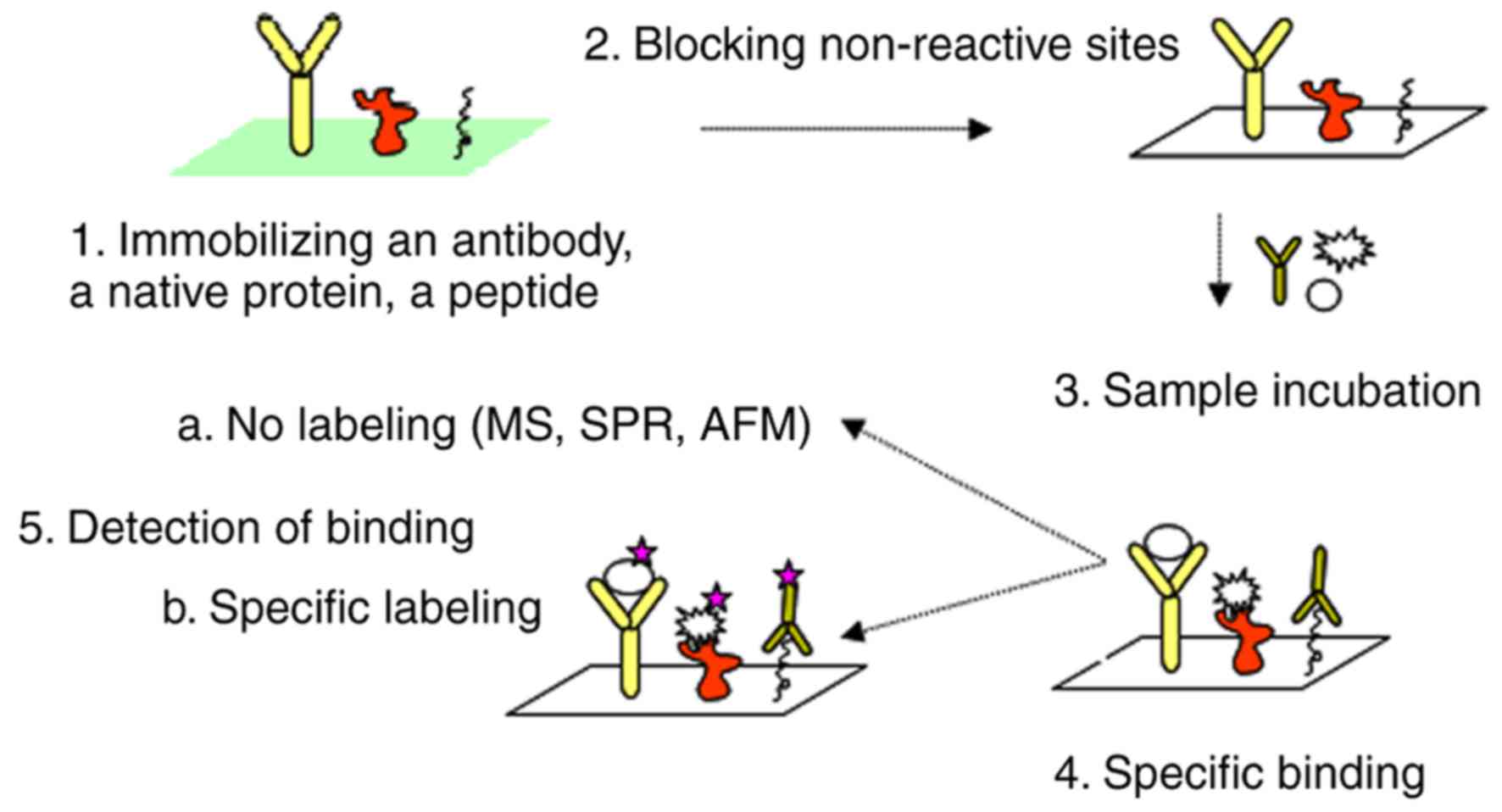
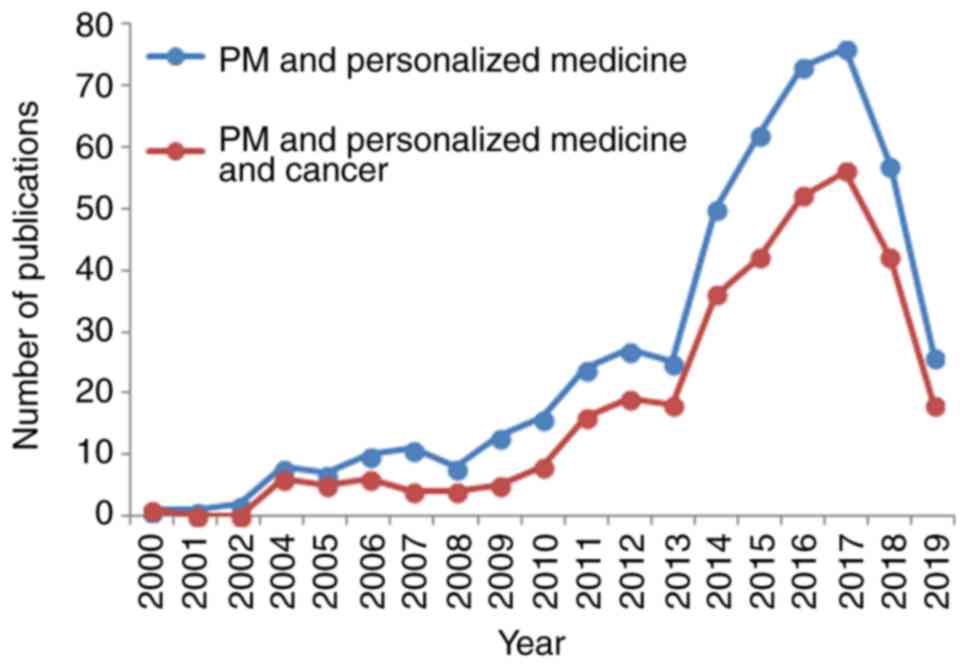
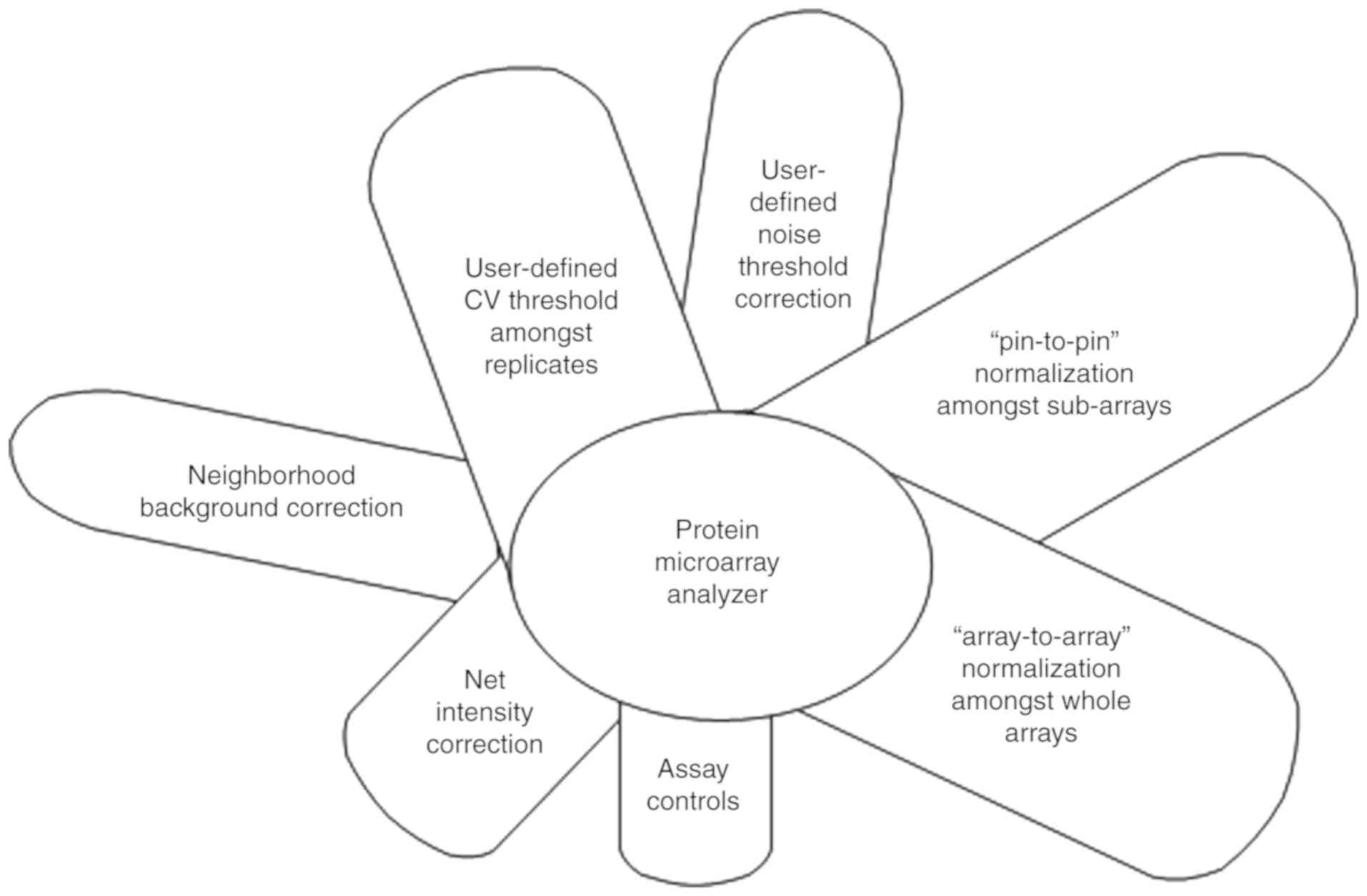
|
1
|
Schena M, Shalon D, Davis RW and Brown PO:
Quantitative monitoring of gene expression patterns with a
complementary DNA microarray. Science. 270:467–470. 1995.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gygi SP, Rochon Y, Franza BR and Aebersold
R: Correlation between protein and mRNA abundance in yeast. Mol
Cell Biol. 19:1720–1730. 1999.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Haab BB: Antibody arrays in cancer
research. Mol Cell Proteomics. 4:377–383. 2005.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chen CS and Zhu H: Protein microarrays.
Biotechniques. 40(423, 425, 427)2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kopf E and Zharhary D: Antibody arrays-An
emerging tool in cancer proteomics. Int J Biochem Cell Biol.
39:1305–1317. 2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Constantin C, Surcel M, Munteanu A and
Neagu M: Protein microarray technology for antibody detection
associated to human pathology. Roman Arch Microbiol Immunol.
77:236–244. 2018.
|
|
7
|
Poetz O, Schwenk JM, Kramer S, Stoll D,
Templin MF and Joos TO: Protein microarrays: Catching the proteome.
Mech Ageing Dev. 126:161–170. 2005.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Speer R, Wulfkuhle J, Espina V, Aurajo R,
Edmiston KH, Liotta LA and Petricoin EF III: Development of reverse
phase protein microarrays for clinical applications and
patient-tailored therapy. Cancer Genomics Proteomics. 4:157–164.
2007.PubMed/NCBI
|
|
9
|
Matei C, Tampa M, Ion RM, Georgescu SR,
Dumitrascu GR, Constantin C and Neagu M: Protein microarray for
complex apoptosis monitoring of dysplastic oral keratinocytes in
experimental photodynamic therapy. Biol Res. 47(33)2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Demyanenko SV, Uzdensky AB, Sharifulina
SA, Lapteva TO and Polyakova LP: PDT-induced epigenetic changes in
the mouse cerebral cortex: A protein microarray study. Biochim
Biophys Acta. 1840:262–270. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sutandy FX, Qian J, Chen CS and Zhu H:
Overview of protein microarrays. Curr Protoc Protein Sci. 27:Unit
27.1. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wilson JJ, Burgess R, Mao YQ, Luo S, Tang
H, Jones VS, Weisheng B, Huang RY, Chen X and Huang RP: Antibody
arrays in biomarker discovery. Adv Clin Chem. 69:255–324.
2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Díez P, Dasilva N, González-González M,
Matarraz S, Casado-Vela J, Orfao A and Fuentes M: Data analysis
strategies for protein microarrays. Microarrays (Basel). 1:64–83.
2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Angenendt P, Kreutzberger J, Glökler J and
Hoheisel JD: Generation of high density protein microarrays by
cell-free in situ expression of unpurified PCR products. Mol Cell
Proteomics. 5:1658–1666. 2006.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Schweitzer B, Predki P and Snyder M:
Microarrays to characterize protein interactions on a
whole-proteome scale. Proteomics. 3:2190–2199. 2003.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Alhamdani MS, Schröder C and Hoheisel JD:
Oncoproteomic profiling with antibody microarrays. Genome Med.
1(68)2009.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Wulfkuhle JD, Aquino JA, Calvert VS,
Fishman DA, Coukos G, Liotta LA and Petricoin EF III: Signal
pathway profiling of ovarian cancer from human tissue specimens
using reverse-phase protein microarrays. Proteomics. 3:2085–2090.
2003.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yuk CS, Lee HK, Kim HT, Choi YK, Lee BC,
Chun BH and Chung N: Development and evaluation of a protein
microarray chip for diagnosis of hepatitis C virus. Biotechnol
Lett. 26:1563–1568. 2004.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gowan SM, Hardcastle A, Hallsworth AE,
Valenti MR, Hunter LJ, de Haven Brandon AK, Garrett MD, Raynaud F,
Workman P, Aherne W and Eccles SA: Application of meso scale
technology for the measurement of phosphoproteins in human tumour
xenografts. Assay Drug Dev Technol. 5:391–401. 2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sigalotti L, Covre A, Fratta E, Parisi G,
Colizzi F, Rizzo A, Danielli R, Nicolay HJ, Coral S and Maio M:
Epigenetics of human cutaneous melanoma: Setting the stage for new
therapeutic strategies. J Transl Med. 8(56)2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Honda K, Ono M, Shitashige M, Masuda M,
Kamita M, Miura N and Yamada T: Proteomic approaches to the
discovery of cancer biomarkers for early detection and personalized
medicine. Jpn J Clin Oncol. 43:103–109. 2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yang L, Guo S, Li Y, Zhou S and Tao S:
Protein microarrays for systems biology. Acta Biochim Biophys Sin
(Shanghai). 43:161–71. 2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
LaBaer J and Ramachandran N: Protein
microarrays as tools for functional proteomics. Curr Opin Chem
Biol. 9:14–19. 2005.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hu S, Xie Z, Qian J, Blackshaw S and Zhu
H: Functional protein microarray technology. Wiley Interdiscip Rev
Syst Biol Med. 3:255–268. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
He M, Stoevesandt O and Taussig MJ: In
situ synthesis of protein arrays. Curr Opin Biotechnol. 19:4–9.
2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tao SC and Zhu H: Protein chip fabrication
by capture of nascent polypeptides. Nat Biotechnol. 24:1253–1254.
2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Miersch S and LaBaer J: Nucleic acid
programmable protein arrays: Versatile tools for array-based
functional protein studies. Curr Protoc Protein Sci Chapter.
27:Unit27.2. 2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wright C, Sibani S, Trudgian D, Fischer R,
Kessler B, LaBaer J and Bowness P: Detection of multiple
autoantibodies in patients with ankylosing spondylitis using
nucleic acid programmable protein arrays. Mol Cell Proteomics.
11:M9 00384. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Neagu M and Constantin C: Immunogenicity
of Stem Cell in Tumorigenesis Versus Regeneration. In: Stem Cells
between Regeneration and Tumorigenesis. Tanase C and Neagu M (eds).
Bentham Pbl. House. pp202–234. 2016. View Article : Google Scholar
|
|
30
|
Chiodoni C, Di Martino MT, Zazzeroni F,
Caraglia M, Donadelli M, Meschini S, Leonetti C and Scotlandi K:
Cell communication and signalling: How to turn bad language into
positive one. J Exp Clin Cancer Res. 38(128)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
James P: Protein identification in the
post-genome era: The rapid rise of proteomics. Q Rev Biophys.
30:279–331. 1997.PubMed/NCBI
|
|
32
|
Joos T and Bachmann J: Protein
microarrays: Potentials and limitations. Front Biosci (Landmark
Ed). 14:4376–4385. 2009.PubMed/NCBI
|
|
33
|
Ramachandran N, Raphael JV, Hainsworth E,
Demirkan G, Fuentes MG, Rolfs A, Hu Y and LaBaer J: Next-generation
high-density self-assembling functional protein arrays. Nat
Methods. 5:535–538. 2008.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Manzano-Romána R and Fuentes M: A decade
of nucleic acid programmable protein arrays (NAPPA) availability:
News, actors, progress, prospects and access. J Proteomics.
198:27–35. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Rivera R, Wang J, Yu X, Demirkan G, Hopper
M, Bian X, Tahsin T, Magee DM, Qiu J, LaBaer J and Wallstrom G:
Automatic identification and quantification of extra-well
fluorescence in microarray images. J Proteome Res. 16:3969–3977.
2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yu X, Petritis B, Duan H, Xu D and LaBaer
J: Advances in cell-free protein array methods. Expert Rev
Proteomics. 15:1–11. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yu X, Song L, Petritis B, Bian X, Wang H,
Viloria J, Park J, Bui H, Li H, Wang J, et al: Multiplexed nucleic
acid programmable protein arrays. Theranostics. 7:4057–4070.
2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Bulman A, Neagu M and Constantin C:
Immunomics in skin cancer-improvement in diagnosis, prognosis and
therapy monitoring. Curr Proteomics. 10:202–217. 2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Neagu M and Constantin C: New Insights in
Cutaneous Melanoma Immune-Therapy-Tackling Immune-Suppression and
Specific Anti-Tumoral Response. In: Melanoma. Murph M (ed).
IntechOpen. pp225–246. 2015. View
Article : Google Scholar
|
|
40
|
Butterfield LH, Ribas A, Dissette VB,
Amarnani SN, Vu HT, Oseguera D, Wang HJ, Elashoff RM, McBride WH,
Mukherji B, et al: Determinant spreading associated with clinical
response in dendritic cell-based immunotherapy for malignant
melanoma. Clin Cancer Res. 9:998–1008. 2003.PubMed/NCBI
|
|
41
|
Yuan J, Wang E and Fox BA: Immune
monitoring technology primer: Protein microarray (‘seromics’). J
Immunother Cancer. 4(2)2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chen YT, Scanlan MJ, Sahin U, Türeci O,
Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M and Old
LJ: A testicular antigen aberrantly expressed in human cancers
detected by autologous antibody screening. Proc Natl Acad Sci USA.
94:1914–1918. 1997.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Fulton KM and Twine SM: Immunoproteomics:
Current technology and applications. Methods Mol Biol. 1061:21–57.
2013.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Sahin U, Tureci O, Schmitt H, Cochlovius
B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I and
Pfreundschuh M: Human neoplasms elicit multiple specific immune
responses in the autologous host. Proc Natl Acad Sci USA.
92:11810–11813. 1995.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Ladd JJ, Chao T, Johnson MM, Qiu J, Chin
A, Israel R, Pitteri SJ, Mao J, Wu M, Amon LM, et al: Autoantibody
signatures involving glycolysis and splicesome proteins precede a
diagnosis of breast cancer among postmenopausal women. Cancer Res.
73:1502–1513. 2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Madoz-Gurpide J, Kuick R, Wang H, Misek DE
and Hanash SM: Integral protein microarrays for the identification
of lung cancer antigens in sera that induce a humoral immune
response. Mol Cell Proteomics. 7:268–281. 2008.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Bouwman K, Qiu J, Zhou H, Schotanus M,
Mangold LA, Vogt R, Erlandson E, Trenkle J, Partin AW, Misek D, et
al: Microarrays of tumour cell derived proteins uncover a distinct
pattern of prostate cancer serum immunoreactivity. Proteomics.
3:2200–2207. 2003. View Article : Google Scholar
|
|
48
|
GuhaThakurta D, Sheikh NA, Fan LQ, Kandadi
H, Meagher T, Hall SJ, Kantoff PW, Higano CS, Small EJ, Gardner TA,
et al: Humoral immune response against nontargeted tumour antigens
after treatment with sipuleucel-T and its association with improved
clinical outcome. Clin Cancer Res. 21:3619–3630. 2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Kwek SS, Dao V, Roy R, Hou Y, Alajajian D,
Simko JP, Small EJ and Fong L: Diversity of antigen-specific
responses induced in vivo with CTLA-4 blockade in prostate cancer
patients. J Immunol. 189:3759–3766. 2012.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Graff JN, Puri S, Bifulco CB, Fox BA and
Beer TM: Sustained complete response to CTLA-4 blockade in a
patient with metastatic, castration-resistant prostate cancer.
Cancer Immunol Res. 2:399–403. 2014.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Gnjatic S, Ritter E, Büchler MW, Giese NA,
Brors B, Frei C, Murray A, Halama N, Zörnig I, Chen YT, et al:
Seromic profiling of ovarian and pancreatic cancer. Proc Natl Acad
Sci USA. 107:5088–5093. 2010.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Abel L, Kutschki S, Turewicz M, Eisenacher
M, Stoutjesdijk J, Meyer HE, Woitalla D and May C: Autoimmune
profiling with protein microarrays in clinical applications. Biomed
Biochim Acta. 1844:977–987. 2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Turewicz M, May C, Ahrens M, Woitalla D,
Gold R, Casjens S, Pesch B, BrüningT Meyer HE, Nordhoff E, et al:
Improving the default data analysis workflow for large autoimmune
biomarker discovery studies with ProtoArrays. Proteomics.
13:2083–2087. 2013.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Zhang DY, Ye F, Gao L, Liu X, Zhao X, Che
Y, Wang H, Wang L, Wu J, Song D, et al: Proteomics, pathway array
and signaling network-based medicine in cancer. Cell Div.
4(20)2009.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Schumacher S, Muekusch S and Seitz H:
Up-to-date applications of microarrays and their way to
commercialization. Microarrays (Basel). 4:196–213. 2015.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Ayoglu B, Schwenk JM and Nilsson P:
Antigen arrays for profiling autoantibody repertoires. Bioanalysis.
8:1105–1126. 2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Stoevesandt O, Taussig MJ and He M:
Protein microarrays: High-throughput tools for proteomics. Expert
Rev Proteomics. 6:145–157. 2009.PubMed/NCBI View Article : Google Scholar
|
|
58
|
No authors listed. The call of the human
proteome. Nat Methods. 7(661)2010.PubMed/NCBI
|
|
59
|
Legrain P, Aebersold R, Archakov A,
Bairoch A, Bala K, Beretta L, Bergeron J, Borchers CH, Corthals GL,
Costello CE, et al: The human proteome project: Current state and
future direction. Mol Cell Proteomics. 10:M111.009993.
2011.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Jantsch MF, Quattrone A, O'Connell M, Helm
M, Frye M, Macias-Gonzales M, Ohman M, Ameres S, Willems L, Fuks F,
et al: Positioning Europe for the EPITRANSCRIPTOMICS challenge. RNA
Biol. 15:829–831. 2018.PubMed/NCBI View Article : Google Scholar
|
|
61
|
González-Gomariz J, Guruceaga E,
López-Sánchez M and Segura V: Proteogenomics in the context of the
human proteome project (HPP). Expert Rev Proteomics. 16:267–275.
2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Tanase C, Albulescu R and Neagu M: Updates
in immune-based multiplex assays. J Immunoassay Immunochem. 40:1–2.
2019.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Popa ML, Albulescu R, Neagu M, Eugen
Hinescu ME and Tanase C: Multiplex assay for multiomics advances in
personalized-precision medicine. J Immunoassay Immunochem. 40:3–25.
2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Cronin KA, Lake AJ, Scott S, Sherman RL,
Noone AM, Howlader N, Henley SJ, Anderson RN, Firth AU, Ma J, et
al: Annual report to the nation on the status of cancer, part I:
National cancer statistics. Cancer. 124:2785–2800. 2018.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Smith ER: A two-tiered health care system:
Is there anything new? Can J Cardiol. 23:915–916. 2007.(In English;
French). PubMed/NCBI
|
|
66
|
Alyass A, Turcotte M and Meyre D: From big
data analysis to personalized medicine for all: Challenges and
opportunities. BMC Med Genomics. 8(33)2015. View Article : Google Scholar
|
|
67
|
Fröhlich H, Balling R, Beerenwinkel N,
Kohlbacher O, Kumar S, Lengauer T, Maathuis MH, Moreau Y, Murphy
SA, Przytycka TM, et al: From hype to reality: Data science
enabling personalized medicine. BMC Med. 16(150)2018.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Wong SH: Pharmacogenomics and personalized
medicine. In: Handbook of drug monitoring methods: Therapeutics and
drugs of abuse. Dasgupta A (ed). Humana Press, New York, NY.
pp211–223. 2007.
|
|
69
|
Yu X, Schneiderhan-Marra N and Joos TO:
Protein microarrays for personalized medicine. Clin Chem.
56:376–387. 2010.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Masuda M and Yamada T: Signalling pathway
profiling by reverse-phase protein array for personalized cancer
medicine. Biochim Biophys Acta. 1854:651–657. 2015.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Rosa M: Advances in the molecular analysis
of breast cancer: Pathway toward personalized medicine. Cancer
Control. 22:211–219. 2015.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Kim DC, Wang X, Yang CR and Gao JX: A
framework for personalized medicine: Prediction of drug sensitivity
in cancer by proteomic profiling. Proteome Sci. 10 (Suppl
1)(S13)2012.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Wistuba II, Gelovani JG, Jacoby JJ, Davis
SE and Herbst RS: Methodological and practical challenges for
personalized cancer therapies. Nat Rev Clin Oncol. 8:135–141.
2011.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Mueller C, Liotta L and Espina V: Reverse
phase protein microarrays advance to use in clinical trials. Mol
Oncol. 4:461–481. 2010.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Kornblau SM, Tibes R, Qiu YH, Chen W,
Kantarjian HM, Andreeff M, Coombes KR and Mills GB: Functional
proteomic profiling of AML predicts response and survival. Blood.
113:154–164. 2009.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Cain JW, Hauptschein RS, Stewart JK, Bagci
T, Sahagian GG and Jay DG: Identification of CD44 as a surface
biomarker for drug resistance by surface proteome signature
technology. Mol Cancer Res. 9:637–647. 2011.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Liotta LA, Espina V, Mehta AI, Calvert V,
Rosenblatt K, Geho D, Munson PJ, Young L, Wulfkuhle J and Petricoin
EF III: Protein microarrays: Meeting analytical challenges for
clinical applications. Cancer Cell. 3:317–325. 2003.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Spurrier B, Ramalingam S and Nishizuka S:
Reverse-phase protein lysate microarrays for cell signaling
analysis. Nat Protoc. 3:1796–1808. 2008.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Kim YB, Yang CR and Gao J: Functional
proteomic pattern identification under low dose ionizing radiation.
Artif Intell Med. 49:177–185. 2010.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Wang X, Dong Y, Jiwani AJ, Zou Y, Pastor
J, Kuro OM, Habib AA, Ruan M, Boothman DA and Yang CR: Improved
protein arrays for quantitative systems analysis of the dynamics of
signaling pathway interactions. Proteome Sci. 9(53)2011.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Laforte V, Lo PS, Li H and Juncker D:
Antibody colocalization microarray for cross-reactivity-free
multiplexed protein analysis. Methods Mol Biol. 1619:239–261.
2017.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Lim MS, Carlson ML, Crockett DK, Fillmore
GC, Abbott DR, Elenitoba-Johnson OF, Tripp SR, Rassidakis GZ,
Medeiros LJ, Szankasi P and Elenitoba-Johnson KS: The proteomic
signature of NPM/ALK reveals deregulation of multiple cellular
pathways. Blood. 114:1585–1595. 2009.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Gnjatic S, Wheeler C, Ebner M, Ritter E,
Murray A, Altorki NK, Ferrara CA, Hepburne-Scott H, Joyce S,
Koopman J, et al: Seromic analysis of antibody responses in
non-small cell lung cancer patients and healthy donors using
conformational protein arrays. J Immunol Methods. 341:50–58.
2009.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Gaudreau PO, Peng D, Rodriguez BL,
Fradette J, Gibson L, Kundu S, Chen L, et al: Co-clinical trials of
MEK inhibitor, anti PD-L1 and anti CTLA-4 combination treatment in
non-small cell lung cancer. J ImmunoTherapy Cancer. 6 (Suppl
1)(S114)2018.
|
|
85
|
Vilgelm AE, Johnson DB and Richmond A:
Combinatorial approach to cancer immunotherapy: Strength in
numbers. J Leukoc Biol. 100:275–290. 2016.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Terracciano R, Pelaia G, Preianò M and
Savino R: Asthma and COPD proteomics: Current approaches and future
directions. Proteomics Clin Appl. 9:203–220. 2015. View Article : Google Scholar
|
|
87
|
Vizcaino JA, Csordas A, Del-Toro N, Dianes
JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F,
Ternent T, et al: 2016 update of the PRIDE database and its related
tools. Nucleic Acids Res. 44(11033)2016.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Hamsten C, Häggmark A, Grundström J, Mikus
M, Lindskog C, Konradsen JR, Eklund A, Pershagen G, Wickman M,
Grunewald J, et al: Protein profiles of CCL5, HPGDS, and NPSR1 in
plasma reveal association with childhood asthma. Allergy.
71:1357–1361. 2016.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Gharib SA, Nguyen EV, Lai Y, Plampin JD,
Goodlett DR and Hallstrand TS: Induced sputum proteome in healthy
subjects and asthmatic patients. J Allergy Clin Immunol.
128:1176–1184, e1176. 2011.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Ohlmeier S, Nieminen P, Gao J, Kanerva T,
Rönty M, Toljamo T, Bergmann U, Mazur W and Pulkkinen V: Lung
tissue proteomics identifies elevated transglutaminase 2 levels in
stable chronic obstructive pulmonary disease. Am J Physiol Lung
Cell Mol Physiol. 310:L1155–L1165. 2016.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Barnes PJ: Corticosteroid resistance in
patients with asthma and chronic obstructive pulmonary disease. J
Allergy Clin Immunol. 131:636–645. 2013.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Levitt JE and Rogers AJ: Proteomic study
of acute respiratory distress syndrome: Current knowledge and
implications for drug development. Expert Rev Proteomics.
13:457–469. 2016.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Priyadharshini VS and Teran LM:
Personalized medicine in respiratory disease: Role of proteomics.
Adv Protein Chem Struct Biol. 102:115–146. 2016.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Foster MW, Morrison LD, Todd JL, Snyder
LD, Thompson JW, Soderblom EJ, Plonk K, Weinhold KJ, Townsend R,
Minnich A and Moseley MA: Quantitative proteomics of
bronchoalveolar lavage fluid in idiopathic pulmonary fibrosis. J
Proteome Res. 14:1238–1249. 2015.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Lavoie JR, Ormiston ML, Perez-Iratxeta C,
Courtman DW, Jiang B, Ferrer E, Caruso P, Southwood M, Foster WS,
Morrell NW and Stewart DJ: Proteomic analysis implicates
translationally controlled tumour protein as a novel mediator of
occlusive vascular remodeling in pulmonary arterial hypertension.
Circulation. 129:2125–2135. 2014.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Abdul-Salam VB, Wharton J, Cupitt J,
Berryman M, Edwards RJ and Wilkins MR: Proteomic analysis of lung
tissues from patients with pulmonary arterial hypertension.
Circulation. 122:2058–2067. 2010.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Kan M, Shumyatcher M and Himes BE: Using
omics approaches to understand pulmonary diseases. Respir Res.
18(149)2017.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Cohen AS, Khalil FK, Welsh EA, Schabath
MB, Enkemann SA, Davis A, Zhou JM, Boulware DC, Kim J, Haura EB and
Morse DL: Cell-surface marker discovery for lung cancer.
Oncotarget. 8:113373–113402. 2017.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Monette A, Bergeron D, Ben Amor A, Meunier
L, Caron C, Mes-Masson AM, Kchir N, Hamzaoui K, Jurisica I and
Lapointe R: Immune-enrichment of non-small cell lung cancer
baseline biopsies for multiplex profiling define prognostic immune
checkpoint combinations for patient stratification. J Immunother
Cancer. 7(86)2019.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Schwill M, Tamaskovic R, Gajadhar AS, Kast
F, White FM and Plückthun A: Systemic analysis of tyrosine kinase
signaling reveals a common adaptive response program in a
HER2-positive breast cancer. Sci Signal. 12:eaau2875.
2019.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Hsu PY, Wu VS, Kanaya N, Petrossian K, Hsu
HK, Nguyen D, Schmolze D, Kai M, Liu CY, Lu H, et al: Dual mTOR
kinase inhibitor MLN0128 sensitizes HR+/HER2+
breast cancer patient-derived xenografts to trastuzumab or
fulvestrant. Clin Cancer Res. 24:395–406. 2018.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Lee J, Geiss GK, Demirkan G, Vellano CP,
Filanoski B, Lu Y, Ju Z, Yu S, Guo H, Bogatzki LY, et al:
Implementation of a multiplex and quantitative proteomics platform
for assessing protein lysates using DNA-barcoded antibodies. Mol
Cell Proteomics. 17:1245–1258. 2018.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Kars MD and Yıldırım G: Determination of
the target proteins in chemotherapy resistant breast cancer stem
cell-like cells by protein array. Eur J Pharmacol. 848:23–29.
2019.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Li M, Li H, Liu F, Bi R, Tu X, Chen L, Ye
S and Cheng X: Characterization of ovarian clear cell carcinoma
using target drug-based molecular biomarkers: Implications for
personalized cancer therapy. J Ovarian Res. 10(9)2017.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Mabuchi S, Sugiyama T and Kimura T: Clear
cell carcinoma of the ovary: Molecular insights and future
therapeutic perspectives. J Gynecol Oncol. 27(e31)2016.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Yanaihara N, Hirata Y, Yamaguchi N,
Noguchi Y, Saito M, Nagata C, Takakura S, Yamada K and Okamoto A:
Antitumor effects of interleukin-6 (IL-6)/interleukin-6 receptor
(IL-6R) signalling pathway inhibition in clear cell carcinoma of
the ovary. Mol Carcinog. 55:832–841. 2016.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Yamashita Y: Ovarian cancer: New
developments in clear cell carcinoma and hopes for targeted
therapy. Jpn J Clin Oncol. 45:405–407. 2015.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Sereni MI, Baldelli E, Gambara G, Deng J,
Zanotti L, Bandiera E, Bignotti E, Ragnoli M, Tognon G, Ravaggi A,
et al: Functional characterization of epithelial ovarian cancer
histotypes by drug target based protein signaling activation
mapping: Implications for personalized cancer therapy. Proteomics.
15:365–373. 2015.PubMed/NCBI View Article : Google Scholar
|
|
109
|
di Martino S, De Luca G, Grassi L,
Federici G, Alfonsi R, Signore M, Addario A, De Salvo L,
Francescangeli F, Sanchez M, et al: Renal cancer: New models and
approach for personalizing therapy. J Exp Clin Cancer Res.
37(217)2018.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Hoff FW, Hu CW, Qiu Y, Ligeralde A, Yoo
SY, Mahmud H, de Bont ESJM, Qutub AA, Horton TM and Kornblau SM:
Recognition of recurrent protein expression patterns in pediatric
acute myeloid leukemia identified new therapeutic targets. Mol
Cancer Res. 16:1275–1286. 2018.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Ragon BK, Odenike O, Baer MR, Stock W,
Borthakur G, Patel K, Han L, Chen H, Ma H, Joseph L, et al: Oral
MEK 1/2 inhibitor trametinib in combination with AKT inhibitor
GSK2141795 in patients with acute myeloid leukemia with RAS
mutations: A phase II study. Clin Lymphoma Myeloma Leuk.
S2152-2650:31542–31548. 2019.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Yang Q, Bavi P, Wang JY and Roehrl MH:
Immuno-proteomic discovery of tumour tissue autoantigens identifies
olfactomedin 4, CD11b, and integrin alpha-2 as markers of
colorectal cancer with liver metastases. J Proteomics. 168:53–65.
2017.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Hagman H, Bendahl PO, Lidfeldt J, Belting
M and Johnsson A: Protein array profiling of circulating
angiogenesis-related factors during bevacizumab containing
treatment in metastatic colorectal cancer. PLoS One.
13(e0209838)2018.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Liu P, Souma T, Wei AZ, Xie X, Luo X and
Jin J: Personalized peptide arrays for detection of HLA
alloantibodies in organ transplantation. J Vis Exp. 2017.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Soni A, Gowthamarajan K and Radhakrishnan
A: Personalized medicine and customized drug delivery systems: The
new trend of drug delivery and disease management. Int J Pharm
Compd. 22:108–121. 2018.PubMed/NCBI
|
|
116
|
Syafrizayanti Lueong SS, Di C, Schaefer
JV, Plückthun A and Hoheisel JD: Personalised proteome analysis by
means of protein microarrays made from individual patient samples.
Sci Rep. 7(39756)2017.PubMed/NCBI View Article : Google Scholar
|