1. Introduction
In eukaryotic cells, genomic DNA is present as chromatin. This complex structuring is achieved as the DNA is bound to DNA-binding proteins, the histones. The complex of DNA and histones is known as a nucleosome and represents the functional unit of chromatin (1). Such a nucleosome includes a DNA region of 145-147 base pairs, which is wound as a superhelix around a histone octamer. The histone group consists of four families: H2A, H2B, H3 and H4. Two histones from each family form a dimer, H2A-H2B and H3-H4, which finally assemble as two tetramers (H2A-H2B)2 and (H3-H4)2 to form the functional histone octamer (1,2). The histones of each of the four families are encoded by several genes and are expressed during DNA synthesis in the S phase of the cell cycle (3).
2. Histone H2A.X and its phosphorylation
The histone family H2A consists of the members, H2A.1, H2A.2, H2A.X and H2A.Z (4). The main part of the histone family is represented by H2A.1 and H2A.2. The two isoforms differ in only a few amino acids and thus far, no differential functions have been detected. In eukaryotes, H2A.Z comprises ~10% of the H2A histones, with H2A.X comprising even up to 25%. In mammals, however, H2A.X is much less expressed and represents only up to 10% of the H2A histones (5).
H2A.X is 124 amino acids in length and differs from the other members of the H2A family by a highly conserved 22 amino acid domain at the C-terminus. The C-terminal motif Ser139-Gln140-Glu141-Tyr142 is used for post-translational modification of the histone, where the protein can be phosphorylated at Ser139 (5,6).
H2A.X, as with all other histones, serves to structure and stabilize the DNA (7). It also has a very specific function in the complex DNA damage detection and repair machinery of higher eukaryotes. The phosphorylation of H2A.X at position Ser139 (γH2A.X) is one of the first signals for the detection of DNA double-strand breaks (DSBs) and an essential step for the initialization of DNA repair (8,9). DSBs are genetic damage events with highest cytotoxicity (10). DSBs occur in human cells ~10 times per day and cell, which impressively reflects the enormous efficiency of the DNA damage detection and repair system of eukaryotic cells (11). The phosphorylation of H2AX serves as one of the first signals for a DSB, and is thus an essential prerequisite for the activation of DNA repair systems, and thus contributes significantly to the stabilization of the genome against genotoxic noxae.
A single DSB leads to the binding of several hundred to thousand γH2A.X proteins to the affected DNA region. γH2A.X-DNA binding extends over a length of 0.5-1.7 megabase pairs, so that the entire environment of the DSB is epigenetically labelled (12). These mechanisms are well studied in radiation-induced DSB. Phosphorylation of H2A.X to γH2A.X occurs within minutes after the damage event and is proportional to the dose of ionizing radiation (biodosimetry). Per gray of radiation, ~1% of the H2A.X protein present in the nucleus is phosphorylated (4).
The phosphorylation of H2A.X is not limited to the effect of ionizing radiation (9,13) (Fig. 1). A number of other exogenous noxae can lead to the formation of γH2A.X. Physical noxae include ultraviolet radiation, low pH, and heat stress (14-17). Chemical factors are DNA-damaging agents, such as bleomycin, doxorubicin and reactive oxygen species (18-20). There are also cellular events that can induce H2A.X phosphorylation. During somatic recombination to ensure antibody variability in B cells and for genome stabilization under replicative stress, phosphorylation of H2A.X occurs (15,21,22). Furthermore, DNA damage, including γH2A.X signals also occurs during aging and apoptosis (23-25). In mammalian germ cells, sex chromosomes X and Y are epigenetically silenced by condensing the chromatin of both chromosomes (XY body). In the regulation of this process, gH2A.X is involved along with a number of other factors of DNA repair (26). Since the phosphorylation of the histone variant sometimes occurs independently of DSBs, the characterization of kinetics, number, size and morphology of detected γH2A.X foci is of great relevance (27). The immunocytochemical γH2A.X detection using a phosphor-specific antibody for the phosphorylation on Ser139 at the C-terminus of H2A.X is based on the assumption that the intensity of immunofluorescence correlates stoichiometrically with the frequency of DSB (14,27,28). The phosphorylated histone isoform can be detected in so-called γH2A.X foci.
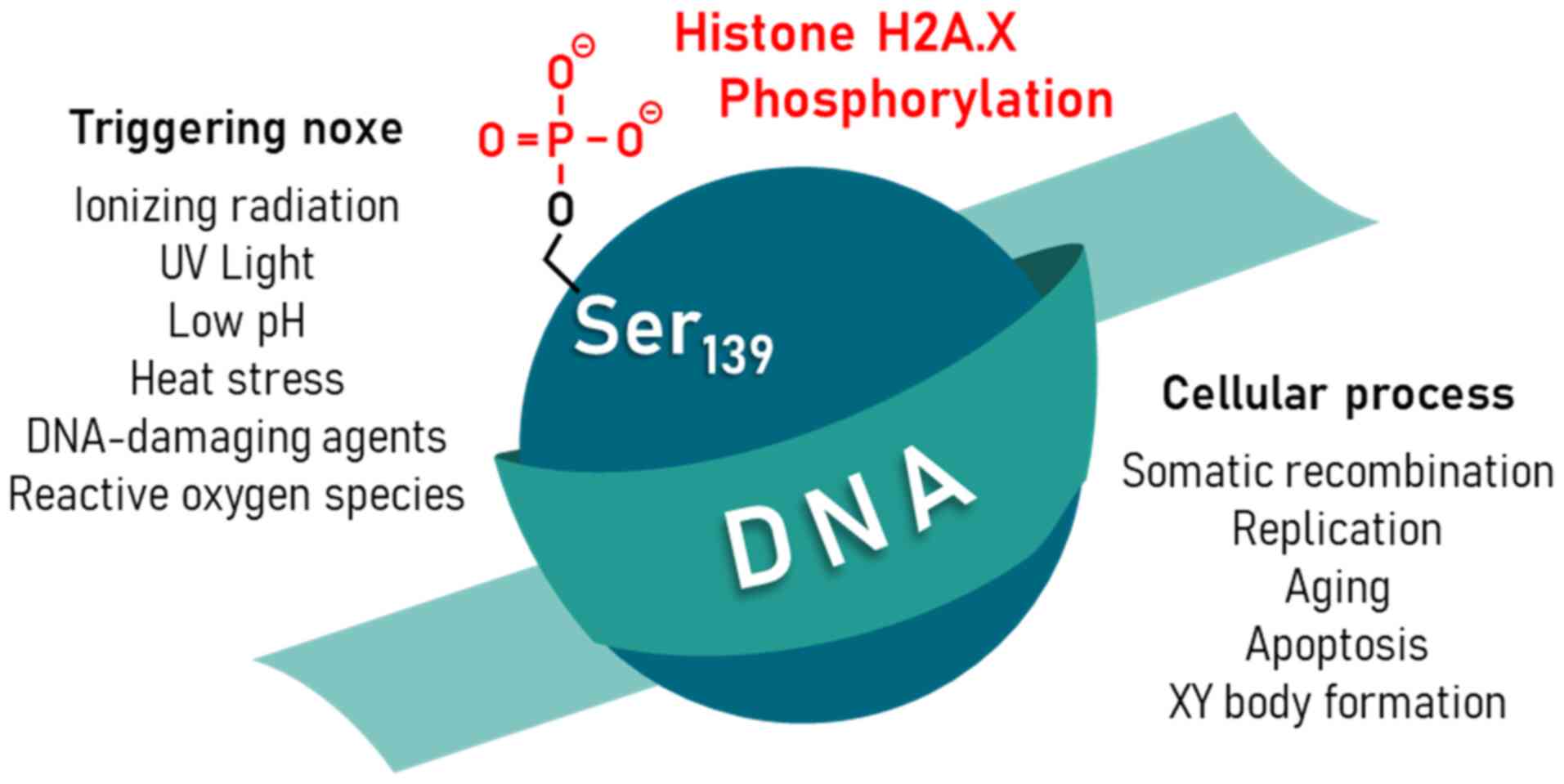 |
Figure 1
Physical and (bio)chemical stress noxae, such as ionizing radiation, UV light, low pH, heat stress, DNA-damaging agents, and reactive oxygen species can lead to the phosphorylation of serine 139 (Ser139) of histone H2A.X in eukaryotic cells. In addition, physiological cellular processes such as somatic recombination, replication, aging, apoptosis, and XY body formation may also result in Ser139-phosphorylated H2A.X. UV, ultraviolet.
|
gH2A.X activity is critical for maintaining genome stability in a wide variety of cellular processes. As with all regulatory processes, however, it must also be possible to switch off the corresponding signals. In the case of H2A.X activity, this is dictated by a phosphorylation-dephosphorylation cycle. Phosphorylation is catalyzed by the kinases ataxia telangiectasia mutated (ATM), ataxia telangiectasia and Rad3-related protein (ATR), DNA-dependent protein kinase (DNA-PK) and mitogen-activated protein (MAP) kinases p38, which can be reversed by the protein phosphatases (PP) PP2A, PP4, PP6 and wild-type p53-induced phosphatase 1 (WIP1) (29-32) (Fig. 2).
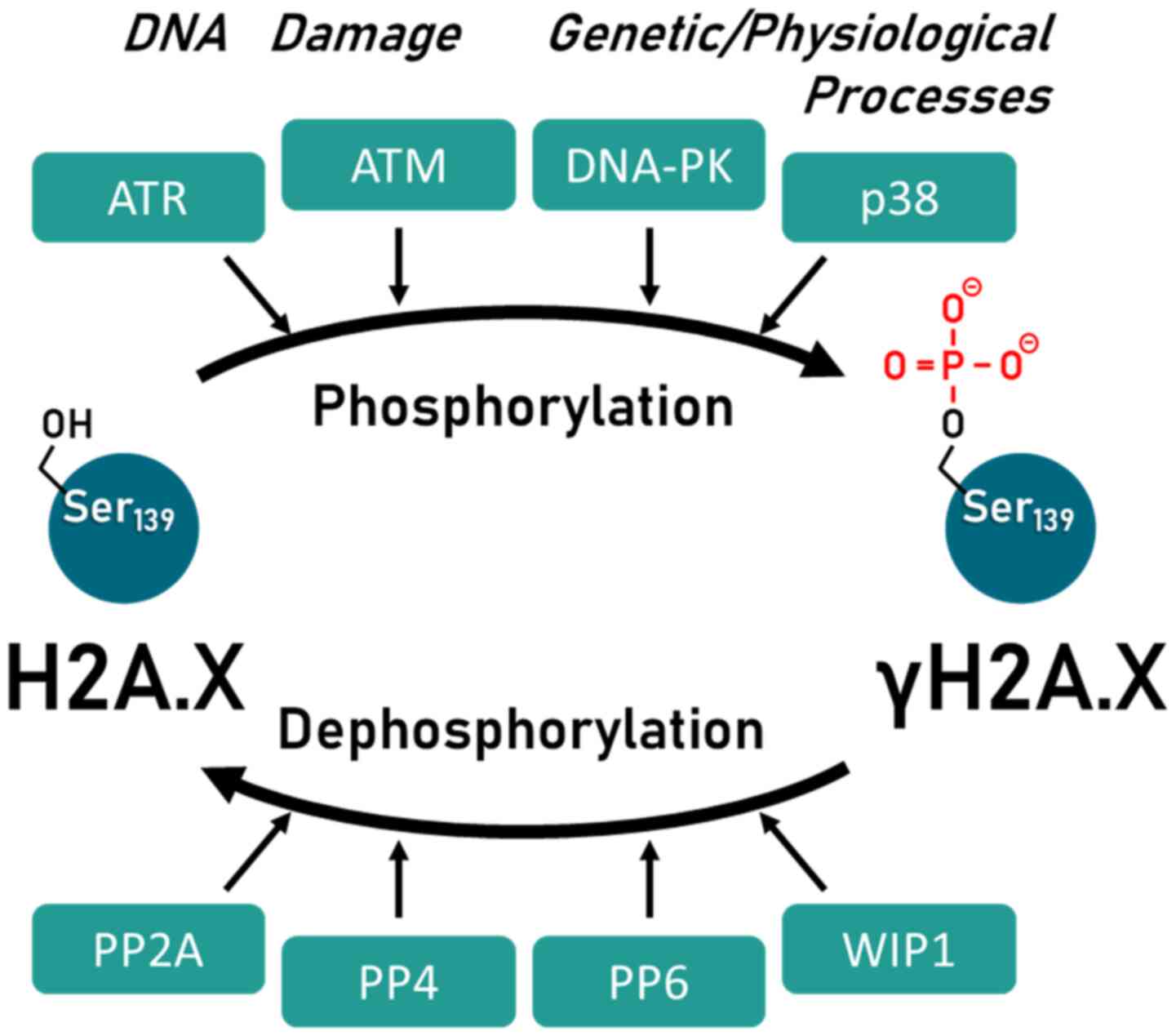 |
Figure 2
Regulation of H2A.X activity by the phosphorylation-dephosphorylation cycle of various kinases and protein phosphatases. Kinases involved are: ATM, ATR, DNA, MAPK p38. Protein phosphatases involved are: PP2A, PP4, PP6, WIP1. The figure has been modified from a previous study (32). ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3-related protein; DNA-PK, DNA-dependent protein kinase; MAPK p38, mitogen-activated protein kinase p38; P2A, protein phosphatase 2A; PP4, protein phosphatase 4; PP6, protein phosphatase 6; WIP1, wild-type p53-induced phosphatase 1.
|
3. Phosphorylation of H2A.X in the DNA damage response
The cellular response to detected DNA damage includes both temporary cellular responses, such as checkpoint control activation, as well as permanent cellular responses such as cell cycle arrest and apoptosis (7).
DNA damage detection and repair constitute a complex cellular event involving the entire histone biochemistry. Both the structure and function of nucleosomes are regulated by post-transcriptional modifications to histone proteins. These include acetylation, methylation, ubiquitination and phosphorylation, which orchestrate the signaling pathways involved in DNA repair and induce structural changes in the DNA histone architecture required for this process (33). A key post-translational modification in the context of DNA damage is the phosphorylation of H2A.X by the kinases, ATM, ATR and DNA-PK (9,15,34-36). As serine/threonine kinases, these enzymes belong to the phosphoinositide 3-kinase (PI3K) family and act as sensors for DNA damage (37). PI3K phosphorylate their substrates on the amino acid motif serine or threonine, glutamine, and glutamic acid (Ser/Thr-Gln-Glu) (38).
During DNA damage detection, γH2A.X and DNA repair proteins, such as p53 binding protein 1 (53BP1) and breast cancer 1 (BRCA1) colocate (39,40). The activation of PI3K is still a general reaction to different DNA damage, and only the activation of H2AX by phosphorylation leads to the mobilization of DSB specific signal and repair proteins (41). However, current investigations indicate that both DSB recognition and the recruitment of repair factors are still feasible without γH2A.X (37).
4. γH2A.X as a protein biomarker
Based on the proportionality of γH2A.X formation and radiation dose as utilized in biodosimetry in radiobiology (4), γH2A.X was previously tested as a direct correlate for physically and chemically induced DSB (6). Furthermore, γH2A.X was also tested as a biomarker for incidences correlating with DSB. It has been used as a biomarker of cell death in the presence of chemotherapeutic agents and to detect the genotoxic effect of tobacco smoke in lung cells (18,42).
In the evaluation of the γH2A.X focus assay with primary mouse embryonic fibroblast cells, the test method was found to be as specific and sensitive as the two genotoxic standard procedures, the micronucleus assay and the comet assay (43). H2A.X phosphorylation has been defined as a genotoxic endpoint along with micronucleus formation and mutation frequency. γH2A.X detection is thus also used for clinical monitoring of DNA damage during chemotherapy (44) and for determining patient radiosensitivity (13,45,46).
In oncology, the quantification of γH2A.X can be used to detect precancerous lesions (47). The level of phosphorylated H2AX correlates with cancer-associated genomic instability of tumor cells and could potentially be used as a biomarker for prediction and recurrence (7,47,48). In addition, the DSB-dependent accumulation of γH2A.X can be used as a biodosimeter to determine age (24). In recent years, it has become increasingly clear that H2AX phosphorylation not only serves to detect DNA damage, but also performs an essential task in the processes of chromatin remodeling and thus DNA repair itself. Therefore, in fractionated radiotherapy, there are approaches to individually determine the time intervals by monitoring the gH2A.X status, so that healthy tissue can recover as completely as possible (Table I) (49,50).
 |
Table I
Current applications of γH2A.X detection in biomedicine.
|
Table I
Current applications of γH2A.X detection in biomedicine.
| γH2A.X application |
(Refs.) |
| Approved biological dosimetry |
(4,6) |
| Approved genotoxicity assay |
(42,44) |
| Cytostatics efficacy assay |
(18) |
| Radiotherapy outcome monitoring |
(13,45,46) |
| Detection of precancerous lesions |
(47) |
| Detection of cancer progression |
(7,48) |
| Tissue regeneration monitoring |
(49,50) |
5. Conclusion and future perspectives
As discussed above, a growing body of literature demonstrates the importance of H2A.X in DNA damage recognition and repair, but also in genome remodeling processes in general. An essential component of this functionality is the activation and inactivation of the histone by phosphorylation and dephosphorylation. The H2A.X phosphorylation status thus represents a central sum parameter for the genome integrity of eukaryotic cells and allows conclusions to be drawn about DNA-associated processes in cells and tissues. This can be of great use for applications in life sciences and medicine. As a biomarker for DNA damage and genotoxicity as well as a clinical marker for radiotherapy outcome, drug efficacy, and tissue regeneration, the H2A.X/γH2A.X status is already used or such applications are emerging. Particularly in the field of biological dosimetry, microscopic routine systems for the detection of γH2A.X are already in use, which allow automated, rapid and cost-effective analysis even of large numbers of samples. The establishment of γH2A.X routine analysis in corresponding specialized treatment centers would therefore be easily feasible.
As with all biomarkers, the specificity of γH2A.X detection is of primary importance. The complex cellular mechanisms in which γH2A.X is involved listed in this review article therefore also highlight potential limitations of practical/clinical use. Countering these is therefore an important goal of future γH2A.X research. For example, the combined detection of γH2A.X and 53BP1 significantly increased the precision of DSB-based biodosimetry following radiation exposure. Combining multiple markers thus represents a promising strategy with which to increase the reliability of biomarkers and thereby bring them into practical application.
Acknowledgements
The author would like to thank Dr Rebekka Einenkel from the Department of Gynecological Endocrinology and Reproductive Medicine (University Hospital Bonn) for her excellent assistance in preparing and styling the figures.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Author's contributions
MBS conceptualized, prepared and drafted the manuscript. MBS confirms the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The author declares that there are no competing interests.
References
|
1
|
Biterge B and Schneider R: Histone variants: Key players of chromatin. Cell Tissue Res. 356:457–466. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Luger K, Mäder AW, Richmond RK, Sargent DF and Richmond TJ: Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 389:251–260. 1997.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Singh RK, Paik J and Gunjan A: Generation and management of excess histones during the cell cycle. Front Biosci. 14:3145–3158. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K and Bonner W: Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 12:162–169. 2002.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kuo LJ and Yang LX: Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo. 22:305–309. 2008.PubMed/NCBI
|
|
6
|
Sharma A, Singh K and Almasan A: Histone H2AX phosphorylation: A marker for DNA damage. Methods Mol Biol. 920:613–626. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Varvara PV, Karaolanis G, Valavanis C, Stanc G, Tzaida O, Trihia H, Patapis P, Dimitroulis D and Perrea D: gamma-H2AX: A potential biomarker in breast cancer. Tumour Biol: Sep 25, 2019 (Epub ahead of print). doi: 10.1177/1010428319878536.
|
|
8
|
Johnson DP, Chandrasekharan MB, Dutreix M and Bhaskara S: Targeting DNA repair and chromatin crosstalk in cancer therapy. Cancers (Basel). 13(381)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Löbrich M and Jeggo PA: ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 64:2390–2396. 2004.PubMed/NCBI View Article : Google Scholar
|
|
10
|
House NC, Koch MR and Freudenreich CH: Chromatin modifications and DNA repair: Beyond double-strand breaks. Front Genet. 5(296)2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ambekar SS, Hattur SS and Bule PB: DNA: Damage and repair mechanisms in humans. Glob J Pharm Pharm Sci. 3(555613)2018.
|
|
12
|
Rogakou EP, Boon C, Redon C and Bonner WM: Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 146:905–916. 1999.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Osipov AN, Pustovalova M, Grekhova A, Eremin P, Vorobyova N, Pulin A, Zhavoronkov A, Roumiantsev S, Klokov DY and Eremin I: Low doses of X-rays induce prolonged and ATM-independent persistence of γH2AX foci in human gingival mesenchymal stem cells. Oncotarget. 6:27275–27287. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Halicka HD, Huang X, Traganos F, King MA, Dai W and Darzynkiewicz Z: Histone H2AX phosphorylation after cell irradiation with UV-B: Relationship to cell cycle phase and induction of apoptosis. Cell Cycle. 4:339–345. 2005.PubMed/NCBI
|
|
15
|
Parsels LA, Parsels JD, Tanska DM, Maybaum J, Lawrence TS and Morgan MA: The contribution of DNA replication stress marked by high-intensity, pan-nuclear γH2AX staining to chemosensitization by CHK1 and WEE1 inhibitors. Cell Cycle. 17:1076–1086. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Xiao H, Li TK, Yang J-M and Liu LF: Acidic pH induces topoisomerase II-mediated DNA damage. Proc Natl Acad Sci USA. 100:5205–5210. 2003.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kaneko H, Igarashi K, Kataoka K and Miura M: Heat shock induces phosphorylation of histone H2AX in mammalian cells. Biochem Biophys Res Commun. 328:1101–1106. 2005.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Banáth JP and Olive PL: Expression of phosphorylated histone H2AX as a surrogate of cell killing by drugs that create DNA double-strand breaks. Cancer Res. 63:4347–4350. 2003.PubMed/NCBI
|
|
19
|
Kiziltepe T, Hideshima T, Catley L, Raje N, Yasui H, Shiraishi N, Okawa Y, Ikeda H, Vallet S, Pozzi S, et al: 5-Azacytidine, a DNA methyltransferase inhibitor, induces ATR-mediated DNA double-strand break responses, apoptosis, and synergistic cytotoxicity with doxorubicin and bortezomib against multiple myeloma cells. Mol Cancer Ther. 6:1718–1727. 2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Guo Z, Kozlov S, Lavin MF, Person MD and Paull TT: ATM activation by oxidative stress. Science. 330:517–521. 2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li Z, Owonikoko TK, Sun SY, Ramalingam SS, Doetsch PW, Xiao ZQ, Khuri FR, Curran WJ and Deng X: c-Myc suppression of DNA double-strand break repair. Neoplasia. 14:1190–1202. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ewald B, Sampath D and Plunkett W: H2AX phosphorylation marks gemcitabine-induced stalled replication forks and their collapse upon S-phase checkpoint abrogation. Mol Cancer Ther. 6:1239–1248. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Williamson J, Hughes CM, Burke G and Davison GW: A combined γ-H2AX and 53BP1 approach to determine the DNA damage-repair response to exercise in hypoxia. Free Radic Biol Med. 154:9–17. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM and Barrett JC: Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol. 6:168–170. 2004.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bekeschus S, Schütz CS, Niessner F, Wende K, Weltmann KD, Gelbrich N, von Woedtke T, Schmidt A and Stope MB: Elevated H2AX phosphorylation observed with kINPen plasma treatment is not caused by ROS-mediated DNA damage but is the consequence of apoptosis. Oxid Med Cell Longev. 2019(8535163)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kulashreshtha M, Mehta IS, Kumar P and Rao BJ: Chromosome territory relocation during DNA repair requires nuclear myosin 1 recruitment to chromatin mediated by γ-H2AX signaling. Nucleic Acids Res. 44:8272–8291. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Goutham HV, Mumbrekar KD, Vadhiraja BM, Fernandes DJ, Sharan K, Kanive Parashiva G, Kapaettu S and Bola Sadashiva SR: DNA double-strand break analysis by γ-H2AX foci: A useful method for determining the overreactors to radiation-induced acute reactions among head-and-neck cancer patients. Int J Radiat Oncol Biol Phys. 84:e607–e612. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sedelnikova OA, Rogakou EP, Panyutin IG and Bonner WM: Quantitative detection of (125)IdU-induced DNA double-strand breaks with gamma-H2AX antibody. Radiat Res. 158:486–492. 2002.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Tomita M: Involvement of DNA-PK and ATM in radiation- and heat-induced DNA damage recognition and apoptotic cell death. J Radiat Res (Tokyo). 51:493–501. 2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Fernandez-Capetillo O, Lee A, Nussenzweig M and Nussenzweig A: H2AX: The histone guardian of the genome. DNA Repair (Amst). 3:959–967. 2004.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Li X, Nan A, Xiao Y, Chen Y and Lai Y: PP2A-B56є complex is involved in dephosphorylation of γ-H2AX in the repair process of CPT-induced DNA double-strand breaks. Toxicology. 331:57–65. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Tu WZ, Li B, Huang B, Wang Y, Liu XD, Guan H, Zhang SM, Tang Y, Rang WQ and Zhou PK: γH2AX foci formation in the absence of DNA damage: Mitotic H2AX phosphorylation is mediated by the DNA-PKcs/CHK2 pathway. FEBS Lett. 587:3437–3443. 2013.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Miller KM and Jackson SP: Histone marks: Repairing DNA breaks within the context of chromatin. Biochem Soc Trans. 40:370–376. 2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhao H, Halicka HD, Garcia J, Li J and Darzynkiewicz Z: ATM activation and H2AX phosphorylation induced by genotoxic agents assessed by flow- and laser scanning cytometry. Methods Mol Biol. 1599:183–196. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hanasoge S and Ljungman M: H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis. 28:2298–2304. 2007.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Park E-J, Chan DW, Park J-H, Oettinger MA and Kwon J: DNA-PK is activated by nucleosomes and phosphorylates H2AX within the nucleosomes in an acetylation-dependent manner. Nucleic Acids Res. 31:6819–6827. 2003.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kinner A, Wu W, Staudt C and Iliakis G: Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36:5678–5694. 2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kim ST, Lim DS, Canman CE and Kastan MB: Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 274:37538–37543. 1999.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhang F and Gong Z: Regulation of DNA double-strand break repair pathway choice: A new focus on 53BP1. J Zhejiang Univ Sci B. 22:38–46. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M and Bonner WM: A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 10:886–895. 2000.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kang J, Ferguson D, Song H, Bassing C, Eckersdorff M, Alt FW and Xu Y: Functional interaction of H2AX, NBS1, and p53 in ATM-dependent DNA damage responses and tumor suppression. Mol Cell Biol. 25:661–670. 2005.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Tanaka T, Huang X, Jorgensen E, Gietl D, Traganos F, Darzynkiewicz Z and Albino AP: ATM activation accompanies histone H2AX phosphorylation in A549 cells upon exposure to tobacco smoke. BMC Cell Biol. 8(26)2007.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Watters GP, Smart DJ, Harvey JS and Austin CA: H2AX phosphorylation as a genotoxicity endpoint. Mutat Res. 679:50–58. 2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al: DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 434:864–870. 2005.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Olive PL and Banáth JP: Phosphorylation of histone H2AX as a measure of radiosensitivity. Int J Radiat Oncol Biol Phys. 58:331–335. 2004.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kawashima S, Kawaguchi N, Taniguchi K, Tashiro K, Komura K, Tanaka T, Inomata Y, Imai Y, Tanaka R, Yamamoto M, et al: γ-H2AX as a potential indicator of radiosensitivity in colorectal cancer cells. Oncol Lett. 20:2331–2337. 2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Gorgoulis VG, Vassiliou L-VF, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B, et al: Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 434:907–913. 2005.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Palla VV, Karaolanis G, Katafigiotis I, Anastasiou I, Patapis P, Dimitroulis D and Perrea D: gamma-H2AX: Can it be established as a classical cancer prognostic factor? Tumour Biol: Mar 29, 2017 (Epub ahead of print). doi: 10.1177/1010428317695931.
|
|
49
|
Baritaud M, Cabon L, Delavallée L, Galán-Malo P, Gilles ME, Brunelle-Navas MN and Susin SA: AIF-mediated caspase-independent necroptosis requires ATM and DNA-PK-induced histone H2AX Ser139 phosphorylation. Cell Death Dis. 3(e390)2012.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Mariotti LG, Pirovano G, Savage KI, Ghita M, Ottolenghi A, Prise KM and Schettino G: Use of the γ-H2AX assay to investigate DNA repair dynamics following multiple radiation exposures. PLoS One. 8(e79541)2013.PubMed/NCBI View Article : Google Scholar
|
















