity are presented as the mean ± SD.
**P<0.01, ***P<0.001 and ****P<0.0001 compared with NC.
miR, microRNA; por cell lines, poorly differentiated cell lines
(KYSE70, KYSE110, TE5) non-por cell lines, well- to moderately
differentiated cell lines (TE4, TE8, TE11). NC, negative
control. |
To investigate the functional roles of these miRNAs
in ESCC progression, cell migration and invasion assays were
conducted using miRNA mimics in KYSE70 cells. Transfection
efficiency for each mimic is shown in Fig. S3A. Cell migration was significantly
inhibited by the miR-203a-3p mimic alone and by the combination of
miR-100-5p and miR-203a-3p mimics compared with the negative
control mimic (Fig. 3B and C). This
result indicates that miR-203a-3p plays a suppressive role in the
migratory ability of ESCC cells. Similarly, cell invasion was
significantly suppressed by the miR-100-5p mimic and by the double
transfection, indicating that miR-100-5p mainly contributes to the
inhibition of invasive potential (Fig.
3D and E). These results suggested that both miRNAs
cooperatively suppress ESCC progression through inhibition of
migration and invasion.
Potential target of miR-100-5p and
miR-203a-3p
FKBP5 was identified as a putative downstream
target of both miR-100-5p and miR-203a-3p using TargetScan Human
8.0 (Fig. S4). To clarify the
functional relevance of FKBP5 in ESCC, comprehensive
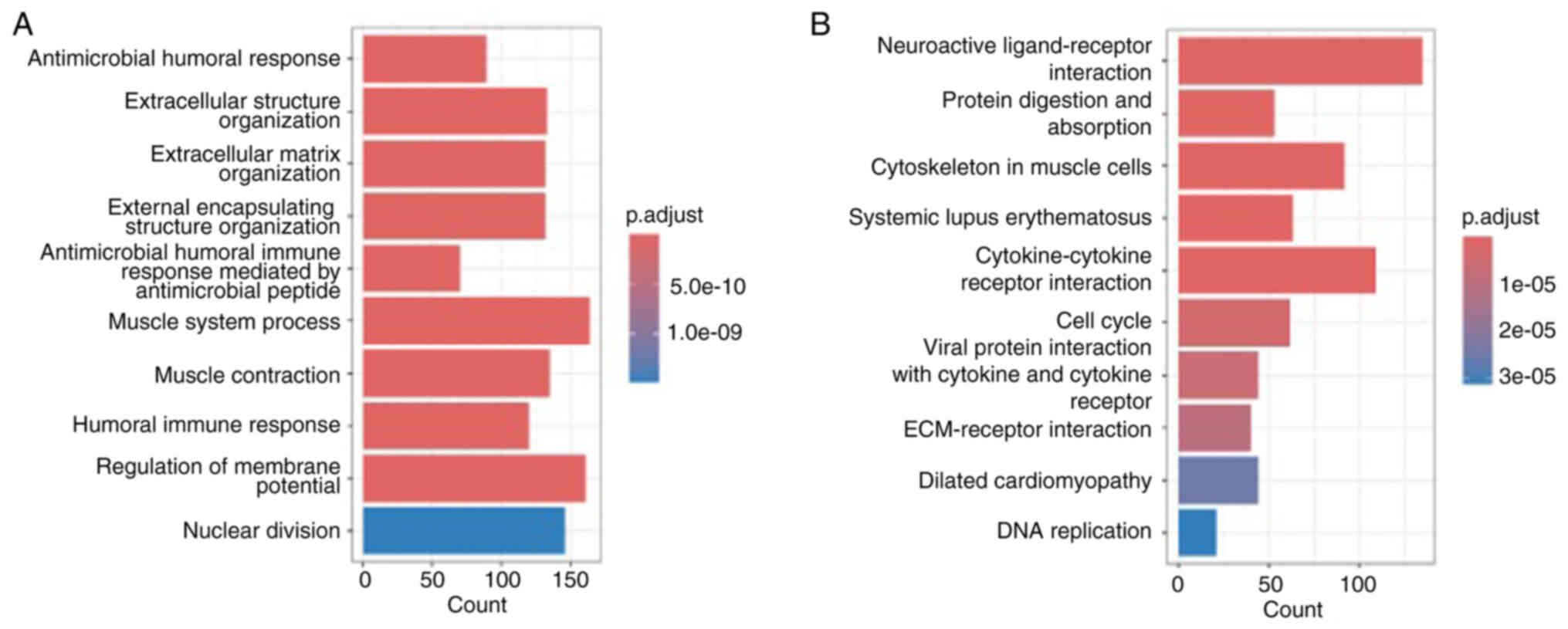
bioinformatics analyses were first performed. GO and KEGG
enrichment of FKBP5-associated genes revealed significant
overrepresentation of pathways involved in cell cycle control,
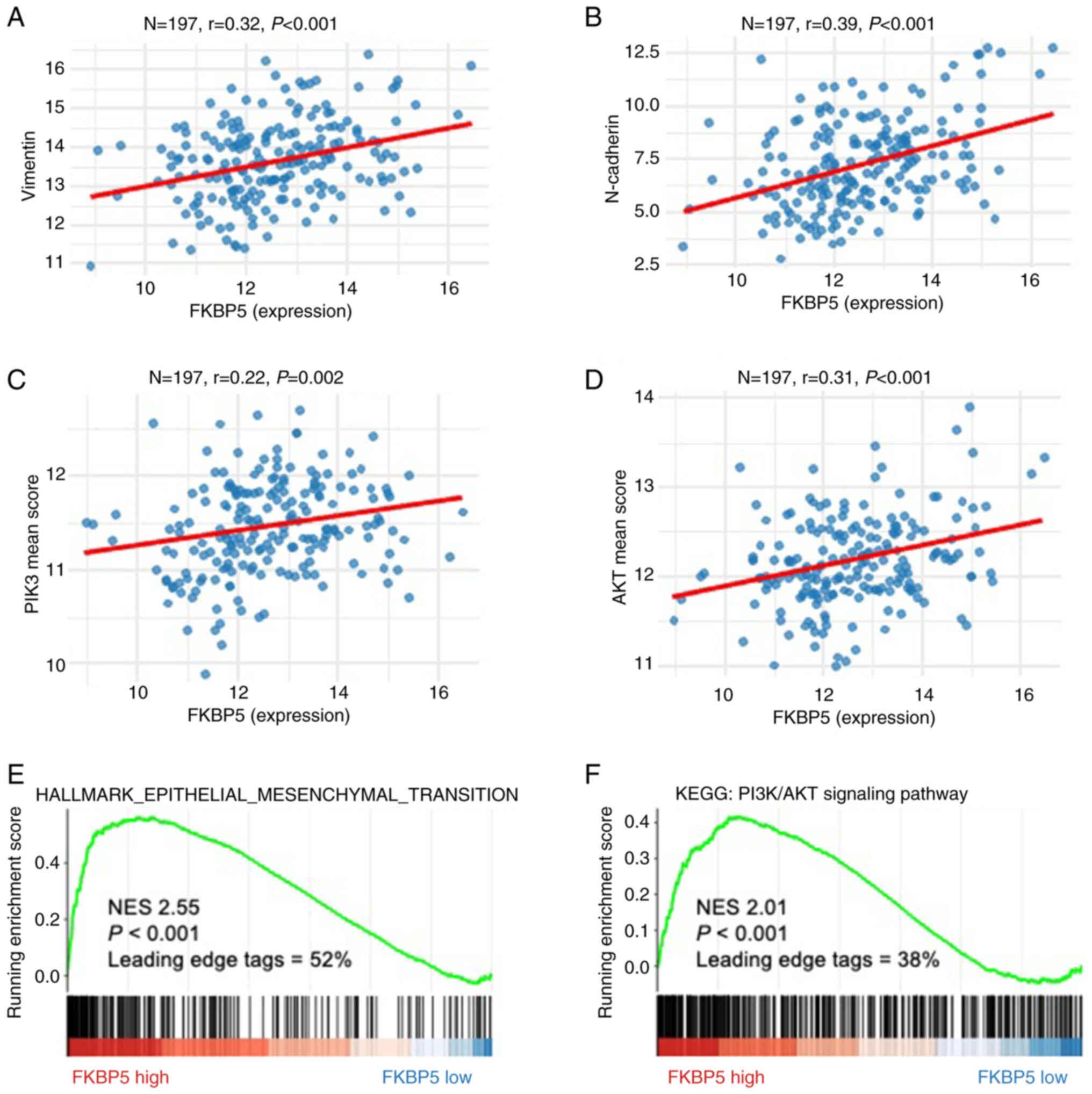
nuclear division and extracellular matrix organization (Fig. 4A and B). Moreover, Pearson
correlation (Fig. 5A-D) and
pre-ranked GSEA (Fig. 5E and F)
demonstrated robust positive associations between FKBP5
expression and both epithelial-mesenchymal transition (EMT) and
PI3K/AKT signalling pathway in ESCC.
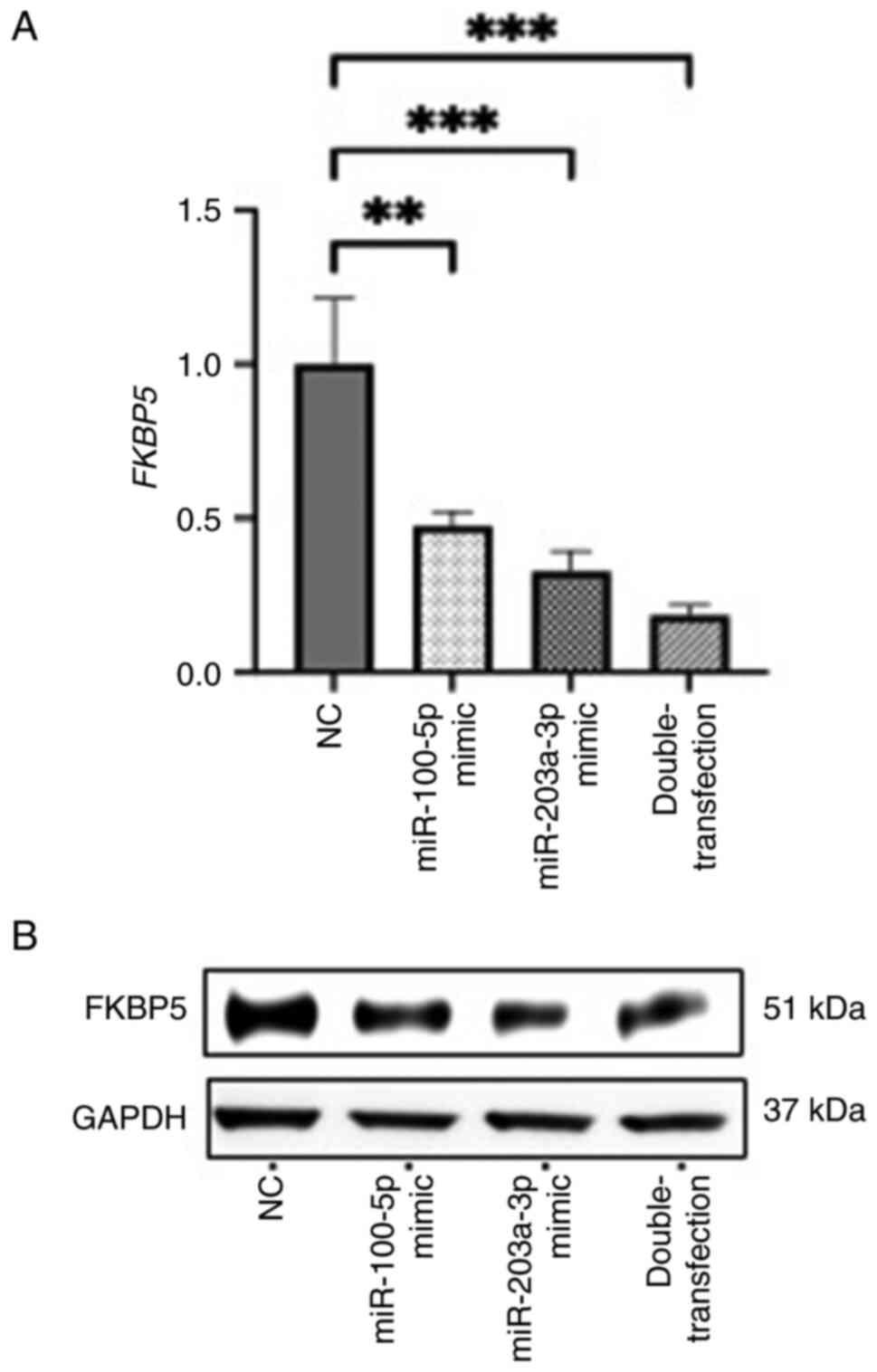
Experimentally, transfection of miR-100-5p and
miR-203a-3p mimics significantly decreased both FKBP5
transcript and protein levels, with the combined transfection of
both miRNAs producing the most pronounced reduction (Fig. 6A and B). These results suggested
that FKBP5 is a downstream target of both miRNAs and that
the two miRNAs may act cooperatively to suppress its expression. To
further confirm the functional involvement of FKBP5 in the
miRNA-mediated suppression of ESCC cell motility, rescue
experiments were performed using the double-transfection of
miR-100-5p and miR-203a-3p mimics with or without FKBP5
siRNA. Adding FKBP5 siRNA to the double-transfection did not
produce additional reductions in migration or invasion compared
with the double-transfection alone, indicating that the effects of
the mimics are largely mediated by FKBP5 suppression
(Fig. S5). In clinical ESCC
samples, the expression levels of miR-100-5p/miR-203a-3p and
FKBP5 protein exhibited a negative correlation trend;
however, this association did not reach statistical significance
(Fig. S6).
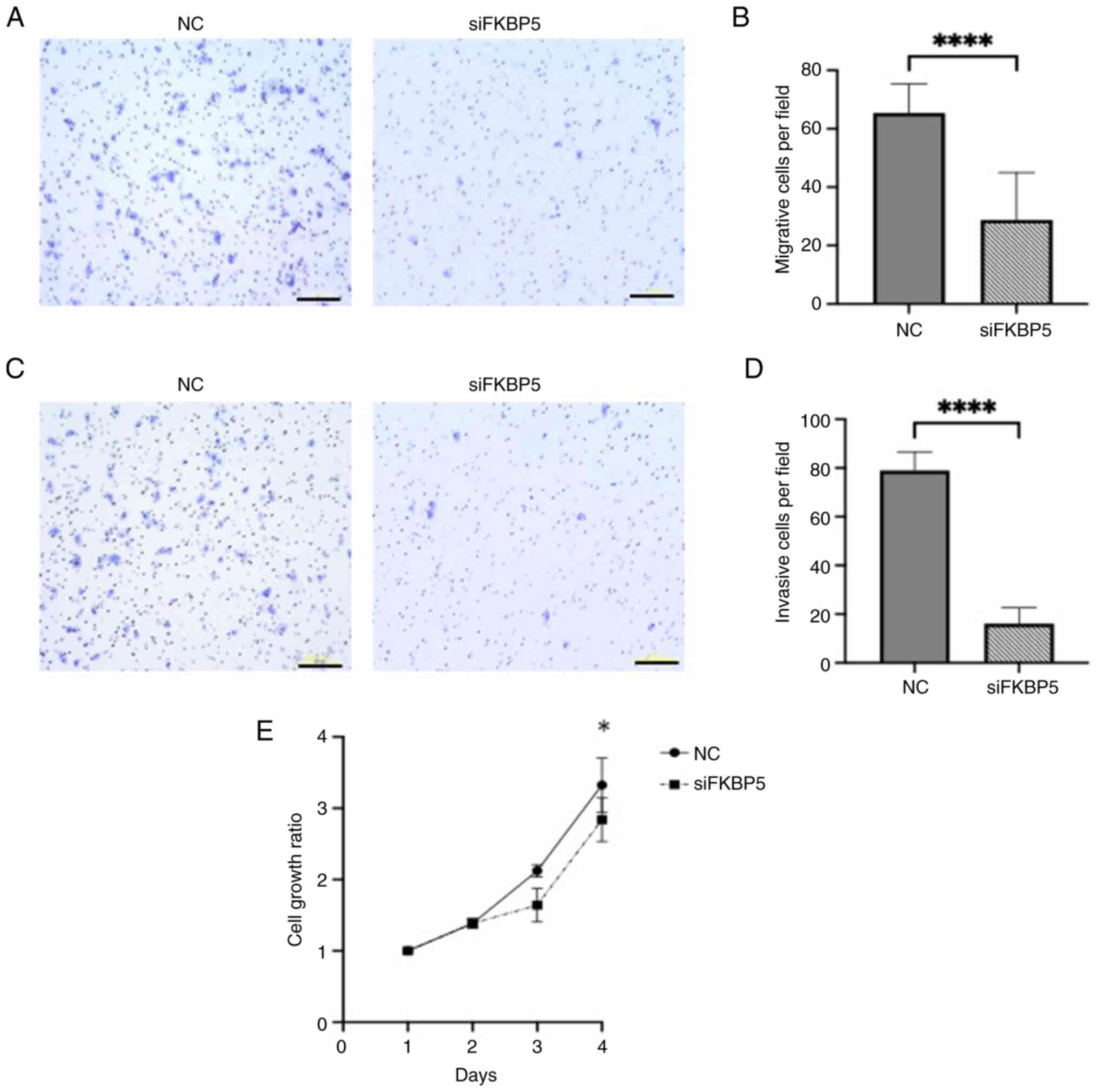
Functional assays further demonstrated that
FKBP5 knockdown, whose efficiency was confirmed by western
blotting (Fig. S3B), significantly
impaired ESCC cell migration, invasion, and increased cytotoxicity
compared with negative controls (Fig.
7A-E), suggesting that FKBP5 promotes malignant
phenotypes in ESCC cells. Additionally, apoptosis assays were
performed. Annexin V/PI flow cytometry showed no significant
differences between FKBP5 siRNA and controls (Fig. S7A). Western blotting revealed
slight changes in Caspase-3/Cleaved Caspase-3 and a marked
reduction in Bcl-2 (Fig. S7B).
These results suggested that FKBP5 knockdown alone did not
provide definitive evidence for apoptosis involvement.
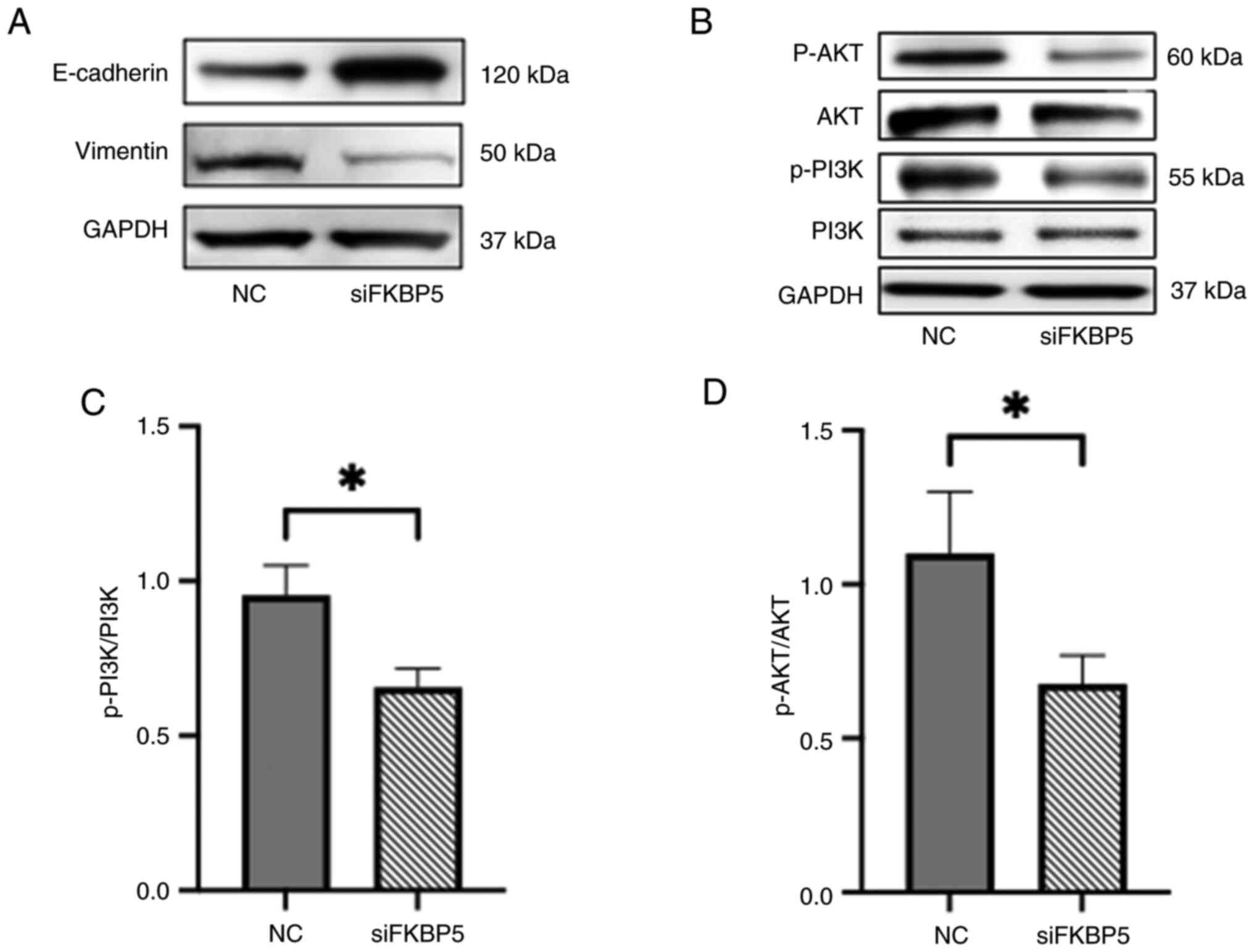
Furthermore, the effect of FKBP5 on EMT and
PI3K/AKT pathway activity was investigated by performing western
blotting for key EMT markers and phosphorylated signaling proteins.
FKBP5 knockdown led to upregulation of the epithelial marker
E-cadherin and downregulation of the mesenchymal marker vimentin
(Fig. 8A). In addition, knockdown
of FKBP5 reduced the levels of p-PI3K and p-AKT, indicating
suppression of the PI3K/AKT signalling pathway (Fig. 8B-D). These findings indicated that
FKBP5 facilitates EMT and activates the PI3K/AKT pathway in
ESCC cells, and that its suppression may help reverse the
mesenchymal phenotype and inhibit oncogenic signaling associated
with invasion and metastasis.
Prognostic impact of FKBP5 expression
in patients with ESCC
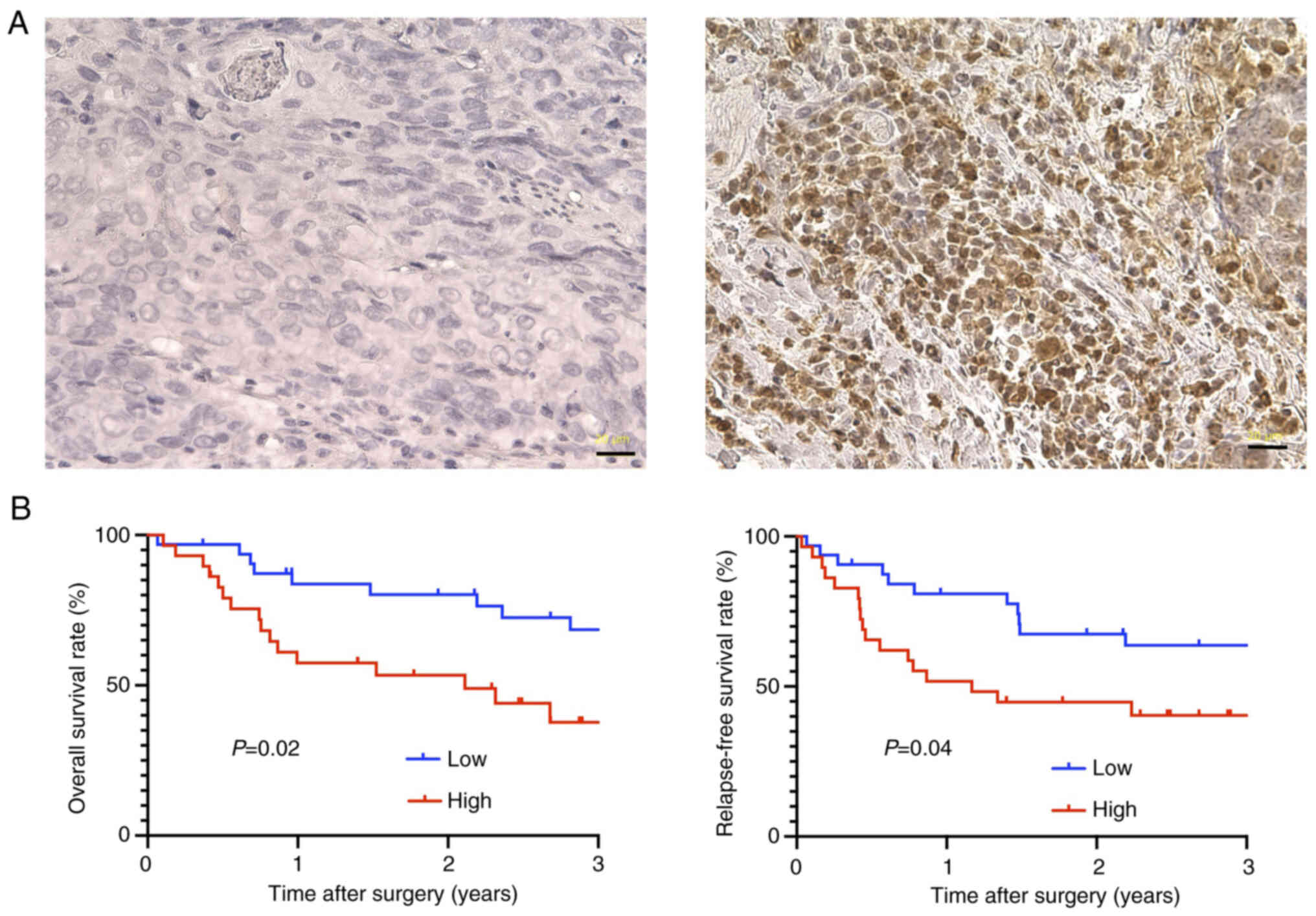
Finally, the prognostic significance of FKBP5
expression was evaluated in patients with ESCC using IHC.
Representative IHC images are shown in Fig. 9A. Based on the median H-score,
patients were categorized into high- and low-FKBP5
expression groups. Survival analysis revealed that patients in the
high FKBP5 expression group exhibited significantly poorer
OS and RFS compared with those in the low expression group
(Fig. 9B). These findings suggested
that elevated FKBP5 expression is associated with adverse
clinical outcomes in ESCC and may serve as a potential prognostic
biomarker.
Discussion
In the present study, it was aimed to elucidate the
role of miRNAs in tumor characteristics within ESCC, specifically
focusing on variations in tumor differentiation. Our findings
strongly suggest that patients with elevated levels of miR-100-5p
and miR-203a-3p have a favorable prognosis. Furthermore, it was
established that these miRNAs inhibit cellular migration and
invasion in ESCC by targeting FKBP5, a promising candidate
for therapeutic intervention in ESCC (Fig. 10).
First, tumor differentiation in ESCC was
investigated. Por ESCC exhibits distinct characteristics compared
with non-por ESCC. Patients with por ESCC have higher rates of
lymph node metastasis and recurrence. Furthermore, the pattern of
lymph node metastasis in por ESCC has been reported to differ
significantly from that in non-por ESCC (5).
Therefore, it was hypothesized that highly malignant
por ESCC could provide insights into identifying molecules that
play a crucial role in the tumor characteristics of ESCC. Through
screening using a comprehensive miRNA array-based approach and
public databases, two miRNAs were identified as potential
candidates.
There are already studies on each of the candidate
miRNAs. Regarding miR-100-5p, it suppresses CXCR7 expression, which
is implicated in initiation, adhesion, angiogenesis and metastasis
(13), and inhibits the activation
of the PI3K/AKT signaling pathway, thereby suppressing
lymphangiogenesis (14). Similarly,
miR-203a-3p suppresses the activation of the PI3K/AKT signaling
pathway, which is associated with tumor initiation and therapy
resistance in ESCC (15).
From a clinical perspective, high expression levels
of these miRNAs are linked to an improved prognosis in ESCC
(16,17). The novelty of the present study lies
in identifying that both miR-100-5p and miR-203a-3p are associated
with tumor differentiation in ESCC using clinical data and cell
lines. Furthermore, the combination of miR-100-5p and miR-203a-3p
synergistically suppressed the malignant behavior of ESCC. In the
present study, miR-100-5p alone did not sufficiently suppress cell
migration, while miR-203a-3p alone was ineffective in suppressing
cell invasion, possibly due to differences in transfection
efficiency or concentration. Our intention was to demonstrate that
simultaneous transfection of both miRNAs more effectively
suppresses both migration and invasion.
FKBP5 was identified as a potential target of
the combined action of miR-100-5p and miR-203a-3p. FKBP5 has
previously been reported as an intracellular protein that promotes
cancer progression and chemoresistance by activating the NF-κB
signaling pathway (18–21). In the present study, western blot
analysis and public database data clearly demonstrated that
FKBP5 also activates the PI3K/AKT signaling pathway, thereby
contributing to enhanced malignancy in ESCC (22). Moreover, siRNA-mediated FKBP5
knockdown significantly affected migration, invasion, cytotoxicity,
and several signaling proteins, whereas no appreciable changes were
detected in apoptosis assays using flow cytometry or western
blotting. These findings suggested that the oncogenic function of
FKBP5 in ESCC may be more strongly associated with cell
motility and survival signaling rather than direct regulation of
apoptosis.
FKBP5 is known to play an oncogenic role in
various malignancies, including thyroid carcinoma, renal clear cell
carcinoma and melanoma (23–25).
In gastrointestinal cancers, similar oncogenic functions have been
reported in esophageal adenocarcinoma, gastric cancer and
colorectal cancer, where elevated FKBP5 expression is
associated with poor prognosis (26–28).
Collectively, these findings suggested that FKBP5
contributes to poor prognosis by promoting EMT and activation of
the PI3K/AKT signaling pathway, which is consistent with our
results. To the best of our knowledge, this is the first study to
demonstrate that FKBP5 promotes tumor aggressiveness through
regulation by miR-100-5p and miR-203a-3p in ESCC.
Based on the results of the present study,
FKBP5 has the potential to become an important therapeutic
target for ESCC in the future. Therefore, suppressing FKBP5
through the administration of both miR-100-5p and miR-203a-3p may
represent an effective treatment option for patients with ESCC.
Further basic and clinical studies are necessary to evaluate the
efficacy of this treatment strategy in suppressing FKBP5 for
ESCC.
The present study has several limitations. It was
conducted at a single institution with a limited number of
patients. In the GEO dataset used, miR-203a-3p was not listed,
which precluded combined analyses of expression arrays and public
databases. While these miRNAs appeared to be associated with tumor
differentiation, it remains unclear whether they and FKBP5
are directly involved in the differentiation process. Furthermore,
dual-luciferase reporter assays were not performed due to the lack
of necessary equipment for gene transfection in our institution. As
a result, the direct interaction between these miRNAs and
FKBP5 was not validated. However, Chen et al
(29) reported such an interaction.
In our functional rescue experiment, adding siFKBP5 to
miRNAs mimic-treated cells did not alter the suppression of
migration and invasion, consistent with the authors' hypothesis
that FKBP5 mediates these effects. Additionally, although a
negative correlation between miR-100-5p/miR-203a-3p and
FKBP5 expression was observed, the association did not reach
statistical significance (Fig.
S6). However, it is considered that a larger cohort study may
confirm a statistically significant negative correlation. Moreover,
cell viability was assessed using the CCK-8 assay, which measures
metabolic activity as an indirect indicator of viable cell number;
therefore, it may not directly reflect cell proliferation rates.
Additional assays based on DNA synthesis or proliferation markers
would provide a more definitive evaluation of proliferation.
Nevertheless, our primary aim was to identify miRNAs associated
with tumor characteristics and potential therapeutic targets in
ESCC, and this objective was achieved. In conclusion, miR-100-5p
and miR-203a-3p inhibited tumor progression by targeting
FKBP5 in ESCC, highlighting FKBP5 as a promising
therapeutic target in ESCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be found
in the Gene Expression Omnibus under accession number GSE43732 or
at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43732.
Authors' contributions
HT, SM and DI conceived and designed the study. HT
and SM confirm the authenticity of all the raw data. HT, SM, YH and
RS performed the experiments and acquired the data. HT, SM, YH, RS,
KT, WI, KeS, SF, YK and HA interpreted the data. SM and DI
supervised the study. HT, SM, YH, TO, KaS, TN and DI drafted the
manuscript. RS, KT, WI, KeS, SF, YK, HA and HK critically revised
the manuscript for important intellectual content. All authors read
and approved the final version of the manuscript and agree to be
accountable for all aspects of the work.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of the University of Yamanashi (approval no. 2723; Chuo,
Japan) for patient experiments. The study adhered to the ethical
standards outlined in The Declaration of Helsinki and its
subsequent amendments. Written informed consent was provided by all
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
por
|
poorly differentiated carcinoma
|
|
non-por
|
well- to moderately differentiated
carcinoma
|
|
ESCC
|
esophageal squamous cell carcinoma
|
|
miRNA or miR
|
microRNA
|
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl.). 135:584–590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brierley JD, Brierley J, Gospodarowicz MK
and Wittekind C: Ebscohost: TNM classification of malignant
tumours. Wiley Blackwell/John Wiley & Sons, Inc; Chichester,
West Sussex: 2017
|
|
4
|
Li J, Wang L, Wang X, Zhao Y, Liu D, Chen
C, Zhang HP and Pan J: Preliminary study of the internal margin of
the gross tumor volume in thoracic esophageal cancer. Cancer
Radiother. 16:595–600. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang J, Liu Y, Che F, Luo Y, Huang W,
Heng X and Li B: Pattern of lymph node metastasis in thoracic
esophageal squamous cell carcinoma with poor differentiation. Mol
Clin Oncol. 8:760–766. 2018.PubMed/NCBI
|
|
6
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang HC and Tang KF: Clinical value of
integrated-signature miRNAs in esophageal cancer. Cancer Med.
6:1893–1903. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Z, Li J, Tian L, Zhou C, Gao Y, Zhou
F, Shi S, Feng X, Sun N, Yao R, et al: MiRNA expression profile
reveals a prognostic signature for esophageal squamous cell
carcinoma. Cancer Lett. 350:34–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shimada Y, Imamura M, Wagata T, Yamaguchi
N and Tobe T: Characterization of 21 newly established esophageal
cancer cell lines. Cancer. 69:277–284. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tanaka H, Shibagaki I, Shimada Y, Wagata
T, Imamura M and Ishizaki K: Characterization of p53 gene mutations
in esophageal squamous cell carcinoma cell lines: Increased
frequency and different spectrum of mutations from primary tumors.
Int J Cancer. 65:372–376. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tanaka H, Shimada Y, Imamura M, Shibagaki
I and Ishizaki K: Multiple types of aberrations in the p16 (INK4a)
and the p15(INK4b) genes in 30 esophageal squamous-cell-carcinoma
cell lines. Int J Cancer. 70:437–442. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou SM, Zhang F, Chen XB, Jun CM, Jing X,
Wei DX, Xia Y, Zhou YB, Xiao XQ, Jia RQ, et al: miR-100 suppresses
the proliferation and tumor growth of esophageal squamous cancer
cells via targeting CXCR7. Oncol Rep. 35:3453–3459. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen C, Yang C, Tian X, Liang Y, Wang S,
Wang X, Shou Y, Li H, Xiao Q, Shu J, et al: Downregulation of
miR-100-5p in cancer-associated fibroblast-derived exosomes
facilitates lymphangiogenesis in esophageal squamous cell
carcinoma. Cancer Med. 12:14468–14483. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Zhang Z, Yu X, Li Q, Wang Q, Chang
A, Huang X, Han X, Song Y, Hu J, et al: SOX9/miR-203a axis drives
PI3K/AKT signaling to promote esophageal cancer progression. Cancer
Lett. 468:14–26. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou S, Yang B, Zhao Y, Xu S, Zhang H and
Li Z: Prognostic value of microRNA-100 in esophageal squamous cell
carcinoma. J Surg Res. 192:515–520. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng Q, Chen L and Ni L: Association of
miR-203 expression with prognostic value in patients with
esophageal cancer: A systematic review and meta-analysis. J Invest
Surg. 36:22857802023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li L, Lou Z and Wang L: The role of FKBP5
in cancer aetiology and chemoresistance. Br J Cancer. 104:19–23.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hähle A, Merz S, Meyners C and Hausch F:
The many faces of FKBP51. Biomolecules. 9:352019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Febbo PG, Lowenberg M, Thorner AR, Brown
M, Loda M and Golub TR: Androgen mediated regulation and functional
implications of fkbp51 expression in prostate cancer. J Urol.
173:1772–1777. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang T, Ma C, Zhang Z, Zhang H and Hu H:
NF-kappaB signaling in inflammation and cancer. MedComm (2020).
2:618–653. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li B, Tsao SW, Li YY, Wang X, Ling MT,
Wong YC, He QY and Cheung AL: Id-1 promotes tumorigenicity and
metastasis of human esophageal cancer cells through activation of
PI3K/AKT signaling pathway. Int J Cancer. 125:2576–2585. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao Z, Yu F, Jia H, Ye Z and Yao S:
FK506-binding protein 5 promotes the progression of papillary
thyroid carcinoma. J Int Med Res. 49:30006052110083252021.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mao S, Zhang D, Chen L, Tan J, Chu Y,
Huang S, Zhou W, Qin H, Xia Q, Zhao Y, et al: FKBP51 promotes
invasion and migration by increasing the autophagic degradation of
TIMP3 in clear cell renal cell carcinoma. Cell Death Dis.
12:8992021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Romano S, Xiao Y, Nakaya M, D'Angelillo A,
Chang M, Jin J, Hausch F, Masullo M, Feng X, Romano MF and Sun SC:
FKBP51 employs both scaffold and isomerase functions to promote
NF-kappaB activation in melanoma. Nucleic Acids Res. 43:6983–6993.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Smith E, Palethorpe HM, Ruszkiewicz AR,
Edwards S, Leach DA, Underwood TJ, Need EF and Drew PA: Androgen
receptor and androgen-responsive gene FKBP5 are independent
prognostic indicators for esophageal adenocarcinoma. Dig Dis Sci.
61:433–443. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu T, Wang C and Xia Z: Overexpressed
FKBP5 mediates colorectal cancer progression and sensitivity to
FK506 treatment via the NF-kappaB signaling pathway. FEBS J.
291:3128–3146. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang RG, Zhang D, Zhao CH, Wang QL, Qu H
and He QS: FKBP10 functioned as a cancer-promoting factor mediates
cell proliferation, invasion, and migration via regulating PI3K
signaling pathway in stomach adenocarcinoma. Kaohsiung J Med Sci.
36:311–317. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen G, Li X, Zhou X, Li Y, Yu H, Peng X,
Bai X, Zhang C, Feng Z, Mei Y, et al: Extracellular vesicles
secreted from mesenchymal stem cells ameliorate renal ischemia
reperfusion injury by delivering miR-100-5p targeting FKBP5/AKT
axis. Sci Rep. 14:67202024. View Article : Google Scholar : PubMed/NCBI
|