Endometriosis is an enigmatic, benign,
hormone-dependent gynecological disorder, defined by the presence
of functional endometrium-like tissue (endometrial glands and
stroma) external to the uterine cavity and affecting ~10% of women
of reproductive age. This condition markedly affects the health of
women, as well as their quality of life (1). Its main clinical symptoms include
chronic pelvic pain, dyspareunia, dysmenorrhea and impaired
fertility, with laparoscopy and biopsy remaining the gold-standard
methods for its diagnosis (2).
Despite significant progress being made in the
understanding of the pathophysiology of endometriosis and the
various theories suggested thus far, its precise etiology remains
unknown. The disease exhibits high complexity, as genetic,
epigenetic and environmental factors appear to interact in order to
formulate the disease phenotype. Furthermore, this multifactorial
nature of endometriosis involves the participation of aberrant
immunological responses, angiogenesis processes and biochemical
alterations. Of note, endometriosis has a strong genetic
predisposition (3,4), as shown by studies conducted on
monozygotic twins and families (5), gene association studies that took
into account candidate genes and single nucleotide polymorphisms
(SNPs) (6), as well as genome-wide
association studies (GWAS) (7).
For example, eight GWAS of women of European and East Asian origin
that have been published to date (2) have identified 19 distinct
disease-associated signals harbored at 14 loci (8). Moreover, a meta-analysis of 15 GWAS
and a new replication analysis, including 58,115 cases and 733,480
controls in total, revealed 27 genetic loci associated with
endometriosis at the genome-wide P-value threshold
(P<5x10-8), 13 of which are novel, while an
additional 8 novel genes identified from gene-based association
analyses (9).
The evidence of an association among genetic
polymorphisms and the risk of developing endometriosis is robust
(10). A number of studies have
reported a nine-fold increase in the risk of developing
endometriosis among women from the East Asian population compared
with the European or American women populations (4,11,12).
Although the risk of endometriosis has been strongly linked to
ethnicity, the main differences identified in population groups
have not been well defined. What remains to be understood is how
risk variation is linked to ethnicity and the minor differences in
SNP variation, as captured in the recent geographical and
evolutionary history of humans. Of note, differences in autoimmune
disease risk variants have also been reported across different
continental populations. In particular, various data have
demonstrated that several systemic lupus erythematosus
susceptibility loci confer risk across multiple ethnicities, while
distinct differences in the risk of developing the disease were
found when populations of European, African, African-American,
Asian and Hispanic ancestry were analyzed (13).
In Africa, ~45,000 to 60,000 years ago, a marked
demographic and geographic spread began that rapidly brought human
presence to almost all habitable areas of the earth. Genetic and
paleoanthropological studies are consistent with the aforementioned
evidence. Genomic data from modern humans suggest that this
expansion was accompanied by a continuous loss of genetic
diversity, a result of what is known as the ‘serial founder effect’
(14). It is generally assumed
that the bottleneck occurred as a small group(s) with an effective
population size of only ~2,000 individuals migrated from the
African continent to the Near East (14-16).
During the great expansion, there was an uninterrupted and
considerable reduction in genetic diversity proportional to the
geographic distance from the African homeland, which is indicated
by the motif of average heterozygosities of contemporary
populations (14). However,
genomes from substructured populations retain a numerous amount of
unique variants. As a result of the relatively profound
substructure within the continent, genetic variation in Africa
varies considerably from region to region. Groups such as the
Khoisan, Hadza, Sandawe and Forest Pygmies have been shown to
maintain extremely high genetic diversity, relative to
out-of-Africa populations, as evidenced by studies on autosomal DNA
polymorphism patterns in present-day African hunter-gatherers
(14-20).
Since nowadays the available genetics data (e.g.
from NGS, GWAS) are in abundance, there is an opportunity for
drawing conclusions, from screening populations for medical genetic
studies, for mapping genotypes to phenotypes and for rendering the
power of natural selection in human history.
However, the gathering of huge data impedes the
identification of such genetic patterns in order to obtain SNPs as
genetic markers for a number of applications such pharmacogenomics,
personalized medicine and genetics. This task requires the
implementation of thousands of SNPs, which are associated with
various clinical disorders in correlation studies. In the present
study, an attempt was made to examine a wide set of genetic targets
that characterize endometriosis through the prism of the genetic
profile of geographically distributed population groups and the
allele frequency of targeted SNPs. To this end, bioinformatics
studies, such as those presented herein, that join the world-wide
genetic information represented by ‘1000 Genomes Project’ with a
disease-related SNP database such as Demetra (21) may alleviate the aforementioned
issue, providing a simple and efficient handling of vast
information and this could lead to the export of results.
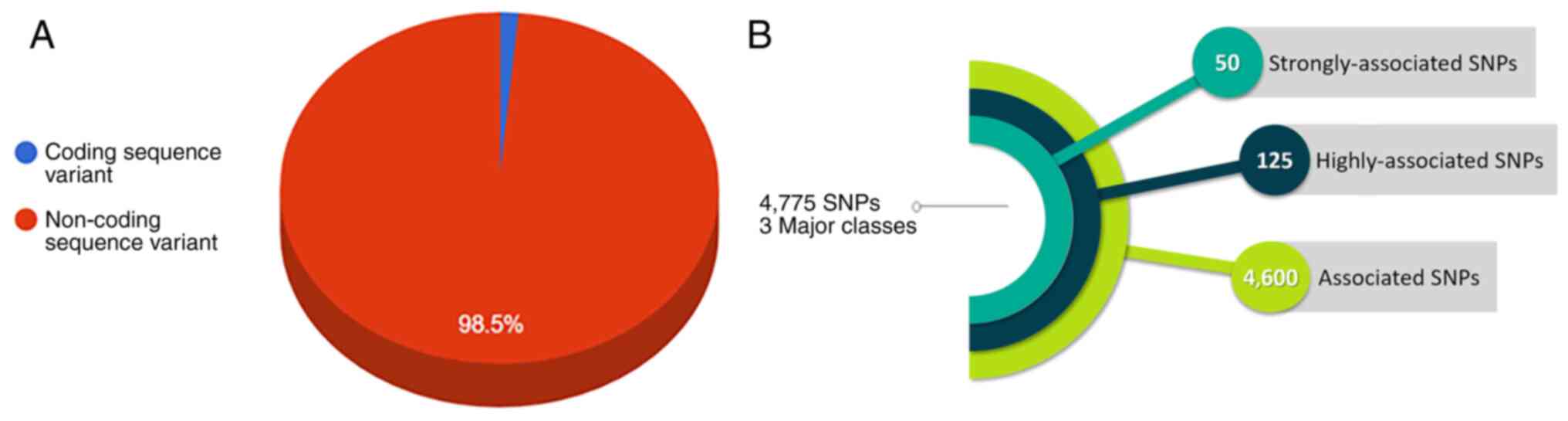
Endometriosis-related SNPs were extracted from the
Demetra Application database (DAD) (21) using the Demetra Application
webserver and have been used in the pipeline of the present study.
The extracted SNPs were labeled with the same classification of the
DAD and contained accompanying information, including the SNP
identification number based on the dbSNP database (22), chromosome name, genetic locus, SNP
type (introns or exons), the name of the responsible gene or the
name of the intragenic region, and the classification based on the
DAD (three different classes, ‘strongly-associated SNPs’,
‘highly-associated SNPs’ and ‘associated SNPs’ with
endometriosis).
In the present study, five geographical population
groups were studied, focusing on Europeans, Africans, Americans,
East Asians and South Asians. Sample sizes and origins of the
individual population samples are provided in the International
Genome Sample Resource (IGSR) (23), which have been developed under the
‘1000 Genomes Project’ (24). The
present study was performed using human genomes that were contained
in the phase three collection of the IGSR on reference assembly
GRCh38 (25,26). Since the ‘1000 Genomes Project’ has
created call sets of sequence variants for each of the different
genomes sequenced, the downloaded data were in multi-individual
variant call format (VCF) (27)
per chromosome, with genotypes listed for each sample (26). The MATLAB Bioinformatics toolbox
(28) was used for pre-analyzing
the extracted data (VCF format) and storing them in a structured
database per chromosome and genetic locus of the identified
variants, and per origin, including using the IGSR directory of the
individuals.
The Python programming language was used to identify
the endometriosis-related SNPs from the DAD, in the structured
database of variants that was developed from the IGSR data. The
selected variants of SNPs were stored in a newly structured
database by combining the information from both of the
aforementioned databases. Each entry in the first columns contains
the information from the DAD and the following columns then contain
the information for the VCF file for each individual. Subsequently,
all the identified variants were pre-analyzed to collect the allele
frequencies for all SNPs, as estimated in the IGSR. Specifically,
for each SNP, information was collected regarding the allele
frequency of appearances in Europeans, Africans, Americans, East
Asians and South Asians.
A specified analysis was performed for drawing the
disease genomic ‘grammar’ (DGG) profiles of endometriosis in the
five major groups of ethnic origins. All the variants were studied
as candidate genetics targets for each group using the Python
programming language. An algorithm was designed to be able to
detect all the allele frequency ‘alerts’ in each studied group. The
algorithm was able to classify each allele frequency in three
clusters including a) ‘Low’, SNP allele frequency ≤0.1; b)
‘Normal’, 0.1 < SNP allele frequency <0.9; and c) ‘High’, SNP
allele frequency ≥0.9. For the purposes of the present study,
‘alerts’ were issued only for cases when the allele frequencies
belong to the ‘Low’ or ‘High’ clusters. Subsequently, by using
specific R programming language packages, such as ‘BioCircos’
(available online at http://bioinfo.ibp.ac.cn/biocircos/), ‘circlize’
(available at http://cran.r-project.org/web/packages/circlize/)
and ‘CMplot’ (available at https://github.com/xiaolei-lab/rMVP), the results were
analyzed to draw the circular visualization and the result
visualization (29-31).
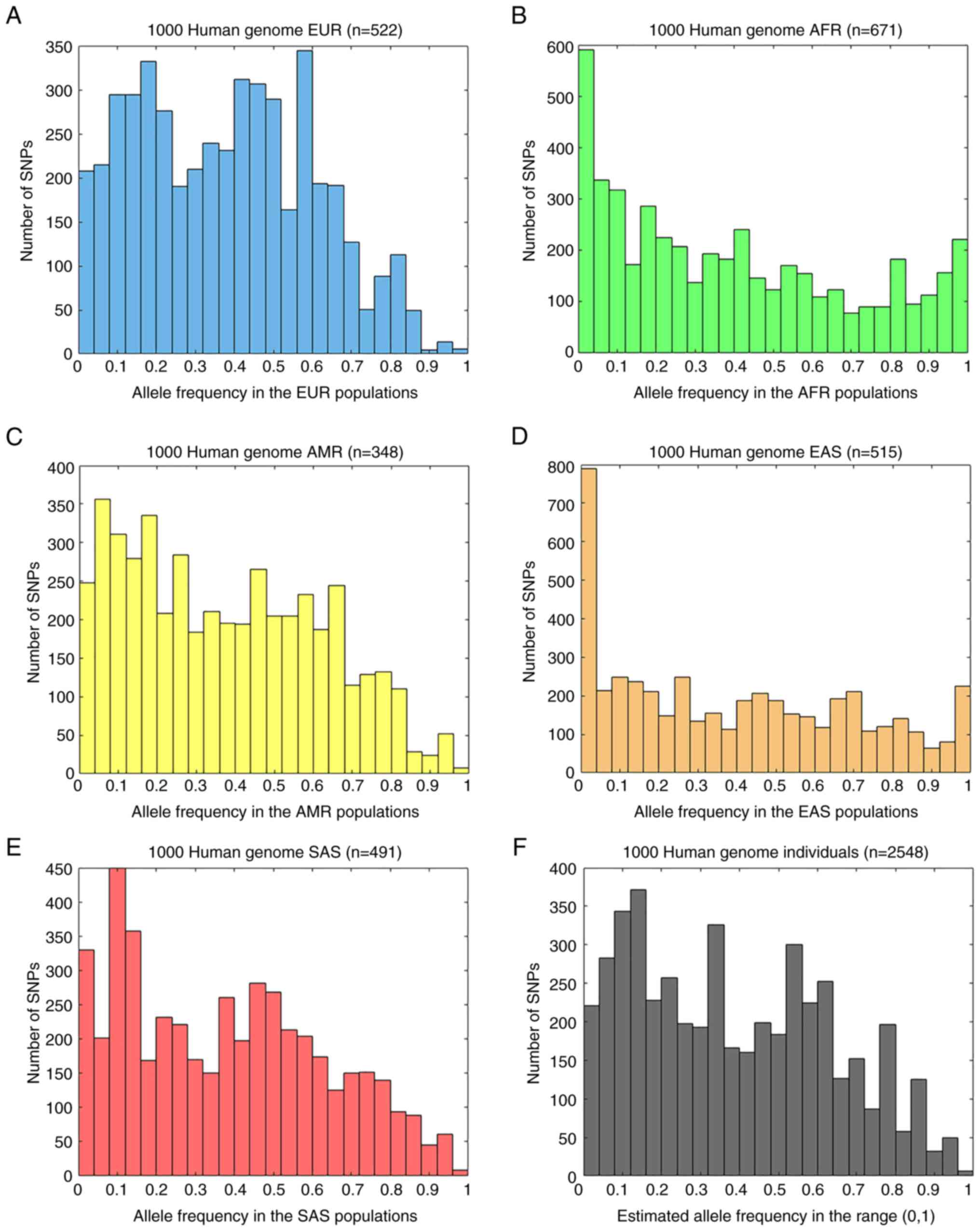
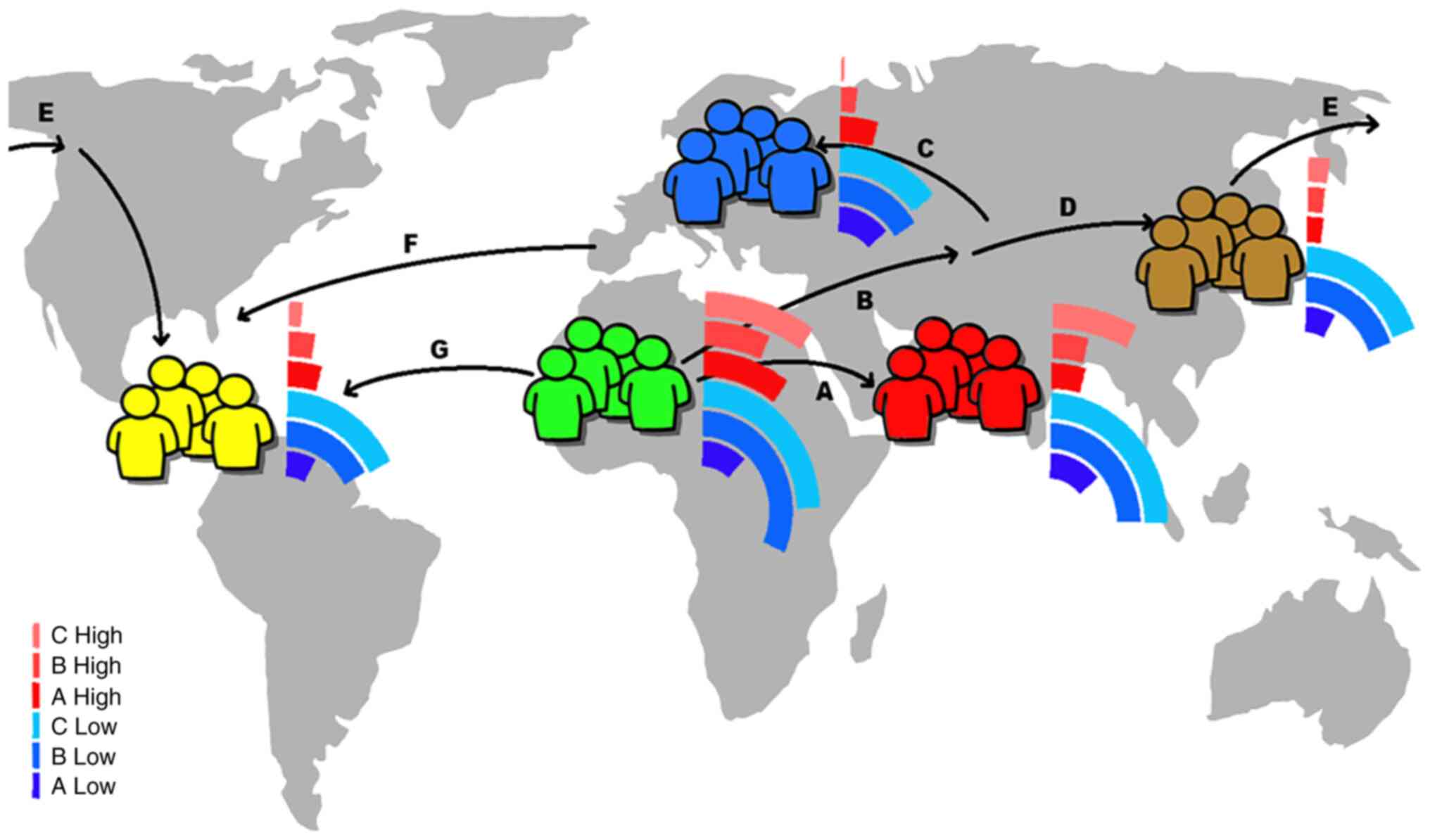
Directional change and reversal in allele
frequencies has been shown in the 4,775 endometriosis-related SNPs
between the individuals of the major five groups. The histogram
analysis of endometriosis-related SNPs using the ‘1000 Genomes
Project’ dataset revealed clearly different distributions in the
five major geographical population groups (Fig. 2). The majority of the groups
exhibited a multimodal distribution, since several peaks are close
together and the top of the distribution forms a plateau (40). Although different allele
frequencies were identified in the studied SNPs, some groups
appeared to have similar distributions, but with different
quantifications, as in between the Africans and East Asians or the
Europeans and the Americans (Fig.
2). The five studied geographical groups accumulated different
SNP totals at the sensitive two ends of the distribution of the
allele frequencies, including the cluster of the ‘low allele
frequencies’ (SNP allele frequency ≤0.1) and the cluster of the
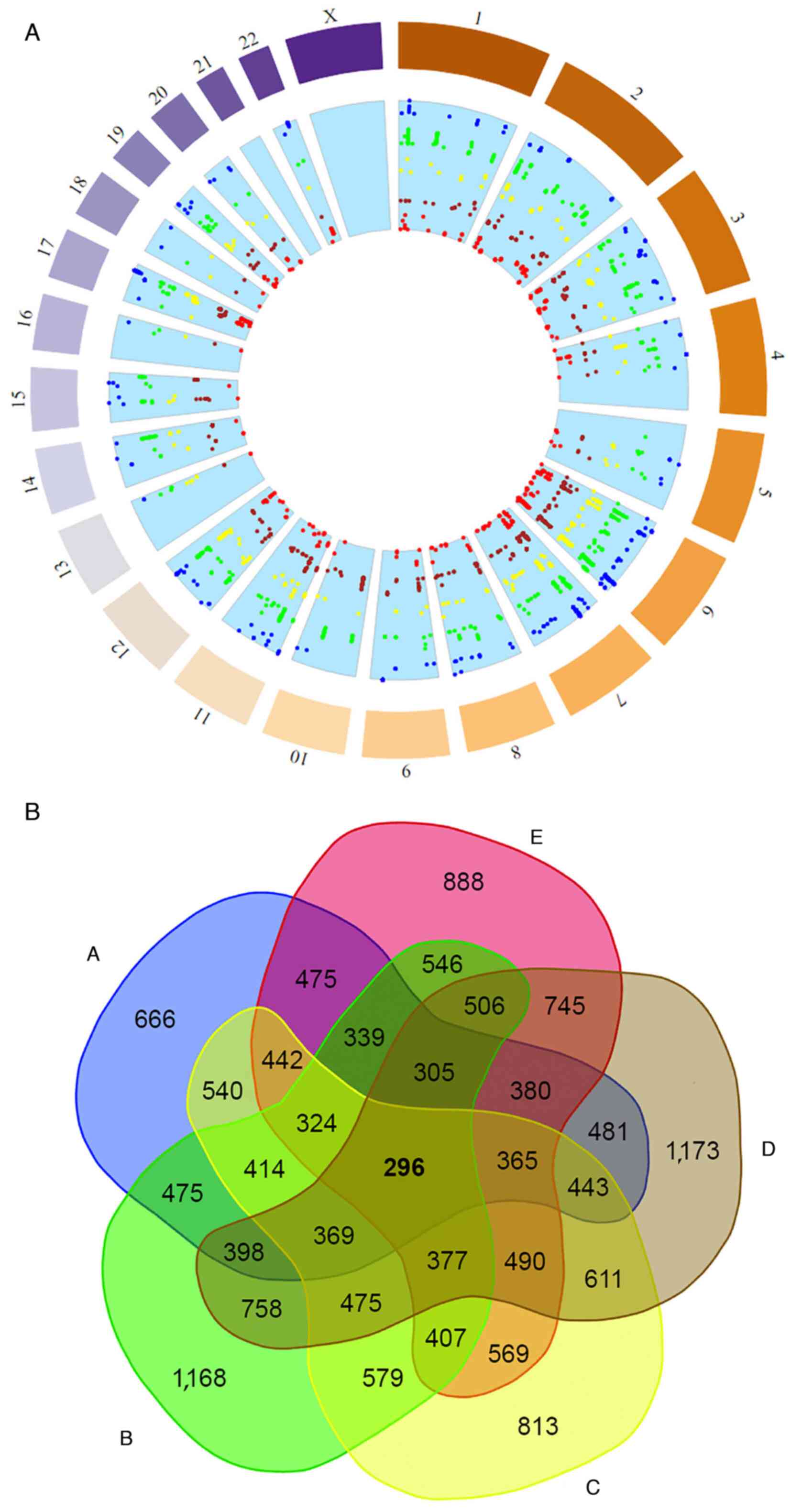
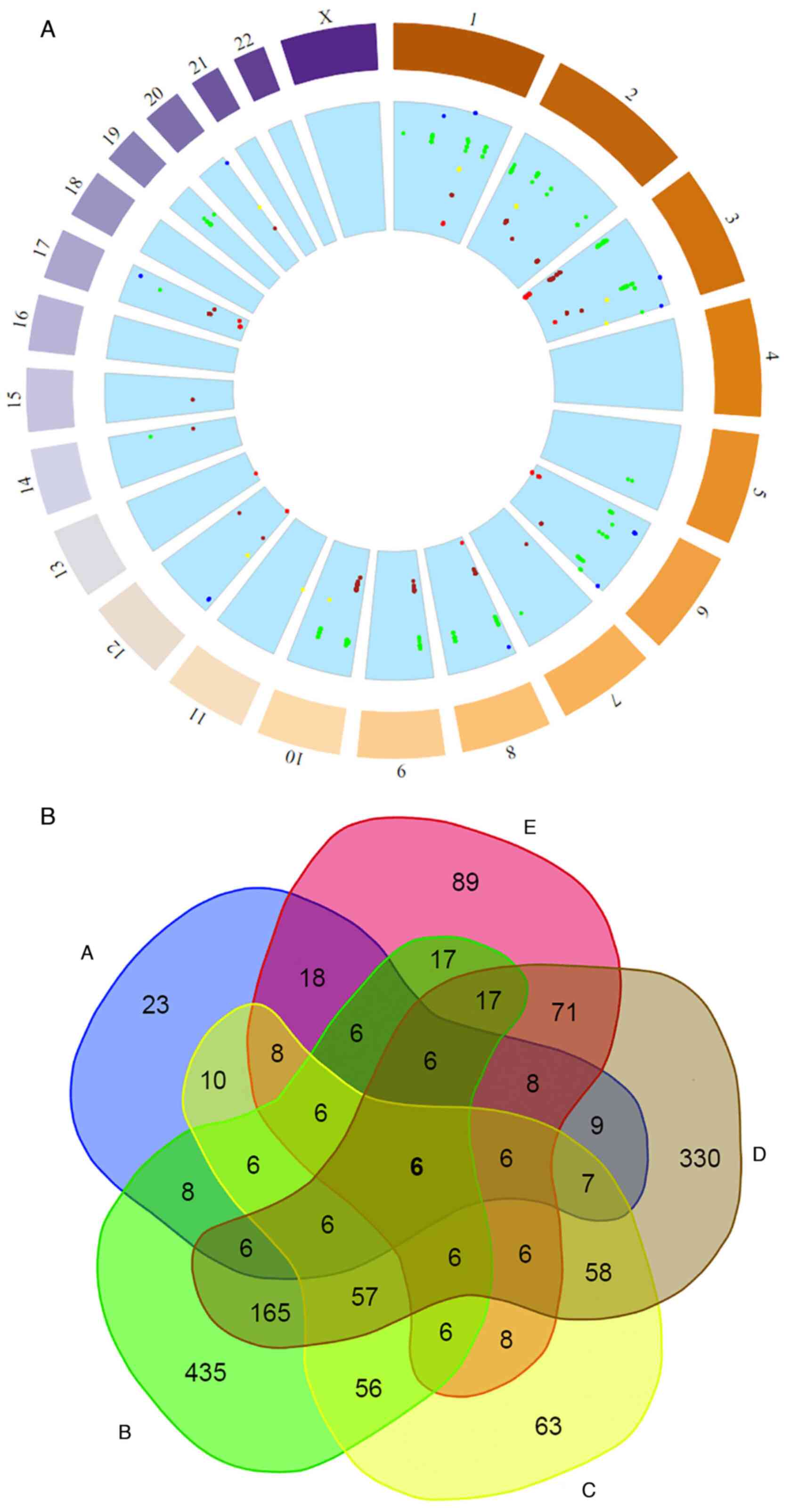
‘high allele frequencies’ (SNP allele frequency ≥0.9) (Figs. 3A and 4A) (41-43).
Different totals and reference SNPs were accumulated
in the low and high clusters among the different population groups
(Figs. 3 and 4). The East Asian group had the largest
sample of SNPs with low allele frequencies, followed by the
Africans, South Asians, Americans and Europeans (Fig. 3). On the other hand, the African
group had the largest sample of different SNPs as expected
(44) with high allele frequencies
followed by the East Asian group (Fig.
4). The South Asian, American and European groups exhibited
markedly fewer totals in SNPs with high allele frequencies
(Fig. 4). Although some population
groups shared some similarities in the identified SNPs in the low
and high clusters, the overall distribution of the
endometriosis-related SNPs and their genetic locus per chromosome
in the five studied population groups revealed clear
differentiation (Figs. 3A and
4A). All the population groups
shared a common genetic background in 296 SNPs with low allele
frequencies and six SNPs with high allele frequencies (Figs. 3B and 4B). These results may be associated with
clinical data in order to understand common or population specific
aspects in endometriosis and form a basis for research towards
understanding the genetic components of the disease that may aid in
improving early diagnosis or directed medical treatment for
patients (45,46).
Allele frequencies in endometriosis-related SNPs
revealed different genetic diversity in the studied geographical
population groups. The genetic SNP distribution for endometriosis
begins from the African population, and follows the Homo
sapiens expansion history, beginning with Africa (47). This scenario is also reflected in
the data of the present study, since in the African population
genomes from the substructured populations of Africa retain an
exceptional number of unique variants. The majority of the
endometriosis-related SNPs with low and high allele frequencies
were identified (Tables I and
II) in Africa. However due to the
founder's effect, there is a loss of genetic diversity in the
selected population groups on endometriosis-associated SNPs
(48). Major common and different
endometriosis-related genetic targets were identified in the five
population groups using the African population group results as a
basis. As Kobayashi et al (49) mentioned, the number of SNPs
detected at a low allele frequency was much higher than the number
of SNPs detected at a higher allele frequency.
The genomic ‘grammar’ of endometriosis has 296
common genetic targets of SNPs with low allele frequencies and six
common genetic targets of SNPs with high allele frequencies in the
five studied population groups (Figs.
3 and 4). Within the Class A
and Class B (strongly- and highly-associated SNPs with
endometriosis) 11 key genetic targets with a low allele frequency
were identified, including SNPs of the MAP3K4, NSD2, MUC2, MUC17,
IL33, KRAS and CEP112 genes (Table
I) (50-57),
as well as two important genetic targets with a high allele
frequency, including the SNPs of the NME7 gene (Table II) (58-61).
A significant differentiation has been identified in SNPs of the
Africans and East Asians populations (62,63).
The African population has 23 unique genetic targets, of which 15
were identified with a low allele frequency and correspond to the
WNT4, TYK2, CDC42, PDLIM5, FEN1 and CDΗ1 genes (56,64-68),
and eight were identified with a high allele frequency and
correspond to the GREB1, MUC4, VEGFA, STXBP4, LILRB2, DNM3 and RNLS
genes (44,69-76)
(Tables I and II). The East Asian population had 11
unique genetic targets, of which ten were identified with a low
allele frequency and correspond to the PACERR, CYP1B1, FAS, IL1A,
ESR, VDR, CYP2C19 and MLLT10 genes (8,77-86),
and one was identified with a high allele frequency and corresponds
to the FN1 gene (8) (Tables I and II). The European population exhibited a
notable differentiation in the IL16(87), MUC4(88) and LAMA5(88) genetic targets, from which MUC4 also
corresponds to the American population and LAMA5 to the American
and East Asian populations (10,88,89).
This is in agreement with the general observation that the genetic
distance between Asia and Africa is shorter than that between
Africa and the other continents, as both Africans and Asians
contributed to the settlement of Europe (90).
The differentiation of endometriosis based on its
genomic ‘grammar’ among population groups is not a hypothetical
scenario. The allele frequency model of the target SNPs related to
endometriosis may explain the degree of differentiation, depending
on the initial genetic background of each population group
(Table I, Table II and Table III and Fig. 5). An even more notable finding is
that this differentiation appears to be consistent with the
continuous loss of genetic diversity along the geographical
expansion of Homo sapiens on earth, and the way that they
have conquered the continents (Fig.
5) (91,92). The African population appears to
have the widest genomic profile for the manifestation of
endometriosis followed by the genomic profile of East Asians,
Europeans, Americans and South Asians (Fig. 5 and Table III). Almost all genetic variants
that affect the risk of developing endometriosis have
human-specific origins, with ancient roots that often trace back to
the Africans population (93,94).
It is currently believed by scientists that SNPs, biological
pathways and consequently systems that are involved in several
diseases may have more ancient origins and each of these ancient
variations generated the conditions for modern disease (94). Indicatively, endometriosis is a
disease that has been recognized in other mammals, including the
olive baboon (Papio anubis) and guinea pig (Cavia
tschudii) (95,96).
Human populations exhibit differences in the allele
frequency of several common and rare SNPs. These observations are
largely the effect of the diverse environmental, cultural,
demographic and genomic scenarios of modern human populations. High
rates of endometriosis in East Asian and European populations have
been previously reported (4,97,98).
However, to the best of our knowledge, the strong association of
endometriosis with African populations through a specific set of
key genetic targets is presented for the first time herein
(Table I, Table II and Table III and Fig. 5), although studies have begun to
converge in this direction (99,100). This may be due to the lack of
reported cases of endometriosis in the African population (63,101). On the other hand, it should be
considered that African women may experience different epigenetic
factors that act negatively in addition to their burdened genomic
profile for the onset of the disease. Apart from the strong link to
hereditary factors, environmental exposures can result in the
development of endometriosis. An increased risk of developing
endometriosis is associated with plethora of environmental factors,
such as persistent organochlorine pollutants, perfluorochemicals,
elevated levels of phthalate esters and exposure to cigarette smoke
(98).
A set of 13 key genetic targets have been identified
in the genomic ‘grammar’ of endometriosis and are representative of
all the studied populations groups. Based on the findings of the
present study, eleven were identified in low allele frequencies and
two in high allele frequencies (Tables
I and II). Moreover, nine
were identified in gene loci and four in epigenetic-related loci. A
literature review revealed three confirmed genes through
genome-wide genetic analyses, including MAP3K4 (rs144240142)
(50), NSD2 (rs14647) (57) and IL33 (rs146597587) SNPs (102,103). It is notable that four
polymorphisms do not involve genes, but epigenetic-associated
targets with endometriosis, including CEP112 (rs76731691) (61), NME7 (rs1209731) (61), Factor V Leiden (F5, rs1894692)
(61) and the unnamed genetic
locus for the rs13177597(61)
SNPs. Indicatively, the polymorphism rs1894692, an intergenic
region between SLC19A2 and F5, has been proven to be a critical
epigenetic target through a GWAS (104) and has been directly linked to
endometriosis (86). The
polymorphisms involving the MUC17 (rs74974199) (52), MUC2 (rs11245936, rs7103978)
(21,51), KRAS (rs61764370) (85,105) and TYK2 (rs34536443, rs12720356)
(106-108)
genes, although they have been confirmed to be associated with
endometriosis in separated subgroups of the population, are
proposed for study in genome-wide genetic analyses based on the
results of the present study.
A total of 36 polymorphisms involving 35 genes and
one epigenetic target [rs7412010(109)], were found to markedly differ in
high and low allele frequencies, and appear to be representative in
one of the studied populations (23 in the African population, 11 in
the East Asian population, one in the South Asian population and
one in the American population, Tables
I and II). All the other
polymorphisms were identified in more than one population groups.
The genomic ‘grammar’ of endometriosis in Africans and East Asians
markedly differs and contains the majority of the SNPs in the two
sensitive clusters (low and high) than the other population groups
(Figs. 3B and 4B). The African population has a markedly
different number of sensitive polymorphisms related to
endometriosis in low and high allele frequencies, including the
genes TYK2 (rs2304256 and rs12720270), WNT4 (rs2235529, rs12037376,
rs61768001, rs3820282, rs56318008 and rs55938609), CDC42
(rs12038474 and rs10917151), PDLIM5 (rs2510770), VEGFA (rs1570360),
LILRB2 (rs383369), CDH1 (rs4783689) and GREB1 (rs13394619)
(56). GWAS performed confirm
these findings for the genes TYK2 (110,111) LILRB2(112) and CDC42 (56,113). The study performed by Välimäki
et al (65) demonstrated
the more frequent occurrence of uterine leiomyoma (UL) in women of
African origin, and suggested the WNT4 as a candidate
predisposition gene that may play a critical role in uterine
pathology and in particular endometriosis. However only two
sensitive polymorphisms of the WNT4 gene have been studied,
including rs3820282 and rs3820282. Although CDH1, PDLIM5, VEGFA,
LILRB2, SYNE1 and GREB1 gene-related polymorphisms are directly
associated with endometriosis and endometrial cancer, to the best
of our knowledge, they have never been studied as sensitive key
genetic targets in women with endometriosis in the African
population (61,72,114). The specialized genomic ‘grammar’
of endometriosis in East Asians includes gene-related polymorphisms
in low and high allele frequencies, including PACERR (rs20417),
CYP1B1 (rs1056836), FAS (rs4064), IL1A (rs2856836, rs1304037,
rs17561), ESR1 (rs9322331), VDR (rs1544410), CYP2C19 (rs11592737),
MLLT10 (rs1802669) and FN1 (rs1250248). GWAS performed confirm
these findings for the genes FN1(7), MLLT10(9), CYP2C19(115), IL1A (82) and CYP1B1(78). However, the reported SNPs regarding
the VDR, ESR1, FAS and PACERR gene loci have never been studied as
sensitive key genetic targets in women with endometriosis in the
East Asian population, at least to the best of our knowledge.
The present study revealed notable ‘key’ genetic
targets in the genomic ‘grammar’ of endometriosis, with the
evidence of population-based heterogeneity. Several of the results
obtained herein appear to be confirmed through a number of
scientific publications. On the other hand, some other ‘key’
genetic targets which are presented herein may be worthy of further
investigations in order to understand the specialized endometriosis
genomic ‘grammar’ of each population group in the future. Moreover,
further analysis is required to indicate how the common genes
within the population groups presented in the present study, or the
unique genes in a specific group, are involved in one or more
biological pathways, resulting in disease development, aiming to
provide personalized medicine and preventive measures for different
population groups.
In conclusion, the study of the genomic ‘grammar’ of
a given disease using GWAS data from several human population
groups may allow for the understanding of the multifunctional
biological pathways that exist and their diversity observed among
populations. The variation of 302 common genetic targets for
endometriosis is analyzed on a set of geographically separated
populations using GWAS information from the ‘1000 Genomes Project’.
Using the allele frequencies, the genetic differentiation among the
five population groups is shown and the common low and high
frequency SNP targets are located. By associating specific SNPs
clustering with low and high frequencies among the population
groups selected, conclusions are drawn to the effect of population
geographical translocation, epigenetic and environmental factors
and regionality of the disease. It remains to be elucidated whether
patients with endometriosis of different ancestral backgrounds have
clear differences in clinical presentation and disease course.
Thus, a more in-depth understanding of some of the molecular
differences between populations, both at the genetic and protein
expression levels may prove valuable, as it may suggest strategies
through which treatment could be personalized. As a consequence, it
may then be possible to determine the unique characteristics of the
patient's disease and select the most beneficial treatment based
upon a patient's individual biology.
Not applicable.
Funding: No funding was received.
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
LP was involved in the conceptualization and
methodology of the study, in obtaining resources, data curation and
investigation, formal analysis, and validation as well as in the
writing of the original draft, and in the reviewing and editing of
the manuscript. AA was involved in data curation, investigation,
validation and formal analysis, as well as in the writing of the
original draft, and in the reviewing and editing of the manuscript.
MZ was involved in data validation and formal analysis, and in the
writing of the original draft. DV was involved in data validation,
in the provision of resources and in data curation. GNG was
involved in the provision of resources, in data validation, as well
as in the writing of the original draft. EE was involved in the
conceptualization and methodology of the study, in data curation,
investigation, validation and formal analysis, in the provision of
resources and project administration, in the writing of the
original draft and in the reviewing and editing of the manuscript,
as well as in the study supervision. LP and EE confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Halis G and Arici A: Endometriosis and
inflammation in infertility. Ann N Y Acad Sci. 1034:300–315.
2004.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zondervan KT, Becker CM, Koga K, Missmer
SA, Taylor RN and Vigano P: Endometriosis. Nat Rev Dis Primers.
4(9)2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Falcone T and Flyckt R: Clinical
management of endometriosis. Obstet Gynecol. 131:557–571.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Dai Y, Li X, Shi J and Leng J: A review of
the risk factors, genetics and treatment of endometriosis in
Chinese women: A comparative update. Reprod Health.
15(82)2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Saha R, Pettersson HJ, Svedberg P,
Olovsson M, Bergqvist A, Marions L, Tornvall P and Kuja-Halkola R:
Heritability of endometriosis. Fertil Steril. 104:947–952.
2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Rahmioglu N, Montgomery GW and Zondervan
KT: Genetics of endometriosis. Womens Health (Lond). 11:577–586.
2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Rahmioglu N, Nyholt DR, Morris AP, Missmer
SA, Montgomery GW and Zondervan KT: Genetic variants underlying
risk of endometriosis: Insights from meta-analysis of eight
genome-wide association and replication datasets. Hum Reprod
Update. 20:702–716. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sapkota Y, Steinthorsdottir V, Morris AP,
Fassbender A, Rahmioglu N, De Vivo I, Buring JE, Zhang F, Edwards
TL, Jones S, et al: Meta-analysis identifies five novel loci
associated with endometriosis highlighting key genes involved in
hormone metabolism. Nat Commun. 8(15539)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nilufer R, Karina B, Paraskevi C, Rebecca
D, Genevieve G, Ayush G, Stuart M, Sally M, Yadav S, Andrew SJ, et
al: Large-scale genome-wide association meta-analysis of
endometriosis reveals 13 novel loci and genetically-associated
comorbidity with other pain conditions. BioRxiv. 7(406967)2018.
|
|
10
|
Ran S, He X, Jiang ZX, Liu Y, Zhang YX,
Zhang L, Gu GS, Pei Y, Liu BL, Tian Q, et al: Whole-exome
sequencing and genome-wide association studies identify novel
sarcopenia risk genes in Han Chinese. Mol Genet Genomic Med.
8(e1267)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hemmert R, Schliep KC, Willis S, Peterson
CM, Louis GB, Allen-Brady K, Simonsen SE, Stanford JB, Byun J and
Smith KR: Modifiable life style factors and risk for incident
endometriosis. Paediatr Perinat Epidemiol. 33:19–25.
2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Williams C, Long AJ, Noga H, Allaire C,
Bedaiwy MA, Lisonkova S and Yong PJ: East and South East Asian
ethnicity and moderate-to-severe endometriosis. J Minim Invasive
Gynecol. 26:507–515. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Goulielmos GN, Zervou MI, Vazgiourakis VM,
Ghodke-Puranik Y, Garyfallos A and Niewold TB: The genetics and
molecular pathogenesis of systemic lupus erythematosus (SLE) in
populations of different ancestry. Gene. 668:59–72. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Henn BM, Cavalli-Sforza LL and Feldman MW:
The great human expansion. Proc Natl Acad Sci USA. 109:17758–17764.
2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
McEvoy BP, Powell JE, Goddard ME and
Visscher PM: Human population dispersal ‘Out of Africa’ estimated
from linkage disequilibrium and allele frequencies of SNPs. Genome
Res. 21:821–829. 2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Amos W and Hoffman JI: Evidence that two
main bottleneck events shaped modern human genetic diversity. Proc
Biol Sci. 277:131–137. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Laval G, Patin E, Barreiro LB and
Quintana-Murci L: Formulating a historical and demographic model of
recent human evolution based on resequencing data from noncoding
regions. PLoS One. 5(e10284)2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Fagundes NJ, Ray N, Beaumont M,
Neuenschwander S, Salzano FM, Bonatto SL and Excoffier L:
Statistical evaluation of alternative models of human evolution.
Proc Natl Acad Sci USA. 104:17614–17619. 2007.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gravel S, Henn BM, Gutenkunst RN, Indap
AR, Marth GT, Clark AG, Yu F and Gibbs RA: 1000 Genomes Project;
Bustamante CD. Demographic history and rare allele sharing among
human populations. Proc Natl Acad Sci USA. 108:11983–11988.
2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li H and Durbin R: Inference of human
population history from individual whole-genome sequences. Nature.
475:493–496. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Papageorgiou L, Zervou MI, Vlachakis D,
Matalliotakis M, Matalliotakis I, Spandidos DA, Goulielmos GN and
Eliopoulos E: Demetra application: An integrated genotype analysis
web server for clinical genomics in endometriosis. Int J Mol Med.
47(115)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Smigielski EM, Sirotkin K, Ward M and
Sherry ST: dbSNP: A database of single nucleotide polymorphisms.
Nucleic Acids Res. 28:352–355. 2000.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fairley S, Lowy-Gallego E, Perry E and
Flicek P: The international genome sample resource (IGSR)
collection of open human genomic variation resources. Nucleic Acids
Res. 48:D941–D947. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
1000 Genomes Project Consortium. Auton A,
Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL,
McCarthy S, McVean GA and Abecasis GR: A global reference for human
genetic variation. Nature. 526:68–74. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zheng-Bradley X, Streeter I, Fairley S,
Richardson D, Clarke L and Flicek P: 1000 Genomes Project
Consortium. Alignment of 1000 genomes project reads to reference
assembly GRCh38. Gigascience. 6:1–8. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lowy-Gallego E, Fairley S, Zheng-Bradley
X, Ruffier M, Clarke L and Flicek P: 1000 Genomes Project
Consortium. Variant calling on the GRCh38 assembly with the data
from phase three of the 1000 genomes project. Wellcome Open Res.
4(50)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Danecek P, Auton A, Abecasis G, Albers CA,
Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST,
et al: The variant call format and VCFtools. Bioinformatics.
27:2156–2158. 2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sobie EA: An introduction to MATLAB. Sci
Signal. 4(tr7)2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cui Y, Chen X, Luo H, Fan Z, Luo J, He S,
Yue H, Zhang P and Chen R: BioCircos.js: An interactive Circos
JavaScript library for biological data visualization on web
applications. Bioinformatics. 32:1740–1742. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Gu Z, Gu L, Eils R, Schlesner M and Brors
B: Circlize Implements and enhances circular visualization in R.
Bioinformatics. 30:2811–2812. 2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yin L, Zhang H, Tang Z, Xu J, Yin D, Zhang
Z, Yuan X, Zhu M, Zhao S, Li X and Liu X: rMVP: A memory-efficient,
visualization-enhanced, and parallel-accelerated tool for
genome-wide association study. Genomics Proteomics Bioinformatics.
19:619–628. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Gazal S, Sahbatou M, Babron MC, Genin E
and Leutenegger AL: High level of inbreeding in final phase of 1000
genomes project. Sci Rep. 5(17453)2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang W, Ng HW, Shu M, Luo H, Su Z, Ge W,
Perkins R, Tong W and Hong H: Comparing genetic variants detected
in the 1000 genomes project with SNPs determined by the
international HapMap consortium. J Genet. 94:731–740.
2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Reisberg S, Galwey N, Avillach P,
Sahlqvist AS, Kolberg L, Magi R, Esko T, Vilo J and James G:
Comparison of variation in frequency for SNPs associated with
asthma or liver disease between Estonia, HapMap populations and the
1000 genome project populations. Int J Immunogenet. 46:49–58.
2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Chen N, Juric I, Cosgrove EJ, Bowman R,
Fitzpatrick JW, Schoech SJ, Clark AG and Coop G: Allele frequency
dynamics in a pedigreed natural population. Proc Natl Acad Sci USA.
116:2158–2164. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lohmueller KE, Albrechtsen A, Li Y, Kim
SY, Korneliussen T, Vinckenbosch N, Tian G, Huerta-Sanchez E, Feder
AF, Grarup N, et al: Natural selection affects multiple aspects of
genetic variation at putatively neutral sites across the human
genome. PLoS Genet. 7(e1002326)2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Taioli E, Pedotti P and Garte S:
Importance of allele frequency estimates in epidemiological
studies. Mutat Res. 567:63–70. 2004.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Adrianto I and Montgomery C: Estimating
allele frequencies. Methods Mol Biol. 850:59–76. 2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Gautier M, Foucaud J, Gharbi K, Cezard T,
Galan M, Loiseau A, Thomson M, Pudlo P, Kerdelhué C and Estoup A:
Estimation of population allele frequencies from next-generation
sequencing data: Pool-versus individual-based genotyping. Mol Ecol.
22:3766–3779. 2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Astrom F, Felsberg M and Scharr H (eds):
Adaptive sharpening of multimodal distributions. Colour and Visual
Computing Symposium (CVCS). 2015.
|
|
41
|
Eberle MA, Rieder MJ, Kruglyak L and
Nickerson DA: Allele frequency matching between SNPs reveals an
excess of linkage disequilibrium in genic regions of the human
genome. PLoS Genet. 2(e142)2006.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Norton N, Williams NM, Williams HJ,
Spurlock G, Kirov G, Morris DW, Hoogendoorn B, Owen MJ and
O'Donovan MC: Universal, robust, highly quantitative SNP allele
frequency measurement in DNA pools. Hum Genet. 110:471–478.
2002.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Fredman D, Sawyer SL, Stromqvist L,
Mottagui-Tabar S, Kidd KK, Wahlestedt C, Chanock SJ and Brookes AJ:
Nonsynonymous SNPs: Validation characteristics, derived allele
frequency patterns, and suggestive evidence for natural selection.
Hum Mutat. 27:173–186. 2006.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Painter JN, Nyholt DR, Krause L, Zhao ZZ,
Chapman B, Zhang C, Medland S, Martin NG, Kennedy S, Treloar S, et
al: Common variants in the CYP2C19 gene are associated with
susceptibility to endometriosis. Fertil Steril. 102:496–502 e5.
2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Gerlinger C, Faustmann T, Hassall JJ and
Seitz C: Treatment of endometriosis in different ethnic
populations: A meta-analysis of two clinical trials. BMC Womens
Health. 12(9)2012.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Bougie O, Yap MI, Sikora L, Flaxman T and
Singh S: Influence of race/ethnicity on prevalence and presentation
of endometriosis: A systematic review and meta-analysis. BJOG.
126:1104–1115. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Nielsen R, Akey JM, Jakobsson M, Pritchard
JK, Tishkoff S and Willerslev E: Tracing the peopling of the world
through genomics. Nature. 541:302–310. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Huang W, Haubold B, Hauert C and Traulsen
A: Emergence of stable polymorphisms driven by evolutionary games
between mutants. Nat Commun. 3(919)2012.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Kobayashi Y, Yang S, Nykamp K, Garcia J,
Lincoln SE and Topper SE: Pathogenic variant burden in the ExAC
database: An empirical approach to evaluating population data for
clinical variant interpretation. Genome Med. 9(13)2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Uimari O, Rahmioglu N, Nyholt DR, Vincent
K, Missmer SA, Becker C, Morris AP, Montgomery GW and Zondervan KT:
Genome-wide genetic analyses highlight mitogen-activated protein
kinase (MAPK) signaling in the pathogenesis of endometriosis. Hum
Reprod. 32:780–793. 2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Chang CY, Chen Y, Lin WC, Chen CM, Chen
CP, Lee SC, Sheu JJC and Tsai FJ: MUC2 polymorphisms are associated
with endometriosis development and infertility: A case-control
study. BMC Med Genet. 13(15)2012.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Yang CW, Chang CY, Lai MT, Chang HW, Lu
CC, Chen Y, Chen CM, Lee SC, Tsai PW, Yang SH, et al: Genetic
variations of MUC17 are associated with endometriosis development
and related infertility. BMC Med Genet. 16(60)2015.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Sun S, Huang DW, Huo LT and Li PL: The
role of NSD2 and EZH2 in the pathogenesis of endometrial carcinoma.
J Biol Regul Homeost Agents. 33:1233–1239. 2019.PubMed/NCBI
|
|
54
|
Miller JE, Monsanto SP, Ahn SH, Khalaj K,
Fazleabas AT, Young SL, Lessey BA, Koti M and Tayade C:
Interleukin-33 modulates inflammation in endometriosis. Sci Rep.
7(17903)2017.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Christensen BC, Moyer BJ, Avissar M,
Ouellet LG, Plaza SL, McClean MD, Marsit CJ and Kelsey KT: A let-7
microRNA-binding site polymorphism in the KRAS 3' UTR is associated
with reduced survival in oral cancers. Carcinogenesis.
30:1003–1007. 2009.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Lalami I, Abo C, Borghese B, Chapron C and
Vaiman D: Genomics of endometriosis: From genome wide association
studies to exome sequencing. Int J Mol Sci. 22(7297)2021.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Zhao ZZ, Croft L, Nyholt DR, Chapman B,
Treloar SA, Hull ML and Montgomery GW: Evaluation of polymorphisms
in predicted target sites for micro RNAs differentially expressed
in endometriosis. Mol Hum Reprod. 17:92–103. 2011.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Maekawa R, Taketani T, Mihara Y, Sato S,
Okada M, Tamura I, Jozaki K, Kajimura T, Asada H, Tamura H, et al:
Thin endometrium transcriptome analysis reveals a potential
mechanism of implantation failure. Reprod Med Biol. 16:206–227.
2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Erikson DW, Barragan F, Piltonen TT, Chen
JC, Balayan S, Irwin JC and Giudice LC: Stromal fibroblasts from
perimenopausal endometrium exhibit a different transcriptome than
those from the premenopausal endometrium. Biol Reprod. 97:387–399.
2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Duan R, Wang Y, Lin A, Lian L, Cao H, Gu
W, Li T and Sun Q: Expression of nm23-H1, p53, and integrin beta1
in endometriosis and their clinical significance. Int J Clin Exp
Pathol. 13:1024–1029. 2020.PubMed/NCBI
|
|
61
|
Montgomery GW, Mortlock S and Giudice LC:
Should genetics now be considered the pre-eminent etiologic factor
in endometriosis? J Minim Invasive Gynecol. 27:280–286.
2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Yen CF, Kim MR and Lee CL: Epidemiologic
factors associated with endometriosis in East Asia. Gynecol Minim
Invasive Ther. 8:4–11. 2019.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Kyama CM, Mwenda JM, Machoki J, Mihalyi A,
Simsa P, Chai DC and D'Hooghe TM: Endometriosis in African women.
Womens Health (Lond). 3:629–635. 2007.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Albertsen HM, Chettier R, Farrington P and
Ward K: Genome-wide association study link novel loci to
endometriosis. PLoS One. 8(e58257)2013.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Valimaki N, Kuisma H, Pasanen A,
Heikinheimo O, Sjoberg J, Butzow R, Sarvilinna N, Heinonen HR,
Tolvanen J, Bramante S, et al: Genetic predisposition to uterine
leiomyoma is determined by loci for genitourinary development and
genome stability. Elife. 7(e37110)2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Gelernter J, Kranzler HR, Sherva R, Almasy
L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu
D, et al: Genome-wide association study of alcohol dependence:
Significant findings in African- and European-Americans including
novel risk loci. Mol Psychiatry. 19:41–49. 2014.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Flynn K, Feben C, Lamola L, Carstens N,
Krause A and Lombard Z: for DDD-Africa as members of the H3Africa
Consortium. Ending a diagnostic odyssey-The first case of
Takenouchi-Kosaki syndrome in an African patient. Clin Case Rep.
9:2144–2148. 2021.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Takeshita H, Fujihara J, Ueki M, Iida R,
Koda Y, Soejima M, Yuasa I, Kato H, Nakajima T, Kominato Y and
Yasuda T: Nonsynonymous single-nucleotide polymorphisms of the
human apoptosis-related endonuclease-DNA fragmentation factor beta
polypeptide, endonuclease G, and Flap endonuclease-1-genes show a
low degree of genetic heterogeneity. DNA Cell Biol. 31:36–42.
2012.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Sapkota Y, Vivo I, Steinthorsdottir V,
Fassbender A, Bowdler L, Buring JE, Edwards TL, Jones S, Dorien O,
Peterse D, et al: Analysis of potential protein-modifying variants
in 9000 endometriosis patients and 150000 controls of European
ancestry. Sci Rep. 7(11380)2017.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Chou YC, Chen MJ, Chen PH, Chang CW, Yu
MH, Chen YJ, Tsai EM, Tsai SF, Kuo WS, Tzeng CR, et al: Integration
of genome-wide association study and expression quantitative trait
locus mapping for identification of endometriosis-associated genes.
Sci Rep. 11(478)2021.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Chang CY, Chang HW, Chen CM, Lin CY, Chen
CP, Lai CH, Lin WY, Liu HP, Sheu JJC and Tsai FJ: MUC4 gene
polymorphisms associate with endometriosis development and
endometriosis-related infertility. BMC Med. 9(19)2011.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Lulinska-Kuklik E, Leznicka K,
Huminska-Lisowska K, Moska W, Michalowska-Sawczyn M, Ossowski Z,
Maculewicz E, Cięszczyk P, Kaczmarczyk M, Ratkowski W, et al: The
VEGFA gene and anterior cruciate ligament rupture risk in the
Caucasian population. Biol Sport. 36:3–8. 2019.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Yang JW, Hutchinson IV, Shah T, Fang J and
Min DI: Gene polymorphism of vascular endothelial growth
factor-1154 G>A is associated with hypertensive nephropathy in a
Hispanic population. Mol Biol Rep. 38:2417–2425. 2011.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Rahim M, El Khoury LY, Raleigh SM, Ribbans
WJ, Posthumus M, Collins M and September AV: Human genetic
variation, sport and exercise medicine, and achilles tendinopathy:
Role for angiogenesis-associated genes. OMICS. 20:520–527.
2016.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Hirayasu K, Ohashi J, Tanaka H, Kashiwase
K, Ogawa A, Takanashi M, Satake M, Jia GJ, Chimge NO, Sideltseva
EW, et al: Evidence for natural selection on leukocyte
immunoglobulin-like receptors for HLA class I in Northeast Asians.
Am J Human Genet. 82:1075–1083. 2008.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Borghese B, Tost J, de Surville M, Busato
F, Letourneur F, Mondon F, Vaiman D and Chapron C: Identification
of susceptibility genes for peritoneal, ovarian, and deep
infiltrating endometriosis using a pooled sample-based genome-wide
association study. Biomed Res Int. 2015(461024)2015.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Chen S, Chen L, Tan Y and Wang J:
Association between rs20417 polymorphism in cyclooxygenase-2 and
gastric cancer susceptibility: Evidence from15 case-control
studies. Medicine (Baltimore). 98(e15468)2019.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Liu F, Luo LM, Wei YG, Li B, Wang WT, Wen
TF, Yang JY, Xu MQ and Yan LN: Polymorphisms of the CYP1B1 gene and
hepatocellular carcinoma risk in a Chinese population. Gene.
564:14–20. 2015.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Gao W, Tan J, Huls A, Ding A, Liu Y,
Matsui MS, Vierkötter A, Krutmann J, Schikowski T, Jin L and Wang
S: Genetic variants associated with skin aging in the Chinese Han
population. J Dermatol Sci. 86:21–29. 2017.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Pissetti CW, Tanaka S, Hortolani ACC and
Marqui ABT: Gene polymorphisms in FAS (Rs3740286 and Rs4064) are
involved in endometriosis development in Brazilian women, but not
those in CASP8 (rs13416436 and rs2037815). Rev Bras Ginecol Obstet.
40:450–457. 2018.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Sawyer SL, Mukherjee N, Pakstis AJ, Feuk
L, Kidd JR, Brookes AJ and Kidd KK: Linkage disequilibrium patterns
vary substantially among populations. Eur J Hum Genet. 13:677–686.
2005.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Hata Y, Nakaoka H, Yoshihara K, Adachi S,
Haino K, Yamaguchi M, Nishikawa N, Kashima K, Yahata T, Tajima A,
et al: A nonsynonymous variant of IL1A is associated with
endometriosis in Japanese population. J Hum Genet. 58:517–520.
2013.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Zhao X, Zong LL, Wang YF, Mao T, Fu YG,
Zeng J and Rao XQ: Association of single nucleotide polymorphism in
CYP17 and ERalpha genes with endometriosis risk in southern Chinese
women. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 28:304–307.
2011.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Yin X, Wang H, Guo J, Zhang L, Zhang Y, Li
L and Hou S: Association of vitamin D receptor BsmI rs1544410 and
ApaI rs7975232 polymorphisms with susceptibility to adolescent
idiopathic scoliosis: A systematic review and meta-analysis.
Medicine (Baltimore). 97(e9627)2018.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Angioni S, D'Alterio MN, Coiana A, Anni F,
Gessa S and Deiana D: Genetic characterization of endometriosis
patients: Review of the literature and a prospective cohort study
on a mediterranean population. Int J Mol Sci.
21(1765)2020.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Zubrzycka A, Zubrzycki M, Perdas E and
Zubrzycka M: Genetic, epigenetic, and steroidogenic modulation
mechanisms in endometriosis. J Clin Med. 9(1309)2020.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Matalliotakis M, Zervou MI, Eliopoulos E,
Matalliotaki C, Rahmioglu N, Kalogiannidis I, Zondervan K,
Spandidos DA, Matalliotakis I, Goulielmos GN, et al: The role of
IL16 gene polymorphisms in endometriosis. Int J Mol Med.
41:1469–1476. 2018.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Vassilopoulou L, Matalliotakis M, Zervou
MI, Matalliotaki C, Krithinakis K, Matalliotakis I, Spandidos DA
and Goulielmos GN: Defining the genetic profile of endometriosis.
Exp Ther Med. 17:3267–3281. 2019.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Christofolini DM, Mafra FA, Catto MC,
Bianco B and Barbosa CP: New candidate genes associated to
endometriosis. Gynecol Endocrinol. 35:62–65. 2019.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Cavalli-Sforza LL: Genes, peoples, and
languages. Proc Natl Acad Sci USA. 94:7719–7724. 1997.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Chakravarti A: Perspectives on human
variation through the lens of diversity and race. Cold Spring Harb
Perspect Biol. 7(a023358)2015.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Templeton AR: Biological races in humans.
Stud Hist Philos Biol Biomed Sci. 44:262–271. 2013.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Choudhury A, Aron S, Botigue LR, Sengupta
D, Botha G, Bensellak T, Wells G, Kumuthini J, Shriner D, Fakim YJ,
et al: High-depth African genomes inform human migration and
health. Nature. 586:741–748. 2020.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Benton ML, Abraham A, LaBella AL, Abbot P,
Rokas A and Capra JA: The influence of evolutionary history on
human health and disease. Nat Rev Genet. 22:269–283.
2021.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Baldi A, Lanza A, Menicagli F, Signorile
PG and Spugnini EP: Histological and immunohistochemical
characterization of a case of endometriosis in a Guinea Pig
(Cavia tschudii). Case Rep Vet Med.
2017(4594510)2017.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Braundmeier AG and Fazleabas AT: The
non-human primate model of endometriosis: Research and implications
for fecundity. Mol Hum Reprod. 15:577–586. 2009.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Eisenberg VH, Weil C, Chodick G and Shalev
V: Epidemiology of endometriosis: A large population-based database
study from a healthcare provider with 2 million members. BJOG.
125:55–62. 2018.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Terzic M, Aimagambetova G, Kunz J,
Bapayeva G, Aitbayeva B, Terzic S and Laganà AS: Molecular basis of
endometriosis and endometrial cancer: Current knowledge and future
perspectives. Int J Mol Sci. 22(9274)2021.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Crespi B: Variation among human
populations in endometriosis and PCOS A test of the inverse
comorbidity model. Evol Med Public Health. 9:295–310.
2021.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Mecha EO, Njagi JN, Makunja RN, Omwandho
COA, Saunders PTK and Horne AW: Endometriosis among African women.
Reprod Fertil. 3:C40–C43. 2022.
|
|
101
|
Kyama MC, D'Hooghe TM, Debrock S, Machoki
J, Chai DC and Mwenda JM: The prevalence of endometriosis among
African-American and African-indigenous women. Gynecol Obstet
Invest. 57:40–42. 2004.PubMed/NCBI
|
|
102
|
Van Hout CV, Tachmazidou I, Backman JD,
Hoffman JD, Liu D, Pandey AK, Gonzaga-Jauregui C, Khalid S, Ye B,
Banerjee N, et al: Exome sequencing and characterization of 49,960
individuals in the UK Biobank. Nature. 586:749–756. 2020.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Mousas A, Ntritsos G, Chen MH, Song C,
Huffman JE, Tzoulaki I, Elliott P and Psaty BM: Blood-Cell
Consortium. Auer PL, et al: Rare coding variants pinpoint genes
that control human hematological traits. PLoS Genet.
13(e1006925)2017.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Johnson AD, Bhimavarapu A, Benjamin EJ,
Fox C, Levy D, Jarvik GP and O'Donnell CJ: CLIA-tested genetic
variants on commercial SNP arrays: Potential for incidental
findings in genome-wide association studies. Genet Med. 12:355–363.
2010.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Gallegos-Arreola MP, Verdin PM,
Magana-Torres MT, Figuera LE, Zuniga-Gonzalez GM, Rosales-Reynoso
MA, Gómez-Meda BC and Puebla-Pérez AM: Association between
rs61764370, rs9266, and rs140080026 polymorphisms of the KRAS gene
and breast cancer risk in a Mexican population. Eur Rev Med
Pharmacol Sci. 25:6454–6464. 2021.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Staels F, Collignon T, Betrains A, Gerbaux
M, Willemsen M, Humblet-Baron S, Liston A, Vanderschueren S and
Schrijvers R: Monogenic adult-onset inborn errors of immunity.
Front Immunol. 12(753978)2021.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Kerner G, Laval G, Patin E, Boisson-Dupuis
S, Abel L, Casanova JL and Quintana-Murci L: Human ancient DNA
analyses reveal the high burden of tuberculosis in Europeans over
the last 2,000 years. Am J Hum Genet. 108:517–524. 2021.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Peluso C, Christofolini DM, Goldman CS,
Mafra FA, Cavalcanti V, Barbosa CP and Bianco B: TYK2 rs34536443
polymorphism is associated with a decreased susceptibility to
endometriosis-related infertility. Hum Immunol. 74:93–97.
2013.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Masuda T, Low SK, Akiyama M, Hirata M,
Ueda Y, Matsuda K, Kimura T, Murakami Y, Kubo M, Kamatani Y and
Okada Y: GWAS of five gynecologic diseases and cross-trait analysis
in Japanese. Eur J Hum Genet. 28:95–107. 2020.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Wang YF, Zhang Y, Lin Z, Zhang H, Wang TY,
Cao Y, Morris DL, Sheng Y, Yin X, Zhong SL, et al: Identification
of 38 novel loci for systemic lupus erythematosus and genetic
heterogeneity between ancestral groups. Nat Commun.
12(772)2021.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Li Z, Rotival M, Patin E, Michel F and
Pellegrini S: Two common disease-associated TYK2 variants impact
exon splicing and TYK2 dosage. PLoS One.
15(e0225289)2020.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Bylinska A, Wilczynska K, Malejczyk J,
Milewski L, Wagner M, Jasek M, Niepiekło-Miniewska W, Wiśniewski A,
Płoski R, Barcz E, et al: The impact of HLA-G, LILRB1 and LILRB2
gene polymorphisms on susceptibility to and severity of
endometriosis. Mol Genet Genomics. 293:601–613. 2018.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Edwards TL, Giri A, Hellwege JN, Hartmann
KE, Stewart EA, Jeff JM, Bray MJ, Pendergrass SA, Torstenson ES,
Keaton JM, et al: A trans-ethnic genome-wide association study of
uterine fibroids. Front Genet. 10(511)2019.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Geng YH, Wang ZF, Jia YM, Zheng LY, Chen
L, Liu DG, Li XH, Tian XX and Fang WG: Genetic polymorphisms in
CDH1 are associated with endometrial carcinoma susceptibility among
Chinese Han women. Oncol Lett. 16:6868–6878. 2018.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Lagana AS, Sturlese E, Retto G, Sofo V and
Triolo O: Interplay between misplaced mullerian-derived stem cells
and peritoneal immune dysregulation in the pathogenesis of
endometriosis. Obstet Gynecol Int. 2013(527041)2013.PubMed/NCBI View Article : Google Scholar
|