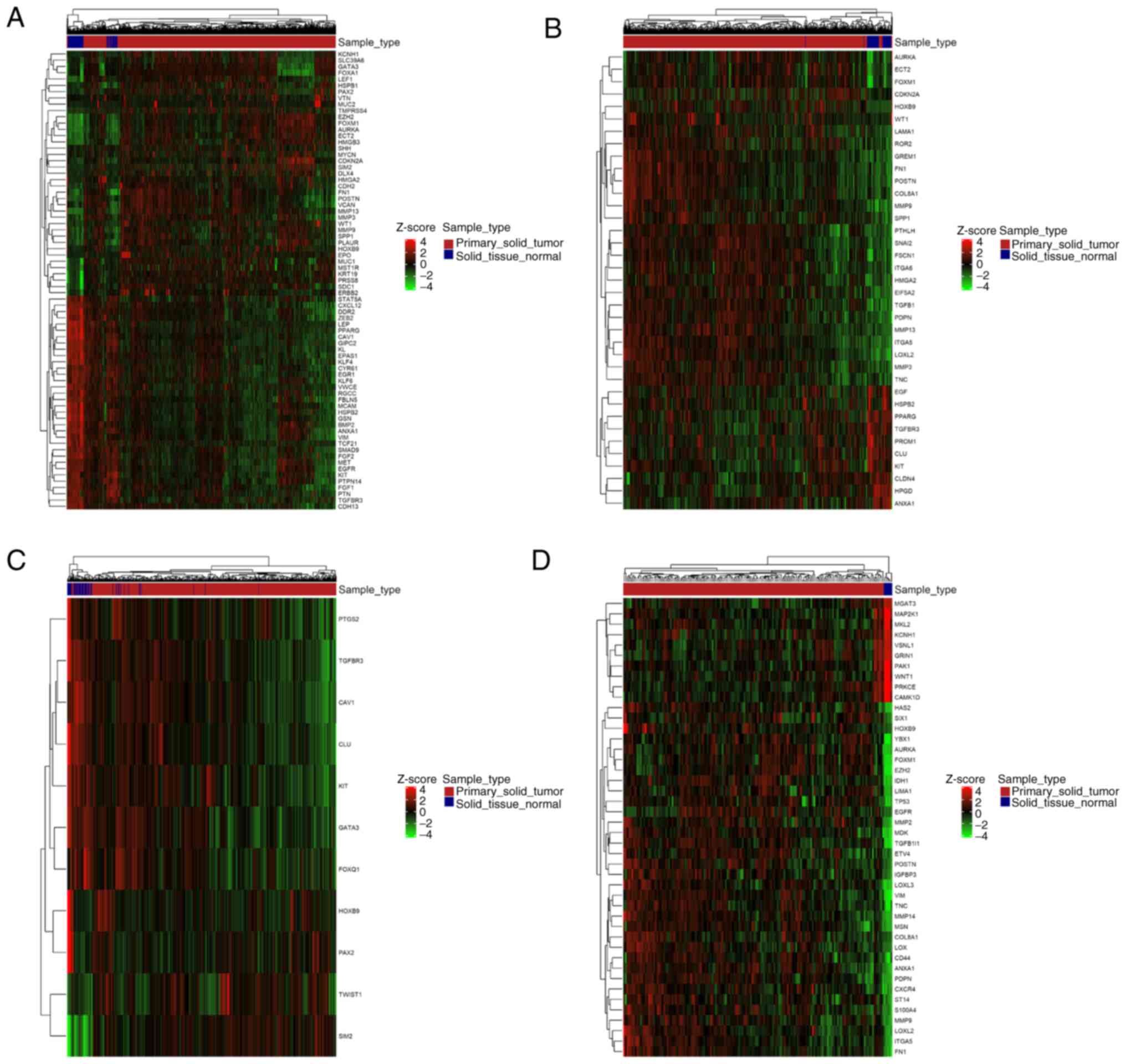
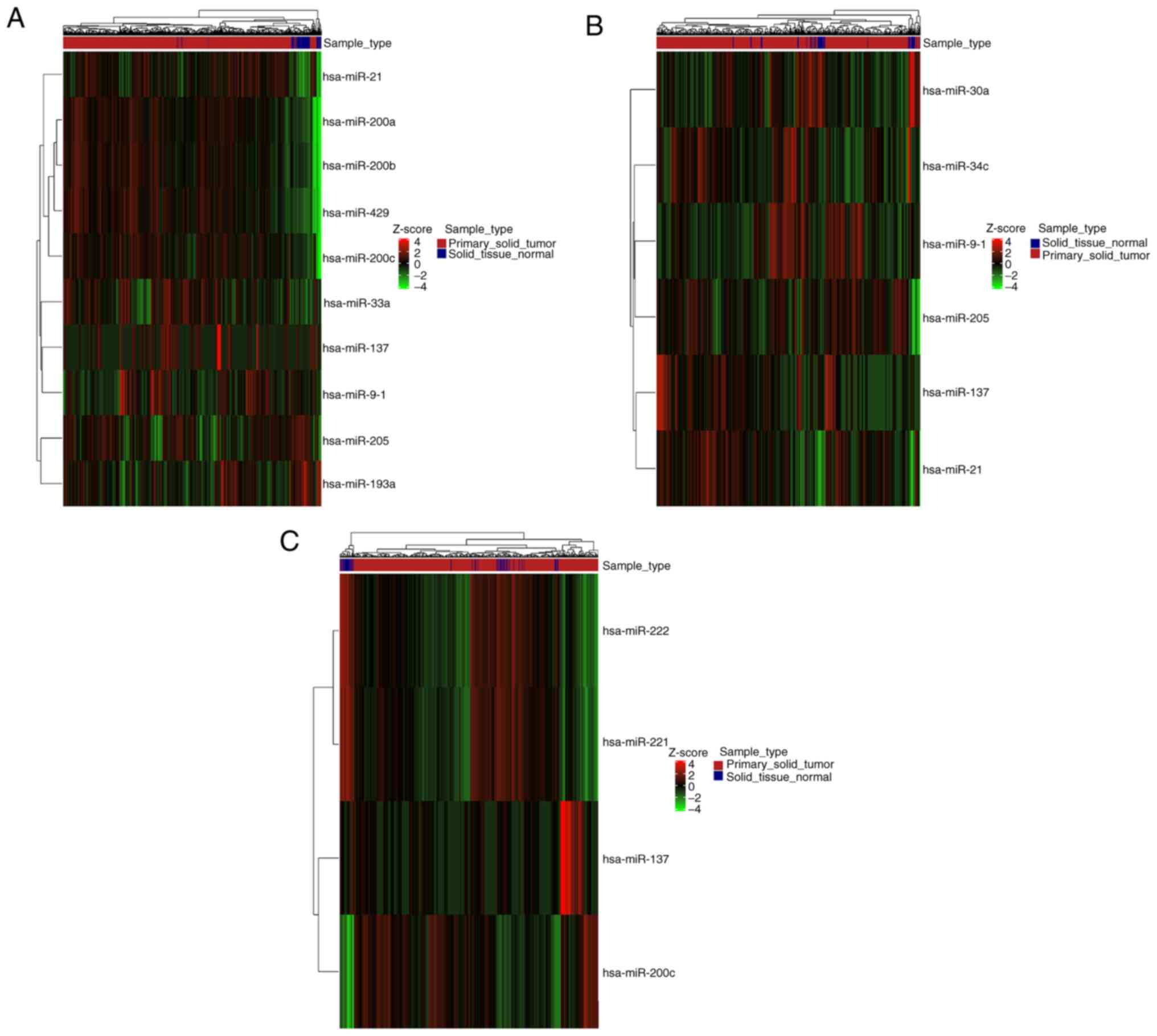
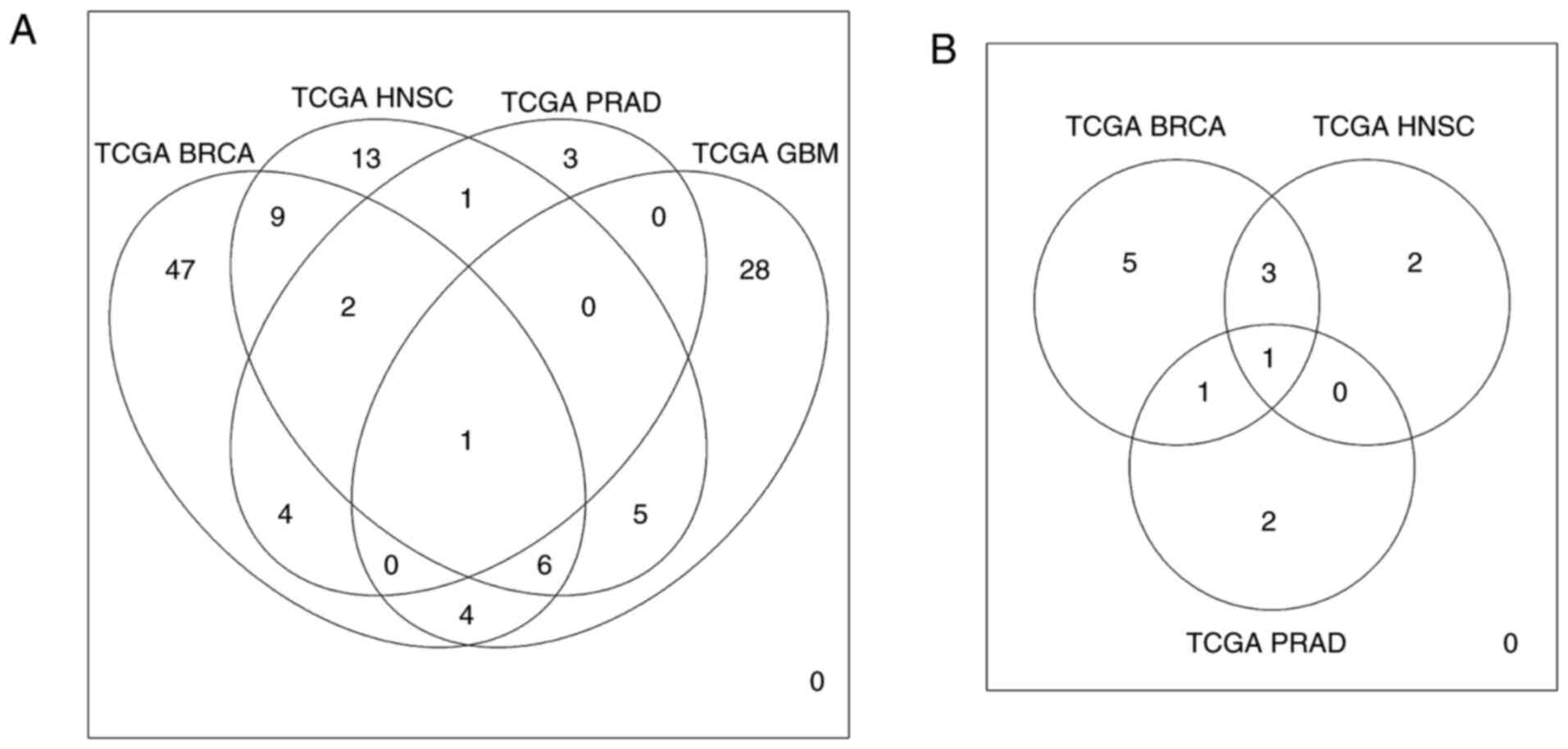
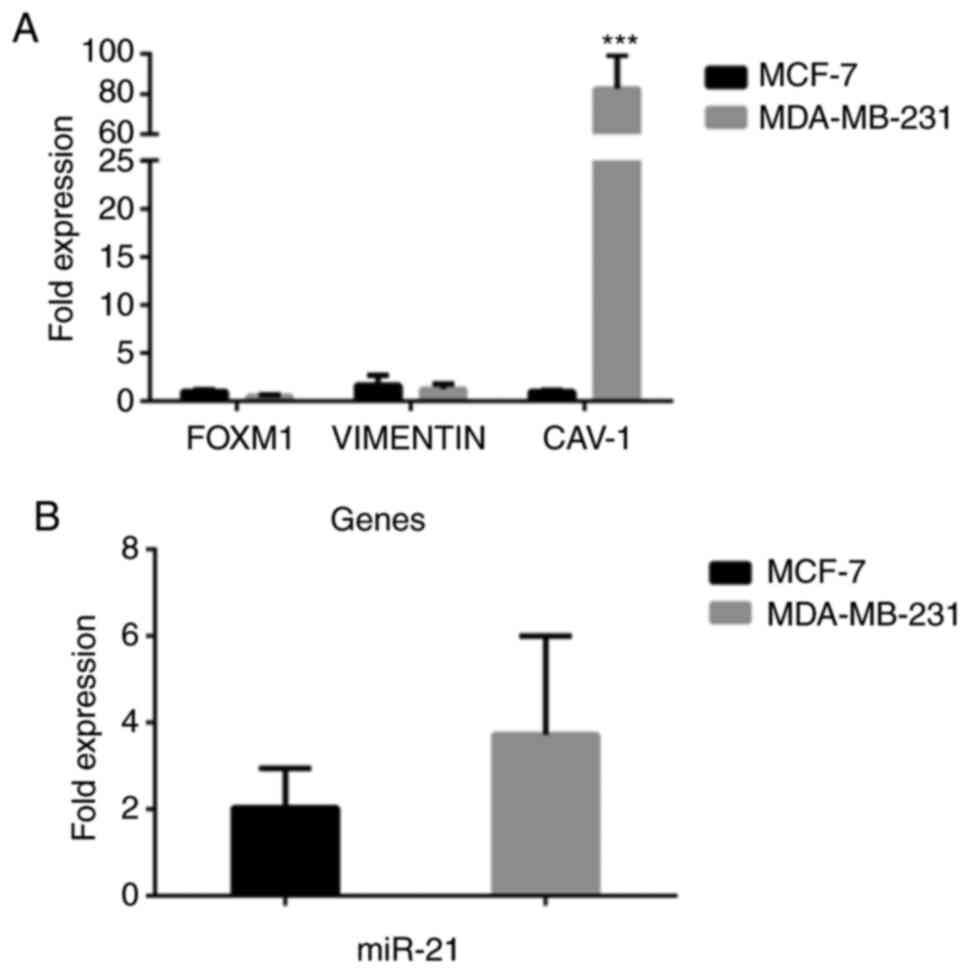
|
1
|
Bocci F, Gearhart-Serna L, Boareto M,
Ribeiro M, Ben-Jacob E, Devi GR, Levine H, Onuchic JN and Jolly MK:
Toward understanding cancer stem cell heterogeneity in the tumor
microenvironment. Proc Natl Acad Sci USA. 116:148–157.
2019.PubMed/NCBI View Article : Google Scholar
|
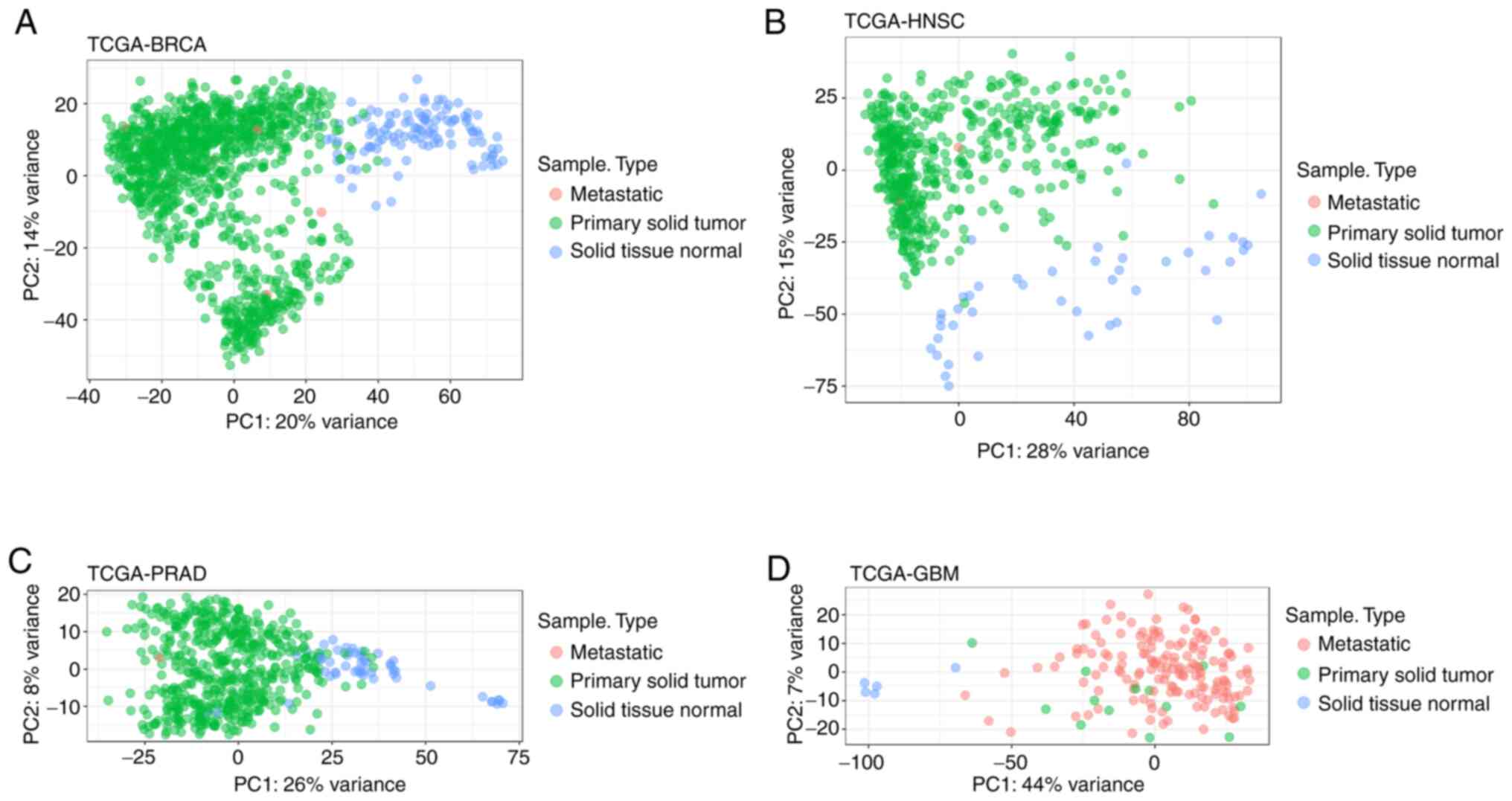
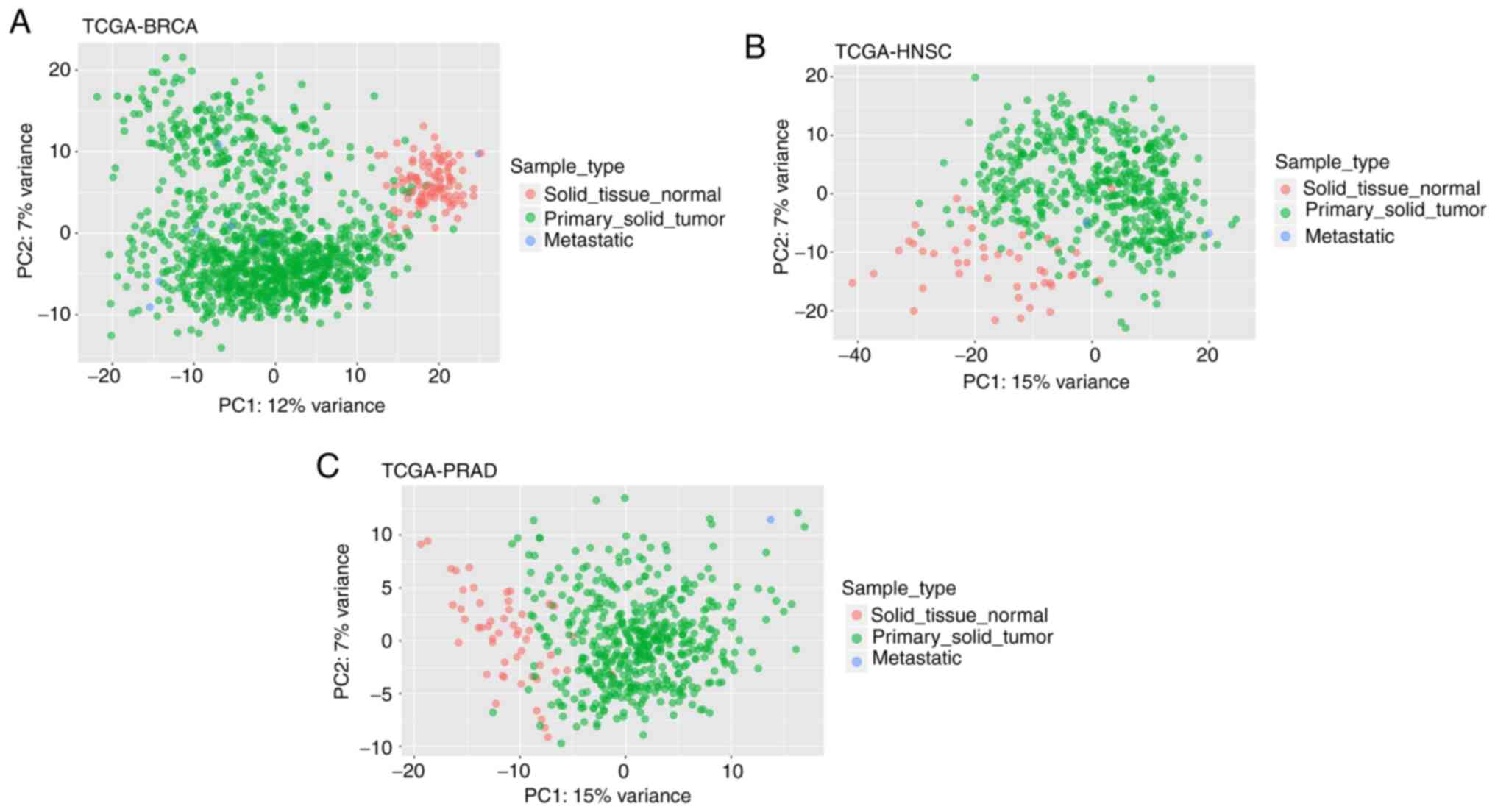
|
2
|
Vizirianakis IS, Miliotou AN, Mystridis
GA, Andriotis EG, Andreadis II, Papadopoulou LC and Fatouros DG:
Tackling pharmacological response heterogeneity by PBPK modeling to
advance precision medicine productivity of nanotechnology and
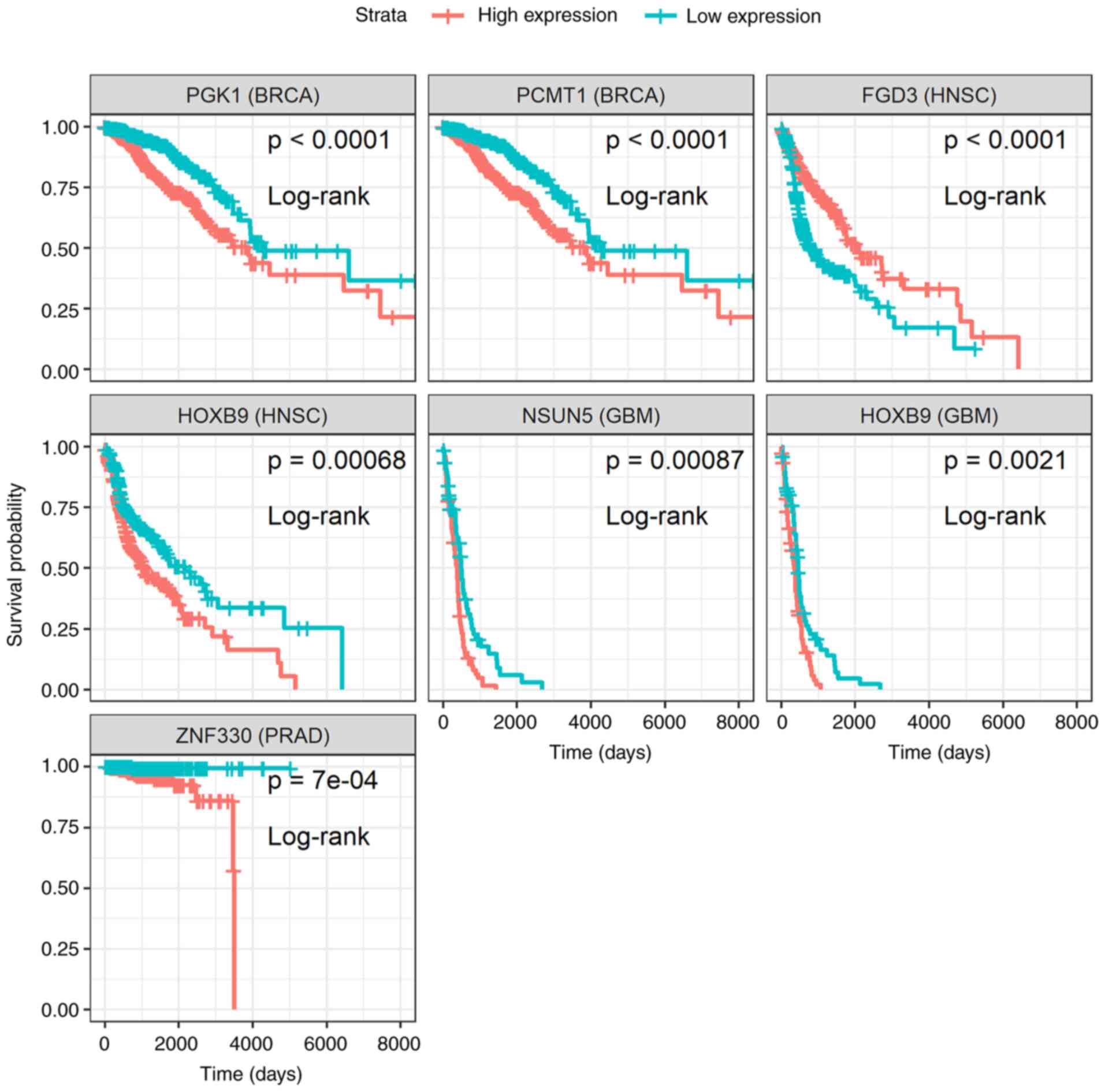
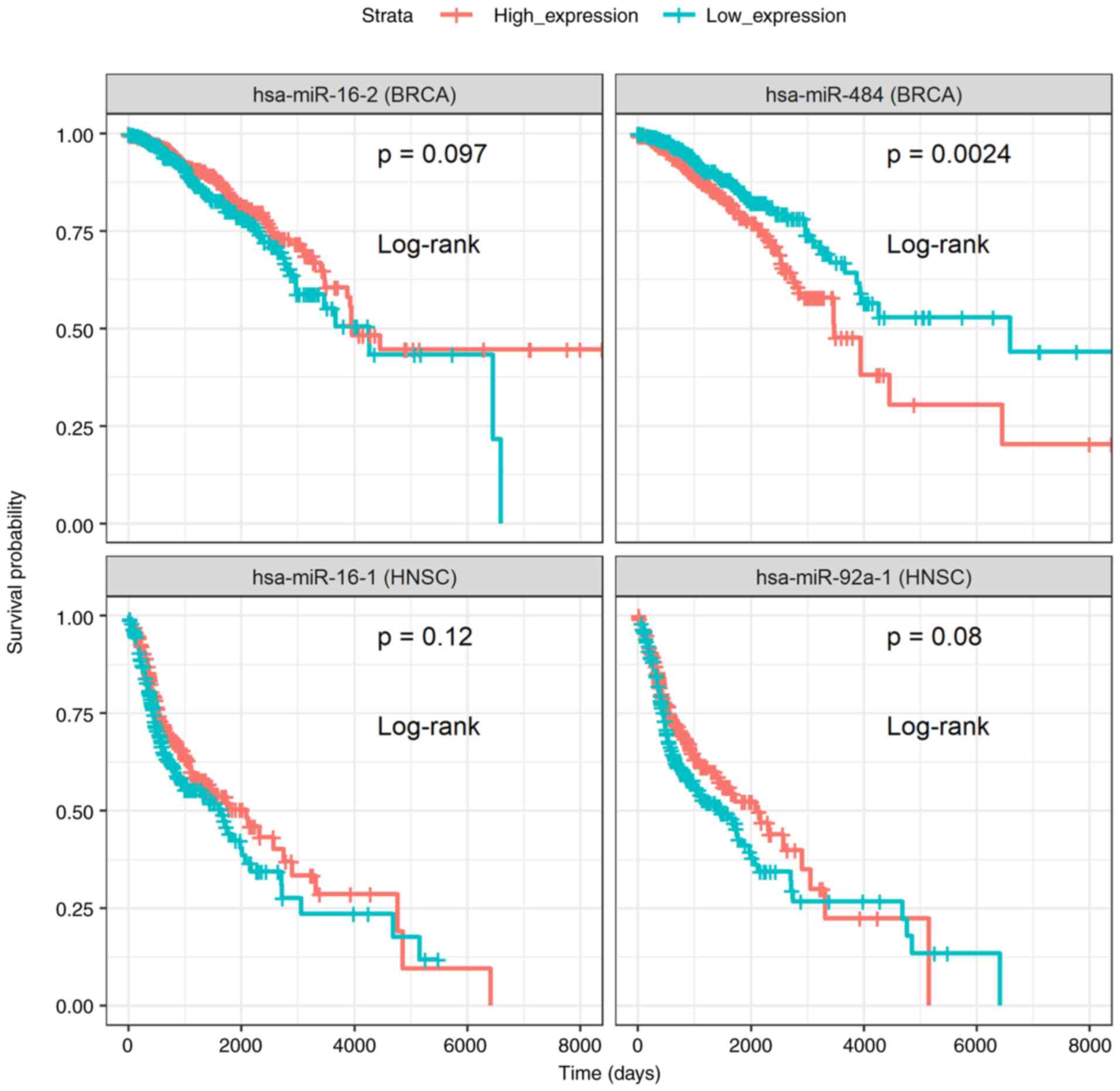
genomics therapeutics. Expert Rev Precis Med Drug Dev. 4:139–151.
2019.
|
|
3
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Roche J: The epithelial-to-mesenchymal
transition in cancer. Cancers (Basel). 10(52)2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Culig Z: Epithelial mesenchymal transition
and resistance in endocrine-related cancers. Biochim Biophys Acta
Mol Cell Res. 1866:1368–1375. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lu W and Kang Y: Epithelial-mesenchymal
plasticity in cancer progression and metastasis. Dev Cell.
49:361–374. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Williams ED, Gao D, Redfern A and Thompson
EW: Controversies around epithelial-mesenchymal plasticity in
cancer metastasis. Nat Rev Cancer. 19:716–732. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kyrodimou M, Andreadis D, Drougou A,
Amanatiadou EP, Angelis L, Barbatis C, Epivatianos A and
Vizirianakis IS: Desmoglein-3/γ-catenin and E-cadherin/ß-catenin
differential expression in oral leukoplakia and squamous cell
carcinoma. Clin Oral Investig. 18:199–210. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Cancer Genome Atlas Research Network.
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 45:1113–1120.
2013.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Zhao M, Kong L, Liu Y and Qu H: dbEMT: An
epithelial-mesenchymal transition associated gene resource. Sci
Rep. 5(11459)2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/Bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res.
44(e71)2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Anders S, Pyl PT and Huber W: HTSeq-a
Python framework to work with high-throughput sequencing data.
Bioinformatics. 31:166–169. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Shaffer JP: Multiple hypothesis testing.
Annu Rev Psychol. 46:561–584. 1995.
|
|
15
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Therneau TM and Grambsch PM: Modeling
survival data: Extending the Cox model. Springer-Verlag New York,
2000.
|
|
17
|
Therneau T: A package for survival
analysis in R. R package version. 3:2–7. 2020.
|
|
18
|
Kassambara A, Kosinski M and Biecek P:
survminer: Drawing survival curves using‘ggplot2’, 2017.
|
|
19
|
Tseligka ED, Rova A, Amanatiadou EP,
Calabrese G, Tsibouklis J, Fatouros DG and Vizirianakis IS:
Pharmacological development of target-specific delocalized
lipophilic cation-functionalized carboranes for cancer therapy.
Pharm Res. 33:1945–1958. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Akrivou MG, Demertzidou VP, Theodoroula
NF, Chatzopoulou FM, Kyritsis KA, Grigoriadis N, Zografos AL and
Vizirianakis IS: Uncovering the pharmacological response of novel
sesquiterpene derivatives that differentially alter gene expression
and modulate the cell cycle in cancer cells. Int J Oncol.
53:2167–2179. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
357(eaan2507)2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liang H, Fu Z, Jiang X, Wang N, Wang F,
Wang X, Zhang S, Wang Y, Yan X, Guan WX, et al: miR-16 promotes the
apoptosis of human cancer cells by targeting FEAT. BMC Cancer.
15(448)2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Qu Y, Liu H, Lv X, Liu Y, Wang X, Zhang M,
Zhang X, Li Y, Lou Q, Li S and Li H: MicroRNA-16-5p overexpression
suppresses proliferation and invasion as well as triggers apoptosis
by targeting VEGFA expression in breast carcinoma. Oncotarget.
8:72400–72410. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ruan L and Qian X: MiR-16-5p inhibits
breast cancer by reducing AKT3 to restrain NF-κB pathway. Biosci
Rep. 39(BSR20191611)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Nilsson S, Möller C, Jirström K, Lee A,
Busch S, Lamb R and Landberg G: Downregulation of miR-92a is
associated with aggressive breast cancer features and increased
tumour macrophage infiltration. PLoS One. 7(e36051)2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zearo S, Kim E, Zhu Y, Zhao JT, Sidhu SB,
Robinson BG and Soon PS: MicroRNA-484 is more highly expressed in
serum of early breast cancer patients compared to healthy
volunteers. BMC Cancer. 14(200)2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Volinia S and Croce CM: Prognostic
microRNA/mRNA signature from the integrated analysis of patients
with invasive breast cancer. Proc Natl Acad Sci USA. 110:7413–7417.
2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ye FG, Song CG, Cao ZG, Xia C, Chen DN,
Chen L, Li S, Qiao F, Ling H, Yao L, et al: Cytidine deaminase axis
modulated by miR-484 differentially regulates cell proliferation
and chemoresistance in breast cancer. Cancer Res. 75:1504–1515.
2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Thompson EW, Reich R, Shima TB, Albini A,
Graf J, Martin GR, Dickson RB and Lippman ME: Differential
regulation of growth and invasiveness of MCF-7 breast cancer cells
by antiestrogens. Cancer Res. 48:6764–6768. 1988.PubMed/NCBI
|
|
31
|
Gjerdrum C, Tiron C, Høiby T, Stefansson
I, Haugen H, Sandal T, Collett K, Li S, McCormack E, Gjertsen BT,
et al: Axl is an essential epithelial-to-mesenchymal
transition-induced regulator of breast cancer metastasis and
patient survival. Proc Natl Acad Sci USA. 107:1124–1129.
2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Theodossiou TA, Ali M, Grigalavicius M,
Grallert B, Dillard P, Schink KO, Olsen CE, Wälchli S, Inderberg
EM, Kubin A, et al: Simultaneous defeat of MCF7 and MDA-MB-231
resistances by a hypericin PDT-tamoxifen hybrid therapy. NPJ Breast
Cancer. 5(13)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sha L, Dong L, Lv L, Bai L and Ji X: HOXB9
promotes epithelial-to-mesenchymal transition via transforming
growth factor-β1 pathway in hepatocellular carcinoma cells. Clin
Exp Med. 15:55–64. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Bhatlekar S, Fields JZ and Boman BM: Role
of HOX genes in stem cell differentiation and cancer. Stem Cells
Int. 2018(3569493)2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hayashida T, Takahashi F, Chiba N,
Brachtel E, Takahashi M, Godin-Heymann N, Gross KW, Vivanco Md,
Wijendran V, Shioda T, et al: HOXB9, a gene overexpressed in breast
cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad
Sci USA. 107:1100–1105. 2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
De Braekeleer E, Douet-Guilbert N, Basinko
A, Le Bris MJ, Morel F and De Braekeleer M: Hox gene dysregulation
in acute myeloid leukemia. Futur Oncol. 10:475–495. 2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Eoh KJ, Kim HJ, Lee JY, Nam EJ, Kim S, Kim
SW and Kim YT: Dysregulated expression of homeobox family genes may
influence survival outcomes of patients with epithelial ovarian
cancer: Analysis of data from the cancer genome atlas. Oncotarget.
8:70579–70585. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Song J, Wang T, Xu W, Wang P, Wan J, Wang
Y, Zhan J and Zhang H: HOXB9 acetylation at K27 is responsible for
its suppression of colon cancer progression. Cancer Lett.
426:63–72. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhan J, Niu M, Wang P, Zhu X, Li S, Song
J, He H, Wang Y, Xue L, Fang W and Zhang H: Elevated HOXB9
expression promotes differentiation and predicts a favourable
outcome in colon adenocarcinoma patients. Br J Cancer. 111:883–893.
2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chiba N, Comaills V, Shiotani B, Takahashi
F, Shimada T, Tajima K, Winokur D, Hayashida T, Willers H, Brachtel
E, et al: Homeobox B9 induces epithelial-to-mesenchymal
transition-associated radioresistance by accelerating DNA damage
responses. Proc Natl Acad Sci USA. 109:2760–2765. 2012.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Carbone C, Piro G, Simionato F, Ligorio F,
Cremolini C, Loupakis F, Alì G, Rossini D, Merz V, Santoro R, et
al: Homeobox B9 mediates resistance to anti-VEGF therapy in
colorectal cancer patients. Clin Cancer Res. 23:4312–4322.
2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zhang G, Fan E, Yue G, Zhong Q, Shuai Y,
Wu M, Feng G, Chen Q and Gou X: Five genes as a novel signature for
predicting the prognosis of patients with laryngeal cancer. J Cell
Biochem, Oct 31, 2019 (Online ahead of print).
|
|
43
|
Xu H, Wu S, Shen X, Wu D, Qin Z, Wang H,
Chen X and Sun X: Silencing of HOXB9 suppresses cellular
proliferation, angiogenesis, migration and invasion of prostate
cancer cells. J Biosci. 45(40)2020.PubMed/NCBI
|
|
44
|
Fu D, He C, Wei J, Zhang Z, Luo Y, Tan H
and Ren C: PGK1 is a potential survival biomarker and invasion
promoter by regulating the HIF-1α-mediated epithelial-mesenchymal
transition process in breast cancer. Cell Physiol Biochem.
51:2434–2444. 2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
He Y, Luo Y, Zhang D, Wang X, Zhang P, Li
H, Ejaz S and Liang S: PGK1-mediated cancer progression and drug
resistance. Am J Cancer Res. 9:2280–2302. 2019.PubMed/NCBI
|
|
46
|
Sambri I, Capasso R, Pucci P, Perna AF and
Ingrosso D: The microRNA 15a/16-1 cluster down-regulates protein
repair isoaspartyl methyltransferase in hepatoma cells:
Implications for apoptosis regulation. J Biol Chem.
286:43690–43700. 2011.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Dong L, Li Y, Xue D and Liu Y: PCMT1 is an
unfavorable predictor and functions as an oncogene in bladder
cancer. IUBMB Life. 70:291–299. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Schosserer M, Minois N, Angerer TB, Amring
M, Dellago H, Harreither E, Calle-Perez A, Pircher A, Gerstl MP,
Pfeifenberger S, et al: Methylation of ribosomal RNA by NSUN5 is a
conserved mechanism modulating organismal lifespan. Nat Commun.
6(6158)2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Heissenberger C, Liendl L, Nagelreiter F,
Gonskikh Y, Yang G, Stelzer EM, Krammer TL, Micutkova L, Vogt S,
Kreil DP, et al: Loss of the ribosomal RNA methyltransferase NSUN5
impairs global protein synthesis and normal growth. Nucleic Acids
Res. 47:11807–11825. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
de Melo IS, Iglesias C, Benítez-Rondán A,
Medina F, Martínez-Barberá JP and Bolívar J: NOA36/ZNF330 is a
conserved cystein-rich protein with proapoptotic activity in human
cells. Biochim Biophys Acta. 1793:1876–1885. 2009.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Willis S, Sun Y, Abramovitz M, Fei T,
Young B, Lin X, Ni M, Achua J, Regan MM, Gray KP, et al: High
expression of FGD3, a putative regulator of cell morphology and
motility, is prognostic of favorable outcome in multiple cancers.
JCO Precis Oncol 1: PO.17.00009, 2017.
|
|
52
|
Renda I, Bianchi S, Vezzosi V, Nori J,
Vanzi E, Tavella K and Susini T: Expression of FGD3 gene as
prognostic factor in young breast cancer patients. Sci Rep.
9(15204)2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Vidigal JA and Ventura A: The biological
functions of miRNAs: Lessons from in vivo studies. Trends Cell
Biol. 25:137–147. 2015.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Peng Y and Croce CM: The role of microRNAs
in human cancer. Signal Transduct Target Ther.
1(15004)2016.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Bartel DP: Metazoan MicroRNAs. Cell.
173:20–51. 2018.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Michael IP, Saghafinia S and Hanahan D: A
set of microRNAs coordinately controls tumorigenesis, invasion, and
metastasis. Proc Natl Acad Sci USA. 116:24184–24195.
2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Cherone JM, Jorgji V and Burge CB:
Cotargeting among microRNAs in the brain. Genome Res. 29:1791–1804.
2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Dong J, Xiao D, Zhao Z, Ren P, Li C, Hu Y,
Shi J, Su H, Wang L, Liu H, et al: Epigenetic silencing of
microRNA-137 enhances ASCT2 expression and tumor glutamine
metabolism. Oncogenesis. 6(e356)2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Zhang Z, Liu R, Shuai Y, Huang Y, Jin R,
Wang X and Luo J: ASCT2 (SLC1A5)-dependent glutamine uptake is
involved in the progression of head and neck squamous cell
carcinoma. Br J Cancer. 122:82–93. 2020.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Wu P, Xiao Y, Guo T, Wang Y, Liao S, Chen
L and Liu Z: Identifying miRNA-mRNA pairs and novel miRNAs from
hepatocelluar carcinoma mirnomes and TCGA database. J Cancer.
10:2552–2559. 2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Nuzzo S, Catuogno S, Capuozzo M, Fiorelli
A, Swiderski P, Boccella S, de Nigris F and Esposito CL:
Axl-targeted delivery of the oncosuppressor miR-137 in
non-small-cell lung cancer. Mol Ther Nucleic Acids. 17:256–263.
2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Drago-García D, Espinal-Enríquez J and
Hernández-Lemus E: Network analysis of EMT and MET micro-RNA
regulation in breast cancer. Sci Rep. 7(13534)2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Schneider E, Pochert N, Ruess C, MacPhee
L, Escano L, Miller C, Krowiorz K, Delsing Malmberg E,
Heravi-Moussavi A, Lorzadeh A, et al: MicroRNA-708 is a novel
regulator of the Hoxa9 program in myeloid cells. Leukemia.
34:1253–1265. 2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Looney AM, Nawaz K and Webster RM:
Tumor-agnostic therapies. Nat Rev Drug Discov. 19:383–384.
2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Astras G, Papagiannopoulos CI, Kyritsis
KA, Markitani C and Vizirianakis IS: Pharmacogenomic testing to
guide personalized cancer medicine decisions in private oncology
practice: A case study. Front. Oncol. 10(521)2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Gu Z, Eils R and Schlesner M: Complex
heatmaps reveal patterns and correlations in multidimensional
genomic data. Bioinformatics. 32:2847–2849. 2016.PubMed/NCBI View Article : Google Scholar
|