Introduction
The transcription factors that form the nuclear receptor (NR) superfamily are either ligand-activated or orphan proteins that regulate numerous physiological processes, such as metabolism, reproduction and homeostasis in humans, as well as in other organisms of the animal kingdom (1,2). The dysregulation of their signaling pathways can lead to the development of severe health abnormalities, such as cancer, organ dysfunction (3), osteoporosis (4), metabolic disorders (1), etc. Due to their ability to bind to small lipophilic molecules and regulate the expression of target genes, NRs are considered ideal drug targets for the confrontation of several of these diseases and are even considered promising targets for the development of personalized medicine (5-7). However, further research is required on the role of NRs as regulators, as their very complex signaling pathways and the number of different genes under their control can lead to the development of drugs with several undesired side-effects (1,8). The distinct subset of the NRs which is referred to as ‘orphans’ lack a defined endogenous ligand and present an excellent opportunity for the discovery of novel promising treatments for several human diseases (9). The identification of novel new innovative ligands for these receptors and the better understanding of their regulatory systems may lead to the discovery of novel agents for the pharmaceutical industry (10-13).
In order to further elucidate the function of NRs, it is crucial to understand their structure (Fig. 1). The majority of NRs are comprised of five protein domains, including the N- and C-terminal, the DNA binding domain (DBD) and ligand binding domain (LBD), as well as the hinge region, that comes between the latter two. The two principal domains are the DBD and LBD, which respectively control the binding of the receptor to specific target genes and the binding of ligands that activate it. Both are highly conserved throughout the evolution of the NR superfamily; however, there is no clear explanation to date as to which one of these domains precedes the other in time. As previously mentioned, it is considered that the ancestral NR may have been an orphan receptor that lacks ligand binding ability (1). Since the evidence to fully support this theory is limited, the present study aimed to further investigate this speculation.
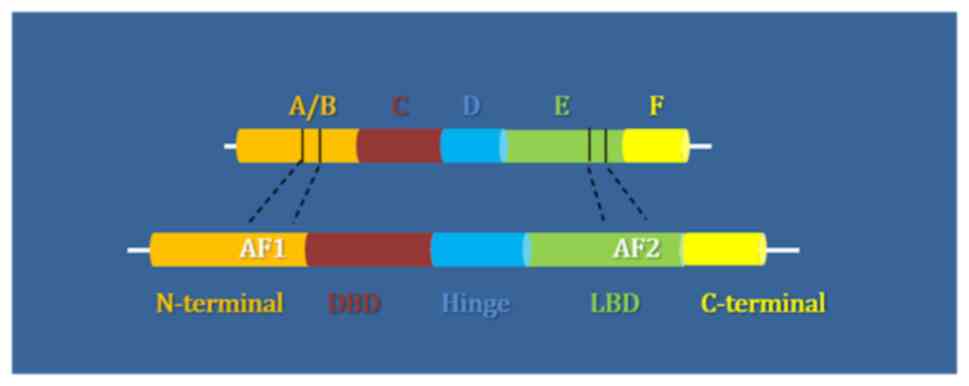 |
Figure 1
Structural domains of the NR superfamily: N-terminal in orange, DNA binding domain (DBD) in red, hinge region in blue, ligand binding domain (LBD) in green and C-terminal domain in yellow. NR, nuclear receptor.
|
The phylogenetic analysis of the NR superfamily has revealed that there is no connection between the chemical nature of a ligand and the evolutionary origin of the corresponding receptor (14,15). This can be easily confirmed by the fact that ligand and orphan receptors are scattered through the evolutionary tree (14). According to Holzer et al (15) and Bridgham et al (16), the NR superfamily is divided into seven subfamilies (NR0-NR6) and subfamily 2, which includes NRs, such as retinoid X receptors (RXRs), hepatocyte nuclear factor 4 (HNF4)s, chicken ovalbumin upstream promoter transcription factors (COUP-TFs) etc., being the one where the whole superfamily originates from (15,16). This subfamily seems to be non-monophyletic, since it does not form a clade where all members originate from the same ancestor, but is rather separated into two groups, one including only HNF4 and the other, the remaining members. Based on this fact, the studies by Holzer et al (15) and Bridgham et al (16) suggested that the ancestral NR was possibly a receptor found in sponges that was similar either to HNF4 or to the rest of the subfamily 2 receptors. Since the NR protein dataset is constantly growing, the aim of the present study was to draw a conclusion regarding the evolution of the NR superfamily, and possibly obtain some new insight on this topic, that may help determine whether any of these NRs or their targets can be used as therapeutic targets.
NR ligands are lipophilic hormones that can enter the cell by passive transport. Once inside the cell, the hormone binds to its congener receptor, which is located in the cytoplasm or nucleus, usually bound to other proteins. Once these proteins are released and the hormone is bound, the receptor is activated, which then binds to the DNA and regulates the transcription of target genes. In the case of cytoplasmic receptors, the binding of the hormone induces their entry into the nucleus, where the hormone-receptor complex acts. NRs bind to DNA in specific sequences known as hormone response elements (HREs). NR targets are genes that are regulated by promoters that contain HREs. The regulation of the transcription of these genes by NRs is usually accomplished by proteins known as co-regulators. These proteins fall into two broad categories: Co-activators, which interact with NRs in the AF-2 region via an LXXLL motif (where L symbolizes leucine and X any amino acid) and help activate gene transcription, and co-repressors which bind to the same region via conserved (L/I) XX (I/V) I or LXXX (I/L) XXX (I/L) motifs (where L denotes leucine, I isoleucine and X any amino acid), and suppress the transcription of target genes (1).
NRs can be categorized based on their mechanisms of action. Category 1 includes homogeneous steroid hormone receptors, which are activated by cholesterol-derived steroid hormones, such as estrogens, androgens, corticosteroids, and progestogens. In the absence of the ligand, these receptors are located in the cytoplasm bound to chaperone proteins; however, following their activation by the ligand, they are released from the chaperone proteins and are transported to the nucleus. In the nucleus, SRs form homodimers and bind to specific DNA sequences (HREs), which consist of two reversed repeats. Category 2 includes RXR-containing heterodimeric receptors, such as RAR and LXR, which often remain in the nucleus, regardless of the presence of a ligand. Following the binding of the ligand, they form heterodimers with RXR receptors and bind to a DNA element with direct repeats of HREs. Category 3 includes homodimeric ‘orphan receptors’. These receptors are named after the fact that their related hormones are unknown. Receptors of this category bind as homodimers to the recognition sequences (HREs), which are arranged as direct repeats. Finally, category 4 comprises the ‘orphan receptor’ monomers, which bind as monomers to asymmetric recognition sequences (HREs) (17).
A NR that has been of considerable interest to the scientific community is the glucocorticoid receptor (GR). This receptor is a ligand-dependent transcription factor, which is activated by glucocorticoid binding and then binds to glucocorticoid response elements (GREs) in the promoter of target genes (18). In this manner, it has the ability to regulate the transcription of genes that respond to glucocorticoids, either positively or negatively. The GR gene is termed NR3C1, is located on chromosome 5q31-32 and contains 10 exons. The alternative splicing of exon 9 results in two isoforms of the human glucocorticoid receptor (hGR), hGRα and hGRβ. When hGRα undergoes alternative translation to exon 2, eight additional isoforms of GR with a smaller N-terminal ends result (GRα-A, GRα-B, GRα-C1, GRα-C2, GRα-C3, GRα-D1, GRα-D2 and GRα-D3). These isoforms exhibit a similar binding affinity to glucocorticoids and subsequently to the GREs of target genes, although they differ in their transcriptional properties and in their subcellular location and expression in tissues (18). From hGRβ, there is a possibility to produce eight more β-isoforms similar to those of hGRα. Each isoform of the above can be further subjected to various post-translational modifications, such as phosphorylation and sumoylation. These modifications affect the stability, transcriptional activity and interaction of GR with other proteins (18,19).
Mutations in GR disrupt glucocorticoid signal transduction, leading to generalized resistance or hypersensitivity to glucocorticoids. A widely studied pathological condition is primary generalized glucocorticoid resistance (PGGR), a rare familial or sporadic disease characterized by partial or general resistance of specific tissues in cortisol. This resistance results in the activation of the hypothalamic-pituitary-adrenal (HPA) axis to repair the reduced activity of glucocorticoids in the target tissues and increased secretion of adrenocorticotropic hormone (ACTH) into the systemic circulation. This excessive secretion of ACTH leads to adrenal hyperfunction, and to the increased secretion of cortisol and other steroid hormones, such as androgens and saline corticosteroids (20).
The molecular basis of Chrousos syndrome has been attributed mainly to mutations occurring in exons 5-9 (regions encoding the LBD region) of the human NR3C1 gene, which affect the mechanisms of action of hGRα (20). In addition to the LBD region, mutations that affect GR activity have been found in the DBD region. These mutations can affect the signaling pathway of glucocorticoids at any stage in the process of activating target gene transcription, such as ligand-receptor binding, DNA binding, and the interaction with co-regulator proteins (21).
Among the 26 mutations studied and associated with loss-of-function mutations, six are in the DBD region and 20 are in the LBD region (22). Some of these mutations, such as I559N, V571A, D641V, V729I, I747M located in the LBD region, appear to adversely affect receptor transcriptional activity, reducing ligand affinity, delaying the displacement of the ligand-receptor complex in the nucleus, and creating problems in its interaction with the GRIP1 co-activator. Moreover, the F774Sfs*24 mutation is a replacement of a phenylalanine by serine. This mutation in the LBD region leads to inability to bind the ligand and therefore, the receptor is not activated and cannot function. Other mutations, such as V423A and R477H, which affect the DBD region, result in a reduction or complete inability of GR to bind to the GREs of the target genes, respectively (20).
Data and methods
Dataset collection, filtering and multiple sequence alignment
Protein sequences of all members of the NR superfamily were extracted from the NCBI protein database (https://www.ncbi.nlm.nih.gov/protein), using the corresponding key words as presented in Table I. Regular expression techniques in the header information, related key words and local alignments with reference sequences were used to remove any protein sequences that did not include members of the NR superfamily, and according to a previously published study by the authors, the final dataset consisted of 333 unique, non-duplicate, representative protein sequences from every phylum and class in Animalia (Table II) (23). Multiple sequence alignment (MSA) was carried out, respectively with the use of the MATLAB Bioinformatics Toolbox (24), as previously described in several studies (25-27), with the neighbor-joining algorithm (NJ), by assuming equal variance and independence of evolutionary distance estimates (28).
 |
Table I
Nuclear receptor superfamily members used in the present study and the subfamilies they belong to.
|
Table I
Nuclear receptor superfamily members used in the present study and the subfamilies they belong to.
| Nuclear receptor |
Abbreviation |
Subfamily |
Groups |
| Thyroid hormone receptor |
TRα |
NR1A1 |
1 |
| |
TRβ |
NR1A2 |
|
| Retinoic acid receptor |
RARα |
NR1B1 |
1 |
| |
RARβ |
NR1B2 |
|
| |
RARγ |
NR1B3 |
|
| Peroxisome proliferator activated receptor |
PPARα |
NR1C1 |
1 |
| |
PPARβ |
NR1C2 |
|
| |
PPARγ |
NR1C3 |
|
| V-ErbA-related protein |
Rev-ErbΑ |
NR1D1 |
1 |
| |
Rev-ErbΒ |
NR1D2 |
|
| Ecdysone-induced protein 78C |
Eip78C |
NR1E1 |
1 |
| RAR related orphan receptor |
RORα |
NR1F1 |
1 |
| |
RORβ |
NR1F2 |
|
| |
RORγ |
NR1F3 |
|
| Liver X receptor |
LXRα |
NR1H2 |
1 |
| |
LXRβ |
NR1H3 |
|
| Farnesoid X receptor |
FXRα |
NR1H4 |
1 |
| |
FXRβ |
NR1H5 |
|
| Vitamin D receptor |
VDR |
NR1I1 |
1 |
| Pregnane X receptor |
PXR |
NR1I2 |
1 |
| Constitutive androstane receptor |
CAR |
NR1I3 |
1 |
| Nuclear receptor HR96, HR8 and HR48 |
HR96 |
NR1J1 |
1 |
| Nuclear receptor HR8 |
HR8 |
NR1J2 |
1 |
| Nuclear receptor HR48 |
HR48 |
NR1J3 |
1 |
| Nuclear receptor HR1 |
HR1 |
NR1K1 |
1 |
| V-erbA-related protein 2 |
EAR-2 |
NR2F3 |
2 |
| Steroid hormone receptor cnr14 |
Cnr14 |
NR1G1 |
2 |
| Estrogen receptor |
ERα |
NR3A1 |
2 |
| |
ERβ |
NR3A2 |
|
| Estrogen related receptor |
ERRα |
NR3B1 |
2 |
| |
ERRβ |
NR3B2 |
|
| |
ERRγ |
NR3B3 |
|
| Glucocorticoid receptor |
GR |
NR3C1 |
2 |
| Mineralocorticoid receptor |
MR |
NR3C2 |
2 |
| Progesterone receptor |
PR |
NR3C3 |
2 |
| Androgen rceptor |
AR |
NR3C4 |
2 |
| Nerve growth factor IB |
NGFIB |
NR4A1 |
2 |
| Nuclear receptor related 1 |
NURR1 |
NR4A2 |
2 |
| Neuron-derived orphan receptor 1 |
NOR-1 |
NR4A3 |
2 |
| Steroidogenic factor 1 |
SF-1 |
NR5A1 |
2 |
| Liver receptor homolog 1 |
LRH-1 |
NR5A2 |
2 |
| Nuclear hormone receptor FTZ-F1 beta |
FTZ-F1β |
NR5B1 |
2 |
| Germ cell nuclear factor |
GCNF |
NR6A1 |
3 |
| Zygotic gap protein knirps |
kni |
NR0A1 |
3 |
| Dosage-sensitive sex reversal |
DSS |
NR0B1 |
3 |
| Small heterodimer partner |
SHP |
NR0B2 |
3 |
| Ecdysone receptor |
EcR |
NR1H1 |
3 |
| Hepatocyte nuclear ractor 4 |
HNF4α |
NR2A1 |
3 |
| |
HNF4γ |
NR2A2 |
|
| Retinoid X receptor |
RXRα |
NR2B1 |
3 |
| |
RXRβ |
NR2B2 |
|
| |
RXRγ |
NR2B3 |
|
| Ultraspiracle |
USP |
NR2B4 |
3 |
| Testicular receptor |
TR2 |
NR2C13 |
|
| |
TR4 |
NR2C2 |
|
| Tailes-related receptor |
TLX |
NR2E1 |
3 |
| Photoreceptor specific nuclear receptor |
PNR |
NR2E2 |
3 |
| COUP transcription factor |
COUP-TF1 |
NR2F1 |
3 |
| |
COUP-TF2 |
NR2F2 |
|
 |
Table II
Eukaryotes in Animalia from which they have been identified NR family members.
|
Table II
Eukaryotes in Animalia from which they have been identified NR family members.
| Domain |
Kingdom |
Phylum |
Class |
| Eukaryotes |
Animals |
Chordates |
Mammals |
| |
|
|
Birds |
| |
|
|
Fish |
| |
|
|
Turtles |
| |
|
|
Amphibians |
| |
|
|
Lizards |
| |
|
Arthropods |
Insects |
| |
|
|
Arachnids |
| |
|
|
Crustaceans |
| |
|
|
Horseshoe crabs |
| |
|
Nematodes |
|
| |
|
Molluscs |
|
| |
|
Flatworms |
|
Phylogenetic analysis
The phylogenetic analysis of the NR superfamily was performed using the MATLAB Bioinfromatics Toolbox (24) utilizing the NJ algorithm and the average distance statistical method with 100 bootstrap replicates (29). The MEGA platform was then used to visualize the constructed phylogenetic tree in a circle form (30). The protein clusters observed, were colored each with a different color using the iTOL platform tools.
Results
Dataset characterization, multiple sequence alignment and protein domain characterization
The primary dataset that was extracted from NCBI included 110,000 protein sequences, related to NRs. Irrelevant, hypothetical, partial, low quality and predicted proteins were eliminated from the dataset. Due to the large amount of data remaining and the presence of duplicate sequences, further filtering and the selection of representative sequences was conducted. The representative protein sequences were selected manually for each class of every NR following multiple alignments and analyses within the family members subgroups. The same species were selected as representative for each class where possible; for example, Homo sapiens was selected as a representative for mammals in the majority of NRs and Xenopus tropicalis for amphibians, etc. Thus, the final dataset consisted of 333 unique, non-duplicate, representative protein sequences for each NR class (Table SI). NRs in genomes of plants, fungi and prokaryotes have yet to be found, since the only ones known belong to the animal kingdom. In the present study, the most ancient NR found was an RXR belonging to the phylum Porifera and the sponge, Suberites domuncula. As previously described, the DBD is the most conserved region amongst the NRs and the amino acids found in this domain are responsible for the identification of the sequences of target genes and binding to it, as well as the dimerization of the receptor itself (23). As regards as the LBD, not only it is important for the binding of ligands, but is also the region where the interaction between the receptor and the coregulator proteins is performed.
In the DBD domain, two highly conserved cystine-rich zinc finger motifs have already been identified: A highly conserved pattern, termed the P-box (located in amino acid positions 908-921 of the MSA), is found in the first zinc finger, whereas another not so highly conserved one known as the D-box, is located in the second zinc finger motif (23). Amino acid residues found in the P-box have been proven to play an important role in the identification and binding of the NR to the target DNA (1), whilst the dimerization of the NRs is known to be affected by the amino acids found in the D-box (31). In the LBD, the LxxLL motif/motif D (where L stands for leucine and x represents any amino acid residue) has been described (23). It is a key regulator of transcription, since amino acids found in this motif are significant for the interaction of the NR with different co-regulators and chromatin-remodeling factors (32). Conserved motifs A and B are also crucial for co-activator function (33) and motif C is critical for ligand binding, as it has been shown that mutations in this region can lead to the complete loss of the ligand binding ability of the NR (33). Based on the above, the conclusion can be drawn that a protein sequence needs to have several specific motifs in order to be considered as a member of the NR superfamily. According to these conserver motifs a NR pedigree can be defined.
Evolutionary findings
The phylogenetic analysis revealed a separation of the NR superfamily members into three major monophyletic branches. Each branch includes one or more NR subfamilies, and as shown in Fig. 2, there are orphan receptors in all three groups. The first group, colored in orange, includes thyroid hormone-like receptors and receptors from subfamily 4, such as nerve growth factor-induced clone B (NGFIB), Nur-related factor 1 (NURR1) and neuron-derived orphan receptor-1 (NOR-1). The second group, depicted in purple, mainly includes steroid hormone receptors, as well as receptors of subfamilies 5 and 6, and the third group is the one that includes RXRs and the orphan receptors, DSS and SHP colored in orange-red. It is apparent that these three clusters that were formed in the phylogenetic tree are based on related NR members and not on classes of the animal kingdom in which they belong. Last but not least, even though only representative protein sequences were used, the separation of the NRs into subfamilies is still preserved.
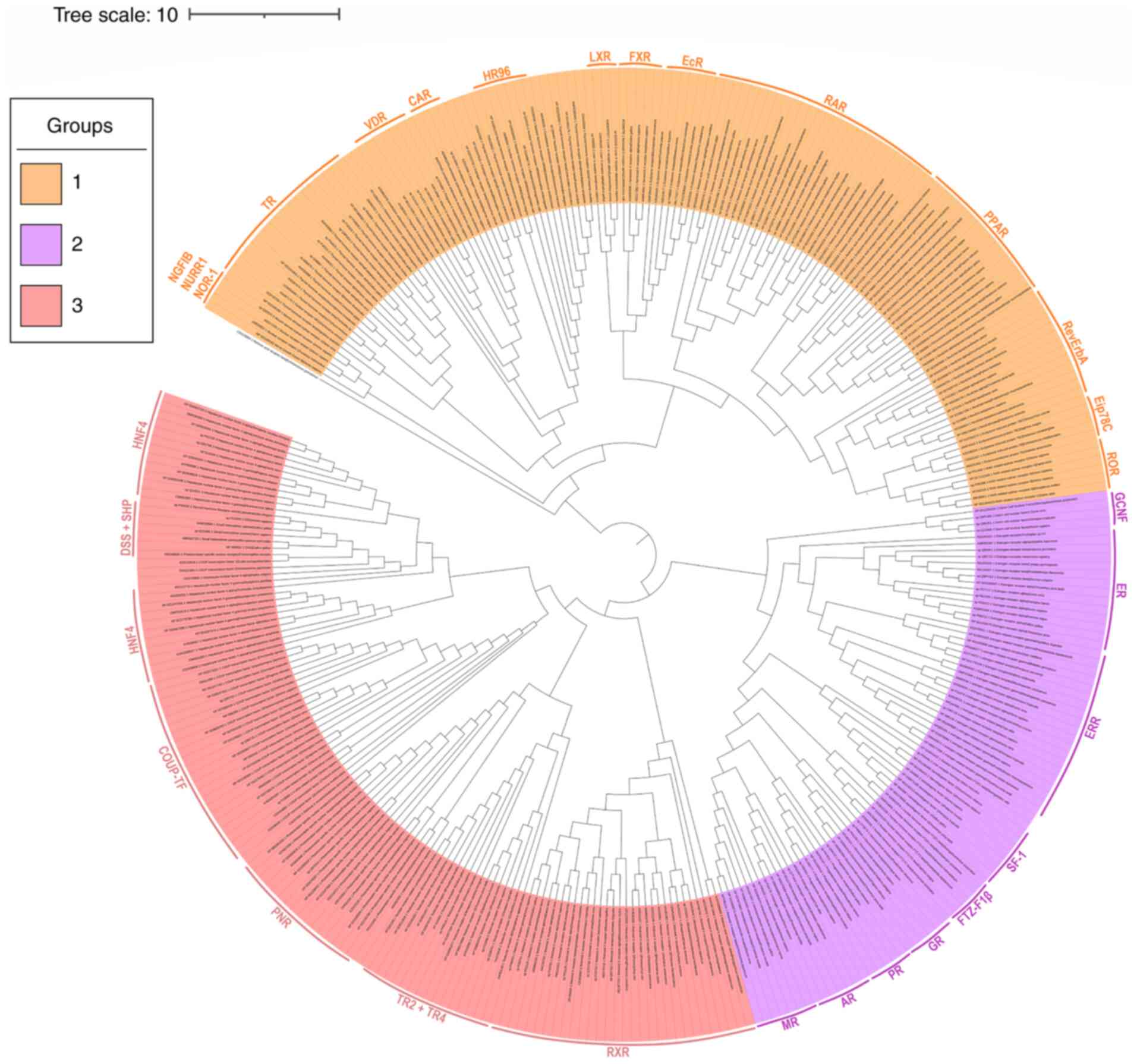 |
Figure 2
Phylogenetic tree of the 333 representative protein sequences of the nuclear receptor superfamily. The tree was calculated with the neighbor-joining method and processed with the MEGA and iTOL programs. The three main groups in which the tree is dived have been colored in orange, purple and orange-red. The receptors that belong to each group are shown on the side with colored stripes.
|
By observing the phylogenetic tree (Fig. 2) it is not possible to pinpoint the exact ancestral receptor of the whole superfamily, although it is clear that the phylogeny of the NR superfamily can be rooted in a location between the two major groups, the first one including the thyroid hormone-like receptors and the second the steroid hormone-like receptors. A possible evolution scenario would be the parallel evolution of a thyroid-like receptor, an RXR-like and a steroid one until their late separation. In order to further understand what really happened, it is crucial to enhance the dataset with more NR protein sequences from various species of the animal kingdom. According to previous studies Holzer et al (15) and Bridgham et al (16), the ancestral NR most likely belongs to the RXR family and is located somewhere between two NRs found in the genome of demosponge Amphimedon queenslandica.
Discussion
NRs are a group of proteins that regulate a large number of biological processes that are important for life. The majority of NRs become activated by the binding of small lipophilic molecules, while for others, the ligands are not yet known. The primary function of NRs is the cell type- and promoter-specific transcriptional regulation of target genes under their control, through the recruitment of negative or positive regulatory proteins, known as co-repressors and co-activators respectively (10,32). Several NR ligands have already been used in order to target a variety of disorders, such as type II diabetes where thiazolidinediones (TZDs), ligands of peroxisome proliferator-activated receptor γ (PPARγ), are widely used (10,34), or corticosteroids which are used in the treatment of asthma and other health associated abnormalities (35). The development of novel drugs targeting NRs that regulate specific genes and also ligand identification for orphan NRs may hold promise for the pharmaceutical industry.
The first NR to be sequenced was the human GR followed by the estrogen receptor (ER). Overall, 48 NR members from the superfamily have been found in humans to date; however, in other organisms of the animal kingdom, >900 have been identified (36). No NR has been found in the plant, fungi or prokaryote kingdoms; however, NR genes are expressed even in the simplest animals, such as those belonging to the Porifera (sponges) and Placozoa phyla (36). As regards the ancestral receptor, two hypothesis are proposed: The first supports the concept of an ancestral receptor lacking ligand-binding ability, whereas the second one suggests that the NR superfamily evolved from a ligand-activated receptor (16). The findings of the present study demonstrated similar conservation in both the DBD and LBD, which indicates the possibility that the two main protein domains of the NRs evolved in parallel, supporting the theory of a possible liganded ancestral receptor.
The evolutionary study of the NR superfamily is crucial, since they play a pivotal role in the regulation of numerous physiological and pathophysiological processes in all organisms of the animal kingdom. In the present study, six conserved motifs were identified and are considered to be to key target regions for the development of novel pharmaceutical agents. A major challenge in this case is accomplishing the desired selectivity within each subfamily of related NRs that bind very similar ligands (22). The equivalence that is visible between the thyroid-like receptor group and the other two, suggests that perhaps the ancestral receptors from each group evolved in a parallel manner. All this information may prove beneficial for the better understanding of the evolutionary pathway of the NRs and the asociation between all the members of the superfamily (37).
A structural analysis of the NR members is required, since the full three-dimensional structure of a NR remains unknown. Difficulties are identified in the crystallization of the DBD of the NRs. A structural analysis of the NRs may provide beneficial knowledge for the NR evolution in the future. However, the structural analysis of the LBD of the NRs by Mitsis et al demonstrated the clear separation of the thyroid hormone-like receptors, the steroid hormone-like receptors and the RXRs (33). Based on the results of the present study, the members of these three major classes have a characteristic LBD, which can be classified in two canonical forms. This evidence also confirms the results of the present evolutionary study. Moreover, the results of the present study are also confirmed from the various studies that have been performed in the response elements of the NRs including primers, promoters and enhancers (38-40). If it is accepted that the evolution of these key sequence regions for the NRs has proceeded in parallel, their separation is also divided into three major classes. Thus, several studies have demonstrated that steroid hormone members share common patterns in their response elements, that are differentiated than the thyroid hormone members response elements (41-43). In the same direction, RXR-like member response elements have been found to share key patterns which differ from the other two major groups.
In conclusion, in the present study, an updated comprehensive sequence analysis of the NR superfamily was performed. A significant amount of sequence data available for the NR superfamily was used by selecting representative protein sequences for every phylum and every class of each member of the superfamily. Thus, through different filtering techniques, a final dataset of 333 unique, non-duplicate, representative protein sequences was formed and used for further research. Considering the important role NRs play in ‘switching on and off’ genes, they present a great potential as innovative drug targets for a variety of diseases, including cancer. In the present study, an updated phylogenetic tree of the NR superfamily was created that provides useful information for the groups formed inside the superfamily and their evolution. This beneficial knowledge may provide the basis towards associating NR members in several aspects, including signaling pathways and biological activities.
Supplementary Material
The 333 representative protein sequences of NRs were used in the present study.
Acknowledgements
Not applicable.
Funding
The authors would like to acknowledge funding from ‘INSPIRED‑The National Re‑search Infrastructures on Integrated Structural Biology, Drug Screening Efforts and Drug Target Functional Characterization; (grant no. MIS 5002550) and by the project: ‘OPENSCREENGR An Open‑Access Research Infrastructure of Chemical Biology and Target‑Based Screening Technologies for Human and Animal Health, Agriculture and the Environment’ (grant MIS 5002691), which are implemented under the Action ‘Reinforcement of the Research and Innovation Infrastructure’, funded by the Operational Program ‘Competitiveness, Entrepreneurship and Innovation’ (NSRF 2014‑2020) and co‑financed by Greece and the European Union (European Regional Development Fund). The authors would also like to acknowledge funding from ‘MilkSafe: A novel pipeline to enrich formula milk using omics technologies’, a research co‑financed by the European Regional Development Fund of the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH‑CREATE‑INNOVATE (project code: T2EDK‑02222).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LP, LS, KP, EP, SM, KD, NCN, AG, FB, GPC, EE and DV contributed to the conceptualization and design of the study, as well as in the writing, drafting, revising, editing and reviewing of the manuscript. All authors confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
GPC is an Editorial Advisor of the journal, but had no personal involvement in the reviewing process, or any influence in terms of adjudicating on the final decision, for this article. The other authors declare that they have no competing interests.
References
|
1
|
Weikum ER, Liu X and Ortlund EA: The nuclear receptor superfamily: A structural perspective. Protein Sci. 27:1876–1892. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Robinson-Rechavi M, Garcia HE and Laudet V: The nuclear receptor superfamily. J Cell Sci. 116:585–586. 2003.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Porter BA, Ortiz MA, Bratslavsky G and Kotula L: Structure and function of the nuclear receptor superfamily and current targeted therapies of prostate cancer. Cancers (Basel). 11(1852)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zuo H and Wan Y: Nuclear receptors in skeletal homeostasis. Curr Top Dev Biol. 125:71–107. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Karki K, Li X, Jin UH, Mohankumar K, Zarei M, Michelhaugh SK, Mittal S, Tjalkens R and Safe S: Nuclear receptor 4A2 (NR4A2) is a druggable target for glioblastomas. J Neurooncol. 146:25–39. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Bakke D and Sun J: Ancient nuclear receptor VDR with new functions: Microbiome and inflammation. Inflamm Bowel Dis. 24:1149–1154. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Cecchin E, De Mattia E and Toffoli G: Nuclear receptors and drug metabolism for the personalization of cancer therapy. Expert Opin Drug Metab Toxicol. 12:291–306. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Schierle S and Merk D: Development of nuclear receptor modulators. Methods Mol Biol. 1824:245–260. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Safe S, Jin UH, Hedrick E, Reeder A and Lee SO: Minireview: Role of orphan nuclear receptors in cancer and potential as drug targets. Mol Endocrinol. 28:157–172. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shi Y: Orphan nuclear receptors in drug discovery. Drug discovery today. 12:440–445. 2007.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Blumberg B and Evans RM: Orphan nuclear receptors-new ligands and new possibilities. Genes Dev. 12:3149–3155. 1998.PubMed/NCBI View Article : Google Scholar
|
|
12
|
De Bosscher K, Desmet SJ, Clarisse D, Estebanez-Perpina E and Brunsveld L: Nuclear receptor crosstalk-defining the mechanisms for therapeutic innovation. Nat Rev Endocrinol. 16:363–377. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vlachakis D and Kossida S: Molecular modeling and pharmacophore elucidation study of the classical swine fever virus helicase as a promising pharmacological target. PeerJ. 1(e85)2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Gronemeyer H, Gustafsson JA and Laudet V: Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 3:950–964. 2004.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Holzer G, Markov GV and Laudet V: Evolution of nuclear receptors and ligand signaling: Toward a soft key-lock model? Curr Top Dev Biol. 125:1–38. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bridgham JT, Eick GN, Larroux C, Deshpande K, Harms MJ, Gauthier MEA, Ortlund EO, Degnan BM and Thornton JW: Protein evolution by molecular tinkering: Diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 8(e1000497)2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Beato M, Chávez S and Truss M: Transcriptional regulation by steroid hormones. Steroids. 61:240–251. 1996.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Nicolaides NC, Chrousos G and Kino T: Glucocorticoid Receptor. In: Endotext. Feingold KR, Anawalt B, Boyce A, Chrousos G, de Herder WW, Dhatariya K, Dungan K, Grossman A, Hershman JM, Hofland J, Kalra S, Kaltsas G, KochC, Kopp P, Korbonits M, Kovacs CS, Kuohung W, Laferrere B, McGee EA, McLachlan R, Morley JE, New M, Purnell J, Sahay R, Singer F, Stratakis CA, Trence DL and Wilson DP (eds). MDText.com, Inc., South Dartmouth, MA, 2000.
|
|
19
|
Vandevyver S, Dejager L and Libert C: Comprehensive overview of the structure and regulation of the glucocorticoid receptor. Endocr Rev. 35:671–693. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Charmandari E: Primary generalized glucocorticoid resistance and hypersensitivity: The end-organ involvement in the stress response. Sci Signal. 5(pt5)2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Nicolaides NC, Kino T, Chrousos G and Charmandari E: Primary generalized glucocorticoid resistance or chrousos syndrome. In: Endotext. Feingold KR, Anawalt B, Boyce A, Chrousos G, de Herder WW, Dhatariya K, Dungan K, Grossman A, Hershman JM, Hofland J, Kalra S, Kaltsas G, Koch C, Kopp P, Korbonits M, Kovacs CS, Kuohung W, Laferrere B, McGee EA, McLachlan R, Morley JE, New M, Purnell J, Sahay R, Singer F, Stratakis CA, Trence DL and Wilson DP (eds). MDText.com, Inc., South Dartmouth, MA, 2000.
|
|
22
|
Papageorgiou L, Papakonstantinou E, Salis C, Raftopoulou S, Mitsis T, Nicolaides N, Hagidimitriou M, Eliopoulos E, Charmandari E, Chrousos GP and Vlachakis D: An updated evolutionary study in glucocorticoid receptors; insights from a comprehensive phylogenetic, snp's and mutation's analysis of the nuclear receptors family. 57th Annual ESPE Meeting, Vol. 89, 2018. https://abstracts.eurospe.org/hrp/0089/hrp0089LB-P4.
|
|
23
|
Papageorgiou L, Shalzi L, Efthimiadou A, Bacopoulou F, Chrousos GP, Eliopoulos E and Vlachakis D: Conserved functional motifs of the nuclear receptor superfamily as potential pharmacological targets. Int J Epigen. 1(3)2021.
|
|
24
|
Sobie EA: An introduction to MATLAB. Sci Signal. 4(tr7)2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Vlachakis D, Papageorgiou L, Papadaki A, Georga M, Kossida S and Eliopoulos E: An updated evolutionary study of the Notch family reveals a new ancient origin and novel invariable motifs as potential pharmacological targets. PeerJ. 8(e10334)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Papageorgiou L, Loukatou S, Sofia K, Maroulis D and Vlachakis D: An updated evolutionary study of flaviviridae NS3 helicase and NS5 RNA-dependent RNA polymerase reveals novel invariable motifs as potential pharmacological targets. Mol Biosyst. 12:2080–2093. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Amidi S, Amidi A, Vlachakis D, Paragios N and Zacharaki EI: Automatic single- and multi-label enzymatic function prediction by machine learning. PeerJ. 5(e3095)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Papageorgiou L, Megalooikonomou V and Vlachakis D: Genetic and structural study of DNA-directed RNA polymerase II of Trypanosoma brucei, towards the designing of novel antiparasitic agents. PeerJ. 5(e3061)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gibbons FD and Roth FP: Judging the quality of gene expression-based clustering methods using gene annotation. Genome Res. 12:1574–1581. 2002.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kumar S, Stecher G and Tamura K: MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Pawlak M, Lefebvre P and Staels B: General molecular biology and architecture of nuclear receptors. Curr Top Med Chem. 12:486–504. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Novac N and Heinzel T: Nuclear receptors: Overview and classification. Curr Drug Targets Inflamm Allergy. 3:335–346. 2004.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Mitsis T, Papageorgiou L, Efthimiadou A, Bacopoulou F, Vlachakis D, Chrousos GP and Eliopoulos E: A comprehensive structural and functional analysis of the ligand binding domain of the nuclear receptor superfamily reveals highly conserved signaling motifs and two distinct canonical forms through evolution. World Acad Sci J. 1:264–274. 2020.
|
|
34
|
Mirza AZ, Althagafi II and Shamshad H: Role of PPAR receptor in different diseases and their ligands: Physiological importance and clinical implications. Eur J Med Chem. 166:502–513. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Abaya R, Jones L and Zorc JJ: Dexamethasone compared to prednisone for the treatment of children with acute asthma exacerbations. Pediatr Emerg Care. 34:53–58. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Mazaira GI, Zgajnar NR, Lotufo CM, Daneri-Becerra C, Sivils JC, Soto OB, Cox MB and Galigniana MD: The nuclear receptor field: A historical overview and future challenges. Nucl Receptor Res. 5(101320)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Vlachakis D, Nicolaides NC, Papageorgiou L, Lamprokostopoulou A and Charmandari E: Tracing the Glucocorticoid Receptor Evolutionary Pedigree: Insights from a Comprehensive Phylogenetic Analysis of the Full NR Super-Family. 55th ESPE Meeting, Paris, France, Vol 82: 2016. https://abstracts.eurospe.org/hrp/0086/hrp0086rfc1.1.
|
|
38
|
Penvose A, Keenan JL, Bray D, Ramlall V and Siggers T: Comprehensive study of nuclear receptor DNA binding provides a revised framework for understanding receptor specificity. Nat Commun. 10(2514)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Khorasanizadeh S and Rastinejad F: Nuclear-receptor interactions on DNA-response elements. Trends Biochem Sci. 26:384–390. 2001.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lisse TS, Hewison M and Adams JS: Hormone response element binding proteins: Novel regulators of vitamin D and estrogen signaling. Steroids. 76:331–339. 2011.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ding XF, Anderson CM, Ma H, Hong H, Uht RM, Kushner PJ and Stallcup MR: Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): Multiple motifs with different binding specificities. Mol Endocrinol. 12:302–313. 1998.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kumar R and McEwan JI: Allosteric modulators of steroid hormone receptors: Structural dynamics and gene regulation. Endocr Rev. 33:271–299. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Klinge C and Rao C: The steroid hormone receptors. Glob. libr. women's med. ISSN: 1756-2228, 2008. DOI 10.3843/GLOWM.10281.
|
















