1. Introduction
The current COVID-19 pandemic caused by severe acute
respiratory syndrome coronavirus-2 (SARS-CoV-2) has markedly
affected the world population (1).
Epidemiology has encountered several pathogenic viral outbreaks in
the past (2), and of these, the
1918 ‘Spanish Flu’ or ‘La Gripe Espanola’ pandemic had exerted the
worst toll on human health and the global economy (3,4).
Viruses are complex ‘biocapsules’ with genomic material (DNA or
RNA), wrapped in a protein coat. They are inherently obligate
intracellular parasites, and must invade live cells and hijack host
machinery to complete their life cycle. According to the
International Committee on Taxonomy of Viruses (ICTV), all known
viruses are classified under 3 orders, 56 families, 9 subfamilies
and 233 genera of >1,550 species (5). Current knowledge is limited when it
comes to their natural ‘reservoirs’ among farm animals, pets,
poultry and wildlife, and subsequent zoonosis to humans. Recent
examples of pathogenic virus outbreaks, including influenza virus,
Hendra virus, Ebola virus, Nipah virus, Zika virus, hantavirus and
coronavirus, have been linked to zoonosis (3).
The newly identified SARS-CoV-2 originated in
December, 2019 in Wuhan, China (6). It is the third highly pathogenic
human coronavirus (HCoV) after the 2002-3 SARS-CoV (renamed,
SARS-CoV-1) and the 2012-13 Middle-East respiratory syndrome CoV
(MERS-CoV) outbreaks (7). Similar
to MERS-CoV infection, SARS-CoV-2 has an incubation period of 2-14
days with symptoms of fever, cough and breathlessness, which may
manifest from mild pneumonia to severe illness and even subsequent
death (1,7,8). The
present 2019-20 SARS-CoV-2 pandemic closely mirrors the events in
‘Contagion’, a 2011 Hollywood movie about a deadly virus outbreak
in the USA, which originated in Hong Kong, and resulted in social
disorder and chaos in healthcare system until a vaccine was
introduced. Currently, several repurposed drugs, such as
hydroxychloroquine, remdesivir, azithromycin and dexamethasone, as
well as some vaccine candidates, are undergoing phase I/II clinical
trials. The present review article presents a sincere effort
towards updating the current understanding of the emergence,
pandemic status, pathogenesis and containment measures of
COVID-19.
2. Infection and epidemiology
The impact of a pandemic depends upon the number of
individuals infected, and countries affected by its
transmissibility, severity and spectrum of clinical manifestations.
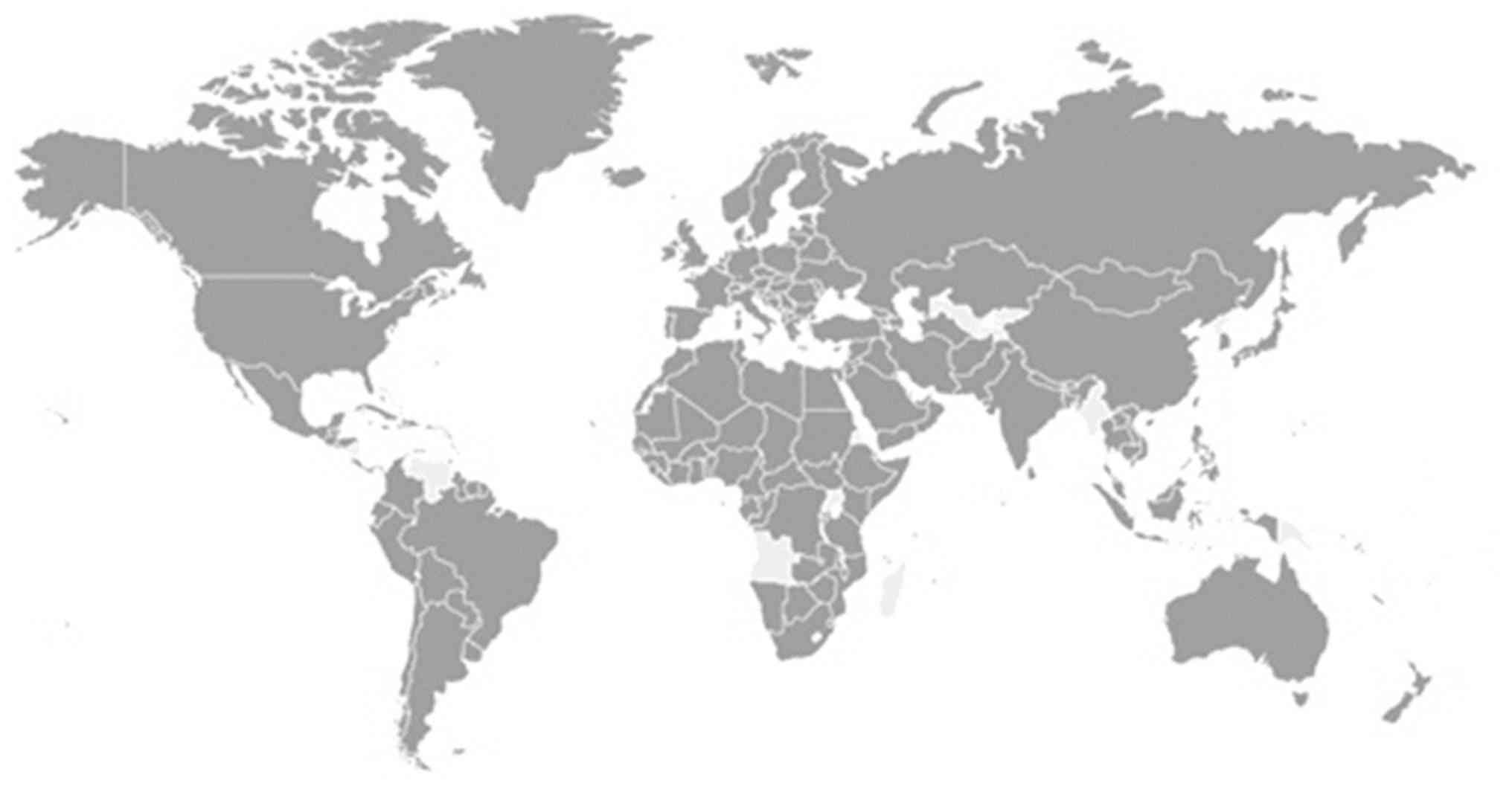
To date, SARS-CoV-2 has affected >200 countries and territories
(Fig. 1). As of June 20, 2020, the
global confirmed cases of COVID-19 have spiked to >8.6 million,
including 460,080 deaths. This includes 285,648 from Africa with
most cases noted in South Africa (87,715); 1,788,752 from Asia with
most cases in India (395,048), Iran (200,262) and China (80,012);
4,282,308 from the Americas, with most cases in the USA (1,133,069)
and Brazil (1,032 913); 2,268,266 cases from Europe, with most
cases in Russia (569,063), UK (301, 815), Spain (245,575) and Italy
(238,011); and 8,905 cases from Oceania, with most cases in
Australia (7,409) (9).
After six months of the first emergence of
SARS-CoV-2 in December, 2019 and its rapid global spread in the
northern and southern hemispheres, seasonal variation has not
significantly affected its transmissibility. SARS-CoV-2 has a high
basic reproductive number (R0), ranging between 2.0 to 2.5 days
(9). The R0 measures the average
number of infections that can result from one infected individual
in a susceptible population (10).
R0 has been, however, estimated with varying results and
interpretations. The current estimate of the mortality rate for
COVID-19 is 3.4%, which is significantly higher than that of
seasonal flu (0.02%), but lower than that of SARS-COV-1 (9.6%) and
MERS-CoV (34%). Affected individuals, mostly males in the elderly
population or those with underlying medical conditions, such as
hypertension, diabetes, or chronic respiratory, renal and hepatic
issues, have exhibited a higher mortality rate (11).
3. Virus biology
SARS-CoV-2 is classified together with SARS-CoV-1,
MERS-CoV, HCoV-OC43 and HCoV-HKU1 within the genus
Betacoronavirus of the Coronaviridae family, and has
a positive sense, single-strand RNA (~29.9 kb) genome (11,12).
The viral RNA is 5'capped and consists of 13 active open reading
frames (ORFs) that encode a total of 27 proteins, i.e., 16
non-structural, 4 structural and 7 accessory proteins (13). However, in a recent sequence
analysis of Betacoronavirus, SARS-CoV-2 has shown that
nonsense mutations introduced 6-stop codons in the accessory coding
region, which could fail to translate ‘3b’ (Padhan and Parvez,
unpublished data). This observation has consolidated the number of
accessory proteins to 6 instead of 7, and the total proteins to 26
in SARS-CoV-2.
The first SARS-CoV-2 RNA sequence (GenBank accession
no. MN908947) was reported using metagenomic sequencing
technologies (14). Phylogenetic
analysis of the viral genome has indicated its close similarity
(~96% identity) with two bat-SARS-like coronaviruses (SL-CoV) viz.,
bat-SL-CoVZC45 and bat-SL-CoVZXC21, but its distinction from
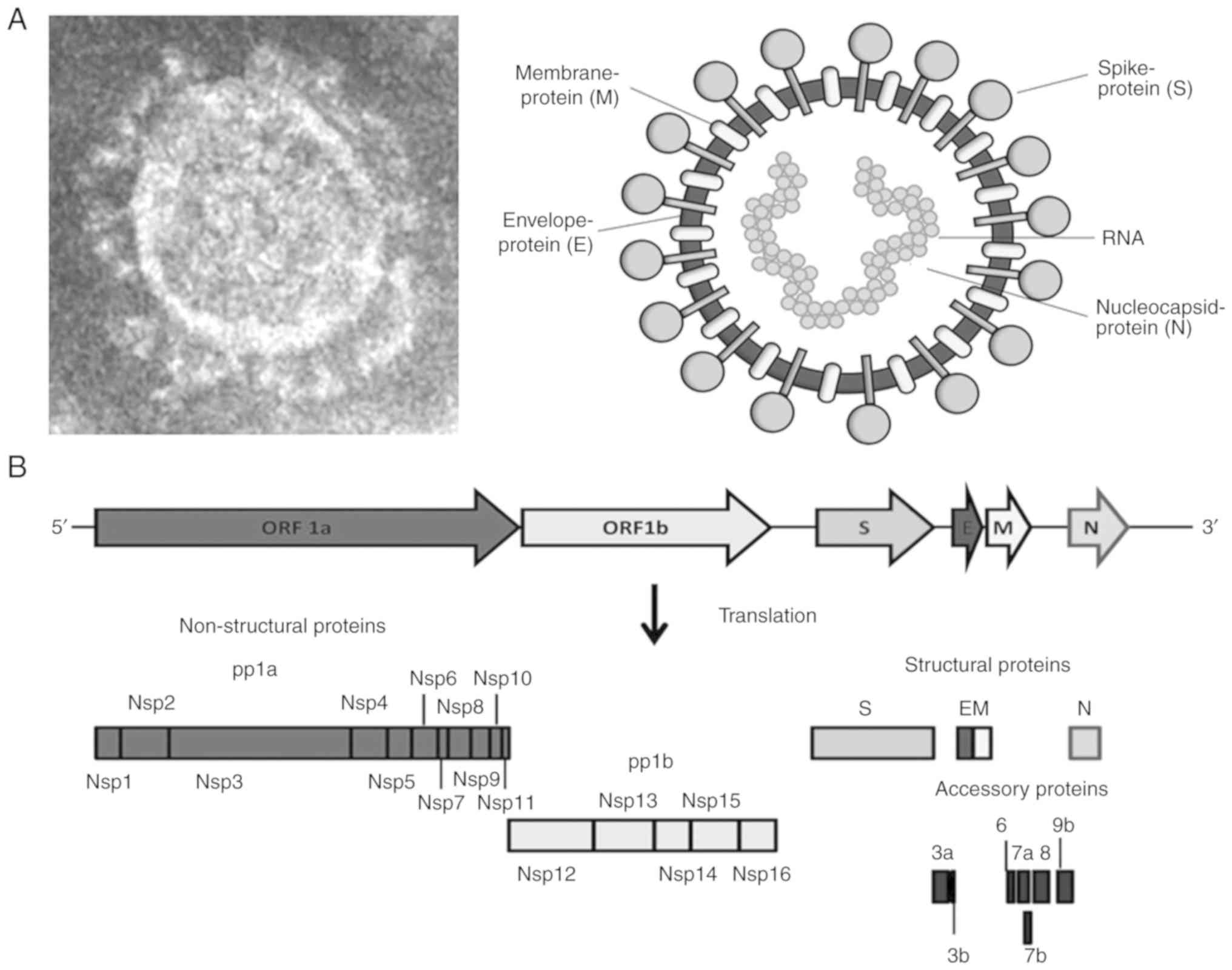
SARS-CoV-1 (~79% similarity) and MERS-CoV (15). SARS-CoV-2 has 4 structural
proteins: Crown-like spike (S), envelope (E), membrane (M) and
nucleocapsid (N) (Fig. 2A). The
‘S’ protein is a type I transmembrane glycoprotein that shares ~76%
sequence identity with that of SARS-CoV-1 and ~80% identity with
bat-SL-CoV (16-18).
‘S’ has two structural subunits (S1 and S2), of which the ‘S1’
subunit contains the human cell-receptor angiotensin-converting
enzyme-2 (ACE2) receptor-binding domain (RBD). The ‘S2’ subunit
contains the structural elements required for membrane fusion.
While the amino acid sequence of the ‘S1’ subunit is highly
variable (~70% sequence identity with bat-SL-CoV and SARS-CoV-1),
‘S2’ sequence is highly conserved and shares ~99% identity with
both bat-SL-CoV and SARS-CoV-1 (17,19).
Notably, of 6 six amino acid residues of RBD, 5 differ between
SARS-CoV-2 and SARS-CoV-1, suggesting the strong binding of the
SARS-CoV-2 spikes with ACE2 receptor and high infectivity (20,21).
The ‘M’ is a trans-membrane glycoprotein, crucial for virion's
fusion with the host cell membrane, whereas the ‘E’ protein is
necessary for the assembly and morphogenesis of nascent virions
(22). In the case of SARS-CoV-1
infection, the ‘N’ protein is highly antigenic, which triggers the
production of SARS-CoV antibodies in approximately 89% of infected
patients and is used as a serological marker (23).
The SARS-CoV-2 non-structural replicase proteins,
pp1a (nsp1-nsp11) and pp1b (nsp12-nsp16), are involved in viral RNA
transcription and replication (Fig.
2B), as well as in modulating host-innate immunity. SARS-CoV-2
varies in its conserved aggregation motif of the ‘3a’ accessory
protein with SARS-CoV-1, as well as civet-SL-CoV (paguma-SARS-CoV)
and bat-SL-CoV (YNLF_31C and NLF_34C) (24).
4. Clinical presentation and
immuno-pathobiology
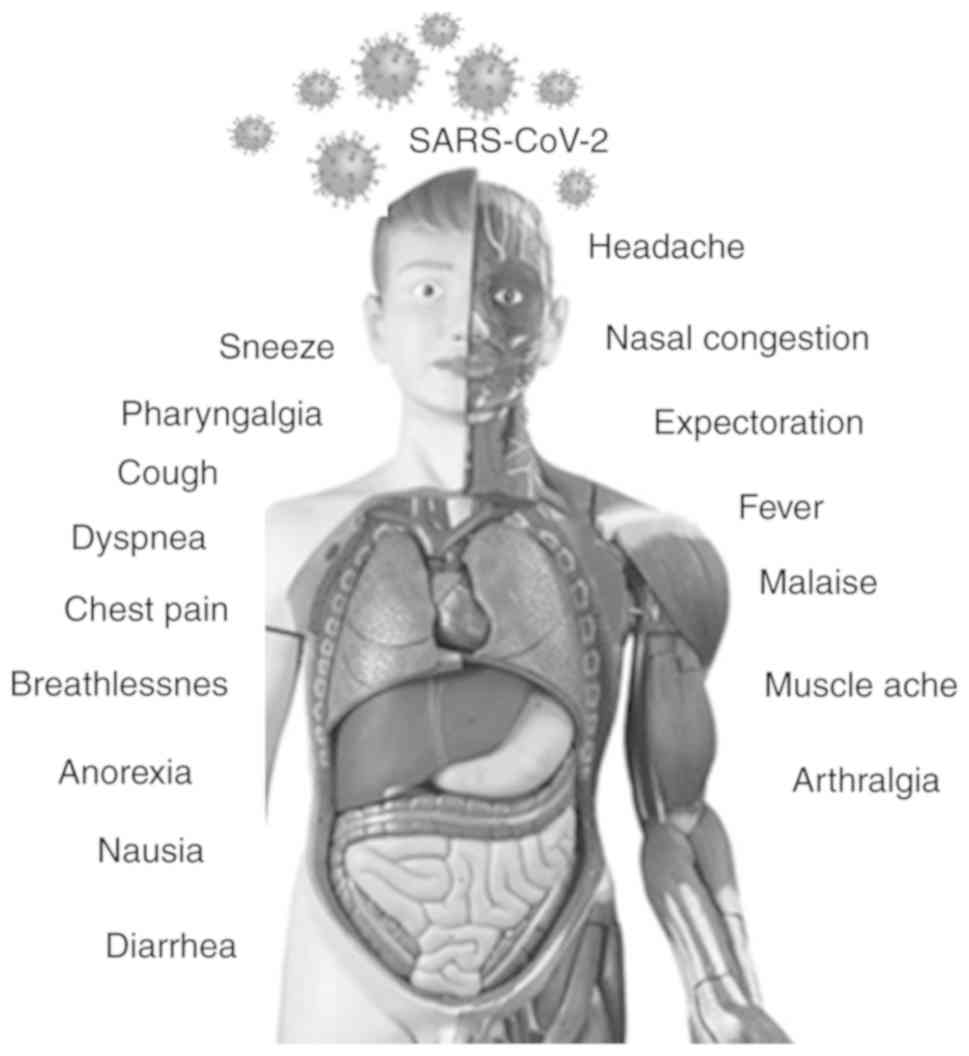
The incubation period for SARS-CoV-2 varies from
2-14 days with a mean incubation period of 6.4 days. This is higher
than that of seasonal flu (2 days), swine flu (1-4 days), MERS-CoV
(2-14 days) and SARS-CoV-1 (2-7 days) (25). Almost 80% of infections with
SARS-CoV-2 remain asymptomatic or exhibit very mild flu-like
symptoms and can recover at home. However, the severe cases (15%)
exhibit high fever, pneumonia and breathlessness, thus requiring
hospitalization. In addition, 5% of cases develop respiratory
failure, septic shock and multi-organ failure (Fig. 3). Patients infected with COVID-19
with severe pneumonia present ground-glass opacity and lung
consolidation features. In >70% of COVID-19 suspected cases, the
disease was apparent in all the 5 lobes of the lung (26). In addition to the ground-glass
opacity, lung consolidation has also been reported with peripheral
(41%), lower zone (50%) and bilateral involvements (50%) (27).
Human betacoronaviruses exhibit high
species-specificity; however, subtle genetic changes can
significantly alter their tissue-tropism, host-barrier and
pathogenicity, as observed with SARS-CoV-1 and MERS-CoV. The
bat-SL-CoVs are known to use their ‘S’ protein (RBD) to bind to
civet and horseshoe bat ACE2 receptors (28). Similarly, the SARS-CoV-2 ‘S’
protein also binds to the ACE2 of the airway epithelium and
alveolar type-2 pneumocytes, pulmonary cells that synthesize
pulmonary surfactant (29), where
the ‘M’ fusion protein facilitates cell entry (21).
When SARS-CoV-2-contaminated droplets are inhaled,
the virus first becomes attached to the inner linings of the throat
and larynx, and remains there for few days. Though generally mild,
SARS-CoV-2 can cause severe symptoms when it travels down the
respiratory tract and infects the lungs, which are even more
abundant in ACE2. As a result, a number of the pulmonary cells are
damaged. In more severe cases, the immune system of the infected
individual goes into ‘overdrive’, attracting immune cells to the
lungs to attack the virus, resulting in the inflammation of the
lungs. In most severe cases, more immune cells rush in, and the
inflammation becomes more severe; this process is known as a
‘cytokine-storm’ and may lead to death. Ample studies have
demonstrated that patients with severe pneumonia may rapidly
progress to acute respiratory distress syndrome (ARDS), septic
shock or multiple organ failure and death (11). As ACE2 is abundantly present on
ciliated cells of the airway epithelium and lung alveolar type-2
cells, ARDS progression, and extensive lung damage in patients
infected with COVID-19 are inevitable (30). However, the mechanism through which
SARS-CoV-2 is able to inhibit or evade host-innate immune responses
to initiate severe pathogenesis remain unclear. Given that
SARS-CoV-2 has similar clinical manifestations as SARS-CoV-1 and
MERS-CoV, it may have a common mechanism of etiology (8). Furthermore, certain individuals have
genetic variants of ACE2 that are slightly more vulnerable to
SARS-CoV-2 than those of the majority in the population. In
addition, individuals with diabetes or hypertension have
significantly increased levels of ACE2, which renders them more
susceptible to the virus (31).
In general, the type-I interferon (IFN)-induced
expression of IFN-stimulated genes (ISGs) significantly inhibits
viral replication when challenged by infection. Given this cellular
antiviral activity, SARS-CoV-2-encoded non-structural and accessory
proteins are suggested to modulate the induction of IFN and
cytokines, and evade the ISG response (32). In addition, the host-immune
responses through inflammatory and cytotoxic lymphocyte (CTL)
activities are critical to inhibiting viral replication and
dissemination. Therefore, the immune overdrive, together with the
cytolytic effects of the virus, results in disease severity. In
addition, certain respiratory viruses, including HCoV, also induce
an increase in the levels of liver function biomarkers, very likely
related to liver inflammation or damage as a result of
IL-6-triggered CTL and Kupffer cell activities (33). Similarly, in some patients with
COVID-19, cases of hepatitis have also been observed (34).
5. Non-respiratory manifestations
A good proportion of COVID-19 patients have
exhibited evidence of gastrointestinal symptoms in response to
SARS-CoV-2 infection (34). In a
recent study from Wuhan, approximately 10% of hospitalized patients
with COVID-19 presented with diarrhea, nausea, vomiting and
abdominal pain within 1-2 days prior to the onset of COVID-19
symptoms, such as fever and dyspnea (34). Although liver function indices are
a noticeable feature of COVID-19 pathology, they are currently not
considered a ‘prominent feature’ (35). Nonetheless, COVID-19 has been
linked to mild to moderate liver injury, as revealed by elevated
levels of serum aminotransferases, bilirubin, hypoproteinemia and
prothrombin time prolongation, supported by liver histopathology
(33,36-38).
Single-cell RNA sequencing data from 2 distinct cohorts of patients
with COVID-19 have demonstrated the higher expression of ACE2 in
cholangiocytes than in hepatocytes, indicating that SARS-CoV-2 may
directly affect intrahepatic bile ducts (39). Taken together, SARS-CoV-2 is
proposed to induce viral hepatitis, while inducing a dysregulated
innate immune response. In general, patients with COVID-19
presenting with digestive issues before respiratory problems have a
higher risk of mortality compared to those without digestive
symptoms.
Cardiac comorbidity has also been reported among a
proportion of patients with COVID-19. Underlying cardiac disease,
arrhythmia and hypertension have been observed twice as often among
critical cases compared to non-critical patients (36,40,41).
Two pathological indicators of cardiac injury, i.e., elevations in
myoglobin (15-17%) and cardiotroponin (8-12%) levels, have been
reported in patients with COVID-19(42). COVID-19 disease severity was also
demonstrated to be positively associated with significantly higher
values of troponin and creatine kinase compared to those of less
severe cases. Moreover, a previous meta-regression analysis
revealed an association of the disease severity with hypertension
(43). These clinical observations
highlight the importance of the correct and timely diagnosis and
treatment of non-respiratory symptoms along with pneumonia in order
to reduce the case fatality rate.
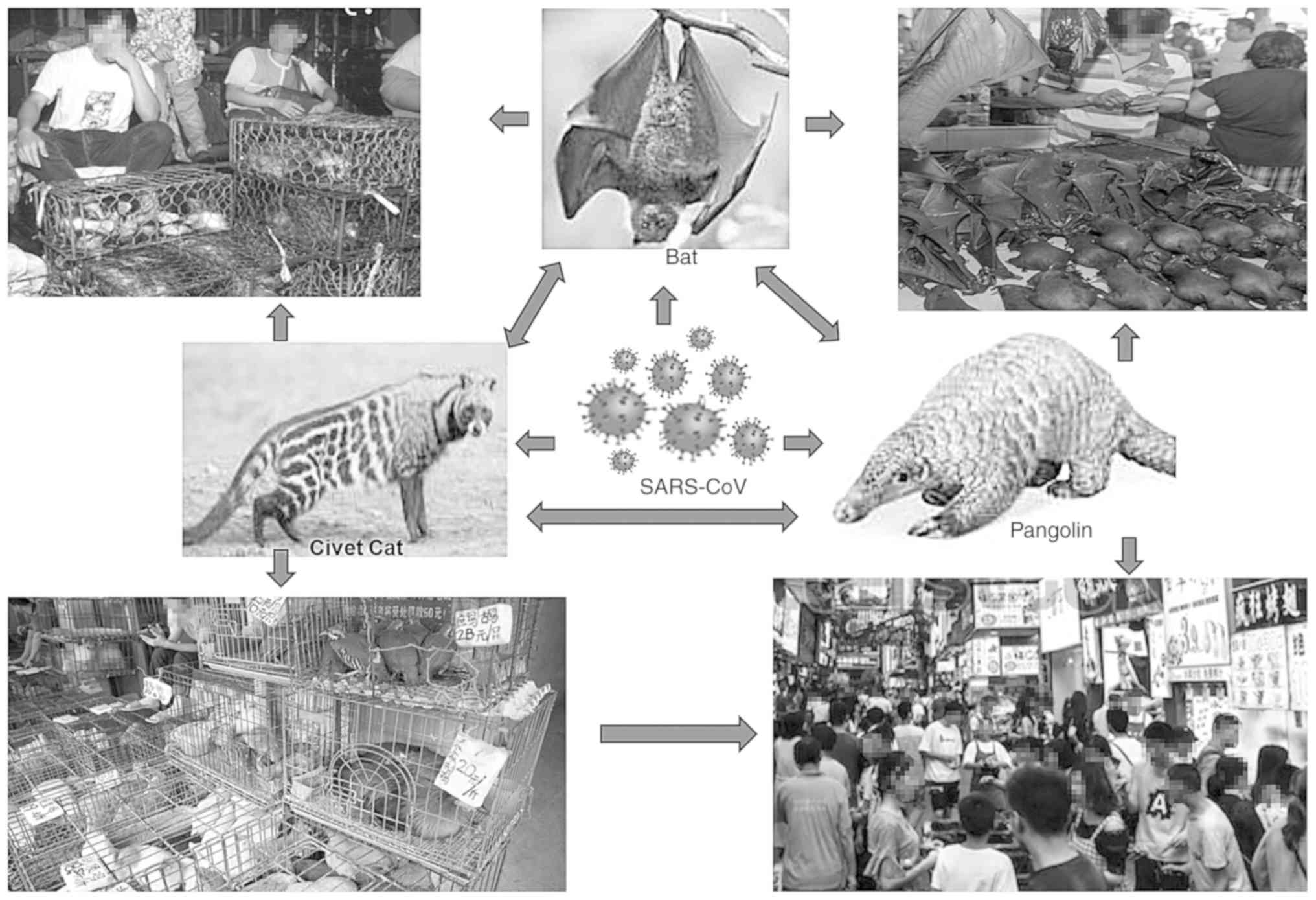
6. Source of origin
The risk of infection is high when viruses transmit
into humans from non-human primates than those from bovine,
porcine, feline, or rodent mammals. For instance, SARS-CoV-1 has
been previously detected in masked-palm or gem-faced civet cats
sold at Chinese wildlife/wet markets (44,45).
The first source of origin, high transmission and mechanisms of the
severity of SARS-CoV-2 in humans are hitherto not clearly
established. However, the virus certainly originated in bats and
transferred to other mammals, such as the pangolin, and
subsequently to its handler or ‘patient zero’ at the Wuhan market
(Fig. 4). Nonetheless, a recent
data analysis of multiple SARS-CoV genomes suggested the natural
evolution of SARS-CoV-2(46).
Previously, the global outbreaks of SARS-CoV-1 and MERS-CoV were
linked to zoonosis due to their close genetic homology to
bat-SL-CoV, but not to any other known HCoV (47). Bats are known to harbor the most
enormous diversity of CoV, which varies from species-to-species and
region-to-region (45,46,48).
Camels are also identified as a potential source of MERS-CoV
transmission to humans (49).
Notably, while zoonosis (anthropozoonosis) of COVID-19 is
established, a few cases of reverse-zoonosis (zooanthroponosis) in
pets and zoo animals are currently being reported (50).
7. Modes of transmission
A number of viruses that affect the respiratory
system, such as the influenza virus, respiratory syncytial virus,
MERS-CoV and SARS-CoV-1, are mainly transmitted when an infected
individual expels virus-loaded water droplets by coughing or
sneezing. The human-to-human direct transmission of COVID-19 has
been confirmed through multiple modes, such as nasal droplets,
aerosols and oral mucus (8).
Recently, anal swabs were shown to contain SARS-CoV-2 RNA in high
amounts, compared to oral swabs, suggesting the possible fecal-oral
transmission of SARS-CoV-2(51).
Furthermore, in pediatric cases of COVID-19, rectal swabs
persistently tested RNA-positive, even though nasopharyngeal tests
were negative (52). In other
studies from China, stool specimens of patients were found
positive, even after viral clearance, presenting evidence of
SARS-CoV-2 shedding in stool (53-55).
Moreover, cell-culture produced SARS-CoV-2 has been shown to
survive in aerosols for 3 h, on copper for 4 h, on cardboard for 24
h, and on plastic and stainless steel surfaces for up to 2-3 days
(56). These results provide vital
information about the environmental stability of SARS-CoV-2 and
suggest potential sources of viral contaminations.
8. Socio-environmental drivers of the
outbreak
Pathogenic viruses introduced into new regions often
cause highly contagious and devastating pandemics, such as
COVID-19. Recurrent outbreaks of novel human viruses suggest the
ability the virus to rapidly adapt compared to the other pathogenic
microbes. There exists a pool of unknown viruses that are likely to
evolve more rapidly over time, of which some tend to disappear in
the course of evolution, while others continue to emerge
aggressively (3). Generally, new
viruses appear when humans are exposed for the first time, to an
evolved virus from other animal hosts. Such viruses may either
become pathogenic in new non-human hosts or may further evolve into
more aggressive strains in humans. Therefore, humans are merely
‘incidental’ or ‘spillover’ hosts (2).
Furthermore, to understand the evolution of novel
viruses is to know the intricate ‘host-pathogen-environment’
interplay. While the emergence of new infections, such as COVID-19
in naïve regions, is caused primarily by human movement, local
emergence is driven by a combination of environmental and
social/traditional changes. Notably, viral transmission rates are
often higher in dense than in sparse populations, and social
contacts greatly enhance the spreading of the virus. Moreover, the
growing human population, global changes in geographical
distribution and the introduction of anthropophilic vectors affect
selective pressure on primary hosts of evolving viruses (3).
Furthermore, there is a key question for individuals
residing in tropical countries, namely that of whether the warm
season would eradicate SARS-CoV-2. Generally, the respiratory, flu,
or pneumonia viruses, including certain CoV lifecycles survive in
cold seasons, and gradually subside when the temperature rises. For
example, the SARS-CoV-1 spread in 2003 was rapidly contained,
leaving little information on the effect of seasonal variations on
disease spread. Since the emergence of SARS-CoV-2 in China in
December, 2019, a number of large-scale outbreaks have been
observed in regions where the weather is cooler, leading to
speculation that the virus would diminish with the arrival of
summer. However, SARS-CoV-2 is too novel to postulate any pattern
of survival as to how it will behave with the season changes. An
unpublished analysis comparing the weather in COVID-19-affected 500
locations suggested a link between its spread and temperature and
relative humidity; however, it was noted that temperature alone
cannot account for the global variation in incidence (12). Although the emergence of SARS-CoV-2
in colder weather indicates for its plausible seasonality, the
arrival of summer in several regions has not significantly affected
the rate of infection.
9. Human adaptation
A zoonotic infection is initially poorly adapted in
a new host, slowly replicated and inefficiently transmitted.
Therefore, its animal-to-human and human-to-human transmission
greatly depends on its evolution to a virulent strain that can well
adapt to the human host. RNA viruses have a much more recent
evolutionary history and ‘human adaptation’ for only thousands of
years as compared to DNA viruses evolving and diversifying for
millions of years (2). Owing to
the high replication-fidelity rate of their
polymerase/reverse-transcriptase enzymes (~10-4
error/site/cycle), RNA viruses are more genetically diversified
than DNA viruses. In the process of evolution and adaptation of RNA
viruses, genetic mutations, re-assortment or virus-host genetic
recombination may lead to the establishment of stable strains or
lineages in human populations (3).
Therefore, it is very much expected that human-adapted novel
viruses, such as SARS-CoV-2, could circulate asymptomatically and
remain undetected until they manifest in the infected population.
However, only a minority of these would persist in specific
populations (endemics), spreading across populations (epidemics) or
globally (pandemics) in the absence of an established
reservoir.
In addition, differential host factors, such as age,
health, physiology, nutritional status, exposure history,
concomitant infections, immuno-competence, underlying comorbidities
and genetics significantly determine the susceptibility of an
individual to a novel infection. As observed with COVID-19
infection, older-aged and immune-compromised individuals are the
most commonly affected population, worldwide. Notably, the severity
of COVID-19 in patients with chronic disease with poor immunity
becomes more inevitable compared to other critically ill patients
with pneumonia.
10. Diagnosis and treatment options
While COVID-19 is rapidly spreading along with other
respiratory viruses in circulation, proper screening and sensitive
diagnostic tools are required to control its further spread. A
chest X-ray and CT scan are the clinical methods of non-invasive
pulmonary assessment. Nasopharyngeal and oropharyngeal swabs, as
well as sputum, tracheal aspirate, or bronchoalveolar lavage are
the recommended specimens for viral RNA molecular (RT-PCR) testing
(57). In the cases of digestive
issues, the testing of rectal swabs and stool samples of patients
with COVID-19 is warranted (34).
Pan-coronavirus based serological or antibody test kits are now
being extensively used in several countries, in order to assess the
protective immunity in patients who recovered from COVID-19.
Although the acquired immunity and longevity against SARS-CoV-2
remain poorly understood, these antibodies could be used as part of
a broader range of care, such as plasma therapy. At this moment,
the test kits that are largely produced in China, are being
reported for inaccurate and unreliable results in countries, such
as the UK, France and India. Currently, few laboratories outside
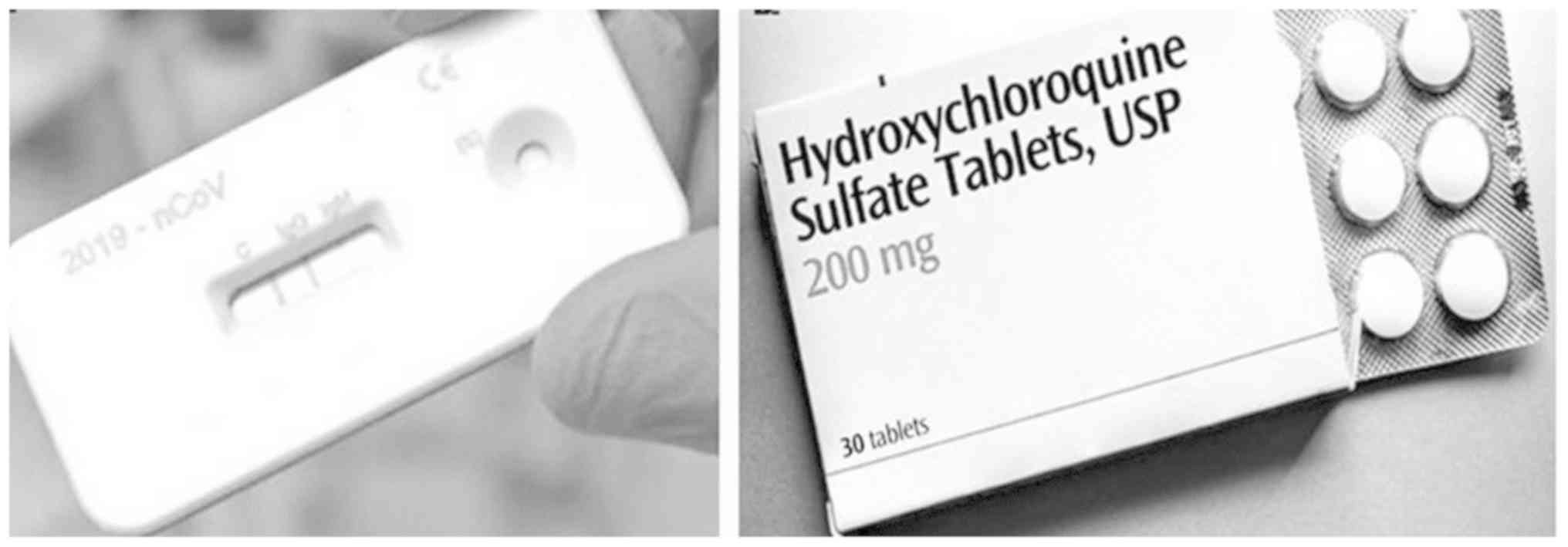
China have also begun the production of rapid COVID-19 antibody
test kits (Fig. 5, left
panel).
The WHO has recommended case definitions for
COVID-19(1) that can however, vary
in countries or even within a given region over time. Suspected
cases of COVID-19 are those with severe acute respiratory
infections requiring hospitalization, and thoroughly explaining the
clinical presentation and a history of visiting China or any
infected population, during the 14 days prior to symptom onset. In
either case, contact with a confirmed or suspected case or working
in or shared a healthcare center where patients with COVID-19 were
treated. Therefore, probable cases are those for whom the COVID-19
test is inconclusive or those tested SARS-CoV-2-positive, and
negative for laboratory evidence of other respiratory viruses. As
per the WHO, a positive case is one with a laboratory confirmation
of SARS-CoV-2 infection, irrespective of clinical
manifestations.
At present, there is no specific treatment regimen
available for COVID-19. As part of the ‘Solidarity Trial’ for
hundreds of COVID-19 treatments and interventions, the
anti-malarial drug, hydroxychloroquine (Fig. 5, right panel), the anti-Ebola drug,
remdesivir, the broad-spectrum antibiotic, azithromycin, including
the anti-retroviral drug, lopinavir, in combination with ritonavir
and IFN-α-1a are currently in advanced stages of clinical
investigations (https://covid19responsefund.org). Of these, remdesivir has
recently been approved for ‘emergency use’ by the US Food and Drug
Administration. In addition, while the anti-influenza drug,
favipiravir, has completed the phase III trials in Japan and
entered the US phase II investigation, it has now been approved in
Russia for hospitalized patients. Thus far, there is no consensus
clinical guidance available on the use, dosing or duration of
treatment in patients with COVID-19(7). There are however, safety concerns
that some of these drugs may cause cardiotoxicity with prolonged
use in patients with pre-existing chronic conditions, such as renal
failure and hepatic disease (58-60).
Nonetheless, though a small sample size, case
studies of patients with COVID-19 with liver issues suggest
focusing on modulating innate-immune dysfunction besides antiviral
trials (35). Recently,
tocilizumab and sarilumab, IL-6 receptor antagonists, have been
approved for phase II/III trials in hospitalized patients with
COVID-19 with severe pneumonia (https://clinicaltrials.gov/ct2/). Moreover, several
other pharmacophores and chemotherapies, such as camostat mesylate
and mefloquine, are undergoing clinical investigations in some
countries (61).
11. Vaccine and preventive measures
Developing a vaccine is the optimal preventive
measure; however, it is also the most time-consuming and most
complex process, which may require significant amounts of time
ranging from 24-30 months. Nonetheless, rapid initiatives have been
already taken, and over 8 promising vaccines candidates, notably
mRNA-1273 (Moderna, Inc.), Ad5-nCoV (CanSino Biologics),
ChAdOx1-nCoV-19 (The Oxford Group); INO-4800 (Inovio
Pharmaceuticals Inc.), and LV-SMENP-DC (Shenzhen Geno-immune
Medical Institute) are currently undergoing phase I/II trials
(62,63). Large scale phase III trials are,
therefore, warranted to determine their optimal required dose,
efficacies in elderly individuals and minimal side-effects.
Additionally, it should be determined whether any additional
adjuvant is required to further boost their effectiveness, before
approval and licensing.
Moreover, reasonably high levels of neutralizing
antibodies termed ‘herd immunity’ are produced in patients with
COVID-19 that maybe protective against future infections. ‘Plasma
therapy’ with significant outcomes has therefore been adopted in
some countries. However, this will not last for >2 years, as
observed with other HCoV infections, where even if the majority of
the population do eventually become exposed, the virus is still
likely to become endemic. The SARS-CoV-2 will be circulating among
us for some time and may become less pathogenic or may mutate to
become more lethal. In summary, COVID-19 does what past flu
pandemics have done, leaving behind enough immune survivors and
searching for viable targets.
In the present situation, the first and most
important measure is to immediately quarantine COVID-19-positive or
suspected individuals, while enforcing public health safety
guidelines on social distancing, the use of sanitizers, protective
masks and gloves. Given the scarcity of medical facilities,
clinical equipment and the lack of treatment options, COVID-19 has
forced several countries to resort to extreme public health
measures, such as the lockdown of cities, suspending domestic and
international travel, and sealing international borders, never
before observed over the past century. A quantitative investigation
on the impact of the travel ban has revealed a significant positive
association between population movement and controlling the spread
of COVID-19 in mainland China (64,65).
12. Mathematical model of assessing viral
evolution and epidemics
Patterns of pathogen evolution and spread are
similar in the way that information or rumors propagate in the
public domain, while editing or morphing occur, particularly
through social media. The mathematical-computational models of
epidemics are currently well known, which can be used to predict or
back-predict the spread of infections accurately. One such
phylodynamic study that combined a modeling framework for host,
epidemiological and molecular data, particularly for RNA viruses,
such as SARS-CoV-2 demonstrated particular promise for
understanding the patterns of viral evolution during epidemics or
pandemics (3). An example of this
is the first case-clusters of the SARS-CoV-1 outbreak and the
subsequent global spread, including the country-by-country
distribution of human cases (66,67).
Notably, the phylodynamic analysis estimated the median time to
most recent common ancestor (TMRCA) as November 17, 2019 and a
genetic mutation rate of 0.8x10-3 (95% CI,
0.14x10-3 to 1.31x10-3) (46). In addition, that study predicted
the doubling time of viral infections each week (7.2 days).
However, it is essential to note that mutation rates differ from
viral protein-to-protein and study-to-study due to the rapid
increase in sequencing data. A recently proposed mathematical
theory highlighted the emergence phenomena of COVID-19 or similar
infections and revealed the effects of evolutionary adaptations on
spreading processes in complex networks (Fig. 6), overcoming the flaws of classical
epidemic models that do not capture evolution (68). This model fully characterizes the
process, accurately predicts the epidemics threshold, the expected
epidemic size, and the expected fraction of individuals infected by
each pathogenic strain. Moreover, another recent algorithm based on
a bacterial protein model could also help understand the
evolutionary pathway of SARS-CoV-2 that evades the immune system or
those viruses who develop drug-resistance in due course of time
(69).
13. Risk factors
It is now well known that emerging human viruses can
be transmitted to humans via close contact with virus-hosting
animals and the consumption of infected meat or meat products,
including freshwater and seafood products (2). The new approach to food safety and
protecting humans against food-borne infections is the most
effective method with which to control human illnesses associated
with zoonosis. However, the vast majority of such infections remain
under-reported, and therefore, novel viral pathogens remain
unidentified and continue to circulate in the general population.
As zoonosis is linked to the majority of the pathogenic viruses,
including SARS-CoV-1 and SARS-CoV-2, precautions and proper care
are required, while selecting, purchasing, cooking, consuming meat
or seafood and avoiding high-risk animals is also mandatory.
Globally, the current bio-/food-security measures appear to be more
successful in constraining bacterial and fungal pathogens that have
been less effective for water-/food-borne viruses (2).
Recent clinical reports on gastrointestinal and
stool shedding of SARS-CoV-2 suggest potential routes of
transmission of COVID-19. Therefore, occurrences of SARS-CoV-2 in
human feces and water sources under poor sanitation conditions may
further aggravate the fecal-oral spread of COVID-19 in
healthcare-deficient nations (70). Nonetheless, further studies are
warranted in order to determine whether infectious and
transmittable amounts of SARS-CoV-2 can be found in water sources.
Moreover, the surveillance of emerging human viruses, such as
SARS-CoV-2 must, therefore, include livestock, wild animals,
potential vectors and their environments at an internationally
coordinated level. Furthermore, animal handlers, such as livestock
herders, hunters, sellers, forest rangers, zookeepers, wildlife
rangers and veterinarians working with potential reservoir or
high-risk animals must take hygienic measures, as well as undergo
routine serological tests.
14. Conclusion and future perspectives
The majority of novel human-adapted viruses with
known origin fall into the category of ‘crowd diseases’ that
require relatively high host-densities for rapid spread and are
greatly enhanced by air travel. Notably, recent incidences of
COVID-19, such as pandemics mark the Asia-Pacific region as the
global hot-spot for emerging novel pathogenic viruses. There is a
growing understanding of COVID-19 pathobiology, epidemiology and
clinical management strategies. However, as evidenced by recent
clinical studies, asymptomatic, as well as discharged patients, can
still remain viremic. The presence of SARS-CoV-2 RNA in uncommon
areas of infection, such as the rectum, despite the absence of
viral load in swabs from regularly affected areas, highlights the
shortcomings in screening, diagnosis and understanding the spread.
The viral shedding in such specimens thereby provides a cautionary
warning that COVID-19 may be transmitted through the fecal-oral
route in developing countries with inadequate sanitization. Most
importantly, the detection of SARS-CoV-2 in rectal and fecal
samples strongly endorses its digestive etiology, directly or
indirectly, in higher-risk patients with impaired immunity.
Worldwide, government authorities, healthcare
providers and non-profit organizations are enforcing safety
guidelines, and providing diagnostics and treatments to the best of
their capacities. Unfortunately, by the time an effective COVID-19
vaccine is available or the tempting ‘herd immunity’ scenario is
expedited, millions of lives could be lost, and there would be
deleterious effects on the health system and economy worldwide.
However, the mass production and open-sharing of sensitive and
cost-effective test kits may be a rapid tool with which to identify
infected and asymptomatic carriers and save the healthy population.
Moreover, epidemiologists and statisticians can adopt the recently
introduced mathematical models to predict further and estimate the
future COVID-19 spread and control measures. Nonetheless, the real
impact of COVID-19 will be known only after the pandemic is
over.
This is the time when international collaborations
and co-ordinations are most needed towards the better utilization
of available resources. Nonetheless, few key issues must be
addressed sincerely, as for instance, ensuring that the preset
standard protocols are appropriately followed amid the rush of
assessing and approving new COVID-19 drugs, as recently experienced
with the hydroxychloroquine controversy. The care of other
infectious or chronic diseases should not be compromised, resulting
in a significant increase in mortality. The mental health of
patients with COVID-19 and their families, healthcare providers,
local administrative or security officials, and underprivileged
individuals must be appropriately taken into consideration in order
to prevent long-term psychological disorders. Moreover, lastly but
importantly, the UNO and WHO must manage the existing distrust and
isolated actions among politically distanced countries to prevent
any future escalation in the crisis. In addition, it becomes the
civic responsibility of academics and scientists to proactively
reach out to the common man to educate on the current understanding
of disease via social media. Therefore, the biomedical,
socio-economical and geopolitical forces of the world must work
together to end the COVID-19 emergency soon. On a positive note,
treated patients are recovering, and the virus is being contained.
The future is not dismal. Importantly, with a united effort, the
indomitable spirit of humanity can remain undefeated.
Acknowledgements
MKP gratefully acknowledges his virology mentors, Dr
Shahid Jameel (CEO, Welcome-Trust India Alliance, Hyderabad,
India); Dr Syed S. Hasnain (VC, Hamdard University, New Delhi,
India); Dr Shiv K. Sarin (Director, Institute of Liver and Biliary
Sciences, New Delhi, India); Dr Robert H. Purcell and Dr Suzanne U.
Emerson (Chief Scientists, National Institutes of Health, Bethesda,
USA); and Dr Robert C. Gallo (Director, Institute of Human
Virology, Baltimore, USA) for providing education and inspiration
in the field.
Funding
No funding received.
Availability of data and materials
Not applicable.
Authors' contributions
MKP conceptualized, planned and designed the study,
performed the literature search, and drafted, edited and prepared
the final version of the manuscript. RMJ, RSP, SKSV, VA, JK and NT
conceptualized and participated in the design, literature search,
and writing and drafting of the manuscript. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Health Organization (WHO):
Coronavirus disease (COVID-19) outbreak situation. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
Accessed June 20, 2020.
|
|
2
|
Parvez MK: Emerging and re-emerging viral
diseases: Controls and preventions. In: Microbial pathogens and
strategies for combating them science, technology and education.
Méndez-Vilas A (ed). Formatex Research Center, Badajoz, 2013.
|
|
3
|
Parvez MK and Parveen S: Evolution and
emergence of pathogenic viruses: Past, present, and future.
Intervirology. 60:1–7. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wever PC and van Bergen L: Death from 1918
pandemic influenza during the first world war: A perspective from
personal and anecdotal evidence. Influenza Other Respir Viruses.
8:538–546. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
International Committee on Taxonomy of
Viruses (ICTV): Virus taxonomy: The classification and nomenclature
of viruses. https://talk.ictvonline.org/ictv-reports/ictv_online_report/.
Accessed June 20, 2020.
|
|
6
|
Ren LL, Wang YM, Wu ZQ, Xiang ZC, Guo L,
Xu T, Jiang YZ, Xiong Y, Li YJ, Li XW, et al: Identification of a
novel coronavirus causing severe pneumonia in human: A descriptive
study. Chin Med J (Engl). 133:1015–1024. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
World Health Organization (WHO):
Director-General's opening remarks at the media briefing on
COVID-19. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-18-march-2020.
Accessed June 20, 2020.
|
|
8
|
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu
Y, Zhang L, Fan G, Xu J, Gu X, et al: Clinical features of patients
infected with 2019 novel coronavirus in Wuhan, China. Lancet.
395:497–506. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kissler SM, Tedijanto C, Goldstein E, Grad
YH and Lipsitch M: Projecting the transmission dynamics of
SARS-CoV-2 through the postpandemic period. Science. 368:860–868.
2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bauch CT and Oraby T: Assessing the
pandemic potential of MERS-CoV. Lancet. 382:662–664.
2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen Y, Liu Q and Guo D: Emerging
coronaviruses: Genome structure, replication, and pathogenesis. J
Med Virol. 92:418–423. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chen B, Liang H, Yuan X, Hu Y, Xu M, Zhao
Y, Zhang B, Tian F and Zhu X: Roles of meteorological conditions in
COVID-19 transmission on a worldwide scale. medRxiv: https://doi.org/10.1101/2020.03.16.20037168.
|
|
13
|
Cárdenas-Conejo Y, Liñan-Rico A,
García-Rodríguez DA, Centeno-Leija S and Serrano-Posada H: An
exclusive 42 amino acid signature in pp1ab protein provides
insights into the evolutive history of the 2019 novel
human-pathogenic coronavirus (SARS-CoV-2). J Med Virol. 92:688–692.
2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song
ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al: A new coronavirus
associated with human respiratory disease in China. Nature.
579:265–269. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Coronaviridae Study Group of the
International Committee on Taxonomy of Viruses: The species severe
acute respiratory syndrome-related coronavirus: Classifying
2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5: 536-544,
2020.
|
|
16
|
Kumar S, Maurya VK, Prasad AK, Bhatt MLB
and Saxena SK: Structural, glycosylation and antigenic variation
between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus
(SARS-CoV). Virus Disease. 31:13–21. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chan JF, Kok KH, Zhu Z, Chu H, Kai-Wang K,
Yuan S and Yuen KY: Genomic characterization of the 2019 novel
human-pathogenic coronavirus isolated from a patient with atypical
pneumonia after visiting Wuhan. Emerg Microbes Infect. 9:221–236.
2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Walls AC, Park YJ, Tortorici MA, Wall A,
McGuire AT and Veesler D: Structure, function, and antigenicity of
the SARS-CoV-2 spike glycoprotein. Cell. 181:281–292.e286.
2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Coutard B, Valle C, de Lamballerie X,
Canard B, Seidah NG and Decroly E: The spike glycoprotein of the
new coronavirus 2019-nCoV contains a furin-like cleavage site
absent in CoV of the same clade. Antiviral Res.
176(104742)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wan Y, Shang J, Graham R, Baric RS and Li
F: Receptor recognition by the novel coronavirus from Wuhan: An
analysis based on decade-long structural studies of SARS
coronavirus. J Virol. 94:e00127–e00120. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wrapp D, Wang N, Corbett KS, Goldsmith JA,
Hsieh CL, Abiona O, Graham BS and McLellan JS: Cryo-EM structure of
the 2019-nCoV spike in the prefusion conformation. Science.
367:1260–1263. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Leung DT, Tam FC, Ma CH, Chan PK, Cheung
JL, Niu H, Tam JS and Lim PL: Antibody response of patients with
severe acute respiratory syndrome (SARS) targets the viral
nucleocapsid. J Infect Dis. 190:379–386. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
European Center for Disease Prevention and
Control (ECDC): COVID-19 situation update worldwide, as of 6 July
2020. https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases.
Accessed June 20, 2020.
|
|
24
|
Schoeman D and Fielding BC: Coronavirus
envelope protein: Current knowledge. Virol J. 16(69)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kakodkar P, Kaka N and Baig MN: A
comprehensive literature review on the clinical presentation, and
management of the pandemic coronavirus disease 2019 (COVID-19).
Cureus. 12(e7560)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li Y and Xia L: Coronavirus disease 2019
(COVID-19): Role of chest CT in diagnosis and management. AJR Am J
Roentgenol. 214:1280–1286. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wong HYF, Lam HYS, Fong AH, Leung ST, Chin
TW, Lo CS, Lui MM, Lee JC, Chiu KW, Chung T, et al:
Frequency and distribution of chest radiographic findings in
COVID-19 positive patients. Radiology 2011: 60, 2019 (Online ahead
of print).
|
|
28
|
Ge XY, Li JL, Yang XL, Chmura AA, Zhu G,
Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, et al: Isolation and
characterization of a bat SARS-like coronavirus that uses the ACE2
receptor. Nature. 503:535–538. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li W, Moore MJ, Vasilieva N, Sui J, Wong
SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough
TC, et al: Angiotensin-converting enzyme 2 is a functional receptor
for the SARS coronavirus. Nature. 426:450–454. 2003.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hamming I, Timens W, Bulthuis ML, Lely AT,
Navis G and van Goor H: Tissue distribution of ACE2 protein, the
functional receptor for SARS coronavirus. A first step in
understanding SARS pathogenesis. J Pathol. 203:631–637.
2004.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fang L, Karakiulakis G and Roth M: Are
patients with hypertension and diabetes mellitus at increased risk
for COVID-19 infection? Lancet Respir Med. 8(e21)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Totura AL and Baric RS: SARS coronavirus
pathogenesis: Host innate immune responses and viral antagonism of
interferon. Curr Opin Virol. 2:264–275. 2012.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Adams DH and Hubscher SG: Systemic viral
infections and collateral damage in the liver. Am J Pathol.
168:1057–1059. 2006.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Gu J, Han B and Wang J: COVID-19:
Gastrointestinal manifestations and potential fecal-oral
transmission. Gastroenterology. 158:1518–1519. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang C, Shi L and Wang FS: Liver injury
in COVID-19: Management and challenges. Lancet Gastroenterol
Hepatol. 5:428–430. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J,
Wang B, Xiang H, Cheng Z, Xiong Y, et al: Clinical characteristics
of 138 hospitalized patients with 2019 novel coronavirus-infected
pneumonia in Wuhan, China. JAMA. 323:1061–1069. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He
JX, Liu L, Shan H, Lei CL, Hui DSC, et al: Clinical characteristics
of coronavirus disease 2019 in China. N Engl J Med. 382:1708–1720.
2020.
|
|
38
|
Shi H, Han X, Jiang N, Cao Y, Alwalid O,
Gu J, Fan Y and Zheng C: Radiological findings from 81 patients
with COVID-19 pneumonia in Wuhan, China: A descriptive study.
Lancet Infect Dis. 20:425–434. 2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Qi F, Qian S, Zhang S and Zhang Z: Single
cell RNA sequencing of 13 human tissues identify cell types and
receptors of human coronaviruses. Biochem Biophys Res Commun.
526:135–140. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Li B, Yang J, Zhao F, Zhi L, Wang X, Liu
L, Bi Z and Zhao Y: Prevalence and impact of cardiovascular
metabolic diseases on COVID-19 in China. Clin Res Cardiol.
109:531–538. 2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Long B, Brady WJ, Koyfman A and Gottlieb
M: Cardiovascular complications in COVID-19. Am J Emerg Med.
38:1504–1507. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lippi G and Plebani M: Laboratory
abnormalities in patients with COVID-2019 infection. Clin Chem Lab
Med. 58:1131–1134. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Li JW, Han TW, Woodward M, Anderson CS,
Zhou H, Chen YD and Neal B: The impact of 2019 novel coronavirus on
heart injury: A systemic review and meta-analysis. Prog Cardiovasc
Dis: April 16, 2020 (Epub ahead of print).
|
|
44
|
Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang
ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YL, et al: Isolation
and characterization of viruses related to the SARS coronavirus
from animals in southern China. Science. 302:276–278.
2003.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Li W, Shi Z, Yu M, Ren W, Smith C, Epstein
JH, Wang H, Crameri G, Hu Z, Zhang H, et al: Bats are natural
reservoirs of SARS-like coronaviruses. Science. 310:676–679.
2005.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Andersen KG, Rambaut A, Lipkin WI, Holmes
EC and Garry RF: The proximal origin of SARS-CoV-2. Nat Med.
26:450–452. 2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Corman VM, Eckerle I, Bleicker T, Zaki A,
Landt O, Eschbach-Bludau M, van Boheemen S, Gopal R, Ballhause M,
Bestebroer TM, et al: Detection of a novel human coronavirus by
real-time reverse-transcription polymerase chain reaction. Euro
Surveill. 17(20285)2012.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Banerjee A, Kulcsar K, Misra V, Frieman M
and Mossman K: Bats and Coronaviruses. Viruses.
11(41)2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Reusken CB, Raj VS, Koopmans MP and
Haagmans BL: Cross host transmission in the emergence of MERS
coronavirus. Curr Opin Virol. 16:55–62. 2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Laguipo A: Tiger with SARS-CoV-2 infection
demostrates reverse zoonosis. News-Medical, 2020. https://www.news-medical.net/news/20200406/Tiger-with-SARS-CoV-2-infection-demostrates-reverse-zoonosis.aspx.
Accessed April 6, 2020.
|
|
51
|
Zhang W, Du RH, Li B, Zheng XS, Yang XL,
Hu B, Wang YY, Xiao GF, Yan B, Shi ZL and Zhou P: Molecular and
serological investigation of 2019-nCoV infected patients:
Implication of multiple shedding routes. Emerg Microbes Infect.
9:386–389. 2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Xu Y, Li X, Zhu B, Liang H, Fang C, Gong
Y, Guo Q, Sun X, Zhao D, Shen J, et al: Characteristics of
pediatric SARS-CoV-2 infection and potential evidence for
persistent fecal viral shedding. Nat Med. 26:502–505.
2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Holshue ML, DeBolt C, Lindquist S, Lofy
KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural
A, et al: First case of 2019 novel coronavirus in the United
States. N Engl J Med. 382:929–936. 2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Tang A, Tong ZD, Wang HL, Dai YX, Li KF,
Liu JN, Wu WJ, Yuan C, Yu ML, Li P and Yan JB: Detection of novel
coronavirus by RT-PCR in stool specimen from asymptomatic child,
China. Emerg Infect Dis. 26:1337–1339. 2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Young BE, Ong SWX, Kalimuddin S, Low JG,
Tan SY, Loh J, Ng OT, Marimuthu K, Ang LW, Mak TM, et al:
Epidemiologic features and clinical course of patients infected
with SARS-CoV-2 in Singapore. JAMA. 323:1488–1494. 2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
van Doremalen N, Bushmaker T, Morris DH,
Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL,
Thornburg NJ, Gerber SI, et al: Aerosol and surface stability of
SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med.
382:1564–1567. 2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Jin YH, Cai L, Cheng ZS, Cheng H, Deng T,
Fan YP, Fang C, Huang D, Huang LQ, Huang Q, et al: A rapid advice
guideline for the diagnosis and treatment of 2019 novel coronavirus
(2019-nCoV) infected pneumonia (standard version). Mil Med Res.
7(4)2020.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Borba MGS, Val FFA, Sampaio VS, Alexandre
MAA, Melo GC, Brito M, Mourão MPG, Brito-Sousa JD, Baía-da-Silva D,
Guerra MVF, et al: Chloroquine diphosphate in two different
dosages as adjunctive therapy of hospitalized patients with severe
respiratory syndrome in the context of coronavirus (SARS-CoV-2)
infection: Preliminary safety results of a randomized,
double-blinded, phase IIb clinical trial (CloroCovid-19 Study).
medRxiv: doi: https://doi.org/10.1101/2020.04.07.20056424.
|
|
59
|
Chorin E, Dai M, Shulman E, Wadhwani L,
RoiBar-Cohen, Barbhaiya C, Aizer A, Holmes D, Bernstein S, Spinelli
M, et al: The QT interval in patients with SARS-CoV-2
infection treated with hydroxychloroquine/azithromycin. medRxiv:
doi: https://doi.org/10.1101/2020.04.02.20047050.
|
|
60
|
Roden DM, Harrington RA, Poppas A and
Russo AM: Considerations for drug interactions on QTc in
exploratory COVID-19 (coronavirus disease 2019) treatment. Heart
Rhythm. 17:e231–e232. 2020.
|
|
61
|
Huang J, Song W, Huang H and Sun Q:
Pharmacological therapeutics targeting RNA-dependent RNA
polymerase, proteinase and spike protein: From mechanistic studies
to clinical trials for COVID-19. J Clin Med. 9(1131)2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Center for Disease control (CDC):
Information for Clinicians on Investigational Therapeutics for
Patients with COVID-19. https://www.cdc.gov/coronavirus/2019-ncov/hcp/therapeutic-options.html.
Accessed April 25, 2020.
|
|
63
|
Akst J: COVID-19 Vaccine Frontrunners. The
scientist, 2020. https://www.the-scientist.com/news-opinion/covid-19-vaccine-frontrunners-67382.
Accessed June 20, 2020.
|
|
64
|
Zhang C, Chen C, Shen W, Tang F, Lei H,
Xie Y, Cao Z, Tang K, Bai J, Xiao L, et al: Impact of population
movement on the spread of 2019-nCoV in China. Emerg Microbes
Infect. 9:988–990. 2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Chinazzi M, Davis JT, Ajelli M, Gioannini
C, Litvinova M, Merler S, Piontti APY, Mu K, Rossi L, Sun K, et al:
The effect of travel restrictions on the spread of the 2019 novel
coronavirus (COVID-19) outbreak. Science. 368:395–400.
2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Anderson RM, Fraser C, Ghani AC, Donnelly
CA, Riley S, Ferguson NM, Leung GM, Lam TH and Hedley AJ:
Epidemiology, transmission dynamics and control of SARS: The
2002-2003 epidemic. Philos Trans R Soc Lond B Biol Sci.
359:1091–1105. 2004.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Hufnagel L, Brockmann D and Geisel T:
Forecast and control of epidemics in a globalized world. Proc Natl
Acad Sci USA. 101:15124–15129. 2004.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Eletreby R, Zhuang Y, Carley KM, Yagan O
and Poor HV: The effects of evolutionary adaptations on spreading
processes in complex networks. Proc Natl Acad Sci USA.
117:5664–5670. 2020.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Zhou J and McCandlish DM: Minimum
epistasis interpolation for sequence-function relationships. Nat
Commun. 11(1782)2020.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Parvez MK: Gastrointestinal and
hepatobiliary manifestations of coronavirus disease-19: Potential
implications for healthcare resource-deficient countries.
Gastroenterol Heptal Lett. 2:7–11. 2020.
|