Introduction
Autism spectrum disorder (ASD) is a broad range of
neurodevelopmental disorders that is generally manifested during
early childhood. ASD can be presented by a lack of social
interaction and communication skills, repetitive and stereotyped
behaviour, restricted interests and activities. ASD is complex and
may be associated with other comorbidities (1).
It is estimated that one to two percent of the
general population is affected by ASD (2), and the condition is four-fold more
common among males than females (3). The recorded increase in the
prevalence of ASD over the past 50 years may be attributed to
changes in the case definitions, increasing awareness, the
availability of specialised centres, earlier detection, improved
reporting and diagnostic substitution. Therefore, the true increase
cannot yet be estimated (4,5).
However, demographic and geographical variables, as well as the
availability of resources are considered to influence the
prevalence of ASD (6).
ASD has a multifactorial aetiology, which is not yet
fully understood. Epigenetic interactions between genetic and
environmental factors are considered as key elements involved in
the onset of this disorder (7).
Notably, in recent research, gastrointestinal (GI) symptoms have
been shown to have a high incidence and association with the
severity of ASD (8). Research into
the functions and alterations in the microbiota-gut-brain axis has
become a promising field which may aid in the understanding of ASD
(9).
The gut-brain axis is a bidirectional communication
pathway of the intestines and central nervous system, whereby the
microbiota-immune axis functions as a central mediator in their
communication (10). It has been
shown that alterations in the microbiome-gut-brain axis may trigger
neuroinflammation, myelination, microglia maturation and the
modulation of complex behaviours, such as anxiety (11). For example, according to previous
studies, increased gut permeability, or ‘leaky gut syndrome’ plays
a crucial role in modification of the normal function of the
gut-brain axis (12). With the
impaired intestinal luminal barrier, bacterial metabolites, such as
short-chain fatty acids, increasingly pass the membrane, enter the
bloodstream and reach the brain. They can alter the levels of the
cytokine expression, triggering the immune response and the
production of neurotransmitters, such as dopamine and serotonin
(12,13). In the cases of ASD, it has been
demonstrated that the concentration of Zonulin, the protein that
modulates gut permeability, is higher in children with ASD compared
to healthy controls; furthermore, the severity of the behavioural
symptoms has been found to be associated with an increase in
Zonulin levels (14). Therefore,
alterations in the gut microbiota may have a tangible effect on the
function of the gut-brain axis in individuals with ASD.
The human gut microbiota is a dynamic and complex
system. The development and fluctuations of this ecosystem can be
affected by an individual's genetics, birth and infant feeding,
diet, medication intake, the environment and geography,
comorbidities, ageing, lifecycle and stress exposure (15). On the other hand, the gut
microbiota itself is considered to influence the individual's
metabolism, nutritional preferences, the physiology of the GI and
immune systems, as well as the neurophysiological and behavioural
functions of the body (16).
Considering the unique homeostasis and composition of the ‘normal’
microbiota in each individual, the state of dysbiosis is generally
difficult to define, Petersen and Round (17). The most common approach used to
investigate dysbiosis in a cohort is to perform a case-controlled
study.
According to a recent review article, a number of
studies have reported an association between gut permeability and
an increase or deprivation of several intestinal microbial genera
and ASD symptoms (8). However,
different literature sources refer to various findings. For
example, a group of scientists (18) found lower levels of
Enterococcus and Bifidobacterium and higher levels of
Clostridium, Bacteroides, Porphyromonas, Prevotella
and Enterobacteria in the composition of the gut microbiome
of children with ASD. A recent review article summarised the
results of nine studies including 254 patients with ASD (19). That study, based on a
meta-analysis, revealed significant differences in abundance of
Akkermansia, Bifidobacterium, Bacteroides, Escherichia
coli (E. coli), and Lactobacillus in the total
detected faecal microbiota of children with ASD compared to the
controls (19). Other researchers
refer to certain intestinal bacteria that may be involved in the
pathogenesis of ASD, including members of the Clostridium
genus (20). It has been shown
that faecal samples from children with ASD have higher levels of
Clostridium bolteae, Clostridium histolyticum or
Clostridium perfringens (21-23).
Some species of Clostridium can produce neurotoxins and may
exert systemic effects that can hypothetically influence the
behavioural patterns of individuals with ASD. Moreover, in a
previous study, following treatment with vancomycin, the reduction
of Clostridium yields was found to be associated with
short-term behavioural improvements in children with
regressive-onset autism (24).
Another study, based on a mouse model, suggested a potential direct
link between Clostridioides (C.) difficile
(previously known as Clostridium difficile) infection, gut
and serum p-cresol levels, dopamine-β-hydroxylase activity and
alterations in dopaminergic activity in the brain, that have direct
implications in the pathogenesis of ASD (25).
C. difficile is an emerging nosocomial
pathogen, common in cases of antibiotic-associated diarrhoea and
pseudomembranous colitis. However, C. difficile is naturally
resistant to ampicillin, amoxicillin, cephalosporins, clindamycin
and fluoroquinolones. The limited number of antibiotics that are
active against C. difficile, such as metronidazole,
vancomycin and fidaxomicin become increasingly less effective, due
to the accumulation of resistance in the C. difficile
population (26). Due to various
reasons, patients with ASD often undergo intensive antibiotic
treatment courses that can lead to acute C. difficile
infections (27). C.
difficile can produce two large toxins: ToxinA (308 kDa) and
ToxinB (270 kDa), which are glucosyltransferases and can inactivate
Rho, Rac and Cdc42 (small GTPases) within target cells. This leads
to the ability of the toxin to disrupt the tight junctions of
epithelial barriers and increase gut permeability (28). Therefore, the prevalence of C.
difficile in the intestinal biome of individuals with ASD has
become a main topic of research.
The first epidemiological study of autism in Georgia
was performed between the years 2007-2009. That analysis was based
on tests and questionnaires. According to the collected data, one
out of 110 children suffered from ASD. This frequency was
approximating the statistical average of Europe at that time
(29). In another study in
2017(30), the levels of the
bacterial metabolite, p-cresol, were measured in the urine
of children with ASD, epilepsy and healthy controls, and were found
to be elevated in the both study groups compared to the controls.
However, the diversity or presence of particular bacterial species
in the gut microbiota of Georgian children with ASD has not been
investigated to date, at least to the best of our knowledge.
The present study aimed to accomplish a preliminary
bacteriological investigation of the gut microbiota and its
association with the intestinal health of children with ASD. In
particular, the present study focused on the prevalence of A/B
toxins producing C. difficile and the abundance of
Bifidobacteria, Lactobacteria, E. coli and
Enterococcus, as well as antibiotic resistant pathogens in
the stool samples collected from an urban population with ASD and
neurotypical children from Tbilisi, Georgia. Since phages are
considered as alternatives to antibiotics, as well as a promising
tool for the re-instatement of disturbed microbiotas, antibiotic
resistance and phage susceptibility profiles of the isolated
intestinal pathogens were defined simultaneously. The present study
was performed in a case-control manner.
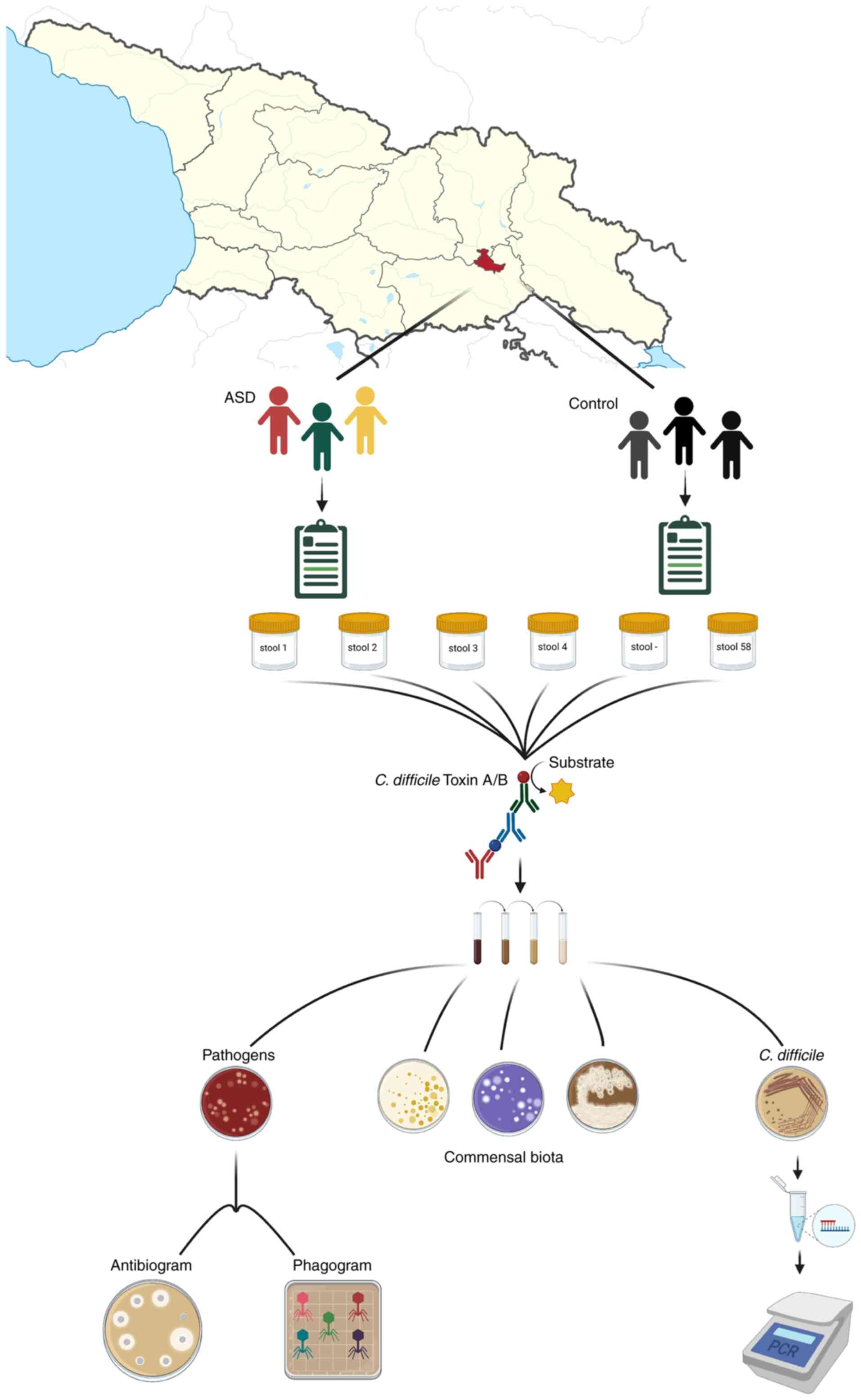
Patients and methods
Study design
The study included two cohorts, one composed of 30
children diagnosed with ASD and a control group of 27 neurotypical
children, all aged between 2 to 17 years and permanently residing
in Tbilisi, Georgia. The enrolment criteria for the patient group
included the following: A previous diagnosis of with ASD in
accordance to DSM-5 (https://psychiatry.org/Psychiatrists/Practice/DSM),
the permanent residence of the family in the Tbilisi urban area and
an age not >17 years; applicants with neurotypical siblings in
the same age groups were prioritized. The present study was
approved by the Scientific Research Ethics Committee of the Ilia
State University, Tbilisi, Georgia (issued on October, 2019).
Parental oral consents were obtained prior to the initiation of the
study.
The participants of the ASD cohort were assessed via
the autistic diagnostic observation schedule (ADOS) (https://www.pearsonclinical.com.au/products/view/502).
Dedicated questionnaires were designed and information concerning
birth type, infant feeding, diet, GI symptoms, antibiotic treatment
history and other medically relevant data were collected from the
parents of the children in the ASD and the control group.
Samples collection and bacteriological
analysis
Fresh faecal samples were collected in sterile
containers and immediately transferred to the Eliava Bacteriophage
Analytical-Diagnostic Canter (BADC), Tbilisi, Georgia. A total of
57 faecal samples were collected for the study.
RIDASCREEN C. difficile Toxin A/B (C0801,
R-biopharm AG) enzyme immunoassay was used for the qualitative
determination of the C. difficile toxins A and B in the
fresh stool specimens. The test was performed according to the
manufacturer's instructions.
Routine bacteriological examination of the same
stool samples was performed according to the Eliava BADC Standard
Operational Procedures for the investigation of dysbiosis. The
enumeration of Bifidobacteria, Lactobacteria, E.
coli and Enterococcus spp., and the detection of
facultative and aerobic enteric pathogens, such as haemolytic E.
coli, Salmonella spp., Shigella spp.,
Klebsiella spp., Citrobacter spp., Morganella
morganii, as well as Acinetobacter baumannii,
Pseudomonas aeruginosa and coccoid species, including
Staphylococcus spp. and Streptococcus spp. was
performed as described below. A total of 1 g of faecal sample was
added to 9 ml sterile saline solution and mixed to obtain a
suspension. Further 10-fold serial dilutions were performed. A
total of 0.1 ml from the dilutions: 10-1,
10-3 and 10-5 were plated or inoculated in
enrichment (brain hart infusion (BHI), 5% sheep blood agar),
selective (brilliant green bile broth, MRS, casein yeast mannitol
salt, phenylethanol, Endo, Sabouraud and triple sugar iron agars)
or differential (MacConkey, SS, cetrimide, bile esculin azide and
Herella agar) medias. All media listed above were produced by
Eliava Media Production Ltd. In particular, BHI agar (Eliava Media
Production Ltd.) plates were used for determining the total
bacterial counts, 5% sheep blood agar for the detection of α- and
β-haemolysis, mannitol salt agar for the selection of
Staphylococcus spp., phenylethanol agar for
Streptococcus spp., brilliant green bile broth, Endo, triple
sugar iron agar and MacConkey agar for Enterobacteriaceae
(E. coli, Klebsiella, etc.), SS agar for the
detection of Salmonella and Shigella species,
cetrimide agar for Pseudomonas aeruginosa and Herellea agar
for Acinetobacter baumannii; bile esculin azide agar was
used to enumerate Enterococcus spp. and MRS medium for
Lactobacillus spp., Sabouraud agar for yeasts, and casein
yeast soft agar (0.7%) with supplement for the cultivation of
Bifidobacteriaceae. The plates and tubes were incubated in
optimal growth conditions of the target species, with anaerobic
environment of <0.1% of oxygen, >15% of CO2 where
needed. The combination of microscopy and biochemical tests were
implemented for further identification of the isolates. The strains
exhibiting α- or β-hemolysis and isolates identified as pathogens
underwent antibiotic and phage susceptibility testing. Antibiotic
susceptibility was determined using the Kirby-Bauer disk method
(31) and interpreted according to
the Clinical and Laboratory Standards Institute criteria (32). Phage susceptibility testing was
conducted using the spot test assay (33) using five commercially available
polyvalent phage preparations: PYO, INTESTI, FERSIS, SES and ENKO
bacteriophages (Eliava Biopreparations, Tbilisi, Georgia) including
the components against Staphylococcus spp,
Streptococcus spp., Enterococcus spp., E.
coli, Pseudomonas aeruginosa and Proteus spp.
TheiIsolation of presumptive C. difficile was
performed at the in-house laboratory of the Eliava Institute of
Bacteriophages, Microbiology and Virology (Tbilisi, Georgia).
Loop-fulls of the same stool sample dilutions 10-1,
10-3, 10-5 were cultured on C.
difficile selective agar (CCFA; Biolife Italiana), differential
chrome-agar (CHROMagar, DRG International, Inc.) and a blood agar
base with 5% of sheep blood (Eliava Media Production Ltd.). The
plates were incubated in an anaerobic jar with a gas-pack and
anaerobic indicator pad (GasPak, Becton, Dickinson and Company (BD)
BBL) at 37˚C for 48 h. C. difficile ATCC 43255 and ATCC
43593 (American Type Culture Collection) were cultured along with
the samples as a positive control for growth conditions. The
selected colonies were Gram-stained (Gram staining kit, Carl Roth
GmbH + Co. KG) and examined for catalase activity by applying 3%
hydrogen peroxide solution (Imedi) to the smear of bacterial
colony.
Phenotypically C. difficile isolates were
identified as follows: Gram-positive and catalase-negative bacilli
with endospores, displaying γ-haemolytic grey colonies with
umbonate edges on 5% sheep blood agar, fluorescence under
ultraviolet light on chrome-agar and circular, raised, opaque grey
colonies, 4 to 6 mm in diameter on CCFA. Furthermore, presumptive
C. difficile isolates were grown on a chrome-agar medium for
genome extraction. DNA was isolated using the UltraClean Microbial
DNA Isolation kit (15800-250, MO BIO Laboratories, Inc.) according
to the manufacturer's instructions. End-Point PCR was performed
using the C. difficile Detection kit (TM37100, Norgen Biotek Corp.)
as per the manufacturer's instructions. The gel electrophoresis of
20 µl PCR products was conducted at 150 V for 30 min on a 1.4%
(w/v) agarose gel, with 10 µl ethidium bromide. The results were
visualised using a transilluminator (Bio-Rad Laboratories, Inc.).
An overview of the study design is presented in Fig. 1.
Statistical analysis
The exposure status was measured using standardised
questionnaires and biological samples. The measure of association
between control and study group was expressed as odds ratios (ORs).
Post-hoc statistical evaluation of odds data was performed using
the GIGAcalculator (www.gigacalculator.com), in a 2-by-2 table, with
two-sided P-value and confidence interval at the level of 95%. Data
were visualised using Datawrapper software (www.datawrapper.de). Recall bias was excluded due to
non-ASD symptom-specific questionnaires.
Results
General group data
The ASD group was formed by all males, including one
pair of siblings. The age range in this group varied from 4 to 17
years, with a mean age of 10.8±3.2 years. The control group
included six pairs of neurotypical siblings and 5 children were
siblings of the ASD group participants. Sex distribution was as
follows: 19 (70.4%) males and eight (29.6%) females. The mean age
was 5.8±2.7 years, ranging from 2 to 15 years (Table I). All participants from the both
groups originated and resided in the same geographical area
(Tbilisi, Georgia).
 | Table IAge, sex and distribution of
gastrointestinal symptoms between the study groups. |
Table I
Age, sex and distribution of
gastrointestinal symptoms between the study groups.
| Group | Average age
(years) | Sex | Exhibiting GI
symptoms | Variety of GI
symptoms | % Individuals | No. of
individuals |
|---|
| ASD | 10.8±3.2 | 30/30 Male | 70% | Constipation | 33.3 | 10 |
| | | 0/30 Female | | Diarrhoea | 23.3 | 7 |
| | | | | Eructation | 26.7 | 8 |
| | | | | Flatulence | 40 | 12 |
| | | | | Heartburn | 10 | 3 |
| | | | | Exhibiting >1
symptom | 30 | 9 |
| Control | 5.8±2.7 | 19/27 Male | 37% | Constipation | 14.8 | 4 |
| | | 8/27 Female | | Diarrhoea | 3.7 | 1 |
| | | | | Eructation | 11.1 | 3 |
| | | | | Flatulence | 25.9 | 7 |
| | | | | Heartburn | 0 | 0 |
| | | | | Exhibiting >1
symptom | 14.8 | 4 |
Nutrition and comorbidities
The average maternal age was the same for both
groups, with 30.3±6.5 for the ASD and 30.3±4.3 for the control
group. In the ASD group, 18 children (60%) were delivered vaginally
and 25 (83.3%) were breastfed, eight participants (26.6%) had a
restricted diet, 19 (63.3%) were fed without restrictions and 3
children (10%) had mild aversions. Another 3 children (10%) had
endocrine comorbidities, 14 (46.6%) demonstrated other types of
comorbidities and 10 (33.3%) had a reported history of various
allergic reactions. In the control group, 11 children (40.7%) were
delivered naturally, 18 (66.6%) were breastfed; no dietary
restrictions were reported. Only two cases of comorbidities were
reported, and 7 participants (25.9%) reported allergies. The
detailed information related to infant feeding for both groups is
presented in Fig. 2.
GI symptoms
At the time of the examination, in the ASD group,
70% of the participants had at least one GI symptom (constipation,
diarrhoea, eructation, flatulence or heartburn). The most frequent
symptom was constipation (33.3%, 10 participants), followed by
diarrhoea (23.3%, 7 participants) and the least frequent was
heartburn affecting (only 10%, 3 participants). It should be noted
that 30% (9 participants) exhibited more than one GI symptom
simultaneously. In the control group, only 37% of the participants
noted at least one GI symptom, with most and least frequent being
flatulence (25.9%, 7 children) and diarrhoea (3.7%, 1 child),
respectively. The details regarding age, sex and GI symptoms are
summarised in Table I.
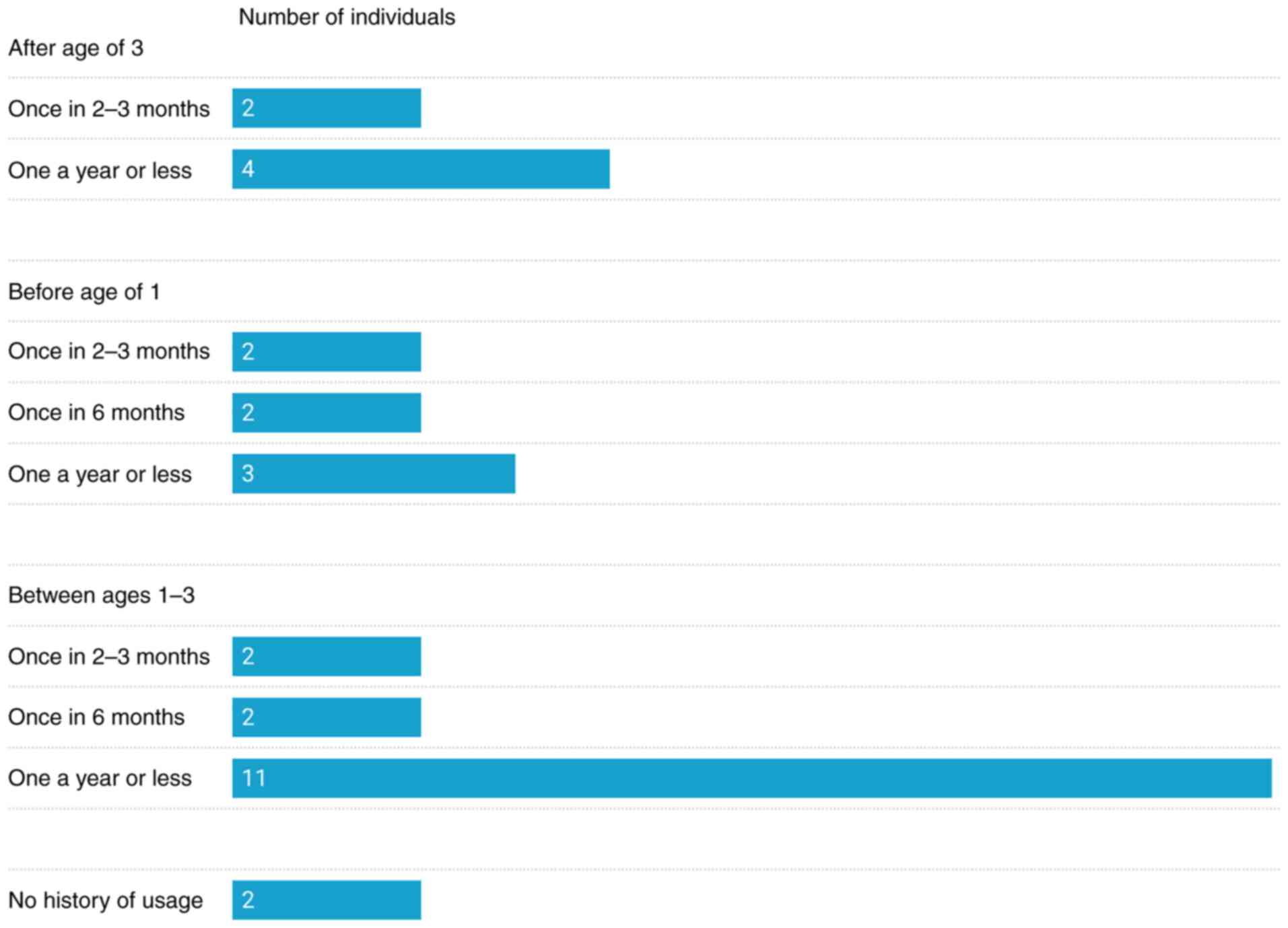
History of antibiotic use
According to the data collected through the
questionnaires, in the ASD group, 93% of the participants underwent
antibiotic therapy between the ages of 0 to 3 years. In total, the
parents of 7 participants (23.3%) reported the usage of penicillin,
cephalosporin or macrolides before the age of one, and 15
participants (50%) between the ages of 1 and 3 years. The parents
of 6 participants (20%) recalled the usage of penicillin,
cephalosporin, quinolones or aminoglycosides after the age of
three. In addition, 9 children (30%) had a history of antibiotic
usage and deficiency in normal bacterial composition (as diagnosed
by BADC) even though they did not have recorded GI symptoms. In
total, 2 participants (7%) had no history of antibiotic usage, but
exhibited constipation and deficiency in normal bacterial
composition. In the control group, the parents of 20 participants
(74%) reported usage of antibiotics at some point in the past.
However, only one parent could recall which antibiotic was
prescribed. The details of antibiotic usage in the ASD group are
illustrated in Fig. 3, while the
classes of used antibiotics are presented in Table II.
 | Table IIClasses of antibiotics used in the
ASD group. |
Table II
Classes of antibiotics used in the
ASD group.
| Antibiotic
classes | No. of
individuals | % Individuals |
|---|
| Penicillins | 12 | 40.0 |
| Cephalosporins | 7 | 23.3 |
| Quinolones | 1 | 3.3 |
| Macrolides | 3 | 10.0 |
|
Aminoglycosides | 1 | 3.3 |
| Unknown | 7 | 23.3 |
| Never used | 2 | 6.7 |
Bacteriological analysis
A near detection level of toxin A/B was found in
only one stool sample from the ASD group. A child with the history
of antibiotic therapy with penicillin in early childhood (1 to 3
years) exhibited diarrhoea, belching and flatulence. The parents of
another participant with a history of frequent use of penicillin
drugs and quinolones after the age of three, suffering with the
interchangeable constipation and diarrhoea, bloating and
flatulence, reported of a previously diagnosed C. difficile
infection; however, at the time of the examination, toxin producing
C. difficile was not detected. Otherwise, A/B toxin
producing C. difficile infection was not detected or
reported by any other participants from either group. The
bacteriological examination of the samples and following
species-specific PCR testing of the isolates did not confirm the
presence of C. difficile in any of the provided samples.
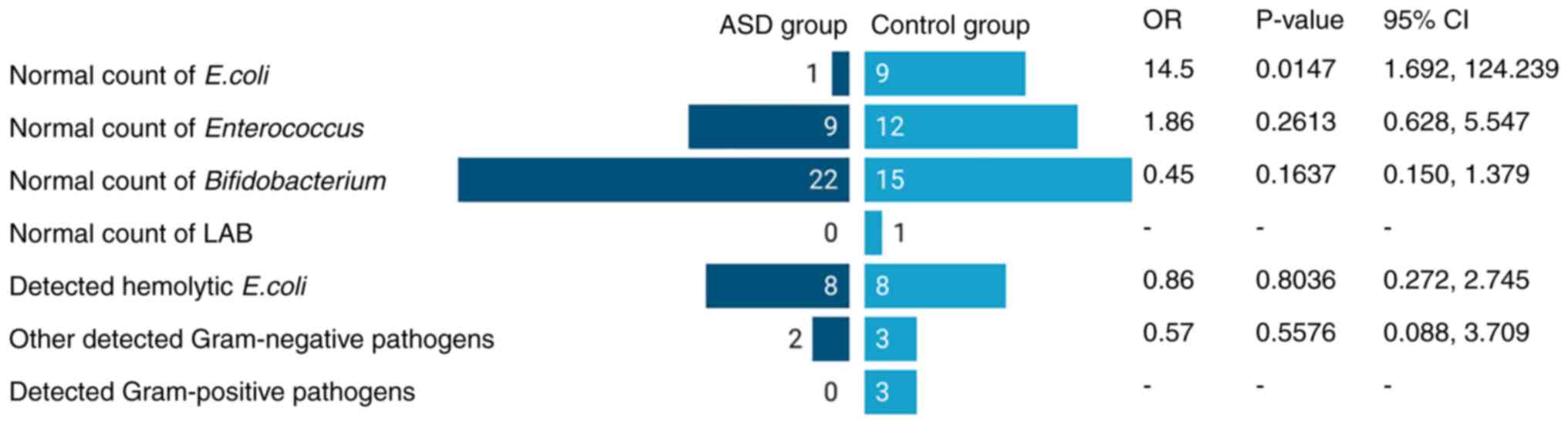
According to the stool analysis performed by the
BADC, no Candida-like fungal growth was detected in any of
the tested samples. All participants, apart from one in the control
group exhibited a deficiency in Lactobacillus spp. counts.
In the ASD group, only 3% of the participants exhibited normal
counts of commensal E. coli, 30% had satisfactory counts of
Enterococcus spp., and 73.3% had regular
Bifidobacterium counts. β-haemolytic E. coli was
isolated from 26.7% of samples, and drug-resistant
Klebsiella spp. and Pseudomonas aeruginosa were found
in two different samples; however, despite this fact, those
participants had not reported any GI symptoms. In the control
group, 33.3% had normal counts of the commensal E. coli,
44.4% had a satisfactory count of Enterococcus spp., and
only 55.6% had regular Bifidobacterium counts. β-haemolytic
E. coli was isolated from 29.6% of samples,
Klebsiella spp. and drug-resistant Pseudomonas
aeruginosa, Morganella morganii and Staphylococcus
aureus were isolated from five different samples as well. The
OR calculation for normal Bifidobacterium counts was 0.45
with significance level of P=0.1637, suggesting that the disruption
in Bifidobacterium biota was more frequent in the control
group than in the ASD group, although at a low level of
significance. For Enterococcus, the analysis revealed an OR
of 1.86 (P=0.2613), suggesting that the disruption in
Enterococcus was 2-fold the odds in the ASD than in the
control group, also with low significance. For the normal E.
coli count, the OR value was 14.5 (P=0.0147), indicating that
the ASD group had 14-fold the odds of a disruption in E.
coli counts than the control. The overview of the
bacteriological analysis is presented in Fig. 4.
Phage and antibiotic
susceptibility
A total of 16 encountered strains of β-haemolytic
E. coli isolated from both groups underwent antibiotic- and
phage-susceptibility tests against five antibiotic classes and five
commercial phage preparations. All strains isolated from the ASD
group demonstrated resistance to penicillin, which was often
associated with resistance to aminoglycoside and in some cases, to
cephalosporin group antibiotics. All isolates obtained from the
control group also revealed resistance to penicillin, often
associated with aminoglycosides and cephalosporins, and rarely to
the group of tetracycline antibiotics (Table SI).
In total, five polyvalent commercial phage
preparations with lines active against E. coli were used on
β-haemolytic E. coli isolates; eight strains out of the 16
tested isolates appeared to be sensitive to at least one
preparation. The overall phage susceptibility of the isolated
pathogens is summarised in Fig.
5.
Discussion
It has been shown that the type of childbirth
delivery, infant feeding and medication intake has a profound
influence on development of an individual's microbiome (34). C-section, perinatal antibiotic
intake and formula feeding are associated with microbiome
perturbation and may influence infants' neurocognitive development
(35). There is still a
controversial view about the link between C-section and the
development of ASD. A 2019 meta-analysis, involving >20 million
individuals, reported that children delivered via C-section were
~30% more likely to be diagnosed with ASD than those born vaginally
(36). At the same time, other
research has indicated that toddlers without breastfeeding in the
first 6 months of life have higher odds of developing ASD when
compared to those who were exclusively breastfed (37). The results of the present study did
not confirm these observations. In particular, the percentage and
duration of breastfeeding, as well as vaginal deliveries were
higher in the ASD group. However, a higher frequency of
early-childhood antibiotic intake and more frequent GI symptoms
were reported in the ASD group.
The present study tried to link the ASD symptoms to
the composition of the gut microbiota and the particular role of
C. difficile in the associated neurological disorders. No
C. difficile was found in the faecal samples of the
participants, which is in agreement with the findings of Khalil
et al (38), whereas no
association between C. difficile and GI manifestation in ASD
was observed.
Some differences in the commensal microbiota counts
were detected between the two groups, with more frequent deficits
in Bifidobacterium in the control group and more frequent
disruptions in Enterococcus and E. coli in the ASD
group. The results of the present study partially correspond to the
data from literature, where the lower abundance of
Bifidobacteria, Enterococcus and E. coli among
children with ASD has been reported (19). In the present study, the slightly
higher incidence of normal counts of Bifidobacteria in the
ASD cohort may be attributed to the frequent administration of
probiotics containing these bacteria. As a popular over-the-counter
supplement in Georgia, probiotics are frequently advised by
pharmacists and doctors for the normalisation of bowel movements.
However, the questions related to probiotic use were not included
in the questionnaire.
Drug-resistant pathogens were encountered in the
samples from both groups, where isolates from the control group
were more diverse than those in the ASD group. The results of
antibiotic sensitivity tests of β-haemolytic E. coli
demonstrated slightly different resistance patterns in the ASD and
control groups. In phage susceptibility testing, 50% of
β-haemolytic E. coli isolates were susceptible to at least
one phage preparation, which is a promising result. Due to the
limited number of the tested strains, it was impossible to identify
significant differences in the phage sensitivity patterns between
the ASD and control groups. At same time, no association between
antibiotic and phage sensitivity patterns was observed.
In order to regulate GI disturbances in patients
with ASD, various personalised dietary approaches, such as the use
of different probiotics and prebiotics, or exclusion diets (gluten-
and casein-free) and microbial transfer therapy have been suggested
and implemented with varying levels of success (1). In some cases, even antibiotic therapy
is prescribed to manage ASD symptoms, which can be devastating for
the commensal microbiota of the individual (39). As an alternative safer treatment
option, phages are gaining increasing momentum. They can be used as
highly specific and natural antibacterial agents that can
specifically eliminate target multi-drug resistant bacteria, or as
a prebiotic agent that can help regulate dysbiosis. As phage
products can be adapted for personalised use, it is possible to
develop phage-based tailored preparations catering to the needs of
individual patients with ASD and thus decrease the overuse of
antibiotics, avoiding antibiotic associated side-effects and
resistant development or offer an option for individuals with
antibiotic-incompatibilities. For example, the present study,
children with β-haemolytic E. coli and at same time, low
commensal E. coli counts, would benefit from highly
specialised, preferably strain specific phages, which only lyse
β-haemolytic E. coli and do not attack commensal strains,
thus ensuring pathogen elimination without further degradation of
commensal microbiota.
Due to the unique nature of human microbiotas, the
further understanding of the functions and alterations of the
microbiome-gut-brain axis and its implications on the ASD aetiology
would benefit from longitudinal observations with a personalized
approach.
Supplementary Material
Antibiotic resistance profiles of the
isolated pathogenic cultures.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by the Shota Rustaveli
National Science Foundation of Georgia (grant no. FR-18-17189).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
All authors (EK, KM, NB, NG, SK, TE, VB, NS, NaC,
EJ, GN, TT, MM, NiC and IA) participated in the conception and
design of the study, revised, edited or reviewed the submitted
versions of the manuscript. In addition, EK, KM, NB, NG, NS, NaC,
EJ, GN, TT, MM contributed to sample collection, processing and
microbiological analysis. SK, TE, VB, NiC and IA participated in
recruitment as well as data collections from the cohorts. NiC, KM
and IA contributed to funding acquisitions and EK, SK and NiC
participated in data analysis, and in the writing and processing of
the manuscript. NiC and IA coordinated the project. All authors
have read and approved the final manuscript. NiC and SK confirmed
the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Scientific
Research Ethics Committee of the Ilia State University, Tbilisi,
Georgia (issued on October, 2019). Parental oral consents were
obtained prior to the initiation of the study and the parents
provided consent for the information of their children to be
published.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Essa MM and Qoronfleh MW (eds):
Personalized food intervention and therapy for autism spectrum
disorder management. Vol. 24. Springer Nature, 2020.
|
|
2
|
Wiśniowiecka-Kowalnik B and Nowakowska BA:
Genetics and epigenetics of autism spectrum disorder-current
evidence in the field. J Appl Genet. 60:37–47. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Loomes R, Hull L and Mandy WPL: What is
the male-to-female ratio in autism spectrum disorder? A systematic
review and meta-analysis. J Am Acad Child Adolesc Psychiatry.
56:466–474. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Fombonne E: Epidemiology of pervasive
developmental disorders. Pediatr Res. 65:591–598. 2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Baxter AJ, Brugha TS, Erskine HE, Scheurer
RW, Vos T and Scott JG: The epidemiology and global burden of
autism spectrum disorders. Psychol Med. 45:601–613. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Global Research on Developmental
Disabilities Collaborators. Developmental disabilities among
children younger than 5 years in 195 countries and territories,
1990-2016: A systematic analysis for the global burden of disease
study 2016. Lancet Glob Health. 6:e1100–e1121. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Muhle RA, Reed HE, Stratigos KA and
Veenstra-VanderWeele J: The emerging clinical neuroscience of
autism spectrum disorder: A review. JAMA Psychiatry. 75:514–523.
2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Oh D and Cheon KA: Alteration of gut
microbiota in autism spectrum disorder: An overview. Soa
Chongsonyon Chongsin Uihak. 31:131–145. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Mayer EA, Padua D and Tillisch K: Altered
brain-gut axis in autism: Comorbidity or causative mechanisms?
Bioessays. 36:933–939. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Fung TC: The microbiota-immune axis as a
central mediator of gut-brain communication. Neurobiol Dis.
136(104714)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sherwin E, Dinan TG and Cryan JF: Recent
developments in understanding the role of the gut microbiota in
brain health and disease. Ann N Y Acad Sci. 1420:5–25.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Borre YE, O'Keeffe GW, Clarke G, Stanton
C, Dinan TG and Cryan JF: Microbiota and neurodevelopmental
windows: Implications for brain disorders. Trends Mol Med.
20:509–518. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Fowlie G, Cohen N and Ming X: The
perturbance of microbiome and gut-brain axis in autism spectrum
disorders. Int J Mol Sci. 19(2251)2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Esnafoglu E, Cırrık S, Ayyıldız SN, Erdil
A, Ertürk EY, Daglı A and Noyan T: Increased serum zonulin levels
as an intestinal permeability marker in autistic subjects. J
Pediatr. 188:240–244. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tiffon C: The impact of nutrition and
environmental epigenetics on human health and disease. Int J Mol
Sci. 19(3425)2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rowland I, Gibson G, Heinken A, Scott K,
Swann J, Thiele I and Tuohy K: Gut microbiota functions: Metabolism
of nutrients and other food components. Eur J Nutr. 57:1–24.
2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Petersen C and Round JL: Defining
dysbiosis and its influence on host immunity and disease. Cell
Microbiol. 16:1024–1033. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
De Angelis M, Piccolo M, Vannini L,
Siragusa S, De Giacomo A, Serrazzanetti DI, Cristofori F, Guerzoni
ME, Gobbetti M and Francavilla R: Fecal microbiota and metabolome
of children with autism and pervasive developmental disorder not
otherwise specified. PLoS One. 8(e76993)2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xu M, Xu X, Li J and Li F: Association
between gut microbiota and autism spectrum disorder: A systematic
review and meta-analysis. Front Psychiatry. 10(473)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li Q, Han Y, Dy ABC and Hagerman RJ: The
gut microbiota and autism spectrum disorders. Front Cell Neurosci.
11(120)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Song Y, Liu C and Finegold SM: Real-time
PCR quantitation of clostridia in feces of autistic children. Appl
Environ Microbiol. 70:6459–6465. 2004.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Parracho HM, Bingham MO, Gibson GR and
McCartney AL: Differences between the gut microflora of children
with autistic spectrum disorders and that of healthy children. J
Med Microbiol. 54:987–991. 2005.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Finegold SM, Summanen PH, Downes J,
Corbett K and Komoriya T: Detection of Clostridium
perfringens toxin genes in the gut microbiota of autistic
children. Anaerobe. 45:133–137. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sandler RH, Finegold SM, Bolte ER,
Buchanan CP, Maxwell AP, Väisänen ML, Nelson MN and Wexler HM:
Short-term benefit from oral vancomycin treatment of
regressive-onset autism. J Child Neurol. 15:429–435.
2000.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Vinithakumari AA, Padhi P, Hernandez B,
Lin SJH, Dunkerson-Kurzhumov A, Showman L, Breitzman MW, Stokes C,
Sulaiman Y, Tangudu CS, et al: Clostridioides difficile
infection increases circulating p-cresol levels and dysregulates
brain dopamine metabolism: linking gut-brain axis to autism
spectrum disorders? bioRxiv, 2021.
|
|
26
|
Huang H, Weintraub A, Fang H and Nord CE:
Antimicrobial resistance in Clostridium difficile. Int J
Antimicrob Agents. 34:516–522. 2009.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kuhn M, Grave S, Bransfield R and Harris
S: Long term antibiotic therapy may be an effective treatment for
children co-morbid with Lyme disease and autism spectrum disorder.
Med Hypotheses. 78:606–615. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Busch C and Aktories K: Microbial toxins
and the glycosylation of rho family GTPases. Curr Opin Struct Biol.
10:528–535. 2000.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Netgazeti Healthcare. Management of autism
spectrum disorder in Georgia: Khachapuridze E. http://netgazeti.ge/life/13498/. Accessed March
6, 2022.
|
|
30
|
Tevzadze G, Shanshiashvilli L and
Mikeladze D: Children with epilepsy and autistic spectrum disorders
show similarly high levels of urinary p-cresol. J Biol Phys Chem.
17:77–80. 2017.
|
|
31
|
Bauer AW, Kirby WM, Sherris JC and Turck
M: Antibiotic susceptibility testing by a standardized single disk
method. Am J Clin Pathol. 45:493–496. 1966.PubMed/NCBI
|
|
32
|
Clinical and Laboratory Standards
Institute. Performance standards for antimicrobial susceptibility
testing, 29th edition. https://clsi.org/standards/products/microbiology/documents/m100/.
Accessed March 6, 2022.
|
|
33
|
Makalatia K, Kakabadze E, Wagemans J,
Grdzelishvili N, Bakuradze N, Natroshvili G, Macharashvili N,
Sedrakyan A, Arakelova K, Ktsoyan Z, et al: Characterization of
Salmonella isolates from various geographical regions of the
caucasus and their susceptibility to bacteriophages. Viruses.
12(1418)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Mueller NT, Bakacs E, Combellick J,
Grigoryan Z and Dominguez-Bello MG: The infant microbiome
development: Mom matters. Trends Mol Med. 21:109–117.
2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Yang I, Corwin EJ, Brennan PA, Jordan S,
Murphy JR and Dunlop A: The infant microbiome: Implications for
infant health and neurocognitive development. Nurs Res. 65:76–88.
2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang T, Sidorchuk A, Sevilla-Cermeño L,
Vilaplana-Pérez A, Chang Z, Larsson H, Mataix-Cols D and
Fernández-de-la-Cruz L: Association of cesarean delivery with risk
of neurodevelopmental and psychiatric disorders in the offspring: A
systematic review and meta-analysis. JAMA Netw Open.
2(e1910236)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Huang S, Wang X, Sun T, Yu H, Liao Y, Cao
M, Cai L, Li X, Lin L, Su X and Jing J: Association of
breastfeeding for the first six months of life and autism spectrum
disorders: A national multi-center study in China. Nutrients.
14(45)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Khalil M, Azouz HG, Ahmed SA, Gad HA and
Omar OM: Sensory processing and gastrointestinal manifestations in
autism spectrum disorders: No relation to Clostridium
difficile. J Mol Neurosci. 71:153–161. 2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ramirez PL, Barnhill K, Gutierrez A,
Schutte C and Hewitson L: Improvements in behavioral symptoms
following antibiotic therapy in a 14-year-old male with autism.
Case Rep Psychiatry. 2013(239034)2013.PubMed/NCBI View Article : Google Scholar
|