Introduction
According to Global Cancer Statistics 2020, breast
cancer is the most prevalent malignancy and is one of the top known
causes of cancer-related mortality among females worldwide
(1). Even in the general Iraqi
population, breast cancer has been the highest-ranked malignancy
since 1986(2), and the latest
Cancer Registry in Iraq has observed 7,515 new breast cancer cases
in 2020, accounting for 37.9% of the total reported cancer cases.
Furthermore, breast cancer is the most common malignant tumor among
Iraqi women (3).
Moreover, well-known risk factors (such as ionizing
radiation, having a family history of cancer, alcoholism, obesity,
hormone therapy during menopause, an older age, etc.), and several
other factors, such as genetic predisposition from parents
(4) along with epigenetic factors,
have been recently reported to be associated with breast cancer
development (5).
Breast carcinogens originate from a small cellular
population known as breast cancer stem cells (BCSCs), that have a
distinct molecular signature (6),
and whose origin continues to be controversial. Some studies have
reported that BCSCs originate from mammary stem cells or progenitor
cells (7-9),
while others have demonstrated that they arise from differentiated
mammary cells (10-12).
A low proportion (5-10%) of breast cancer cases may be associated
with inherited mutations in the BRCA genes, which occur only in
women (13,14).
MicroRNAs (miRNAs/miRs) are small, non-coding,
single stranded (18-22 nucleotides in length) RNA molecules that
function to regulate the post-transcriptional machinery of gene
expression (15). Previous
research has reported that miRNAs are involved in tumorigenesis
(16), playing a critical role in
the genesis and progression of breast cancer (17), potentially through the regulation
of BCSCs (18). miRNAs play a
crucial role in regulating the biological functions of a cell on a
variety of levels. A number of diseases, including cancer, have
been associated with miRNAs. There has been a rapid increase in
interest in miR-146a in particular, as a modulator of
differentiation and function as well as innate and adaptive
immunity. miR-146a has been implicated in regulating a number of
key cellular functions; thus, there are various types of tumors
(papillary thyroid carcinoma, breast cancer and cervical cancer)
with a dysregulated expression of miR-146a (19). Evidently, miR-146 levels have been
shown to be consistently higher in breast cancer cells that exhibit
tumor aggressive characteristics, among a variety of molecular
subcategories of breast malignancy, along with an observed peak
enrichment in BCSCs (20).
As regards the molecular mechanisms of miR-146 in
breast cancer, it was previously demonstrated that miR-146a-5p
overexpression in MCF-7 cells led to an increased proliferation,
and the low expression miR-146a-5p in MCF-7 cells led to a
decreased proliferation (21). By
analyzing bioinformatics data and detecting fluorescent reporter
genes, miR-146a-5p was identified as a gene target for BRCA1.
Breast cancer tissue and MCF-7 cells expressing miR-146a-5p may
regulate the proliferation of the cancer cell line via
BRCA1(21). Another study also
found that the miR-146a expression levels were significantly higher
in breast cancers with various pathological classifications, while
non-metastatic protein 23 H1 (NM23-H1) expression levels were
significantly lower, which were closely correlated (22). In a breast cancer cell line,
miR-146 and NM23-H1 were verified to have target regulatory
associations by double luciferase reporter gene assays. miR-146a
was closely related to the proliferation and metastasis of breast
cancer. Additionally, miR-146a targeted NM23-H1 in vivo to
promote breast cancer growth (22).
Malondialdehyde (MDA) is an organic compound that
occurs naturally and its determination in blood plasma or tissues
functions as a predictive marker of oxidative stress (23,24).
When biomolecule peroxidation occurs, a number of carcinogenic and
mutagenic factors are produced (25). Exploring the potential role of
ectopic gene expression in patients with cancer has recently
attracted the research interests of the authors (26-29).
Despite the fact that a higher expression of miR-146 is associated
with increased levels of MDA in normal tissues (30-32),
to date, at least to the best of our knowledge, there are no
reports available suggesting a similar association in malignant
tissues. Therefore, the present study aimed to investigate the
association between miR-146 expression and oxidative stress, as
indicated by MDA levels, in breast tumorigenesis.
Subjects and methods
Subjects and sampling
Blood samples were collected from patients with
breast cancer (n=30) and healthy women (age-matched controls;
n=20), between January 3 to March 23, 2022. The inclusion criteria
used to recruit individuals in the present study involved female
patients diagnosed with stage I-III breast primary tumors aged
30-70 years. Control individuals included apparently healthy
females with the same age range aforementioned. Males, patients
with breast secondary tumors and those out of the age range
mentioned above were, otherwise, excluded. Relevant ethics approval
was obtained from the Biomedical Research Ethics Committee of the
leading National Cancer Research Center at the University of
Baghdad (reference no. NCRCEC/01/001). All individuals
participating in the study were recruited from the Baghdad Teaching
Hospital and Oncology Hospital, Baghdad, Iraq, after providing
written informed consent. The recruitment process was carried out
based on clinical/laboratory examinations and diagnoses by
specialist doctors in the hospitals. A questionnaire was prepared
to obtain patient information, including name, age and a family
health history. The clinical information of all the study subjects
is presented in Table I.
 | Table IClinical information of the patients
with breast cancer in the present study. |
Table I
Clinical information of the patients
with breast cancer in the present study.
| Subject no. | Age of the healthy
controls, years | Age of the
patients, years | Patient
relative | Tumor marker
(cA15.3) | Tumor stage |
|---|
| 1 | 32 | 70 | Not relative | 20.5 | PT 1 |
| 2 | 44 | 67 | Sister | 19.4 | PT 2 |
| 3 | 52 | 62 | Aunt | 20.6 | PT 1 |
| 4 | 22 | 70 | Not relative | 18.2 | PT 2 |
| 5 | 50 | 57 | Nephew | 13.4 | PT 2 |
| 6 | 38 | 70 | Not relative | 11.2 | PT 1 |
| 7 | 25 | 62 | Not relative | 21.4 | PT 2 |
| 8 | 31 | 60 | Not relative | 17.3 | PT 2 |
| 9 | 34 | 52 | Not relative | 22.2 | PT 1 |
| 10 | 24 | 59 | Daughter | 16.5 | PT 1 |
| 11 | 62 | 56 | Sister | 10.5 | PT 1 |
| 12 | 59 | 50 | Sister | 9.3 | PT 1 |
| 13 | 43 | 55 | Sister | 7.1 | PT 2 |
| 14 | 21 | 59 | Daughter | 20.7 | PT 2 |
| 15 | 30 | 61 | Not relative | 14.9 | PT 1 |
| 16 | 37 | 67 | Not relative | 18 | PT 1 |
| 17 | 70 | 69 | Sister | 28.6 | PT 1 |
| 18 | 58 | 48 | Not relative | 23.8 | PT 1 |
| 19 | 24 | 68 | Not relative | 16.3 | PT 1 |
| 20 | 21 | 55 | Not relative | 8.6 | PT 1 |
| 21 | | 59 | Not relative | 10.2 | PT 1 |
| 22 | | 62 | Not relative | 19.8 | PT 1 |
| 23 | | 45 | Not relative | 12.1 | PT 1 |
| 24 | | 68 | Not relative | 7.5 | PT 1 |
| 25 | | 66 | Not relative | 12.9 | PT 1 |
| 26 | | 43 | Not relative | 14.8 | PT 1 |
| 27 | | 51 | Not relative | 18.2 | PT 1 |
| 28 | | 55 | Not relative | 8.2 | PT 1 |
| 29 | | 49 | Not relative | 13.9 | PT 1 |
| 30 | | 32 | Not relative | 24.2 | PT 1 |
Blood samples were collected in VACUETTE®
tubes, K3EDTA tubes (cat. no. 454021, Greiner Bio One Ltd.) and
allowed to stand for 20 min. RNA was then extracted using TRIzol
reagent (cat. no. T9424, MilliporeSigma) by the addition of 200 µl
blood and 400 µl TRIzol to reagent to each sample. In addition,
blood plasma (serum) was collected from all samples for the MDA
assay.
RNA isolation and cDNA
preparation
Total RNA was extracted from whole blood of patients
with breast cancer and healthy women using the mirVana™ miRNA
Isolation kit (AM1560, Thermo Fisher Scientific, Inc.), following
the manufacturer's protocol. The quantity of miRNA was measured
using a Qubit 4 fluorometer (Thermo Fisher Scientific, Inc.). This
assay is highly selective for the miRNA quantification of other
types of RNA. Following RNA extraction, cDNA was synthesized from
isolated miRNA, through optimized primers, using a Protoscript cDNA
synthesis kit (E6300L, New England BioLabs, Inc.). Briefly, 5 µl of
each RNA sample were added to the Protoscript reaction mix,
containing dNTPs, 10 µl buffer, 2 µl MuLV enzyme and 2 µl specific
primers for each sample. All mixtures were incubated for 1 h at
42˚C in a thermocycler, followed by an incubation at 80˚C to
inactivate the enzyme. The cDNA products were quantified using a
Qubit 4 fluorometer. The products were electrophoresed on a 2%
agarose gel and visualized on a UV transilluminator by ethidium
bromide staining.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was used to detect the expression of miR-146
using the Luna® Universal qPCR Master Mix kit (cat. no.
M3003, New England BioLabs, Inc.). Primers for miR-146 and U6
calibrators were designed by Macrogen, Inc. (Korea), and are
presented in Table II.
 | Table IIPrimers used for the analysis of
miR-146 and U6 gene expression. |
Table II
Primers used for the analysis of
miR-146 and U6 gene expression.
| Primers | Sequence |
|---|
| miR-146 | |
|
Forward |
GGGTGAGAACTGAATTCCA |
|
Reverse |
CAGTGCGTGTCGTGGAGT |
| U6 | |
|
Forward |
CTCGCTTCGGCAGCACA |
|
Reverse |
AACGCTTCACGAATTTGCGT |
Synthesized cDNA from patients and healthy controls
were run simultaneously, including the target miR-146, and the
housekeeping gene, U6 snRNA. The reaction mix consisted of 10 µl
Luna Universal qPCR Master Mix, 1 µl forward primer (10 µM), 1 µl
reverser primer (10 µM), 5 µl template DNA and 3 µl nuclease-free
water. The qPCR program was set up with the indicated thermocycling
protocol, as presented in Table
III. Relative mRNA quantification was performed using the
2-ΔΔCq method (33).
 | Table IIIThermocycling conditions used in
RT-qPCR. |
Table III
Thermocycling conditions used in
RT-qPCR.
| Cycle step | Temperature
(˚C) | Time | Cycles |
|---|
| Initial
denaturation | 95 | 60 sec | 1 |
| Denaturation | 95 | 15 sec | 40-45 |
| Extension | 60 | 30 sec (+ plate
read) | |
| Melting curve | 60-95 | 40 min | 1 |
MDA assay
MDA, a highly reactive an organic compound that
causes toxic stress in cells, is a naturally occurring marker of
oxidative stress (23). In the
present study, to determine whether the MDA capacity of serum
differs between healthy women and patients with breast cancer, the
serum MDA concentrations were detected and its association with
miR-146 gene expression was examined.
The MDA assay kit (cat. no. ab118970; Abcam) was
used in the present study. Serum samples from 30 patients with
breast cancer and 20 healthy women were collected and centrifuged
at 2,500 x g for 10 min at room temperature to remove any residual
cells. Subsequently, 200 µl of each serum sample or standard
solution were mixed with 600 µl thiobarbituric Acid (TBA). All of
the samples were then heated in a 100˚C water bath for 30 min and
cooled down to room temperature 25˚C for 20 min. The total
antioxidant capacity (TAC) was measured using a relevant kit (cat.
no. ab65329; Abcam); 100 µl Cu2+ working solution was
added to 100 µl of each sample or standard solution, followed by
centrifugation at 1,008 x g for 10 min. Finally, the absorbance of
the samples was measured using an Accuris™ SmartReader™ 96
microplate absorbance reader (cat. no. Z742712; Merck KGaA) at 532
and 570 nm, respectively.
Statistical analysis
Comparisons of miRNA gene expression frequencies and
the results of the MAD assay among the study groups were determined
using an unpaired t-test (independent samples t-test). All
measurements were taken from three replicates and are presented as
mean values, from which the standard error (SE) of the mean was
calculated. A value of P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of U6 gene
The results of the expression of the U6 gene
(endogenous control) in healthy women and patients with breast
cancer are presented in Table IV.
The mean Ct values for healthy women and patients with breast
cancer were 20.71 and 20.59, respectively. There were no
significant differences in U6 expression between the healthy and
breast cancer samples.
 | Table IVComparison of the Ct value between
study groups for the U6 gene (mean ± SE). |
Table IV
Comparison of the Ct value between
study groups for the U6 gene (mean ± SE).
| | | Ct mean |
|---|
| Group | No | Statistic | SE |
|---|
| Healthy
controls | 20 | 20.71 | 0.32 |
| Patients | 30 | 20.59 | 0.34 |
| LSD | - | 1.018 (NS) | |
| P-value | - | 0.238 | |
The 2-ΔΔCq values for healthy women and
patients with breast cancer were (0.64E-8) and (0.60E-8),
respectively. There was no significant difference in U6 gene
expression between the two groups. However, there was a decrease in
fold change in patients with breast cancer, when compared with the
healthy controls (0.93 and 1, respectively), as shown in Table V.
 | Table VComparison of U6 gene fold expression
between the study groups. |
Table V
Comparison of U6 gene fold expression
between the study groups.
| Group | Means Ct of U6 |
2-ΔCq | Experimental
group/control group | Fold of gene
expression |
|---|
| Healthy
controls | 20.71 | 0.64E-8 |
0.64E-8/0.64E-8 | 1 |
| Patients | 20.59 | 0.60E-8 |
0.60E-8/0.64E-8 | 0.93 |
| LSD | 1.017 (NS) | - | - | 0.230 (NS) |
| P-value | 0.223 | - | - | 0.087 |
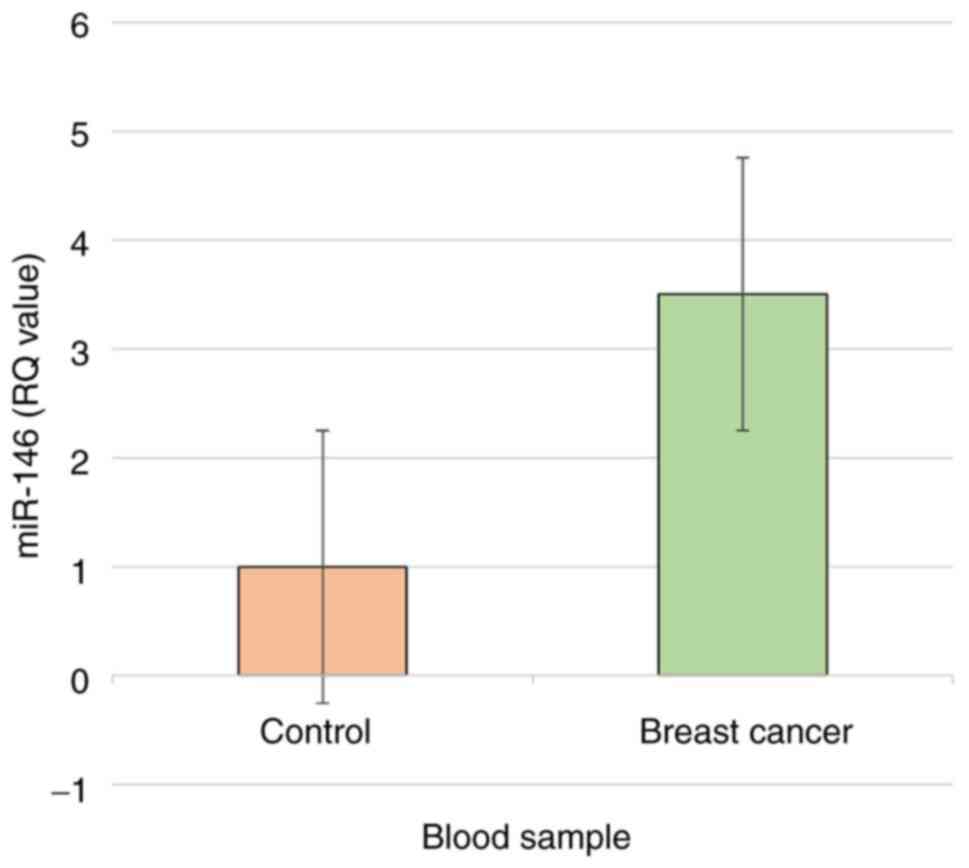
Expression of the miR-146 gene
The values of Ct, ΔCq and 2-ΔCq of the miR-146 gene
in healthy women and breast cancer patients are presented in
Table VI. The Ct values of the
miR-146 gene for healthy women and patients with breast cancer were
29.76 and 28.04, respectively. The ΔCq values of the miR-146 gene
were significantly (P<0.05) lower in patients with breast cancer
compared with healthy women (7.47 and 9.1, respectively). In
contrast, the 2-ΔCq values of miR-146 were significantly
(P<0.05) higher in breast cancer patients than in healthy women
(0.0056 and 0.0018, respectively). The results related the fold
change in miR-146 gene expression, based on the 2-ΔCq
and 2-ΔΔCq values, as presented in Table VI.
 | Table VIComparison between Ct, ΔCq and 2-ΔCq
values of miR-146 in different groups. |
Table VI
Comparison between Ct, ΔCq and 2-ΔCq
values of miR-146 in different groups.
| Group | No | Ct value | ΔCq |
2-ΔCq |
|---|
| Healthy
controls | 20 | 29.76 | 9.1 | 0.0018 |
| Patients | 30 | 28.04 | 7.47 | 0.0056 |
| LSD | - | 1.483 (NS) | 1.761a |
0.00055a |
| P-value | - | 0.0688 | 0.0420 | 0.0495 |
As shown in Table
VII, depending on the 2-ΔCq method, the actual
results were significantly associated with the predictions by the
model (P<0.01); miR-146 expression was higher in patients with
breast cancer compared with healthy women (3.1- and 1-fold,
respectively). In addition, depending on the 2-ΔΔCq
method, the values of ΔΔCq were significantly (P<0.01) lower in
patients with breast cancer than in the healthy subjects (-1.52 and
0.11, respectively). The value of 2-ΔΔCq was
significantly (P<0.01) higher in patients with breast cancer
than in healthy women (2.867 and 0.926, respectively). Therefore,
depending on the 2-ΔΔCq method, miR-146 gene expression
was significantly (P<0.01) higher in patients with breast cancer
than in healthy women (3.096- and 1-fold, respectively) (Fig. 1).
 | Table VIIFold of miR-146 gene expression
depending on the ΔCq and 2-ΔCq methods. |
Table VII
Fold of miR-146 gene expression
depending on the ΔCq and 2-ΔCq methods.
| Parameters | 2-ΔCq
method | |
|---|
| Group | Healthy
controls | Patients | P-value |
|---|
| Ct target | 9.1 | 7.47 | 0.0420a |
| Experimental | 0.0018/0.0018 | 0.0056/0.0018 | - |
| Fold of gene
expression | 1 | 3.1 | 0.0028b |
| Parameters | 2-ΔΔCq
method | |
| Group | Healthy | Patients | |
| Ct calibrator | 8.99 | 8.99 | 1.00 (NS) |
| ΔΔCt | 0.11 | -1.52 | 0.0042b |
|
2-ΔΔCt | 0.926 | 2.867 | 0.0002b |
| Experimental | 0.926/0.926 | 2.867/0.926 | |
| Fold of gene
expression | 1 | 3.096 | 0.0030b |
MDA levels in serum
The serum MDA values in patients with breast cancer
(n=30) and the healthy controls are presented in Table VIII. The MDA levels were
significantly (P<0.01) higher in patients with breast cancer
compared with the healthy controls (Fig. 2).
 | Table VIIISerum MDA values at 532 nm. |
Table VIII
Serum MDA values at 532 nm.
| Subject no. | Healthy women | Breast cancer
patients | P-value |
|---|
| 1 | 1.04±0.08 | 2.87±0.31 | 0.002a |
| 2 | 0.84±0.05 | 2.50±0.22 | 0.002a |
| 3 | 0.31±0.03 | 4.29±0.23 | 0.002a |
| 4 | 1.05±0.11 | 2.92±0.26 | 0.002a |
| 5 | 0.89±0.07 | 5.62±0.09 | 0.002a |
| 6 | 0.68±0.06 | 2.62±0.29 | 0.002a |
| 7 | 0.73±0.05 | 4.07±0.12 | 0.002a |
| 8 | 0.94±0.04 | 2.99±0.82 | 0.002a |
| 9 | 1.07±0.12 | 3.81±0.22 | 0.002a |
| 10 | 0.98±0.07 | 2.92±0.18 | 0.002a |
| 11 | 0.34±0.05 | 2.21±0.22 | 0.002a |
| 12 | 0.69±0.09 | 2.07±0.02 | 0.002a |
| 13 | 1.97±0.07 | 3.62±0.09 | 0.002a |
| 14 | 0.54±0.03 | 4.25±0.27 | 0.002a |
| 15 | 1.24±0.10 | 2.41±0.11 | 0.002a |
| 16 | 0.80±0.02 | 2.04±0.08 | 0.002a |
| 17 | 1.21±0.08 | 2.84±0.05 | 0.002a |
| 18 | 0.73±0.05 | 3.61±0.12 | 0.002a |
| 19 | 0.38±0.07 | 2.05±0.03 | 0.002a |
| 20 | 1.94±0.11 | 3.86±0.46 | 0.002a |
| 21 | | 2.99±0.82 | |
| 22 | | 2.56±0.34 | |
| 23 | | 6.89±0.16 | |
| 24 | | 3.31±0.03 | |
| 25 | | 5.66±0.09 | |
| 26 | | 2.21±0.22 | |
| 27 | | 2.25±0.58 | |
| 28 | | 2.25±0.58 | |
| 29 | | 3.86±0.46 | |
| 30 | | 2.04±0.08 | |
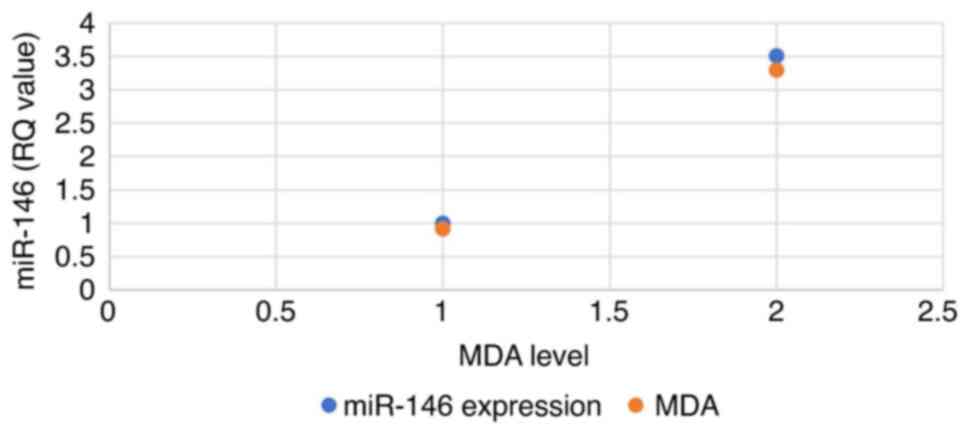
Association between miR-146 expression
and MDA levels
A positive association was found between miR-146
expression and MDA levels in patient serum, where miR-146
expression in normal healthy samples was 0.84, whereas the MDA
level was 0.76. By contrast, the breast cancer samples exhibited a
3-fold increase in miR-146 expression and the MDA levels also
exhibited a similar 2-fold increase (Fig. 3).
Discussion
A small, non-coding, single-stranded RNA that
consists of 20-24 nucleotides, known as miRNA, plays a crucial role
in gene transcription and expression by regulating gene expression
(34). While miRNAs do not code
for proteins, they are capable of directly degrading mRNA or
preventing mRNA translation by creating complete or incomplete
complementary combinations with the target mRNA (35).
A variety of studies have demonstrated that miRNAs
are involved in tumor growth, metastasis and angiogenesis through
the modulation of oncogenesis, migration and other related genes
(36,37). Several miRNAs have also been shown
to be associated with the clinical and pathological aspects of
breast cancer, including the expression of estrogen and
progesterone receptors, as well as vascular invasion. For example,
Blenkiron et al (37)
discovered 133 miRNAs expressed in both normal breast and cancer
tissues; some of these types are associated with the molecular
subtypes of breast cancer.
The aim of the present study was to elucidate the
role of miRNA-146 in the context of oxidative stress in breast
cancer. miRNAs are increasingly being implicated in the development
of breast cancer. It has been found that miR-146a-5p expression is
considerably higher in breast cancer tissues than it is in
paraneoplastic tissue (21). The
study by Gao et al (21)
also reported that the expression of miR-146a-5p was significantly
higher in MCF-7 cells than in control cells, as verified using
RT-qPCR. MCF-7 cells with a high expression of miR-146a-5p
exhibited an increased proliferation, while cells with a low
expression exhibited a decreased proliferation (21). This finding supports the findings
of the present study, indicating high levels of miR-146 expression
in patients with breast cancer.
For a more in-depth understanding of oxidative
stress, the present study measured the MDA levels in patients with
breast cancer and compare these to those of healthy women.
Increased levels of MDA have been reported in breast, ovary,
gastric and lung cancers, as well as in colorectal adenomas
(38-43).
The reaction between polyunsaturated fatty acids and free radicals
can produce MDA, a low-molecular-weight aldehyde. Patients with
breast cancer have been found to have higher plasma levels of MDA
(40). It was demonstrated that
the MDA serum levels were indeed higher in patients with breast
cancer than in healthy individuals (40). The study performed by Bhattacharjee
et al (44) revealed that
the median MDA level for patients with breast cancer was 3.98±0.35
nmol/ml, which was higher than that in controls (3.04±0.36
nmol/ml), with a P-value of 0.001. In the Ropanasuri Specialized
Surgery Hospital, Padang, Indonesia, breast cancer patients and
controls also exhibited significantly different MDA levels
(44). Furthermore, patients with
breast cancer had significantly higher serum MDA levels than those
with benign breast diseases (P=0.042). The MDA concentrations and
age of the patients with breast cancer and lymph node metastases
differed significantly (P=0.006) (45). Similarly, Sahu et al also
observed an increased level of MDA in patients with breast cancer,
with an average value of 5.8±3.2 nmol/ml as compared to the control
group with an average value of 1.9±0.28 nmol/ml, indicating that
there were statistically significant differences between the two
groups (38).
The results support the hypothesis that MDA plays a
causal role in the development of breast cancer. In addition,
malignant tissues contain higher MDA concentrations than normal
tissue samples obtained from healthy individuals (46). The abnormally high levels of MDA in
breast cancer patients can be attributed to excessive reactive
oxygen species reactive oxygen species (ROS) production and a lack
of antioxidant defenses.
The observed increase in MDA levels may be caused by
ROS induction in breast cancer cells leading to oxidative stress
and molecular damage, including lipid peroxidation (47). It is possible that MDA represents a
product of lipid peroxidation, induced by an increase in ROS in the
body, a process that could lead to the development of breast cancer
(48,49). Biological, chemical and physical
carcinogens can induce excessive ROS production. Significantly
higher levels of oxidative stress and lower levels of antioxidants
are associated with increased MDA levels in cancer patients. This
event plays a critical role in the development and pathogenesis of
tumors (50).
Since MDA is one of the most common products of
lipid peroxidation, by interacting with proteins and DNA, it can
lead to gene mutations that increase tumor development, explaining
why increasing MDA levels can act as a marker cancer cell
development (46,47). Previous studies have provided
evidence that ROS plays a critical role in the development and
progression of breast cancer (51). As a result, previous findings
(52), as well as the results of
the present study support the hypothesis that oxidative stress is
prevalent, not only in cancer cells, but also throughout the entire
body affected in cancer patients. In addition, MDA levels increased
with progressing TNM stages in malignant breast cancer tissue
(46).
Cancer progression and treatment resistance are
characterized by ROS accumulation, altered redox balance and
signaling. Oxidative phosphorylation generates ROS, primarily at
the mitochondrial level. It is possible that the increased ROS
levels detected in cancer cells arise due to several factors,
including high metabolic activity, cellular signaling, peroxisomal
activity, mitochondrial dysfunction, oncogene activity and
increased enzyme activity of oxidases, cyclooxygenases,
lipoxygenases, and thymidine phosphorylases (53). A number of antioxidants are
involved in maintaining intracellular homeostasis, including
catalase, superoxide dismutase and glutathione peroxidase.
Furthermore, glutathione, a potent antioxidant, and the
transcription factor, Nrf2, also contribute to balancing oxidative
stress (53). Free radicals,
oxidative stress and lipid peroxidation have been well documented
as factors contributing to the carcinogenesis initiation and the
progression of the process (39).
It has been demonstrated that MDA is a potent marker for evaluating
oxidative stress in patients with breast cancer. An individual's
age and disease stage determine the level of oxidative stress
(39). The levels of MDA can serve
as a marker of an oxidative state. The disease stage and age have
been shown to be associated with higher levels of malondialdehyde,
suggesting a more severe state of oxidative stress (39).
A recent study demonstrated that miR-146a regulates
inflammatory reactions in diseases associated with inflammation and
oxidative stress (54). The
association between miRNA-146a expression and MDA levels were
examined in the present study. NF-κB is a transcription factor
located upstream of the miR-146a promoter that triggers miR-146a
expression in response to pro-inflammatory factors and reactive
oxygen species. In turn, miR-146a can impede NF-κB and mediate
inflammatory processes by inhibiting the expression of some of its
target genes, such as IRAK1 and TRAF6 (55,56).
Further analysis revealed that miR-146a overexpression inhibited
neuronal apoptosis, reduced the production of pro-inflammatory
cytokines, and reduced oxidative stress in ICH mice. miR-146a
appears to function as a protective factor against ICH by
inhibiting inflammatory and oxidative stress (57).
miR-146a levels have recently been found to be
negatively associated with chronic inflammation and oxidative
stress. In 2018, Xie et al (58) found that chronic type 2 diabetes
(cT2DM) rats with elevated inflammation and oxidative stress status
exhibited neurodegenerative disorders that were negatively
correlated with miR-146a levels. miR-146a may therefore serve as a
positive indicator of inflammation and oxidative stress in the
brain of rats with chronic type 2 diabetes. Overall, it has been
demonstrated that increased levels of inflammation and oxidative
stress in cT2DM rats contribute to brain impairment, which is
negatively regulated by miR-146a (58). Furthermore, inflammatory mediators,
such as COX-2, TNF-α and IL-1β, as well as oxidative stress
indicators, such as MDA and p22phox were elevated in the brain
tissues of cT2DM rats and negatively correlated with miR-146a
expression (58). Accordingly, the
present study examined the association between miR-146a expression
and MDA, an oxidative stress indicator, comparing patients with
breast cancer and healthy women. However, no such association was
observed. However, one of the limitations of the present study is
the lack of sampling that affected the sample size. In addition,
due to the difficulty of the survey, data on smoking, alcohol
consumption and body mass index were not included.
miRNAs regulate a wide range of biological processes
within a cell. Cancer is one of the diseases associated with miRNAs
There has been a rapid increase in interest in miR-146a in
particular, as a modulator of differentiation and function, as well
as innate and adaptive immunity (19). Various types of tumors have a
dysregulated expression of miR-146a due to the fact that miR-146a
regulates several important cellular functions (19). Furthermore, to evaluate the
effectiveness of free and total MDA as indicators of oxidative
stress, a more in-depth understanding of the association between
free and total MDA in different biological media is essential
(59).
The mechanisms underlying this observation are not
yet entirely clear; however, miRNAs may influence gene abundance
via several different mechanisms. The mechanism underlying this
phenomenon needs to be examined in more detail in future studies.
Similarly, high levels of 8-hydroxydeoxyguanosine and MDA, and low
superoxide dismutase levels have been shown to increase oxygen
radical activity in certain inflammatory diseases (60). Thus, further studies are warranted
to determine the association of miRNAs with breast cancer in more
detail.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author upon
reasonable request.
Authors' contributions
ASKAK made substantial contributions to the
conception and design of the study; the acquisition, analysis and
interpretation of the data; generated the datasets; and drafted the
work. IMH collected blood samples from the primary breast cancer
patients and healthy individuals, sufficiently participated in the
acquisition, analysis and interpretation of the data, generated the
datasets, and revised the manuscript. MAAN collected blood samples
from primary breast cancer patients and healthy individuals,
performed their laboratory analyses, contributed to the
acquisition, analysis and interpretation of the data, and revised
the manuscript. GOA contributed to the acquisition of data,
performed the data analysis and interpretation, and revised the
manuscript. ASKAK and GOA confirm the authenticity of all the raw
data. All the authors have read and approved the final version of
the manuscript for publication.
Ethics approval and consent to
participate
Ethical approval for the present study was obtained
from the Research Ethics Committee at the University of Baghdad
under the reference no. NCRCEC/01/001. All clinical samples were
collected autonomously from individuals who provided their informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
International Agency for Research on
Cancer: Globocan 2012. World Health Organization, International
Agency for Research on Cancer, Lyon, 2013.
|
|
3
|
Anothaisintawee T, Wiratkapun C,
Lerdsitthichai P, Kasamesup V, Wongwaisayawan S, Srinakarin J,
Hirunpat S, Woodtichartpreecha P, Boonlikit S, Teerawattananon Y
and Thakkinstian A: Risk factors of breast cancer: A systematic
review and meta-analysis. Asia Pac J Public Health. 25:368–387.
2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Shoukry M, Broccard S, Kaplan J and
Gabriel E: The emerging role of circulating tumor DNA in the
management of breast cancer. Cancers (Basel).
13(3813)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Feng Y, Spezia M, Huang S, Yuan C, Zeng Z,
Zhang L, Ji X, Liu W, Huang B, Luo W, et al: Breast cancer
development and progression: Risk factors, cancer stem cells,
signaling pathways, genomics, and molecular pathogenesis. Genes
Dis. 5:77–106. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Butti R, Gunasekaran VP, Kumar TV,
Banerjee P and Kundu GC: Breast cancer stem cells: Biology and
therapeutic implications. Int J Biochem Cell Biol. 107:38–52.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu
Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, et al:
Breast cancer stem cells transition between epithelial and
mesenchymal states reflective of their normal counterparts. Stem
Cell Reports. 2:78–91. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bao L, Cardiff RD, Steinbach P, Messer KS
and Ellies LG: Multipotent luminal mammary cancer stem cells model
tumor heterogeneity. Breast Cancer Res. 17(137)2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sin WC and Lim CL: Breast cancer stem
cells-from origins to targeted therapy. Stem Cell Investig.
4(96)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lagadec C, Vlashi E, Della Donna L,
Dekmezian C and Pajonk F: Radiation-induced reprogramming of breast
cancer cells. Stem Cells. 30:833–844. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chaffer CL, Marjanovic ND, Lee T, Bell G,
Kleer CG, Reinhardt F, D'Alessio AC, Young RA and Weinberg RA:
Poised chromatin at the ZEB1 promoter enables breast cancer cell
plasticity and enhances tumorigenicity. Cell. 154:61–74.
2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Koren S, Reavie L, Couto JP, De Silva D,
Stadler MB, Roloff T, Britschgi A, Eichlisberger T, Kohler H, Aina
O, et al: PIK3CAH1047R induces multipotency and multi-lineage
mammary tumours. Nature. 525:114–118. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Colditz GA, Kaphingst KA, Hankinson SE and
Rosner B: Family history and risk of breast cancer: Nurses' health
study. Breast Cancer Res Treat. 133:1097–1104. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Allison KH: Molecular pathology of breast
cancer: What a pathologist needs to know. Am J Clin Pathol.
138:770–780. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Dykes IM and Emanueli C: Transcriptional
and post-transcriptional gene regulation by long non-coding RNA.
Genomics Proteomics Bioinformatics. 15:177–186. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
O'Rourke JR, Swanson MS and Harfe BD:
MicroRNAs in mammalian development and tumorigenesis. Birth Defects
Res C Embryo Today. 78:172–179. 2006.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Saito Y, Nakaoka T and Saito H:
microRNA-34a as a therapeutic agent against human cancer. J Clin
Med. 4:1951–1959. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tordonato C, Di Fiore PP and Nicassio F:
The role of non-coding RNAs in the regulation of stem cells and
progenitors in the normal mammary gland and in breast tumors. Front
Genet. 6(72)2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Rusca N and Monticelli S: MiR-146a in
immunity and disease. Mol Biol Int. 2011(437301)2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tordonato C, Marzi MJ, Giangreco G, Freddi
S, Bonetti P, Tosoni D, Di Fiore PP and Nicassio F: miR-146
connects stem cell identity with metabolism and pharmacological
resistance in breast cancer. J Cell Biol.
220(e202009053)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gao W, Hua J, Jia Z, Ding J, Han Z, Dong
Y, Lin Q and Yao Y: Expression of miR 146a 5p in breast cancer and
its role in proliferation of breast cancer cells. Oncol Lett.
15:9884–9888. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chen J, Jiang Q, Jiang XQ, Li DQ, Jiang
XC, Wu XB and Cao YL: miR-146a promoted breast cancer proliferation
and invasion by regulating NM23-H1. J Biochem. 167:41–48.
2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Del Rio D, Stewart AJ and Pellegrini N: A
review of recent studies on malondialdehyde as toxic molecule and
biological marker of oxidative stress. Nutr Metab Cardiovasc Dis.
15:316–328. 2005.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Nair V, O'Neil CL and Wang PG:
Malondialdehyde. In: Encyclopedia of Reagents for Organic
Synthesis. John Wiley & Sons, Hoboken, NJ, 2001.
|
|
25
|
Mateos R, Goya L and Bravo L:
Determination of malondialdehyde by liquid chromatography as the 2,
4-dinitrophenylhydrazone derivative: A marker for oxidative stress
in cell cultures of human hepatoma HepG2. J Chromatogr B Analyt
Technol Biomed Life Sci. 805:33–39. 2004.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Al-Khafaji ASK, Pantazi P, Acha-Sagredo A,
Schache A, Risk JM, Shaw RJ and Liloglou T: Overexpression of HURP
mRNA in head and neck carcinoma and association with in vitro
response to vinorelbine. Oncol Lett. 19:2502–2507. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Al-Khafaji AS, Davies MP, Risk JM, Marcus
MW, Koffa M, Gosney JR, Shaw RJ, Field JK and Liloglou T: Aurora B
expression modulates paclitaxel response in non-small cell lung
cancer. Br J Cancer. 116:592–599. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Al-Khafaji ASK, Marcus MW, Davies MPA,
Risk JM, Shaw RJ, Field JK and Liloglou T: AURKA mRNA expression is
an independent predictor of poor prognosis in patients with
non-small cell lung cancer. Oncol Lett. 13:4463–4468.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Tarannum J, Manaswini P, Deekshitha C,
Reddy BP and Sunder AS: Elucidative Histopathological study in
female cancer patients: Histopathology in female cancers. Iraq J
Sci. 61:720–726. 2020.
|
|
30
|
Lei B, Liu J, Yao Z, Xiao Y, Zhang X,
Zhang Y and Xu J: NF-κB-Induced Upregulation of miR-146a-5p
promoted hippocampal neuronal oxidative stress and pyroptosis via
TIGAR in a Model of Alzheimer's Disease. Front Cell Neurosci.
15(653881)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Jin X, Liu J, Chen YP, Xiang Z, Ding JX
and Li YM: Effect of miR-146 targeted HDMCP up-regulation in the
pathogenesis of nonalcoholic steatohepatitis. PLoS One.
12(e0174218)2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Mao H and Xu G: Protective effect and
mechanism of microRNA-146a on ankle fracture. Exp Ther Med.
20(3)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kagiya T: MicroRNAs: Potential biomarkers
and therapeutic targets for alveolar bone loss in periodontal
disease. Int J Mol Sci. 17(1317)2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ohtsuka M, Ling H, Doki Y, Mori M and
Calin GA: MicroRNA processing and human cancer. J Clin Med.
4:1651–1667. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Blenkiron C, Goldstein LD, Thorne NP,
Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE,
Green AR, Ellis IO, et al: MicroRNA expression profiling of human
breast cancer identifies new markers of tumor subtype. Genome Biol.
8(R214)2007.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Sahu A, Varma M and Kachhawa K: A
prognostic study of MDA, SOD and catalase in breast cancer
patients. Int J Sci Res. 4:157–159. 2015.
|
|
39
|
Didžiapetrienė J, Bublevič J, Smailytė G,
Kazbarienė B and Stukas R: Significance of blood serum catalase
activity and malondialdehyde level for survival prognosis of
ovarian cancer patients. Medicina (Kaunas). 50:204–208.
2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Sadati Zarrini A, Moslemi D, Parsian H,
Vessal M, Mosapour A and Shirkhani Kelagari Z: The status of
antioxidants, malondialdehyde and some trace elements in serum of
patients with breast cancer. Caspian J Intern Med. 7:31–36.
2016.PubMed/NCBI
|
|
41
|
Bakan E, Taysi S, Polat MF, Dalga S,
Umudum Z, Bakan N and Gumus M: Nitric oxide levels and lipid
peroxidation in plasma of patients with gastric cancer. Jpn J Clin
Oncol. 32:162–166. 2002.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Gupta A, Srivastava S, Prasad R, Natu SM,
Mittal B, Negi MP and Srivastava AN: Oxidative stress in non-small
cell lung cancer patients after chemotherapy: Association with
treatment response. Respirology. 15:349–356. 2010.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Tao D, Zhou Z, Xu X and Luo H: Lipid
disorders and lipid peroxidation associated with the malignant
transformation of colorectal adenoma. Chin Ger J Clin Oncol.
10:270–273. 2011.
|
|
44
|
Bhattacharjee J, Jogdand S, Shinde RK and
Goswami S: Assessment of oxidative stress in breast cancer
patients: A hospital based study. Int J Basic Clin Pharmacol.
7:966–970. 2018.
|
|
45
|
Gubaljevic J, Srabović N, Jevrić-Čaušević
A, Softić A, Rifatbegović A, Mujanović-Mustedanagić J, Dautović E,
Smajlović A and Mujagić Z: Serum levels of oxidative stress marker
malondialdehyde in breast cancer patients in relation to
pathohistological factors, estrogen receptors, menopausal status,
and age. J Health Sci. 8:154–161. 2018.
|
|
46
|
Sener DE, Gönenç A, Akıncı M and Torun M:
Lipid peroxidation and total antioxidant status in patients with
breast cancer. Cell Biochem Funct. 25:377–382. 2007.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Qebesy HS, Zakhary MM, Abd-Alaziz MA,
Abdel Ghany AA and Maximus DW: Tissue levels of oxidative stress
markers and antioxidants in breast cancer patients in relation to
tumor grade. Al-Azhar Assiut Med J. 13:10–17. 2015.
|
|
48
|
Hauck AK and Bernlohr DA: Oxidative stress
and lipotoxicity. J Lipid Res. 57:1976–1986. 2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Gönenç A, Erten D, Aslan S, Akıncı M,
Şimşek B and Torun M: Lipid peroxidation and antioxidant status in
blood and tissue of malignant breast tumor and benign breast
disease. Cell Biol Int. 30:376–380. 2006.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Kilic N, Yavuz Taslipinar M, Guney Y,
Tekin E and Onuk E: An investigation into the serum thioredoxin,
superoxide dismutase, malondialdehyde, and advanced oxidation
protein products in patients with breast cancer. Ann Surg Oncol.
21:4139–4143. 2014.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Himmetoglu S, Dincer Y, Ersoy YE,
Bayraktar B, Celik V and Akcay T: DNA oxidation and antioxidant
status in breast cancer. J Investig Med. 57:720–723.
2009.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Mena S, Ortega A and Estrela JM: Oxidative
stress in environmental-induced carcinogenesis. Mutat Res.
674:36–44. 2009.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Didžiapetrienė J, Kazbarienė B, Tikuišis
R, Dulskas A, Dabkevičienė D, Lukosevičienė V, Kontrimavičiūtė E,
Sužiedėlis K and Ostapenko V: Oxidant/antioxidant status of breast
cancer patients in pre-and post-operative periods. Medicina
(Kaunas). 56(70)2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wan RJ and Li YH: MicroRNA-146a/NAPDH
oxidase4 decreases reactive oxygen species generation and
inflammation in a diabetic nephropathy model. Mol Med Rep.
17:4759–4766. 2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Li K, Ching D, Luk FS and Raffai RL:
Apolipoprotein E enhances microRNA-146a in monocytes and
macrophages to suppress nuclear factor-κB-driven inflammation and
atherosclerosis. Circ Res. 117:e1–e11. 2015.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Lo WY, Peng CT and Wang HJ:
MicroRNA-146a-5p mediates high glucose-induced endothelial
inflammation via targeting interleukin-1 receptor-associated kinase
1 expression. Front Physiol. 8(551)2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Qu X, Wang N, Cheng W, Xue Y, Chen W and
Qi M: MicroRNA-146a protects against intracerebral hemorrhage by
inhibiting inflammation and oxidative stress. Exp Ther Med.
18:3920–3928. 2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Xie Y, Chu A, Feng Y, Chen L, Shao Y, Luo
Q, Deng X, Wu M, Shi X and Chen Y: MicroRNA-146a: A comprehensive
indicator of inflammation and oxidative stress status induced in
the brain of chronic T2DM rats. Front Pharmacol.
9(478)2018.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Cui X, Gong J, Han H, He L, Teng Y, Tetley
T, Sinharay R, Chung KF, Islam T, Gilliland F and Grady S:
Relationship between free and total malondialdehyde, a
well-established marker of oxidative stress, in various types of
human biospecimens. J Thorac Dis. 10:3088–3097. 2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Canakci CF, Cicek Y, Yildirim A, Sezer U
and Canakci V: Increased levels of 8-hydroxydeoxyguanosine and
malondialdehyde and its relationship with antioxidant enzymes in
saliva of periodontitis patients. Eur J Dent. 3:100–106.
2009.PubMed/NCBI
|