Introduction
In 2020, the incidence of gastric cancer reached
~1.09 million cases, ranking 5th among all malignant tumors and
accounting for 5.6% of all malignant diseases (1,2). The
number of mortalities caused by gastric cancer totaled ~770,000,
ranking fourth and accounting for 7.7% of overall malignant
tumor-related fatalities (1,2). The
high prevalence and poor prognosis of gastric cancer have markedly
impacted the well-being of the population, especially in China
(3). In 2022, China reported
~397,000 new cases of gastric cancer, accounting for 37% of the
global total, with both incidence and mortality rates ranking third
among malignant tumors in China (4,5). East
Asia bears the brunt of the global burden, with ~60% of gastric
cancer cases occurring in this region (6–8). Along
with the health burden comes a pronounced economic loss to
residents and the government due to the diagnosis and treatment of
gastric cancer (9,10).
Early-stage gastric cancer often presents with
inconspicuous symptoms, which leads to late detection, suboptimal
treatment outcomes, high recurrence rates and low survival rates
(6,10–12).
The 5-year survival rate for advanced gastric cancer remains as low
as 5% globally (13–15). Currently, the predominant treatment
strategy for gastric cancer is comprehensive, with a primary
emphasis on surgical intervention (16–18).
Surgical procedures dominate the treatment landscape for
early-stage gastric cancer, while chemotherapy improves survival
and quality of life for locally advanced or metastatic cases (stage
Ib to IIIb) (8,17,19,20).
Despite the mature theory and practice of abdominal anatomy,
coupled with inherent shortcomings of chemotherapy, advances in
surgical and chemotherapeutic approaches to the treatment of
gastric cancer have been limited (8). Overall, the efficacy of gastric cancer
treatment remains unsatisfactory, with only modest improvements in
prognosis (21).
New therapeutic approaches, such as targeted drugs
and immunotherapy, offer hope for patients with gastric cancer
(22,23). Targeting central mechanisms of tumor
development, developing drugs that prevent uncontrolled cell
proliferation or directly inducing apoptosis represents a promising
path to a successful transformation and potentially a cure of
advanced gastric cancer (24).
GPR176, a G protein-coupled receptor located on
15q14-q15.1, belongs to the G protein-coupled receptor family and
functions as a cell surface receptor that responds to hormones,
growth factors and neurotransmitters (25). GPR176 is primarily expressed in the
brain, followed by the gallbladder and testis (25). While earlier studies focused
primarily on the role and mechanisms of GPR176 in circadian rhythms
(25,26,27),
later research recognizes its potential in tumors (28–32).
For example, Tang et al (28) revealed that GPR176 recruits GNAS,
activates the cAMP/PKA/BNIP3L signaling pathway and inhibits
mitochondrial autophagy, which promotes stem cell formation and
proliferation of colorectal adenocarcinoma. Yun et al
(29) demonstrated an association
between the expression of GPR176 and the prognosis of breast
adenocarcinoma. Interfering with GPR176 suppresses the
PI3K/AKT/mTOR signaling pathway, glycolysis, epithelial-mesenchymal
transition and proliferation of breast adenocarcinoma cells. The
prognostic value of GPR176 in esophageal adenocarcinoma has also
been established, indicating an association with prognosis and
resistance of esophageal adenocarcinoma (30). Analyses of publicly available data
have suggested an association between GPR176 and gastric cancer
prognosis, warranting further exploration of underlying mechanisms
(31,32).
PIP5K1A, which encodes the protein
phosphatidylinositol-4-phosphate 5-kinase type 1 α, carries out a
role in several processes, including activation of GTPase activity
(33,34). It serves as an upstream regulator of
the PI3K/AKT/mTOR signaling pathway and genetic or pharmacological
inhibition of PIP5K1A markedly inhibits AKT phosphorylation
(35,36).
Building upon previous research, the present study
first evaluated the expression and prognostic significance of
GPR176 in gastric cancer and further elucidated the mechanism by
which GPR176 promotes cancer cell invasion and its functional
interaction with PIP5K1A, using both in vitro and in
vivo models.
Materials and methods
Data acquisition
RNA-sequencing (RNA-seq) data from 448 gastric
cancer (STAD) samples, including 410 tumor and 38 normal samples,
were collected, along with clinical data from 383 patients with
STAD, sourced from the Cancer Genome Atlas (TCGA) database
(https://portal.gdc.cancer.gov/)
(37). The Limma package in R 4.5.1
(https://bioconductor.org/packages/release/bioc/html/limma.html)
facilitated the standardization of RNA-seq information (38). The transcriptomic details for all
samples were preserved to investigate the nuanced differences
between adenocarcinomatous and adjacent tissues. However, 17
samples with incomplete clinical information were excluded from
subsequent survival analysis.
The RNA-seq data and associated clinical/prognostic
information for 300 patients diagnosed with gastric cancer were
methodically extracted from the GSE66254 dataset in the Gene
Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66254)
(39). The original chip data from
GSE66254 underwent a rigorous annotation and standardization
process in R, utilizing the Limma package.
Patient samples
From the biobank of the Department of
Gastrointestinal and Glandular Surgery at The First Affiliated
Hospital of Guangxi Medical University (Nanning, China), tissue
specimens (including gastric cancer tumor and adjacent non-tumorous
tissues) were collected from 48 patients with gastric cancer who
underwent surgery. Postoperative pathological reports indicated
lymph node metastasis in 24 of these patients. These tissue samples
were subsequently utilized to analyze the correlation between lymph
node metastasis and the activation of the PI3K/AKT/mTOR and EMT
signaling pathways, as well as the expression of GPR176.
Cell culture
The cell lines HGC-27 and NCI-N87 were procured from
the American Type Culture Collection (The Global Bioresource
Center; http://www.atcc.org/) cell repository.
Culturing these cells involved a comprehensive medium consisting of
DMEM (cat. no. 10566016; Gibco, Thermo Fisher Scientific, Inc.),
supplemented with 10% FBS (cat. no. 10099141C; Gibco; Thermo Fisher
Scientific, Inc.), 1% streptomycin and 1% penicillin (cat. no.
P1400; Beijing Solarbio Science & Technology Co., Ltd.). The
controlled environment for these cell cultures was maintained in
incubators with 5% CO2 and 37°C.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
RNA extraction from each sample (gastric cancer
tumor and adjacent non-tumorous tissues) was carried out using the
TRI Reagent™, by following the manufacturer's protocol
(cat. no. AM9738; Invitrogen; Thermo Fisher Scientific, Inc.).
Subsequently, reverse transcription (RT) into cDNA was carried out
according to the manufacturer's instructions for the PrimeScript RT
Reagent Kit with gDNA Eraser (cat. no. RR047B; Takara Bio, Inc.).
The primers were designed using Primer3 (https://primer3.ut.ee/) and are listed in Table SI. qPCR was performed using the
FastStart Universal SYBR® Green Master Mix (cat. no.
06402712001; Roche Diagnostics GmbH) on an Applied Biosystems 7500
(Thermo Fisher Scientific, Inc.) with the following thermocycling
conditions: Initial denaturation at 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 1 min. Gene expression was
analyzed using the 2−∆∆Cq method (40).
Construction of lentivirus and stable
cell lines
Design and packaging of overexpression (OE)/RNA
interfering (RNAi) lentiviruses into cells was carried out by
Shanghai GeneChem Co., Ltd. Lentiviruses for GPR176 overexpression,
GPR176 knockdown (sh-GPR176), PIP5K1A overexpression and PIP5K1A
knockdown (sh-PIP5K1A) were constructed using the wild-type
sequences of GPR176 and PIP5K1A, respectively. Lentiviruses
containing empty vectors were used as the control group for
infection. The infection concentration of all lentiviruses in the
present study was set to 5 multiplicity of infection (MOI), with an
infection duration of 12 h in 37°C cell incubator. Puromycin was
added at a concentration of 10 ng/ml to select cells with
off-target effects and the selection period was 5 days in in 37°C
cell incubator. The GPR176 overexpression lentivirus contained the
wild-type sequence of the GPR176 gene, while the PIP5K1A
overexpression lentivirus contained the wild-type sequence of the
PIP5K1A gene. The functional sequences of sh-GPR176 and sh-PIP5K1A
are provided in Table SII. The
efficacy of these lentiviruses was verified through a using RT-qPCR
assay and western blotting (WB).
WB
Extraction of proteins from cells (HGC-27 and
NCI-N87) and tissues (gastric cancer and adjacent normal gastric)
entailed a combination of RIPA reagent (cat. no. R0010; Beijing
Solarbio Science & Technology Co., Ltd.) and 1% PMSF (cat. no.
P0100; Beijing Solarbio Science & Technology Co., Ltd.).
Quantification of protein concentration was carried out using the
BCA protein assay kit (cat. no. P0009; Beyotime Institute of
Biotechnology). The proteins (2 µg) underwent separation using 10%
SDS-PAGE electrophoresis and were then transferred to PVDF
membranes. Following blocking with 5% skim milk at room temperature
for 30 min, the PVDF membranes were incubated overnight at 4°C with
the diluent for the primary antibody. After two washes with
PBS-Tween (1%), the membranes were incubated with secondary
antibody diluent at 23°C for 1 h. The visualization of protein
bands was carried out using the Bio-Rad ChemiDoc MP Imaging System
(Bio-Rad Laboratories, Inc.). The information and usage
concentrations of the corresponding primary antibodies are listed
in Table SIII. HRP-conjugated Goat
Anti-Mouse IgG (H+L) (1:100; cat. no. SA00001-1, Proteintech Group
Inc.) and HRP-conjugated Goat Anti-Rabbit IgG (H+L) (1:100; cat.
no. SA00001-2, Proteintech Group Inc.) were used as the secondary
antibody.
Immunofluorescence (IF)
IF staining was carried out following standard
protocols (https://www.ptgcn.com/support/protocols/#if). Briefly,
1×105 HGC-27 and NCI-N87 cells were fixed with 4%
paraformaldehyde at room temperature for 15 min, permeabilized with
0.2% Triton X-100 (cat. no. 9002-93-1; Beijing Solarbio Science
& Technology Co., Ltd.) and blocked with 5% normal goat serum
(cat. no. SL038, Beijing Solarbio Science & Technology Co.,
Ltd.) at room temperature for 1 h. Samples were then incubated with
primary antibodies (Vimentin, 1:200; E-cadherin, 1:200) at 4°C
overnight, followed by Multi-rAb® CoraLite®
Plus 488-Goat Anti-Rabbit Recombinant Secondary Antibody (H+L)
(1:500; cat. no. RGAR002; Proteintech Group Inc.) and
Multi-rAb® CoraLite® Plus 594-Goat
Anti-Rabbit Recombinant Secondary Antibody (H+L) (1:500; cat. no.
RGAR004; Proteintech Group Inc.) at room temperature for 1 h and
DAPI (cat. no. 28718-90-3, Beijing Solarbio Science &
Technology Co., Ltd.) nuclear counterstaining at room temperature
for 15 min. Fluorescence images were captured with an Olympus
inverted fluorescence microscope (Olympus IX73; Olympus
Corporation) equipped with appropriate filter sets and a digital
camera and analyzed using Olympus CellSens software 2.3.
Cell invasion assays
The cell suspension was obtained by digesting the
cells with 1 ml trypsin (cat. no. T1321; Beijing Solarbio Science
& Technology Co., Ltd.) for 2 min, adding 5 ml of complete
medium, and then repeatedly pipetting the mixture. Before adding
the cell suspension in 6.5 mm Transwell® with 8.0 µm
Pore Polycarbonate Membrane Inserts (cat. no. 3422; Corning, Inc.),
the Matrigel (cat. no. 356234; Beijing Solarbio Science &
Technology Co., Ltd.) was prepared according to the manufacturer's
instructions. Cells were plated in the upper chambers, and the
culture medium was added to the lower chambers. Specifically, 200
µl of a serum-free cell suspension containing 100,000 cells was
added to the upper Transwell inserts, while 600 µl of DMEM
supplemented with 10% FBS was added to the lower chambers of a
24-well plate. The plate was then incubated for 48 h at 37°C and 5%
CO2. After incubation, the insert was fixed with 4%
paraformaldehyde at room temperature for 30 min and stained with a
1% crystal violet solution at room temperature for 20 min.
Subsequent actions included the erasure of cells on the upper
layer, which were then removed, washed with PBS, and the invaded
cells were observed under an Olympus IX53.
Cell wound healing assays
Cells (HGC-27 cells and NCI-N87) were grouped
according to the regulation of GPR176 and PIP5K1A, with the details
stated in Construction of lentivirus and stable cell lines.
To observe cell proliferation (HGC-27 and NCI-N87), a precise wound
was induced by scratching the plate with a 200 µl suction tip when
the cells covered 90–100% of the culture plate area. After washing
twice with PBS, serum-free medium was added to the plate, and cells
were cultured in incubators with 5% CO2 and 37°C.
Subsequently, the width of the wound was observed and imaged
consecutively under the Olympus IX53 at 0 and 48 h.
Nude-mouse transplanted tumor model
construction
Male nude mice (n=40; 6–8 weeks old; 18–22 g) were
housed under specific pathogen-free conditions with a controlled
temperature of 22±2°C, humidity of 50±10% and a 12-h light/dark
cycle. All animals had ad libitum access to a standard laboratory
diet and autoclaved water. The nude mice were randomly assigned
into four groups (10 per group) and received subcutaneous
injections of HGC-27 cells: sh-NC, sh-GPR-176, Lenti-null and
Lenti-GPR-176, respectively. HGC-27 cells in a stable growth state
were resuspended in pre-cooled PBS at a concentration of
1×107 cells/200 µl. This suspension was aseptically
injected subcutaneously into the right flank of each mouse.
After transplantation, the general health status and
body weight of the mice were monitored daily. Tumor volume was
measured every two days using a digital caliper and calculated
using the formula V=(length × width2 × π)/6. To minimize
observer bias, all measurements were conducted in a blinded
fashion. Humane endpoints were strictly defined as follows: Tumor
burden >1.5 cm in diameter, significant weight loss (>20% of
initial body weight) or signs of severe distress. None of the
animals reached these endpoints prior to the scheduled experimental
conclusion. At the end of the experiment (day 28 post-inoculation),
all mice were euthanized by CO2 asphyxiation with a
chamber displacement rate of 30–50% per min, followed by cervical
dislocation to ensure mortality.
Statistical analysis
Each experiment was carried out to at least three
independent replicates. Data analysis was conducted using IBM SPSS
Statistics software (version 26.0; IBM Corp.). Results are
expressed as the mean ± standard error. Differences between two
groups were assessed using an unpaired Student's t-test, while
multiple comparisons were carried out by one-way or two-way ANOVA.
Linear regression and correlation analysis were employed to assess
the correlation between GPR176 and PIP5K1A expression. P<0.05
was considered to indicate a statistically significant
difference.
Results
GPR176 is associated with the
prognosis and clinicopathologic factors of gastric
adenocarcinomas
Expression matrices and corresponding clinical data
were respectively obtained from TCGA-STAD dataset and GSE66254
dataset (37,39). The expression of GPR176, its
prognostic significance and its association with clinicopathologic
factors were evaluated. Analysis of the pan-cancer data from TCGA
revealed significant differences in GPR176 expression between
cancer and adjacent tissues in cholangiocarcinoma (CHOL), colon
adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck
squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney
renal clear cell carcinoma (KIRC), liver hepatocellular carcinoma
(LIHC), lung adenocarcinoma (LUAD), stomach adenocarcinoma (STAD)
and uterine corpus endometrial carcinoma (UCEC) (Fig. 1A). In the TCGA-STAD dataset, the
expression of GPR176 was significantly higher in gastric cancer
tissues when compared with adjacent tissues (Fig. 1B). Additionally, GPR176 expression
levels were lower in patients with TNM stage I gastric
adenocarcinoma when compared with patients with stage II/III/IV
gastric adenocarcinoma (Fig. 1C),
and GPR176 expression levels were significantly lower in patients
with stage T1 when compared with patients with pathologic stage
T2/T3/T4 (Fig. 1D).
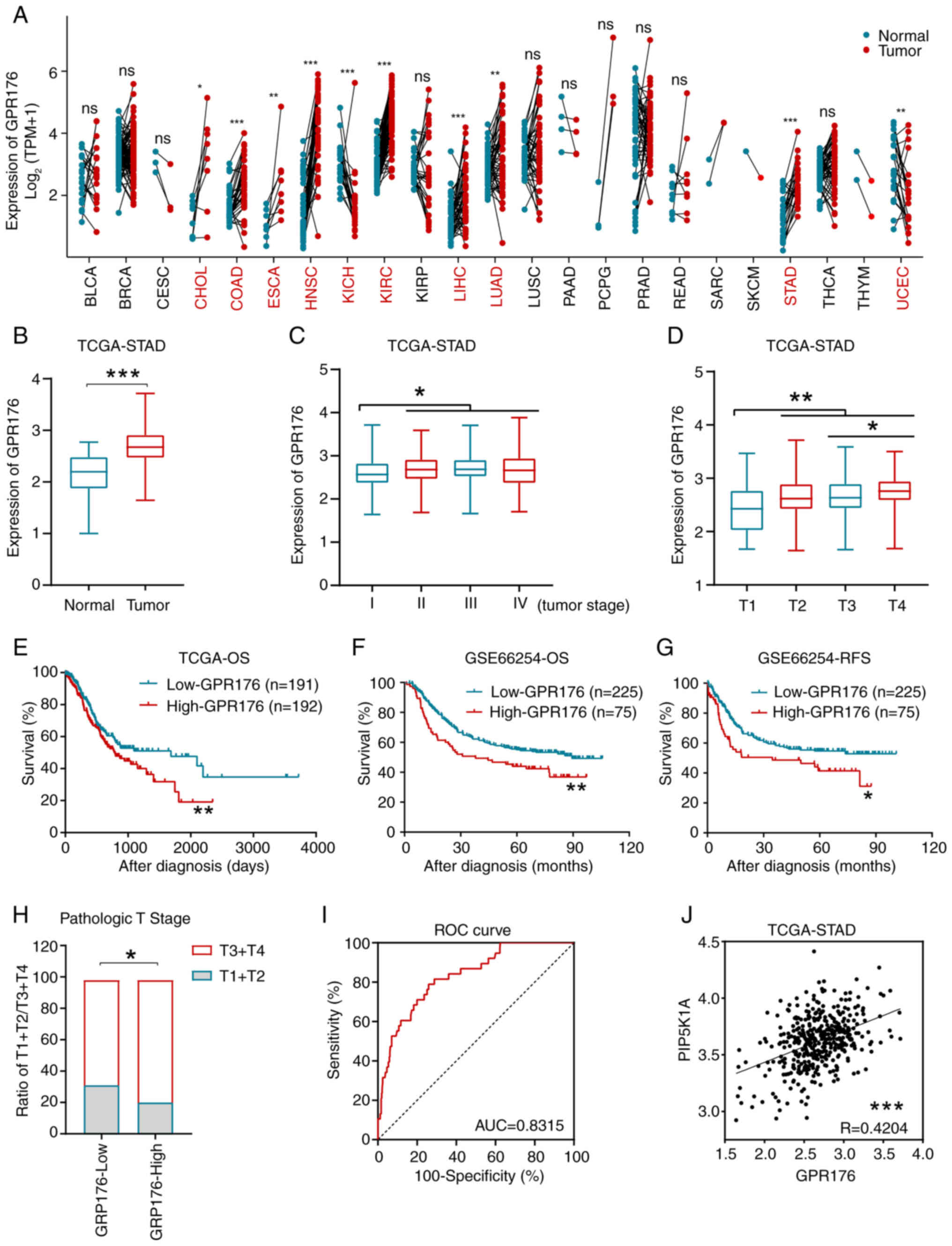 | Figure 1.GPR176 expression levels were
associated with clinicopathological characteristics and prognosis
in STAD patients. (A) GPR176 expression levels in common malignant
tumors and adjacent tissues. (B) Comparison of GPR176 expression
levels in STAD tumor tissues and adjacent normal stomach tissues.
(C) Expression levels of GPR176 mRNA in STAD tumors after TNM
staging (I, II, III and IV). (D) Expression levels of GPR176 mRNA
in STAD tumors at pathologic T1, T2, T3 and T4 stages. (E) Tn the
TCGA-STAD dataset, OS of patients with STAD with GPR176 high and
low expression groups using the median value as a cut-off. (F) In
the GSE66254 dataset, OS of patients with STAD with GPR176 high and
low expression groups using the 75th percentile value as a cut-off.
(G) In the GSE66254 dataset, RFS of patients with STAD with GPR176
high and low expression groups using the 75th percentile as a
cut-off. (H) Association of GPR176 expression with pathologic T
staging in the TCGA-STAD dataset. (I) The ROC curve based on GPR176
plotted for distinguishing between gastric cancer and adjacent
tissues. (J) Significant positive correlation between GPR176 and
PIP5K1A expression in the gastric cancer samples in the TCGA-STAD
dataset. ROC, receiver operating characteristic; TCGA, The Cancer
Genome Atlas; STAD, stomach adenocarcinoma; OS, overall survival;
RFS, Recurrence-free survival. *P<0.05, **P<0.01,
***P<0.001. Unpaired Student's t-test was used in Fig. 1A-D, Survival analysis with
Kaplan-Meier method was used in Fig.
1E-G; Chi-squared test was used in Fig. 1F; Receiver Operating Characteristic
(ROC) was used in Fig. 1G; Linear
regression was used in Fig. 1H. |
Statistical analysis confirmed the association of
GPR176 with tissue type and tumor stage, suggesting its importance
in the occurrence and progression of gastric adenocarcinoma.
Subsequently, in the TCGA-STAD and GSE66254 datasets, significant
association was observed between GPR176 expression levels and
overall survival/recurrence-free survival in gastric
adenocarcinoma, with high GPR176 expression levels associated with
worse prognosis (Fig. 1E-G). Using
the median expression of GPR176 as a cut-off point, patients were
categorized into GPR176-High and GPR176-Low groups. An increased
proportion of patients at T3/T4 in the GPR176-High group was
observed (Fig. 1H). The area under
the receiver operating curve reached 0.8315, indicating GPR176 as a
good biomarker to distinguish gastric adenocarcinoma from normal
gastric tissue (Fig. 1I).
Correlation analysis of GPR176 with PIP5K1A expression levels in
gastric adenocarcinoma tissues using the TCGA-STAD dataset verified
a linear positive association (Fig.
1J).
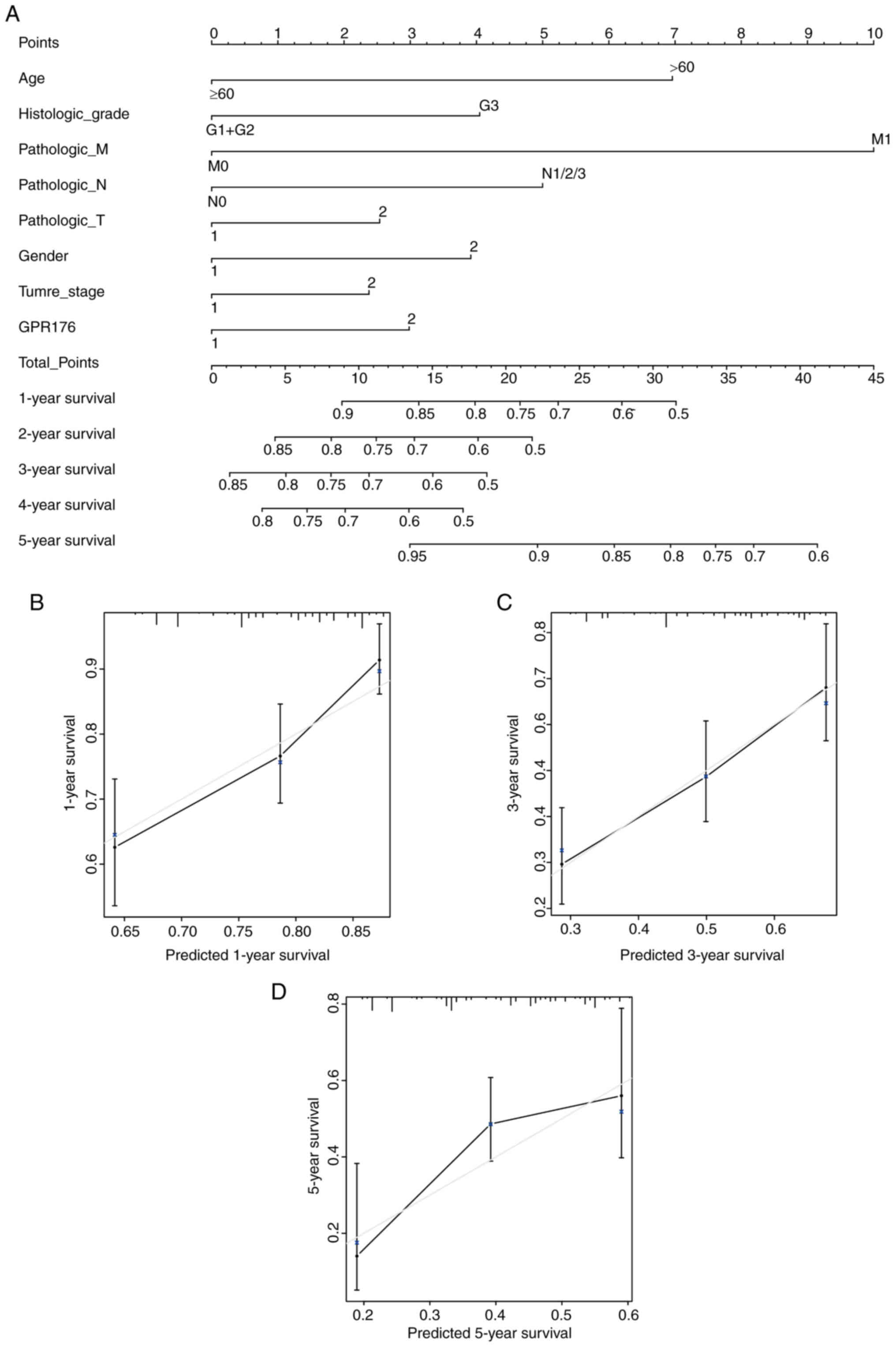
Nomogram construction
Nomogram were created based on GPR176 expression and
clinicopathologic parameters. Univariate Cox regression analysis
and multivariate Cox regression analysis were carried out using
clinical case characteristics such as age, sex, TNM stage, T stage,
N stage, M stage, histologic grade and GPR176 expression (Table SIV). The results of univariate Cox
regression analysis results indicated that age, sex, TNM stage, T
stage, N stage, M stage and histological grade were associated with
the overall survival of patients with gastric adenocarcinoma. In
Multivariate Cox regression analysis, only age, histologic grade
and GPR176 expression were associated with overall survival of
patients with gastric adenocarcinoma. The nomogram graph based on
age, sex, TNM stage, T stage, N stage, M stage, histological grade
and GPR176 expression were constructed to assess the risk of
mortality for specific patients (Fig.
2A). The predictive power of the histogram was evaluated by
comparing the grade between the training group and the validation
group. The nomogram showed a high degree of overlap between the
self-validation cohort and the training group in predicting the 1-,
3- or 5-year prognosis (Fig.
2B-D).
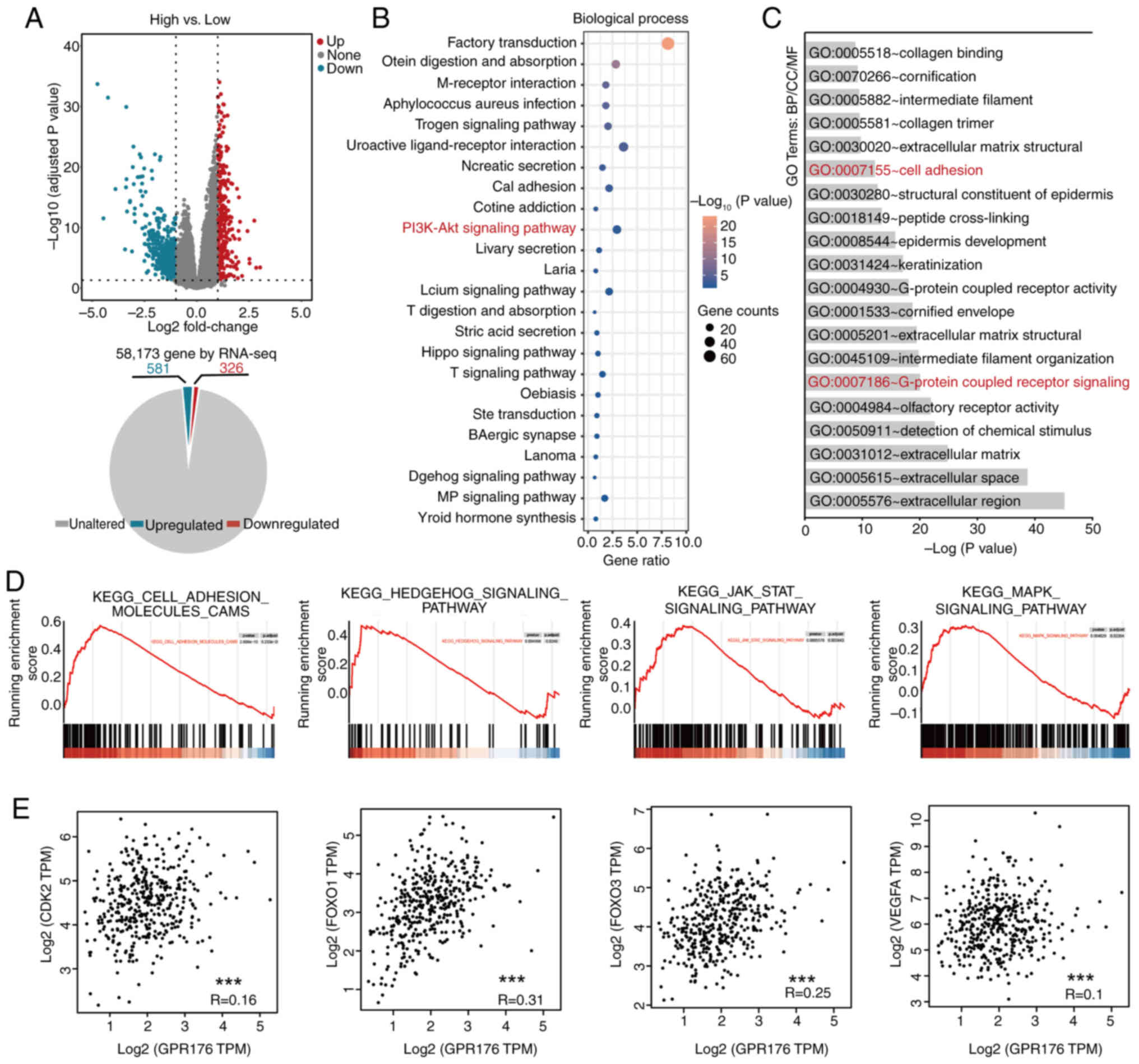
Exploration of the mechanisms of
GPR176 action using bioinformatics tools
Based on the expression levels of GPR176, patients
in the TCGA-STAD dataset were categorized into high and low
expression groups. Differential expression analysis was carried out
using the Limma package for RNA-seq and a volcano plot was
generated (Fig. 3A). The
corresponding pie charts showed that 326 genes were upregulated,
and 581 genes were downregulated. Functional enrichment analysis of
differentially expressed genes associated with GPR176 revealed
enrichment in signaling pathways such as ‘PI3K-Akt signaling
pathway’; Fig. 3B), and cellular
functions such as ‘Cell adhesion’ and ‘G-protein-coupled receptor
signaling’ (Fig. 3C). Gene Set
Enrichment Analysis indicated associations between GPR176 and
tumor-related signaling pathways such as ‘cell adhesion molecules
CAMs’, ‘Hedgehog signaling pathway’, ‘Jak-stat signaling pathway’
and ‘MAPK signaling pathway’ (Fig.
3D). CDK2, FOXO1, FOXO3 and VEGFA are downstream target genes
of the PI3K/AKT/mTOR signaling pathway, reflecting the
activation/inhibition status of the pathway (41). Linear regression analysis using
RNA-seq data from gastric adenocarcinoma tissues in TCGA-STAD
revealed an association between GPR176 and CDK2, FOXO1, FOXO3 and
VEGFA (Fig. 3E).
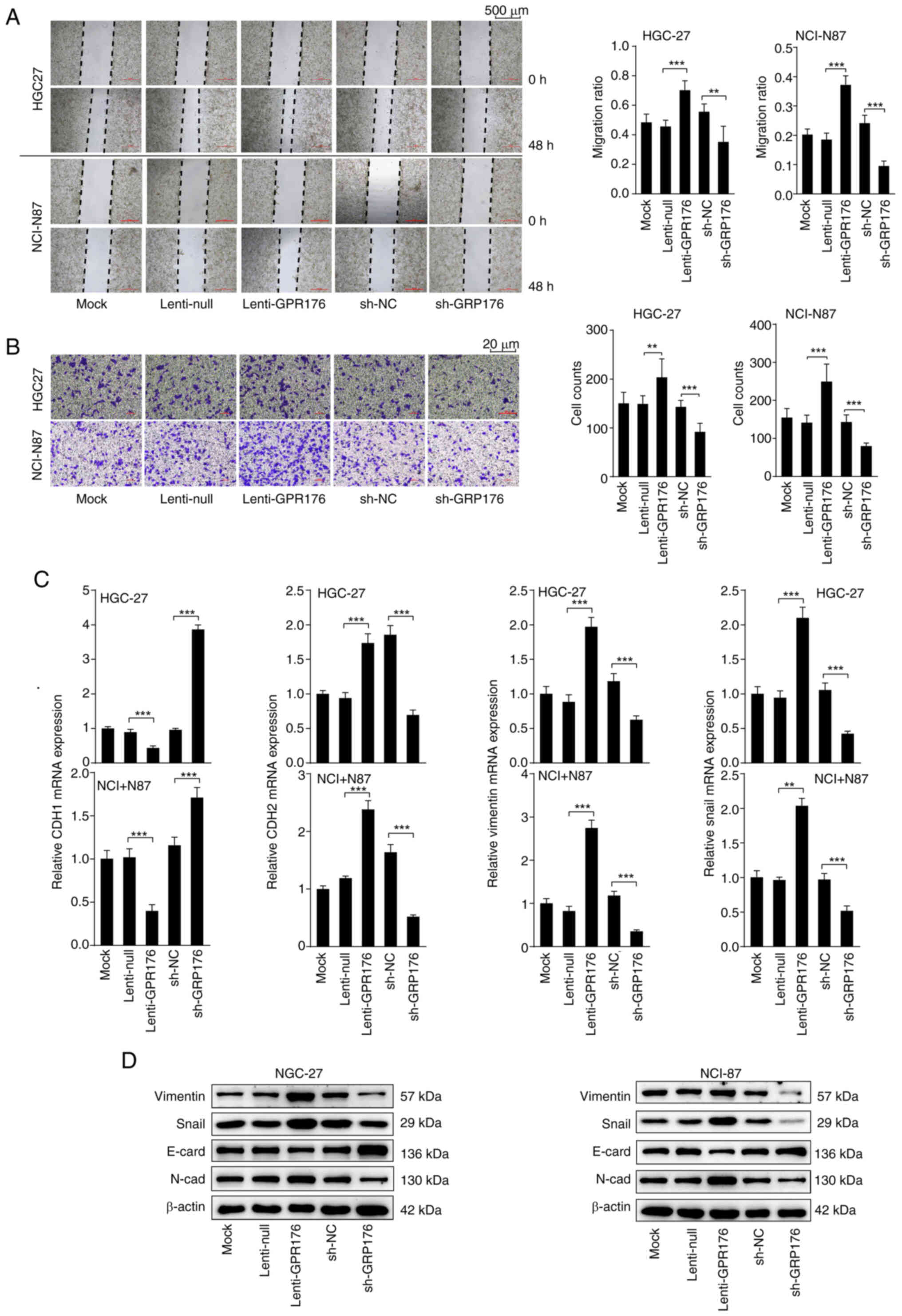
GPR176 enhances the migration and
invasion capabilities of gastric cancer cells and induces EMT
To confirm the effect of GPR176 on the biological
behavior of gastric cancer cells and explore the corresponding
mechanisms, lentiviral vectors were used to overexpress and
knockdown GPR176 expression levels. RT-qPCR and WB confirmed the
satisfactory efficiency of OE lentivirus and RNAi lentivirus in
HGC-27 and NCI-N87 cells (Fig.
S1A). Subsequently, scratch-wound healing and Transwell assays
were carried out to evaluate the effects of modulating GPR176
expression on cell migration and invasion. After upregulation of
GPR176, the migration and invasion ability of cells increased
significantly, while downregulation of GPR176 resulted in a
significant decrease in this ability (Fig. 4A and B). Subsequent RT-qPCR analysis
revealed a significant increase in the mRNA levels of
EMT-activating genes, such as CDH2, VIM and SNAI1, following
upregulation of GPR176 (Fig. 4C).
By contrast, the expression of the gene CDH1 which encodes
E-cadherin, associated with the inhibition of the EMT pathway
(42), significantly decreased
(Fig. 4C). Conversely,
downregulation of GPR176 led to a significant decrease in the mRNA
levels of CDH2, VIM and SNAI1, accompanied by a significant
increase in the mRNA levels of CDH1, associated with the inhibition
of the EMT pathway (Fig. 4C). WB
results corroborated the RT-qPCR results and revealed an increase
in the protein concentration of N-cadherin, vimentin and Snai1, and
a decrease in the protein concentration of E-cadherin after
upregulation of GPR176 (Fig. 4D),
with the corresponding bar chart shown in Fig. S2A and B. IF showed that GPR176
upregulation increased vimentin and reduced E-cadherin, whereas
GPR176 knockdown had the opposite effect (Fig. S1E). The results from RT-qPCR, WB,
and IF experiments showed that GPR176 expression was associated
with the activation of the EMT signaling pathway and increased
migration and invasion of gastric adenocarcinoma cells.
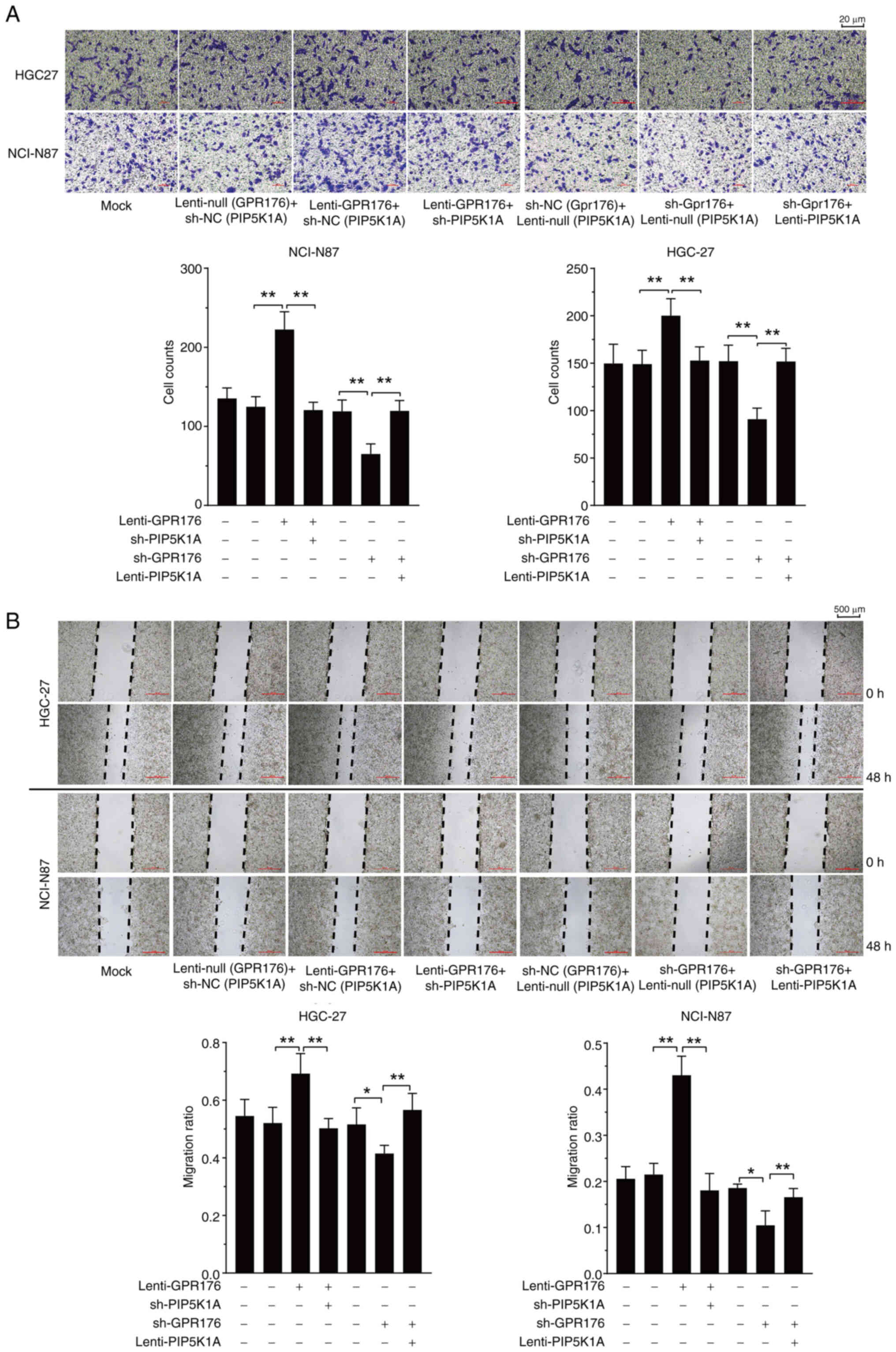
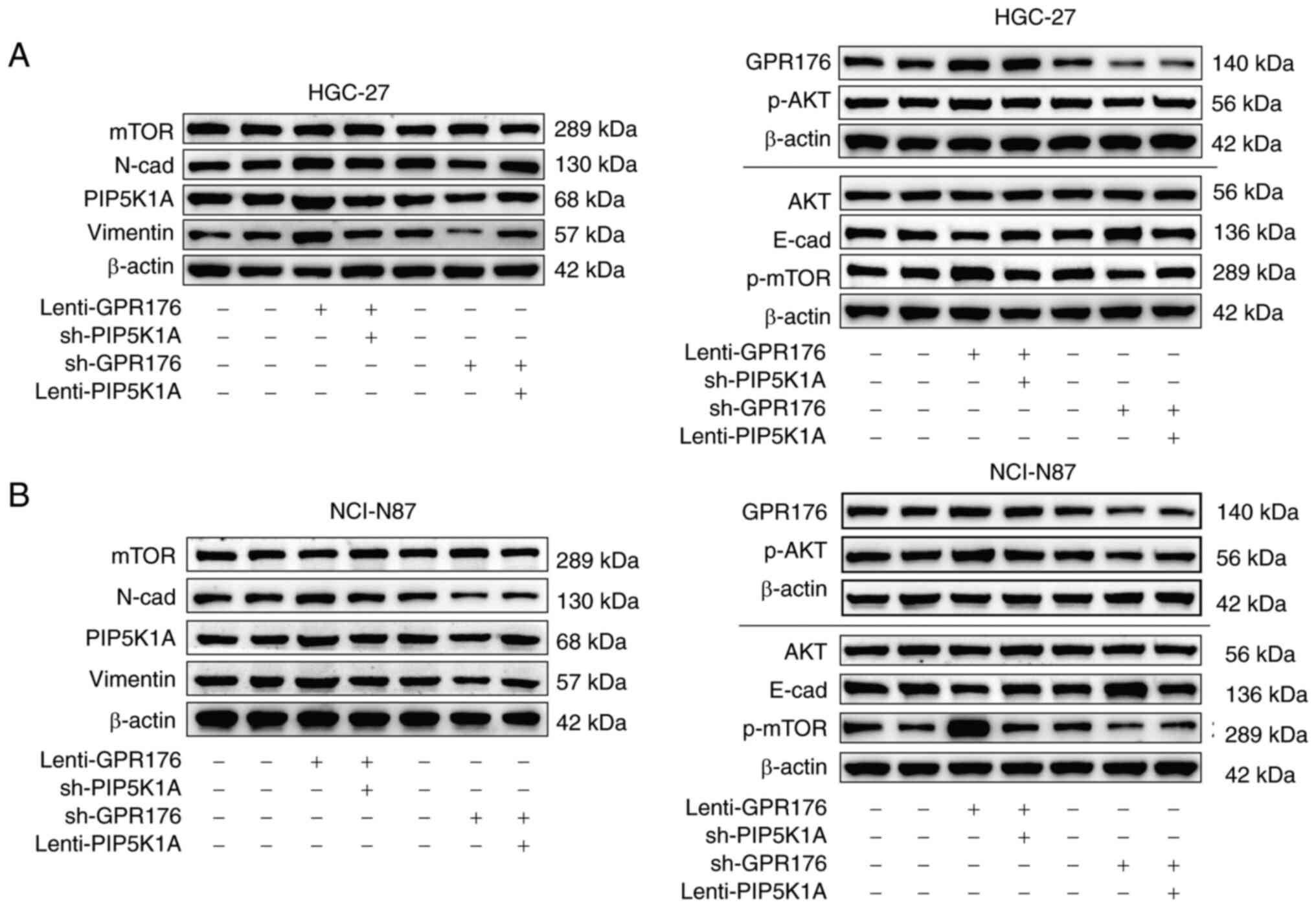
Downregulation of PIP5K1A prevents the
effects of GPR176 on cell migration, invasion, EMT and on the
PI3K/AKT/mTOR pathway
Previous studies have confirmed that PIP5K1A is an
upstream molecule in the PI3K/AKT/mTOR signaling pathway (30–33).
Analysis of RNA-seq data from the TCGA-STAD dataset in the present
study revealed a significant positive association between GPR176
and PIP5K1A expression (Fig. 1J).
Therefore, we hypothesized that GPR176 may activate the
PI3K/AKT/mTOR signaling pathway by inducing upregulation of PIP5K1A
expression. Interference and overexpression experiments of PIP5K1A
were performed based on the overexpression of GPR176 and the
repression of GPR176. Analysis revealed that interference with
PIP5K1A expression significantly prevented the migration and
invasion induced by GPR176 overexpression (Fig. 5A and B). Conversely, downregulation
of PIP5K1A also significantly reversed the slowed migration and
invasion of gastric cancer cells caused by GPR176 knockdown
(Fig. 5A and B). Following this, WB
was carried out to evaluate the phosphorylation levels of molecules
within the PI3K/AKT/mTOR pathway and the EMT signaling pathway,
along with assessing the expression levels of PIP5K1A. The
activation of the PI3K/AKT/mTOR pathway and EMT signaling pathway
after GPR176 upregulation was attenuated by the downregulation of
PIP5K1A. Conversely, the inhibition of the PI3K/AKT/mTOR pathway
and EMT signaling pathway after GPR176 downregulation was
counteracted by the OE of PIP5K1A (Fig.
6A and B), with the corresponding bar chart shown in Fig. S3A and B. The results of the RT-qPCR
assay were in concordance with the aforementioned experiments,
indicating a preventing in changes to the mRNA expression levels of
molecules in the EMT signaling pathway after GPR176 upregulation,
which was suppressed by the downregulation of PIP5K1A in HGC-27 and
NCI-N87 cells; similarly, in HGC-27 and NCI-N87 cells, the
downregulation of GPR176 resulted in prevention of the mRNA
expression levels of molecules in the EMT signaling pathway, under
the upregulating influence of PIP5K1A (Fig. 7A-L).
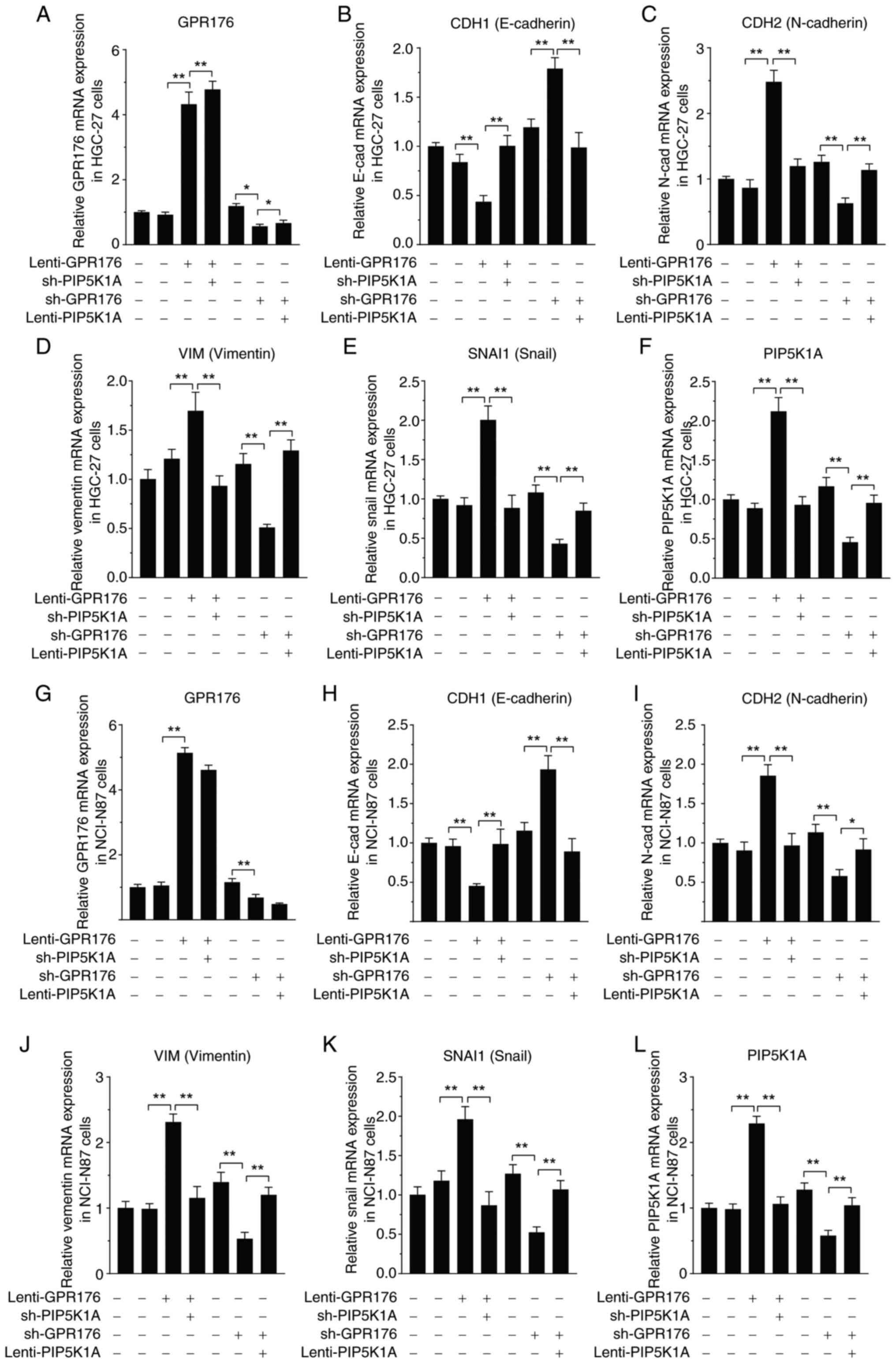 | Figure 7.Genetic regulation of PIP5K1A
counteracts the effects of GPR176 on the expression of EMT pathway
genes at the mRNA level. Combined genetic regulation of GPR176 and
PIP5K1A on the expression of (A) GPR176 (B) CHD1 (E-cadherin), (C)
CHD2 (N-cadherin) (D) VIM (Vimentin), (E) SNAI1 (Snail), (F)
PIP5K1A in in HGC-27 cells. Combined genetic regulation of GPR176
and PIP5K1A on the expression of (G) GPR176, (H) CHD1 (E-cadherin),
(I) CHD2 (N-cadherin), (J) VIM (Vimentin), (K) SNAI1 (Snail), (L)
PIP5K1A in NCI-N87 cells. EMT, epithelial-mesenchymal transition;
sh-, short hairpin-; cad, cadherin. Statistical analysis was
performed using two-way ANOVA. **P<0.01, *P<0.05. |
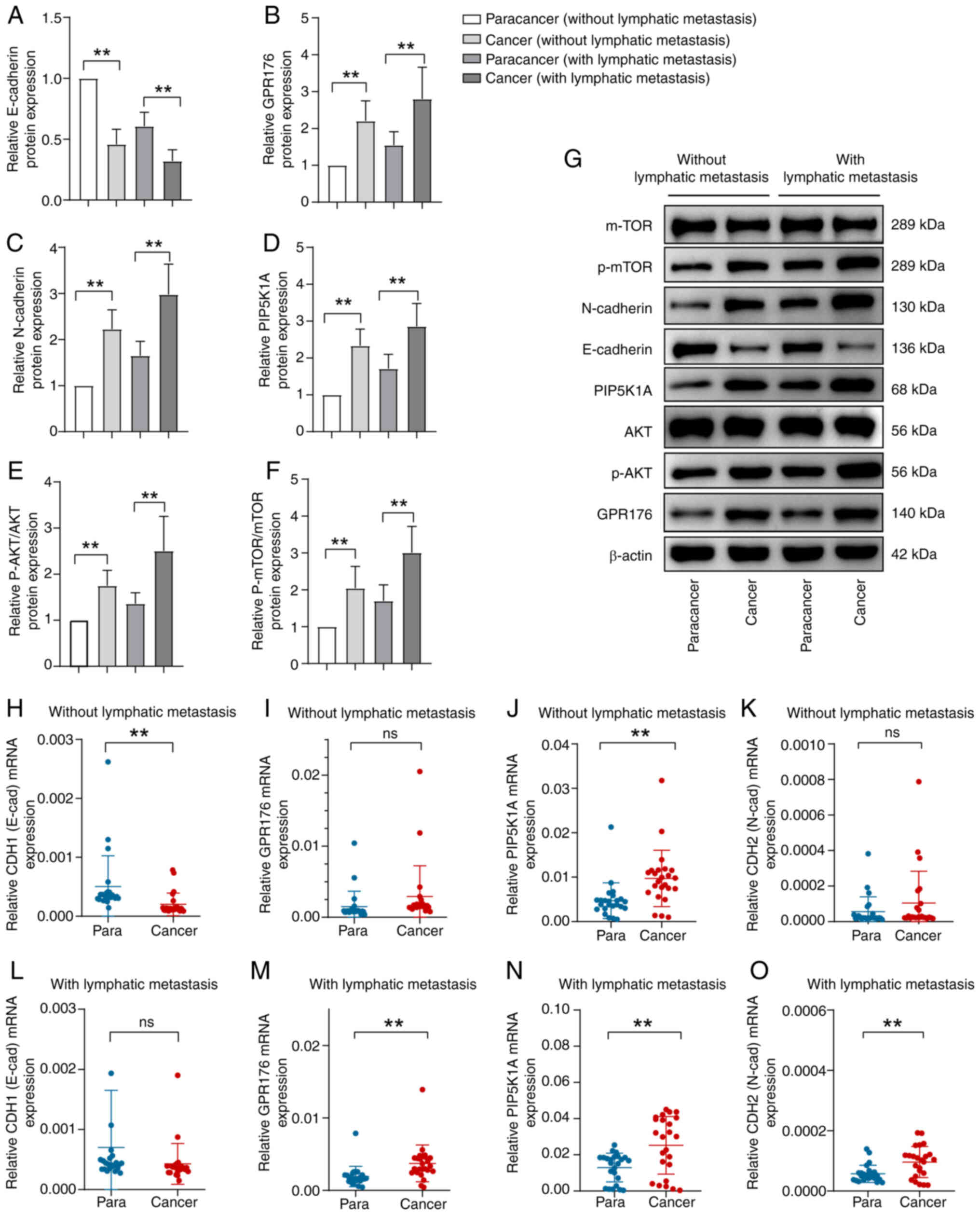
The status of EMT and PI3K/AKT/mTOR
between para-cancer and cancer tissues, as well as the expression
of PIP5K1A and GPR176
Firstly, patients with gastric cancer were
categorized into two groups based on the presence or absence of
lymph node metastasis: Those with lymph node metastasis and those
without. Further classification was made based on histological
type, distinguishing between the para-cancer group and the cancer
group. Initially, WB was employed to assess the protein levels of
E-cadherin, N-cadherin, PIP5K1A, GPR176, AKT, p-AKT, mTOR and
p-mTOR in each group. The findings revealed that irrespective of
lymph node metastasis, the expression of E-cadherin in the cancer
group was significantly reduced compared with the para-cancer
group. Conversely, levels of N-cad, PIP5K1A, GPR176, p-AKT, p-mTOR,
p-AKT/AKT and p-mTOR/mTOR were increased in the cancer group
compared with the para-cancer group (Fig. 8A-F). The representative blots of WB
experiments are referenced in Fig.
8G.
Subsequently, validation of the expression levels of
key genes in the EMT pathway and PI3K/AKT/mTOR signaling pathway,
as well as PIP5K1A and GPR186, was conducted using RT-qPCR. Among
patients without lymph node metastasis, no significant differences
were observed in GPR186 and CDH2 (N-cadherin) expression levels
between the para-cancer and cancer groups (Fig. 8I and K). However, CDH1 (E-cadherin)
expression was significantly reduced in the cancer group compared
with the para-cancer group (Fig.
8H). Conversely, PIP5K1A expression was markedly higher in the
cancer group compared with the para-cancer group (Fig. 8J). In patients with lymph node
metastasis, there were no significant differences in CDH1
expression between the para-cancer and cancer groups (Fig. 8L). Nevertheless, GPR186, CDH2 and
PIP5K1A expression levels were significantly increased in the
cancer group compared with the para-cancer group (Fig. 8M-O). In summary, the expression
levels of PIP5K1A and GPR176 were increased in the cancer group
compared with the para-cancer group. Additionally, activation of
the EMT pathway and PI3K/AKT/mTOR pathway was evident in the cancer
group.
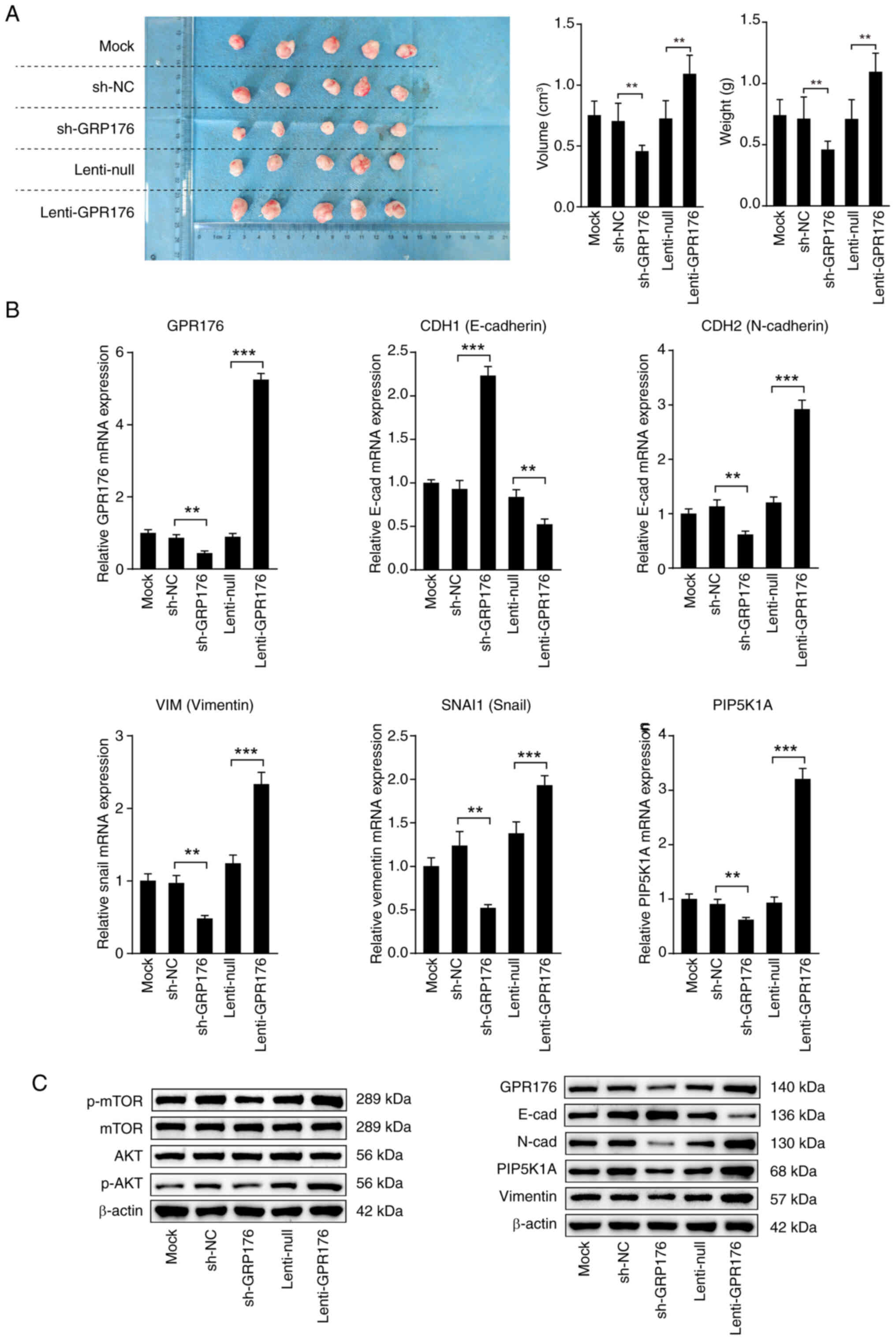
GPR176 promotes the growth of
transplanted tumors by activating the PI3K/AKT/mTOR pathway
In vitro experiments provided evidence that
GPR176 could activate the PI3K/AKT/mTOR pathway, facilitating EMT
and promoting cell proliferation by inducing the overexpression of
PIP5K1A. To further validate the role of GPR176 in gastric cancer,
a nude mouse subcutaneous tumor experiment was designed in the
present study to assess the influence of GPR176 expression on the
PI3K/AKT/mTOR pathway and EMT. In comparison with the Lenti-NC
group, the GPR176 upregulation group exhibited significantly larger
tumor volume and weight. Conversely, relative to the sh-NC group,
the sh-GPR176 group displayed a substantial reduction in tumor
volume and weight (Fig. 9A).
Subsequent examination of the mRNA expression levels of EMT pathway
molecules, consistent with the in vitro results, revealed
activation of the EMT pathway following GPR176 upregulation, while
the EMT signaling pathway was inhibited after GPR176 downregulation
(Fig. 9B). Western blot analysis
suggested the activation of the EMT and PI3K/AKT/mTOR pathways
following GPR176 upregulation, as indicated by increased levels of
key pathway markers. Conversely, GPR176 downregulation resulted in
changes indicative of pathway inhibition. (Fig. 9C), with the corresponding bar chart
shown in Fig. S4A-G.
Discussion
Gastric cancer is a highly malignant tumor
characterized by the absence of typical early symptoms, often
leading to late-stage diagnosis and missing of the optimal
treatment window (4,5). This poses several challenges for the
treatment of gastric cancer (43).
In the advanced stages, gastric cancer has usually spread to lymph
nodes or other organs, making treatment considerably more difficult
(44–46). Due to the different subtypes of
gastric cancer, each with different biological behaviors and
treatment responses, the development of universal treatment plans
is complex and requires more individualized and precise treatment
strategies (47,48). Some patients develop resistance to
conventional chemotherapeutic agents, leading to a decrease in
treatment efficacy and requiring constant adjustment of drug
combinations during treatment to overcome the adaptability of the
tumor (49,50). Given these challenges, there is an
urgent need for further research in the field of gastric cancer
treatment to develop more effective and personalized treatment
strategies.
GPR176 is generally considered to be associated with
circadian rhythms (26). However,
some researchers have begun to recognize its role in tumors. For
example, Tang et al (28)
found that GPR176 interacts with the G protein GNAS to inhibit
mitochondrial autophagy in colorectal cancer cells, thereby
promoting cancer progression. In the present study, compared with
normal gastric tissue, upregulation of GPR176 expression in gastric
cancer was observed, with significant differences in expression
between different TNM stages and tissue types. This heterogeneous
expression suggests that GPR176 may play different biological roles
in different subtypes of gastric cancer.
Through integrated analysis of data from TCGA and
GEO databases, it was clarified that high GPR176 expression is
significantly associated with shorter overall survival and
disease-free survival in patients with gastric cancer. This
suggests that GPR176 could serve as an independent prognostic
marker, providing a new molecular standard for the assessment of
patient survival. Similarly, researchers such as Yun et al
(29) have previously suggested
that GPR176 may also serve as a prognostic biomarker in breast
cancer.
Bioinformatic analysis revealed that the gene
expression profile regulated by the accumulation of GPR176 is
associated with signaling pathways such as PI3K/AKT/mTOR. This
suggests that GPR176 may be involved in the development of gastric
cancer via this signaling pathway. Further experimental
verification demonstrated an association between GPR176 and the
PI3K/AKT/mTOR signaling pathway and provided an experimental basis
for understanding the specific role of GPR176 in cell
proliferation, survival and metastasis.
The PI3K signaling pathway carries out a key role in
the proliferation and progression of various cancer cells,
including gastric cancer (51,52).
Numerous previous studies have indicated that the PI3K signaling
pathway promotes the progression of gastric cancer through various
mechanisms, including inhibiting apoptosis, inducing drug
resistance, facilitating metastasis and promoting angiogenesis
(53–55).
After activation by PI3K and PIP2, AKT kinase
relocates downstream to the cell membrane, triggering its
conformational activation (56).
AKT carries out a key role in activating the PI3K axis and elevated
expression of AKT and p-AKT has been detected in >74% of gastric
cancer cases (57). Aberrant
expression of p-AKT is associated with the overexpression of PI3K
and HER2, and high levels of p-AKT are regarded as indicators of
tumor progression, metastasis, and poor prognosis in gastric cancer
(58).
Analysis of the TCGA molecular subtypes reveals that
the majority of gastric cancer cases studied display varying
degrees of PIK3CA gene mutations, along with amplifications of RTK
genes such as EGFR and HER2 (48).
Genomic amplifications markedly contribute to tumor progression.
Amplification of PIK3CA is associated with tumor progression,
prognosis and the development of gastric cancer resistance
(59). The substantial involvement
of the PI3K/AKT/mTOR signaling pathway in gastric cancer
progression suggests that targeting this signaling axis holds
potential for cancer therapy (60).
The present study identified the upregulation of
GPR176 as a promoter of EMT in gastric cancer cells, thereby
revealing a mechanism that drives tumor invasion and metastasis.
In vitro assays confirmed that GPR176 overexpression
enhances cell migration and invasion, whereas its knockdown
produces the opposite effect. These findings highlight the pivotal
role of GPR176 in regulating EMT and suggest that it may serve as a
novel modulator of gastric cancer progression. Future studies
should further dissect the upstream regulators and downstream
effectors of GPR176 in EMT, as well as its interactions with other
EMT-related transcription factors and regulatory networks.
The present study also identified an association
between GPR176 and PIP5K1A, and that downregulation of PIP5K1A
prevented the effects of GPR176 on migration, invasion, EMT and
activation of the PI3K/AKT/mTOR pathway. This indicates that GPR176
may exert its tumor-promoting function, at least in part, through
synergistic regulation with PIP5K1A. Additional studies employing
gene knock-in models, CRISPR/Cas9-mediated editing or
co-immunoprecipitation assays are warranted to validate the direct
interaction between GPR176 and PIP5K1A and to elucidate their
precise molecular mechanisms. Although the present study
highlighted the involvement of GPR176 in the PI3K/AKT/mTOR
signaling pathway, it is important to note that this pathway
represents a common oncogenic mechanism (61–63).
Future research should therefore investigate the potential upstream
regulators that modulate GPR176 expression, such as transcription
factors, epigenetic modifications or non-coding RNAs, which may
contribute to its dysregulation in gastric cancer. In addition,
downstream interactions of GPR176 beyond PI3K/AKT/mTOR, including
crosstalk with MAPK/ERK or Wnt/β-catenin pathways, may further
shape tumor cell proliferation, survival and metastatic behavior.
Exploring these upstream and downstream aspects will broaden the
understanding of GPR176-mediated oncogenic signaling and may
uncover novel therapeutic opportunities.
As a member of the G protein-coupled receptor
family, GPR176 remains relatively underexplored in both
physiological and pathological contexts. Findings of the present
study expand the current understanding of its role in gastric
adenocarcinoma, particularly in relation to cell signaling,
proliferation, invasion and metastasis. Given the high
heterogeneity of gastric cancer and the variable therapeutic
responses among patients, studying GPR176 expression and function
across tumor subtypes could contribute to the development of more
individualized treatment strategies. Furthermore, the observed
association between GPR176 expression and patient survival
underscores its potential as a prognostic biomarker and as a
candidate for risk stratification in clinical practice.
Furthermore, the findings of the present study
indicate that high GPR176 expression in gastric adenocarcinoma is
associated with tumor invasion, metastasis and poor prognosis,
highlighting its potential translational significance. GPR176 may
serve not only as a prognostic biomarker for assessing patient
survival and recurrence risk but also as a promising therapeutic
target due to its key role in tumor cell proliferation, migration
and activation of the PI3K/AKT/mTOR pathway. Future studies should
further validate the predictive value of GPR176 in independent
patient cohorts and explore the feasibility of targeting GPR176
through small-molecule inhibitors, antibodies or gene-editing
strategies, providing new avenues for personalized therapy and
precision oncology.
Despite these insights, several limitations should
be acknowledged. First, the bioinformatics analyses were based on
retrospective datasets (TCGA-STAD and GSE66254) with limited sample
sizes, which may introduce selection bias. Second, although the
prognostic value and functional role of GPR176 were validated
through in vitro and in vivo experiments, the models
may not fully capture the complexity of the human tumor
microenvironment. Third, the mechanistic investigations mainly
focused on the PI3K/AKT/mTOR signaling pathway and EMT, while other
potential pathways or crosstalk mechanisms remain unexplored.
Finally, the translational relevance of GPR176 as a biomarker or
therapeutic target requires further validation in large-scale,
prospective and independent clinical cohorts. Addressing these
limitations will be key for advancing the clinical application of
GPR176 in gastric cancer.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported in part by the Medical Excellence Award
Funded by the Creative Research Development Grant from the First
Affiliated Hospital of Guangxi Medical University (grant no.
202204), Self-funded Research Project of Health Commission of
Guangxi Zhuang Autonomous Region (grant no. Z-A20220397) and Hubei
Chen Xiaoping Science and Technology Development Foundation (grant
no. CXPJJH122002-027).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
GM conceived and designed the research program; GM
and KL contributed to acquisition of data; CH, HR, DL, GZ and JC
performed data analysis. GM, ZH, HR and KL performed the
experiments. KL wrote the manuscript. GM guided and supervised the
manuscript. All authors read and approved the final manuscript. All
authors made a significant contribution to the work reported,
whether that is in the conception, study design, execution,
acquisition of data, analysis and interpretation, or in all these
areas; took part in drafting, revising or critically reviewing the
article; gave final approval of the version to be published; have
agreed on the journal to which the article has been submitted; and
agree to be accountable for all aspects of the work. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The investigation had been approved by the ethics
committee of Guangxi Medical University the first affiliated
hospital, Nanning, China (approval no. 2023-S243-01)]. All methods
in this research were carried out in accordance with Declaration of
Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GPR176
|
G protein-coupled receptor 176
|
|
PIP5K1A
|
phosphatidylinositol-4-phosphate
5-kinase type 1 α
|
|
DAVID
|
Database for Annotation, Visualization
and Integrated Discovery
|
|
GO
|
gene ontology
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
WB
|
western blotting
|
|
EMT
|
epithelial-mesenchymal transition
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Guo HX, Wang Q, Wang C, Yin QC, Huo HZ and
Yin BH: Secular trends in gastric and esophageal cancer
attributable to dietary carcinogens from 1990 to 2019 and
projections until 2044 in China: Population-based study. JMIR
Public Health Surveill. 9:e484492023. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maomao C, He L, Dianqin S, Siyi H, Xinxin
Y, Fan Y, Shaoli Z, Changfa X, Lin L, Ji P and Wanqing C: Current
cancer burden in China: Epidemiology, etiology, and prevention.
Cancer Biol Med. 19:1121–1138. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Y, Yan Q, Fan C, Mo Y, Wang Y, Li X,
Liao Q, Guo C, Li G, Zeng Z, et al: Overview and countermeasures of
cancer burden in China. Sci China Life Sci. 66:2515–2526. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qiu H, Cao S and Xu R: Cancer incidence,
mortality, and burden in China: A time-trend analysis and
comparison with the United States and United Kingdom based on the
global epidemiological data released in 2020. Cancer Commun (Lond).
41:1037–1048. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sekiguchi M, Oda I, Matsuda T and Saito Y:
Epidemiological trends and future perspectives of gastric cancer in
Eastern Asia. Digestion. 103:22–28. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li GZ, Doherty GM and Wang J: Surgical
management of gastric cancer: A review. JAMA Surg. 157:446–454.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hong J, Tsai Y, Novick D, Hsiao FCH, Cheng
R and Chen JS: The economic burden of advanced gastric cancer in
Taiwan. BMC Health Serv Res. 17:6632017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu H and Li W: Early detection of gastric
cancer in China: Progress and opportunities. Cancer Biol Med.
19:1622–1628. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Machlowska J, Baj J, Sitarz M, Maciejewski
R and Sitarz R: Gastric cancer: Epidemiology, risk factors,
classification, genomic characteristics and treatment strategies.
Int J Mol Sci. 21:40122020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen D, Fu M, Chi L, Lin L, Cheng J, Xue
W, Long C, Jiang W, Dong X, Sui J, et al: Prognostic and predictive
value of a pathomics signature in gastric cancer. Nat Commun.
13:69032022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao R, Gong L and Dong D: Pathological
diagnosis and prognosis of gastric cancer through a multi-instance
learning method. EBioMedicine. 73:1036712021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Y, Chen T and Fang JY: Burden of
gastrointestinal cancers in China from 1990 to 2019 and projection
through 2029. Cancer Lett. 560:2161272023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li H, Zhang H, Zhang H, Wang Y, Wang X and
Hou H; Global Health Epidemiology Reference Group, : Survival of
gastric cancer in China from 2000 to 2022: A nationwide systematic
review of hospital-based studies. J Glob Health. 12:110142022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lordick F, Carneiro F, Cascinu S, Fleitas
T, Haustermans K, Piessen G, Vogel A and Smyth EC; ESMO Guidelines
Committee. Electronic address, : simpleclinicalguidelines@esmo.org:
Gastric cancer: ESMO clinical practice guideline for diagnosis,
treatment and follow-up. Ann Oncol. 33:1005–1020. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guan WL, He Y and Xu RH: Gastric cancer
treatment: Recent progress and future perspectives. J Hematol
Oncol. 16:572023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ajani JA, D'Amico TA, Bentrem DJ, Chao J,
Cooke D, Corvera C, Das P, Enzinger PC, Enzler T, Fanta P, et al:
Gastric cancer, version 2.2022, NCCN clinical practice guidelines
in oncology. J Natl Compr Canc Netw. 20:167–192. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kobayashi D and Kodera Y: Intraperitoneal
chemotherapy for gastric cancer with peritoneal metastasis. Gastric
Cancer. 20 (Suppl 1):S111–S121. 2017. View Article : Google Scholar
|
|
20
|
Boilève J, Touchefeu Y and Matysiak-Budnik
T: Clinical management of gastric cancer treatment regimens. Curr
Top Microbiol Immunol. 444:279–304. 2023.PubMed/NCBI
|
|
21
|
Patel TH and Cecchini M: Targeted
therapies in advanced gastric cancer. Curr Treat Options Oncol.
21:702020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li K, Zhang A, Li X, Zhang H and Zhao L:
Advances in clinical immunotherapy for gastric cancer. Biochim
Biophys Acta Rev Cancer. 1876:1886152021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kawakami H and Okamoto I: MET-targeted
therapy for gastric cancer: The importance of a biomarker-based
strategy. Gastric Cancer. 19:687–695. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Joshi SS and Badgwell BD: Current
treatment and recent progress in gastric cancer. CA Cancer J Clin.
71:264–279. 2021.PubMed/NCBI
|
|
25
|
Doi M, Murai I, Kunisue S, Setsu G, Uchio
N, Tanaka R, Kobayashi S, Shimatani H, Hayashi H, Chao HW, et al:
Gpr176 is a Gz-linked orphan G-protein-coupled receptor that sets
the pace of circadian behaviour. Nat Commun. 7:105832016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakagawa S, Nguyen Pham KT, Shao X and Doi
M: Time-restricted G-protein signaling pathways via GPR176,
Gz, and RGS16 Set the pace of the master circadian clock
in the suprachiasmatic nucleus. Int J Mol Sci. 21:50552020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yamaguchi Y, Murai I, Takeda M, Doi S,
Seta T, Hanada R, Kangawa K, Okamura H, Miyake T and Doi M:
Nmu/Nms/Gpr176 triple-deficient mice show enhanced light-resetting
of circadian locomotor activity. Biol Pharm Bull. 45:1172–1179.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang J, Peng W, Ji J, Peng C, Wang T, Yang
P, Gu J, Feng Y, Jin K, Wang X and Sun Y: GPR176 promotes cancer
progression by interacting with G protein GNAS to restrain cell
mitophagy in colorectal cancer. Adv Sci (Weinh). 10:e22056272023.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yun WJ, Xue H, Yang N, Xiao LJ, Sun HZ and
Zheng HC: Oncogenic roles of GPR176 in breast cancer: A potential
marker of aggressiveness and a potential target of gene therapy.
Clin Transl Oncol. 25:3042–3056. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yun WJ, Li J, Yin NC, Zhang CY, Cui ZG,
Zhang L and Zheng HC: The promoting effects of GPR176 expression on
proliferation, chemoresistance, lipogenesis and invasion of
oesophageal cancer. J Cancer Res Clin Oncol. 149:14641–14655. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Gu X, Zhu F, Li Y, Huang Y and Ju
S: High expression of GPR176 predicts poor prognosis of gastric
cancer patients and promotes the proliferation, migration, and
invasion of gastric cancer cells. Sci Rep. 13:93602023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ni L, Chen S, Liu J, Li H, Zhao H, Zheng
C, Zhang Y, Huang H, Huang J, Wang B and Lin C: GPR176 is a
biomarker for predicting prognosis and immune infiltration in
stomach adenocarcinoma. Mediators Inflamm. 2023:71235682023.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
East MP, Laitinen T and Asquith CRM:
PIP5K1A: A potential target for cancers with KRAS or TP53
mutations. Nat Rev Drug Discov. 19:4362020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ding T, Ji J, Zhang W, Liu Y, Liu B, Han
Y, Chen C and Yu L: The phosphatidylinositol
(4,5)-bisphosphate-Rab35 axis regulates migrasome formation. Cell
Res. 33:617–627. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yin M and Wang Y: The role of PIP5K1A in
cancer development and progression. Med Oncol. 39:1512022.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sarwar M, Syed Khaja AS, Aleskandarany M,
Karlsson R, Althobiti M, Ødum N, Mongan NP, Dizeyi N, Johnson H,
Green AR, et al: The role of PIP5K1α/pAKT and targeted inhibition
of growth of subtypes of breast cancer using PIP5K1α inhibitor.
Oncogene. 38:375–389. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu WT, Li YJ, Feng AZ, Li L, Huang T, Xu
AD and Lyu J: Data mining in clinical big data: The frequently used
databases, steps, and methodological models. Mil Med Res.
8:442021.PubMed/NCBI
|
|
38
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Somerville TDD, Wiseman DH, Spencer GJ,
Huang X, Lynch JT, Leong HS, Williams EL, Cheesman E and
Somervaille TC: Frequent derepression of the mesenchymal
transcription factor gene FOXC1 in acute myeloid leukemia. Cancer
Cell. 28:329–342. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Glaviano A, Foo ASC, Lam HY, Yap KCH,
Jacot W, Jones RH, Eng H, Nair MG, Makvandi P, Geoerger B, et al:
PI3K/AKT/mTOR signaling transduction pathway and targeted therapies
in cancer. Mol Cancer. 22:1382023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wong SHM, Fang CM, Chuah LH, Leong CO and
Ngai SC: E-cadherin: Its dysregulation in carcinogenesis and
clinical implications. Crit Rev Oncol Hematol. 121:11–22. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jim MA, Pinheiro PS, Carreira H, Espey DK,
Wiggins CL and Weir HK: Stomach cancer survival in the United
States by race and stage (2001–2009): Findings from the CONCORD-2
study. Cancer. 123 (Suppl 24):S4994–S5013. 2017. View Article : Google Scholar
|
|
44
|
Budginaite E, Kloft M, van Kuijk SMJ,
Canao PA, Kooreman LFS, Pennings AJ, Magee DR, Woodruff HC and
Grabsch HI: The clinical importance of the host anti-tumour
reaction patterns in regional tumour draining lymph nodes in
patients with locally advanced resectable gastric cancer: A
systematic review and meta-analysis. Gastric Cancer. 26:847–862.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xia X, Zhang Z, Zhu C, Ni B, Wang S, Yang
S, Yu F, Zhao E, Li Q and Zhao G: Neutrophil extracellular traps
promote metastasis in gastric cancer patients with postoperative
abdominal infectious complications. Nat Commun. 13:10172022.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang ZY and Ge HY: Micrometastasis in
gastric cancer. Cancer Lett. 336:34–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cristescu R, Lee J, Nebozhyn M, Kim KM,
Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al: Molecular
analysis of gastric cancer identifies subtypes associated with
distinct clinical outcomes. Nat Med. 21:449–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv
L, Liu J, Xu Y, Shen Y and Yang M: Noncoding RNAs in gastric
cancer: Implications for drug resistance. Mol Cancer. 19:622020.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yuan L, Xu ZY, Ruan SM, Mo S, Qin JJ and
Cheng XD: Long non-coding RNAs towards precision medicine in
gastric cancer: Early diagnosis, treatment, and drug resistance.
Mol Cancer. 19:962020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Willems L, Tamburini J, Chapuis N, Lacombe
C, Mayeux P and Bouscary D: PI3K and mTOR signaling pathways in
cancer: New data on targeted therapies. Curr Oncol Rep. 14:129–138.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhou X, Li T, Xie H, Huang H, Yang K, Zeng
X and Peng T: HBV-induced N6 methyladenosine modification of PARP1
enhanced AFB1-related DNA damage and synergistically contribute to
HCC. Ecotoxicol Environ Saf. 298:1182542025. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li H, Prever L, Hirsch E and Gulluni F:
Targeting PI3K/AKT/mTOR signaling pathway in breast cancer. Cancers
(Basel). 13:35172021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fresno Vara JA, Casado E, de Castro J,
Cejas P, Belda-Iniesta C and González-Barón M: PI3K/Akt signalling
pathway and cancer. Cancer Treat Rev. 30:193–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhou X, Luo J, Xie H, Wei Z, Li T, Liu J,
Liao X, Zhu G and Peng T: MCM2 promotes the stemness and sorafenib
resistance of hepatocellular carcinoma cells via hippo signaling.
Cell Death Discov. 8:4182022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
57
|
Almhanna K, Strosberg J and Malafa M:
Targeting AKT protein kinase in gastric cancer. Anticancer Res.
31:4387–4392. 2011.PubMed/NCBI
|
|
58
|
Zhou XD, Chen HX, Guan RN, Lei YP, Shu X,
Zhu Y and Lv NH: Protein kinase B phosphorylation correlates with
vascular endothelial growth factor A and microvessel density in
gastric adenocarcinoma. J Int Med Res. 40:2124–2134. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shi J, Yao D, Liu W, Wang N, Lv H, Zhang
G, Ji M, Xu L, He N, Shi B and Hou P: Highly frequent PIK3CA
amplification is associated with poor prognosis in gastric cancer.
BMC Cancer. 12:502012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kobayashi I, Semba S, Matsuda Y, Kuroda Y
and Yokozaki H: Significance of Akt phosphorylation on tumor growth
and vascular endothelial growth factor expression in human gastric
carcinoma. Pathobiology. 73:8–17. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang Y, Kwok-Shing Ng P, Kucherlapati M,
Chen F, Liu Y, Tsang YH, de Velasco G, Jeong KJ, Akbani R,
Hadjipanayis A, et al: A Pan-cancer proteogenomic atlas of
PI3K/AKT/mTOR pathway alterations. Cancer Cell. 31:820–832.e3.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Janku F, Yap TA and Meric-Bernstam F:
Targeting the PI3K pathway in cancer: Are we making headway? Nat
Rev Clin Oncol. 15:273–291. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Okkenhaug K, Graupera M and Vanhaesebroeck
B: Targeting PI3K in cancer: Impact on tumor cells, their
protective stroma, angiogenesis, and immunotherapy. Cancer Discov.
6:1090–1105. 2016. View Article : Google Scholar : PubMed/NCBI
|