1. Introduction
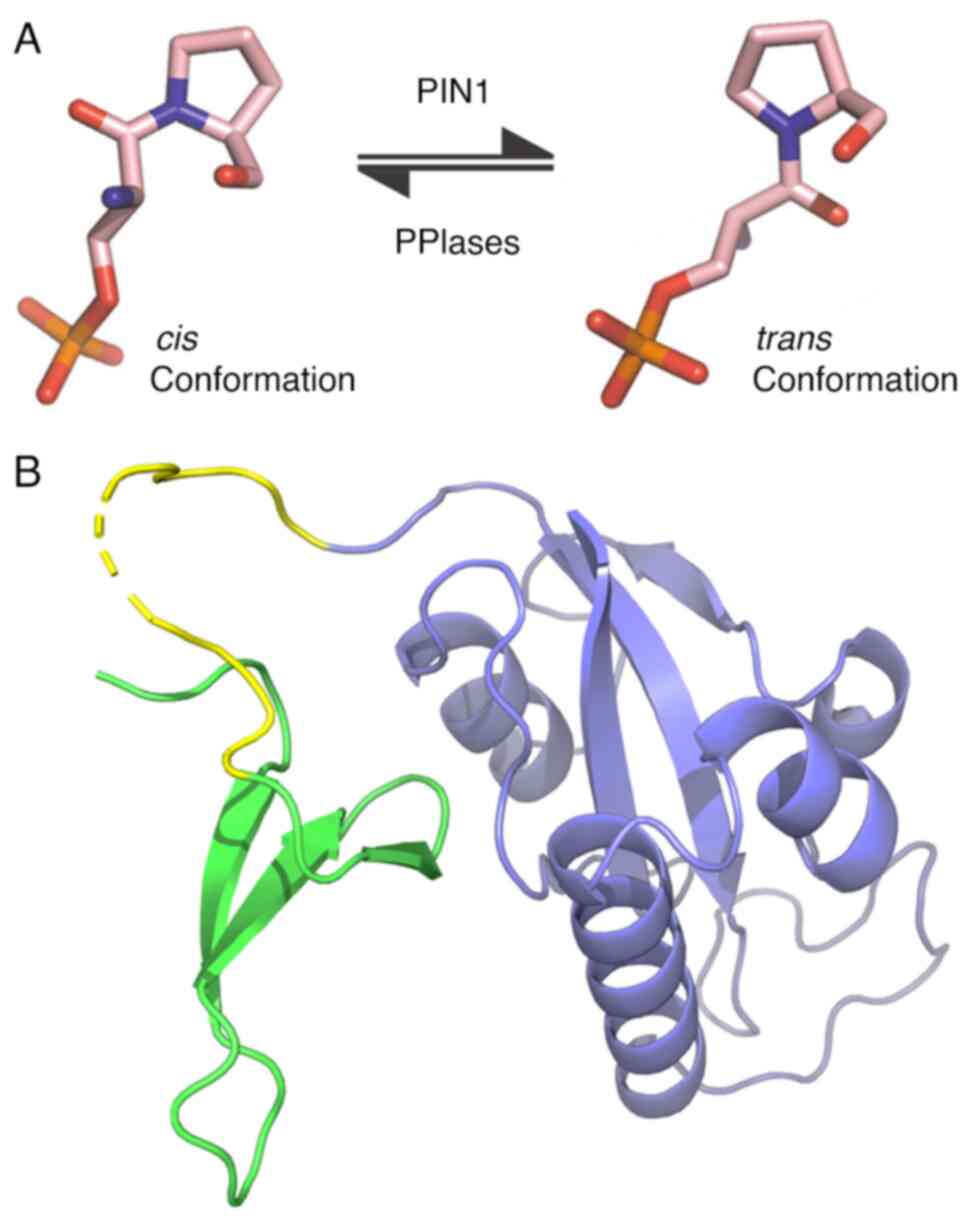
Proline (Pro) is the only non-synthetic amino acid
able to adopt cis or trans conformations. The process
of transitioning between these conformations is known as
isomerization. This process can be catalyzed by a group of proteins
termed peptidyl-prolyl isomerases (PPIases) enzymes. Thus far, the
only enzyme capable of recognizing and catalyzing the
cis-trans isomerization of Pro in phosphorylated
Ser/Thr-Pro motifs (p-Ser/Thr-Pro) is the peptidyl-prolyl
cis-trans isomerase 1 interacting with NIMA (PIN1)
(1) (Fig. 1A). The Ser/Thr-Pro motifs are one
of the most crucial phosphorylation domains present in proteins
(2). The alteration in the
cis-trans conformation of the Pro within these
domains, modulated by PIN1, emerges as another pivotal aspect in
orchestrating intracellular signaling pathways (3).
PIN1 is composed of two domains: The ‘WW’ domain,
responsible for the recognition and binding of PIN1 to the
p-Ser/Thr-Pro motifs of its target proteins, and the PPIase, which
provides PIN1 with its isomerase catalytic activity (4) (Fig.
1B). PIN1 can regulate and modify the destiny of numerous
proteins through these two properties, affecting various signaling
pathways and participating in numerous cellular processes. Such
actions are achieved by influencing target proteins via diverse
mechanisms, such as promoting activation or inhibition, altering
cellular localization, affecting stability, and modifying
interactions with other proteins (5).
This capacity of PIN1 to engage in a broad spectrum
of cellular processes is accompanied by rigorous regulation. Under
normal physiological conditions, PIN1 is tightly regulated at the
transcriptional, post-transcriptional and post-translational levels
(6-11).
This regulatory framework ensures precise control over its activity
and allows PIN1 to orchestrate intricate protein modifications,
thus contributing to the complex network of cellular signaling and
responses.
The expression of PIN1 under normal physiological
conditions is induced during neuronal differentiation. Generally,
the expression level of this enzyme is directly associated with the
replicative potential present in normal cells and gradually
decreases with aging (12,13). Conversely, when some of the
mechanisms that regulate PIN1 are disrupted, the deregulation of
PIN1 can give rise to a range of pathologies. The present review
aimed to systematically outline the diverse information on the
dysregulation of PIN1 activity associated with various disorders,
as enlisted in Fig. 2. By
presenting a comprehensive synthesis of the currently available
knowledge, the present review aimed to shed light on the
multifaceted roles of PIN1 and its potential implications in
disease mechanisms.
2. Metabolic diseases
Metabolic diseases (MetS) represent a constellation
of related metabolic disturbances, such as insulin resistance,
atherogenic dyslipidemia, central adiposity and high blood
pressure. The pathogenesis of MetS involves various genetic and
acquired factors related to insulin resistance and chronic systemic
inflammation. If left unaddressed, MetS are strongly linked to a
heightened susceptibility to developing diabetes and cardiovascular
disorders. Together, MetS are currently known as metabolic syndrome
(14).
Disordered insulin signaling is associated with
various metabolic conditions, such as adiposity, non-alcoholic
steatohepatitis (NASH), and type 2 diabetes (15). In this context, a number of
researchers describe PIN1 as a vital regulator in insulin
signaling. In the study by Nakatsu et al (16), it was demonstrated that PIN1
fosters insulin secretion in islet β-cells by heightening
salt-inducible kinase 2 activity. Indeed, PIN1 encourages cellular
proliferation and transformation by modulating activator protein-1
(AP-1) and ERK1/2 activation induced by insulin through
interactions with p70S6K (16).
Moreover, previous research has shown that PIN1 positively
regulates signaling by promoting insulin receptor substrate 1
phosphorylation, increasing protein stability and the levels of
acetyl-CoA carboxylase 1 and fatty acid synthase, and repressing
AMPK activity in NASH (17). While
the involvement of PIN1 in these metabolic disorders is partially
due to its control of insulin signaling, it also interacts with or
regulates key molecules relevant to other processes (Table I). For instance, in adipogenesis,
PIN1 has been reported to function as an enhancer of adipocyte
differentiation by regulating the function of PPARγ (18).
 | Table IPIN1-related processes involved in
osteoporosis, metabolic and cardiovascular diseases. |
Table I
PIN1-related processes involved in
osteoporosis, metabolic and cardiovascular diseases.
| Metabolic diseases
(Refs.) | Osteoporosis
(Refs.) | Cardiovascular
diseases (Refs.) |
|---|
| Insulin signaling
(16,17) | Osteoclast cell
signaling and fusion (23,28) | Bioavility and
release of nitric oxide (35,36) |
| Adipogenesis
(18,19) | Osteoblast
differentiation (24-26) | Vascular
homeostasis (35,41,42) |
| Lipid accumulation
(20) | Bone resorption and
osteogenesis (22,27) | Apoptosis and
inflammation (25,37,43) |
Additionally, it is involved in the function of
adipocytes through its association with PR/SET domain 16 and
patatin like phospholipase domain containing 2(19). A recent study suggested that an
increase in the expression of PIN1 in liver cells contributes to
lipid accumulation in NASH (20).
All these data lead to a better understanding of the role of PIN1
in MetS and its potential use as a therapeutic target.
3. Osteoporosis
Osteoporosis is a global disease associated in
particular with advancing age, and it occurs when there is a loss
of bone mineral density, leading to an increased predisposition to
fragility fractures (21).
A keystone process that outlines this disease is the
balance between osteoblasts and osteoclasts, which manage bone
restoration and breakdown, respectively (22). In this regard, PIN1 is a key
regulator of numerous pathways involved in osteoblast and
osteoclast cell signaling. A clear indication is that
PIN1-deficient mice exhibit osteoporosis-like characteristics with
a low bone mass and density (23).
In-depth studies reveal that PIN1 promotes osteoblast function and
bone formation by enhancing bone morphogenetic protein signaling
and associating with Runx2, a critical factor for osteoblast
differentiation (24-26).
By binding to Runx2 and Osterix, another essential factor for
osteoblast differentiation, PIN1 boosts their transcriptional
activity, thereby enhancing osteogenesis (27).
Conversely, PIN1 is a negative regulator of
osteoclast fusion, leading to the accumulation of dense bone and
promoting osteoporosis by reducing the activity of the dendritic
cell-specific transmembrane protein, a key fusion-mediating
molecule in osteoclastogenesis (23,28).
In addition, several studies denote that genetics and regulatory
factors regulate osteoporosis through multiple pathways, including
Wnt, Notch and the MAPK signaling pathways (29-32).
Furthermore, as reported by Xu et al (22), the cytokine network plays a crucial
role in bone resorption and formation. Considering the
participation of PIN1 in regulating a broad range of these
signaling pathways (Table I), the
role of this protein in a balance between osteoclasts and
osteoblasts could be of utmost importance. Overall, further
investigations are required to reveal the potential utility of PIN1
stabilizers, inhibitors, or activators for the treatment of
osteoporosis.
4. Cardiovascular diseases
Cardiovascular diseases (CVDs) are a global disease,
mainly related to an increasing age (21), involving a spectrum of pathological
conditions. Atherosclerosis (AS), which underlies the majority of
CVDs, is a persistent ailment that represents the primary cause of
coronary heart disease, cerebral infarction and peripheral vascular
disease (33).
The development of early-stage AS is primarily due
to endothelial dysfunction. To maintain a healthy blood pressure
and prevent AS, endothelial nitric oxide (NO) synthase (eNOS) plays
a key role by producing NO, a vasodilator and a molecule that
protects the vasculature (34). In
this manner, PIN1 has been reported to function as a conformational
switch in the modulation of the bioavailability of NO (35). Kennard et al (36) demonstrated that PIN1 regulated
bovine eNOS activity by interacting with residues Ser116-Pro117 in
a phosphorylation-dependent manner. Additionally, PIN1 has been
identified as a crucial driver of vascular cell proliferation,
apoptosis and inflammation, all implicated in various vascular
diseases, such as AS, hypertension and cardiac hypertrophy
(25,37).
Nonetheless, PIN1 regulates NO release by
interacting with other intracellular factors involved in vascular
homeostasis, such as vascular endothelial growth factor (VEGF) and
transforming growth factor β (TGF-β) (35). TGF-β is a crucial mediator of
endothelial-to-mesenchymal transition, a critical driver of
vascular inflammation and AS (38). Despite the broad array of cellular
activities, the TGF-β signaling route oversees, the mechanism is
relatively straightforward. TGF-β family ligands attach to a type
II receptor, leading to the recruitment and phosphorylation of a
type I receptor. This type I receptor subsequently phosphorylates
receptor-governed SMADs. Therefore, members of the SMAD family
gather in the cell nucleus, functioning as gene regulators and
aiding in the oversight of target gene output (39). Studies have demonstrated that PIN1
accelerates the degradation of SMAD2/3 ubiquitin proteasome brought
about by the E3 ubiquitin-protein ligase Smurf2 and impedes TGF-β
signal transduction, thereby effectively preventing the onset of AS
(35). Furthermore, Kurakula et
al (25) reported that the
inhibition of PIN1 decreased TGF-β/SMAD2/3 signaling in cultured
microvascular endothelial cells, representing a novel therapeutic
strategy with which to reverse the abnormal vascular remodeling in
pulmonary hypertension.
In addition, PIN1 is related to the activation of
the VEGF signaling pathway. This pathway has been described as a
central regulator of eNOS function (40). Regarding this, PIN1 has been
reported as a positive regulator of the transcriptional and protein
levels of VEGF by activating hypoxia-inducible factor-α and AP-1,
promoting endothelial dysfunction and hypertension (41,42).
Lastly, researchers have demonstrated a pivotal role
for PIN1 as a signaling network regulator in cardiac hypertrophy.
In the myocardial context, through AKT and MEK-ERK cascade
regulation, PIN1 activity influences signaling pathways, finally
determining the overall outcome of the heart when challenged by
hypertrophic stimulation (43).
Several studies have demonstrated that PIN1 plays a
critical role in multiple cellular processes that govern CVDs
(Table I). However, further
in-depth investigations are necessary to reveal the specific
functions of PIN1.
5. Cancer
Oncological diseases are among the most extensively
documented pathologies linked to PIN1 dysregulation. PIN1 levels
are markedly elevated in the majority of tumors, and its high
expression is negatively associated with clinical prognosis. As
previously outlined, this upregulation of active PIN1 in tumor
cells invariably results from disruptions of the transcription and
post-translation modifications governing PIN1(44). The transcriptional and
post-translational regulation of PIN1 is perturbed by various
mechanisms that increase its expression and/or hyperactivation in
cancer. Notable mechanisms include PIN1 overexpression mediated by
the aberrant activation of oncogenes, such as E2F and NOTCH1, or
alterations in tumor suppressor genes, such as BCRA1 and
p53(44).
Beyond its clinical associations, PIN1 can bind to
and modulate the fate of an extensive pool of proteins harboring
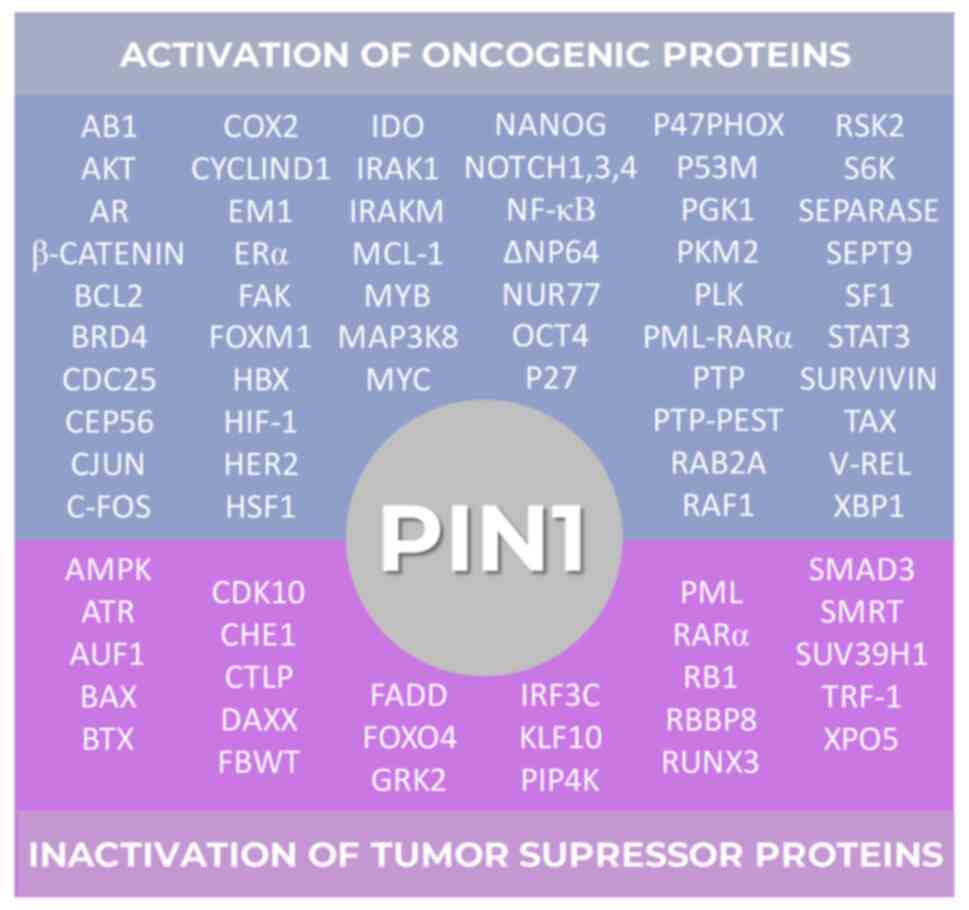
p-Ser/Thr-Pro motifs. Thus, it has been reported that PIN1 enhances
the expression and/or activation of >50 oncogenes, while
inhibiting the expression and/or activation of >25 tumor
suppressor genes (44) (Fig. 3). The intricate network of proteins
and cellular signaling pathways with which PIN1 can interact
substantiates its molecular rationale for involvement in all
pivotal cellular processes associated with tumorigenesis and
progression, as previously described by Hanahan and Weinberg
(45), such as sustained
proliferative signaling, the evasion of growth suppressors, immune
system evasion, replicative immortality, inflammation, invasion,
angiogenesis, genomic instability, resistance to cell death and the
dysregulation of cellular metabolism (46). This compilation of processes,
collectively known as cancer hallmarks, expanded in 2022 to
incorporate epigenetic reprogramming, senescent cell formation,
phenotypic plasticity and polymorphic microbiomes (47). There is suggestive evidence
pointing towards the involvement of PIN1 mainly in two of these
processes, as research has indicated its role in fostering cells
with a stem-like phenotype and in regulating epigenetic
modifications (48,49).
In this manner, this elucidates why PIN1 is
upregulated in various types of human cancer, such as breast
(50), prostate (51), lung (52), ovarian (53), gastric (54), esophageal (55), colorectal (56), cervical (57), melanoma (58) and brain tumors (59), which exhibit a heightened
expression or activation of PIN1 in comparison to their
corresponding normal tissues.
By contrast, single nucleotide polymorphisms in the
promoter region of the PIN1 gene that reduce its expression exhibit
a decrease in the susceptibility to developing multiple types of
tumors (60). Additionally, PIN1
knockout mice exhibited resistance to oncogenesis, even following
overexpression of specific oncogenes, such as HER2 and HRAS
(61) or upon the mutation or
deletion of the tumor suppressor gene TP53 (62,63).
6. Autoimmune diseases
Systemic lupus erythematosus
(SLE)
SLE is a systemic, debilitating autoimmune disease
with a variety of clinical manifestations affecting multiple organs
in the body (64). The Toll-like
receptor 7 (TLR-7)/TLR-9/interleukin (IL)-1 receptor-associated
kinase 1 (IRAK-1)/interferon regulatory factor 7 (IRF-7) pathway
plays a key role in the development and progression of SLE
(65-67).
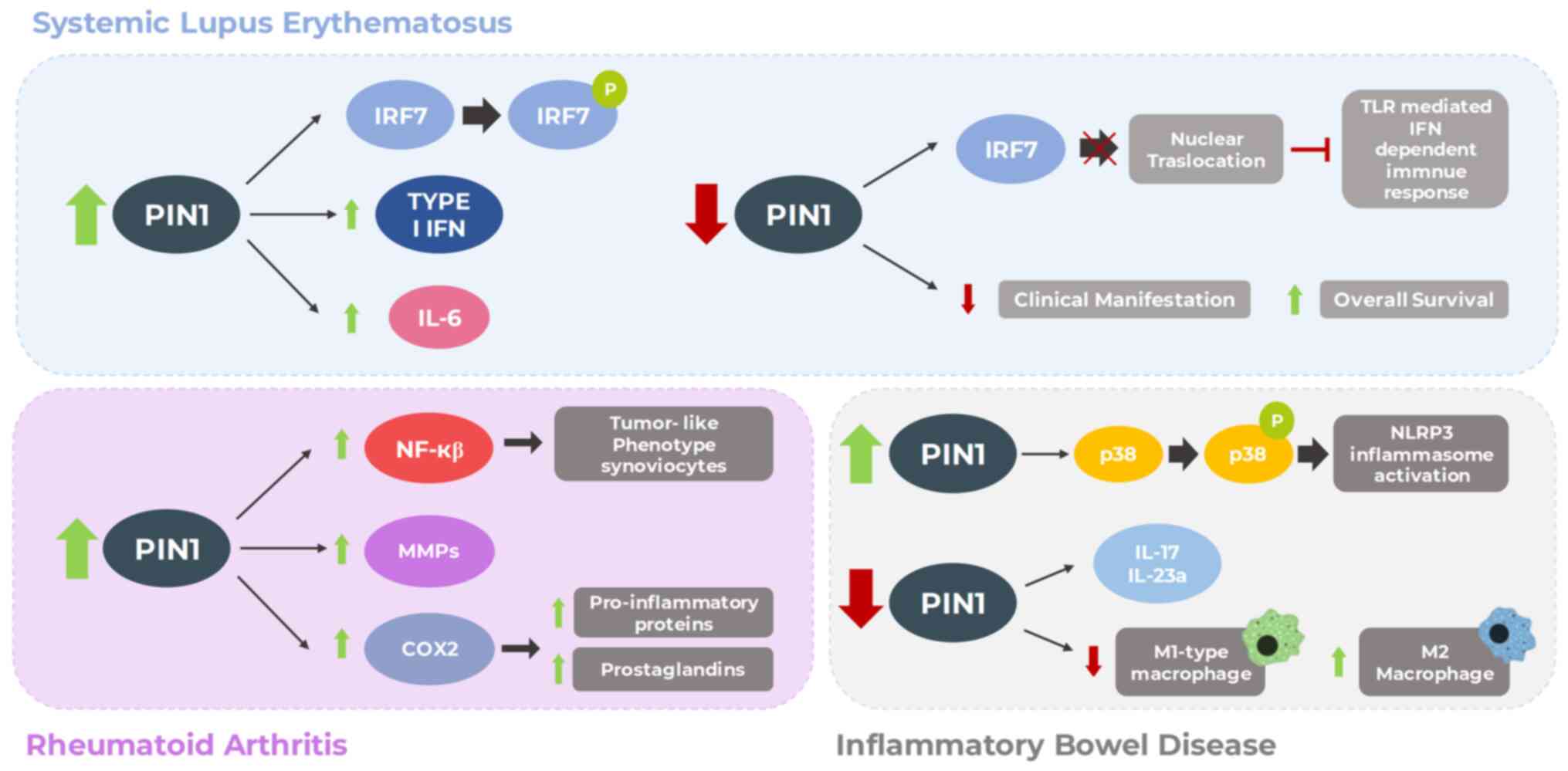
Furthermore, it has been observed that PIN1 activity is upregulated
upon the activation of TLR7 or TLR9 and interacts with IRAK1 in
primary dendritic cells. This interaction is crucial for IRAK1
activation and downstream signaling, including the activation of
IRF7 and the induction of type I interferons. Indeed, PIN1
deficiency impairs the nuclear translocation of IRF7, thereby
hampering TLR-mediated interferon-dependent immune responses
(68). In this context, it has
been demonstrated that PIN1 inhibition reduces SLE manifestations
and improves overall survival in lupus-prone animal models
(69).
In addition, IL-6, a pro-inflammatory cytokine
produced by various cell types, has emerged as a crucial regulator
in SLE. IL-6 affects the function of different cell types,
including T- and B-cells, macrophages and neutrophils by promoting
cell activation and differentiation, leading to systemic
autoimmunity and subsequent inflammatory responses (70). Multiple studies have revealed
elevated IL-6 serum levels in patients with SLE (71). In particular, the IL-6 pathway is
suspected to play an essential role in B-cell hyperactivity and
immunopathology in human SLE, and may directly mediate tissue
damage. Thus, the blockade of IL-6 has been shown to improve the
SLE phenotype in various models (72). Actually, IL-6 signaling is driven
by the IL-6 receptor and the signal transducer and activator of
transcription 3 (STAT3). It has been reported that PIN1 plays a
critical role in inducing IL-6 expression in SLE by interacting
with STAT3, promoting its phosphorylation at Ser727, and enhancing
its transcriptional activity (73)
(Fig. 4). Although further
research is required to fully elucidate the underlying mechanisms
and potential clinical applications, PIN1 inhibition emerges as a
promising novel therapeutic strategy for patients with SLE
(Fig. 4).
Rheumatoid arthritis (RA)
RA is a complex autoimmune disorder characterized by
chronic joint inflammation, the destruction of cartilage and bone,
abnormal synovial cell proliferation, and the formation of an
aggressive tumor-like structure termed pannus (74). Fibroblast-like synoviocyte samples
from patients with RA have been observed to exhibit a neoplastic
phenotype due to the activation of the NF-κB pathway (75). In addition, NF-κB, a critical
player in inflammation and immunity, is pivotal in producing matrix
metalloproteinases (MMPs) (76).
These enzymes contribute to the destructive nature of RA by
participating in cartilage destruction (77,78).
Notably, PIN1 has been reported to upregulate NF-κB activity,
thereby promoting the tumor-like phenotype of RA synoviocytes.
Indeed, an association has been found between the expression of
MMPs, NF-κB subunits and PIN1 in the synovial tissue of patients
with RA (75).
Moreover, the progression of RA is dictated by
inflammatory mediators, such as prostaglandins (PGs) and
pro-inflammatory cytokines, that orchestrate the immune response
(79,80). Cyclooxygenase 2 (COX2) controls PG
production. Notably, in a previous study, both COX2 and PIN1
expression levels were found to be increased in the ankle tissue of
mice with RA following induction with type II collagen (81). The overexpression of PIN1 led to
the increased expression of COX2. Moreover, it was shown that
during the progression of RA, PIN1 induction stimulated the
expression of pro-inflammatory proteins, such as iNOS, TNF-α and
IL-1β by activating NF-κB, CREB and the C/EBP pathways.
Consistently, PIN1 inhibition significantly reduced RA progression
and COX2 expression in ankle tissues (81) (Fig.
4). Given the complex interplay of PIN1 and its impact on RA,
further research is essential to uncover its potential role as a
therapeutic target for the treatment of RA.
Inflammatory bowel diseases (IBDs):
Crohn's disease and ulcerative colitis (UC)
IBDs are chronic relapsing disorders encompassing
Crohn's disease and UC affecting the colon and small intestine.
Symptoms range from chronic diarrhea and abdominal pain to
inflammation and epithelial injury involving the gastrointestinal
tract. Although the etiology of IBDs remains unknown, it has been
proposed that intestinal mucosa damage occurs due to the
dysregulated abnormal immune response against the microorganisms of
the intestinal flora (82).
Matsunaga et al (83) suggested the contribution of PIN1 to
the development and severity of IBDs. Indeed, its expression was
shown to be markedly enhanced in the colons of model mice with
dextran sodium sulfate (DSS)-induced UC. A PIN1 knockout mouse
model consistently exhibited significantly reduced DSS-induced
colitis symptoms and colon tissue damage. In the colons of PIN1
knockout mice, its inhibition decreased M1-type macrophages, known
for their pro-inflammatory cytokine expression (83). Conversely, M2 macrophages, a
producer of anti-inflammatory cytokines, exhibited an increased
percentage. Furthermore, PIN1 inhibition was associated with the
downregulation of IL-17 and IL-23a expression, cytokines implicated
in IBD development (83).
Of note, the NOD-like receptor family pyrin
domain-containing protein 3 (NLRP3) is exclusively expressed in
several inflammatory and autoimmune diseases. This innate immune
receptor triggers the inflammasome complex assembly, which has been
reported to be associated with various inflammation- and
immune-related diseases that are a key factor in the pathogenesis
and progression of UC and Crohn's disease. Indeed, the
overexpression, aberrant activation, polymorphism and
gain-of-function mutation of the NLRP3 inflammasome contribute to
IBD pathogenesis (84).
Additionally, it has been suggested that PIN1 promotes NLRP3
inflammasome activation through the phosphorylation of the p38 MAPK
signaling pathway in macrophages (85) (Fig.
4). A comprehensive understanding of the role of PIN1 in
activating NLRP3 necessitates further research on IBDs.
Nonetheless, additional exploration is necessary to
unveil its potential as a therapeutic focal point for addressing
RA.
7. Viral infections
Viruses are prevalent infectious agents leading to
contagious diseases. Upon a viral attack, the host triggers its
innate immune response to combat or eliminate the infection
(86). However, specific proteins
within the host assist the virus in minimizing the host's
resistance or facilitating the viral infectivity process. In this
framework, PIN1 has been identified as one of these proteins that
share a strong association with viral infections (Table II) (87).
 | Table IIMain PIN1-regulated targets and their
effects on viral infection. |
Table II
Main PIN1-regulated targets and their
effects on viral infection.
| | Target | Effect | (Refs.) |
|---|
| Hepatitis C
virus | p-Ser/Thr-Pro
motifs of NS5A and NS5B. | Enhancing viral
replication and propagation | (89) |
| Hepatitis B
virus | p-Ser41-Pro motif
of HBx. | Promoting
development of HCC | (91,92) |
| Epstein-Barr
virus | p-Thr78-Pro motif
of BALF5. | Enhancing viral DNA
replication | (99) |
| Human T-cell
leukemia virus type I | p-Ser160-Pro motif
of Tax. | NF-кB activation,
promoting cancer progression. | (106) |
| High-risk human
papillomavirus | NF-кB and
STAT3. | Promoting
development of cancer. | (48,111) |
| Human
immunodeficiency virus | p-Ser16-Pro motif
of HIV capsid | Discarding capside
proteins. | (117,118) |
| | protein. Host
protein A3G. p-Ser57-Pro | Enhancing reverse
transcription of | (116) |
| | motif of HIV
integrase. | HIV genome
Promoting HIV cDNA integration. | (119) |
| Severe acute
respiratory syndrome coronavirus-2 | p-Ser79-Pro motif
of the N protein. | Not yet
described. | |
Hepatitis C virus (HCV)
HCV is the primary infectious agent of acute and
chronic hepatitis, which can eventually lead to permanent liver
damage and hepatocellular carcinoma (HCC). HCV, a sheathed RNA
virus, has its reproduction reliant primarily on the host cell
cycle and requires the involvement of host proteins (88). In particular, PIN1 is a cellular
factor required for HCV replication, and it may enhance the viral
infection principally due to its interaction with both the NS5A and
NS5B HCV proteins. Specifically, PIN1 interacts with the
p-Ser/Thr-Pro motifs of NS5A and NS5B, stabilizing them. Notably,
Lim et al (89)
demonstrated that increased levels of PIN1 were associated with
higher levels of intracellular HCV RNA and these viral
proteins.
By contrast, when PIN1 is inhibited, it prevents the
propagation of HCV by disrupting the interaction between PIN1 and
these proteins. Furthermore, NS5B can increase the expression of
PIN1, thus promoting its spread (89). Therefore, PIN1 may be exploited as
a host target to impair HCV reproduction and infection, emerging as
a potential target for antiviral therapies.
Hepatitis B virus (HBV)
HBV is a virus that can lead to chronic hepatitis B,
causing cirrhosis, liver failure and HCC. Its genome in the viral
capsid is relaxed circular DNA, and when it enters the hepatocyte
nucleus, it is converted to covalently closed circular DNA
(cccDNA). The transcription of the viral cccDNA is modulated by the
HBV-encoded protein, HBx (90). Of
note, there is evidence to indicate that PIN1 is overexpressed in
HBV-related HCC and that HBx comprises two p-Ser-Pro motifs that
can serve as potential targets for PIN1. Actually, PIN1 binds to
HBx through its Ser41-Pro motif, enhancing its stability and
transactivation, and thereby promoting hepatocarcinogenesis
(91,92).
HBx-activated AKT and ERKs also phosphorylate and
inactivate glycogen synthase kinase-3β (GSK-3β), leading to the
stabilization of β-catenin and cyclin D1 gene transcription, and
promoting the development of HCC (93). The overexpression of PIN1 not only
elevates the expression of cyclin D1, but also encourages the
intracellular buildup of β-catenin in the Wnt/β-catenin signaling
pathway. These two signaling pathways can trigger the expression of
oncogenes and encourage the occurrence of HCC in HBV infection
(94).
In addition, HBV is formed with a core particle
composed of multiple HBV core protein (HBc) molecules. Notably,
PIN1 has been observed to interact with HBc at the p-Thr160-Pro and
p-Ser162-Pro motifs and promote its stability to sustain efficient
HBV replication (95). Conversely,
Kwon et al (96) recently
reported that PIN1 interacts with HBV core particles, but not HBc
dimer or monomer. Moreover, they posited PIN1 as a positive
regulator of HBV propagation and that the interaction between PIN1
and the core particle may be involved in HBV-associated
hepatocarcinogenesis., supported by the observation that PIN1
overexpression enhances, while its knockdown reduces it (96).
Epstein-Barr virus (EBV)
EBV, a highly prevalent virus, infects 95% of
individuals worldwide at some point. While often asymptomatic, some
develop infectious mononucleosis (97). Additionally, an association exists
between EBV infection and Burkitt lymphoma and T-cell malignancies
(98). Considering the main
processes in EBV infection, one of the interesting aspects to
analyze is the role of PIN1 in replication. By performing PIN1
knockdown, Narita et al (99) reported that PIN1 promoted viral DNA
replication through its interaction with the p-Thr178-Pro motif in
the EBV DNA polymerase subunit.
In addition, EBV infection is closely related to
nasopharyngeal carcinoma (NPC). The majority of cases of NPC
exhibit a distinctive feature: They are nonkeratinizing carcinomas
associated with EBV infection (100). In this context, previous studies
have demonstrated that PIN1 is overexpressed in all EBV-associated
NPC cells, contributing to the growth and aggressiveness of cancer
(101,102).
Human T-cell leukemia virus type
1
Adult T-cell leukemia/lymphoma is an uncommon
malignancy caused by human T-cell lymphotropic virus type I
(HTLV-1) (103). The HTLV-1
encodes for an oncoprotein known as Tax and this plays a crucial
role in viral gene expression. This protein influences NF-κB
signaling pathways, perturbing the normal cell cycle, interfering
with apoptosis and prompting genomic instability (104). Tax interacts with a number of
human cellular proteins to regulate viral gene expression and
foster the pathogenic activation of signaling pathways, such as
NF-κB (105). Notably, PIN1 is
highly expressed in adult T-cell leukemia cells expressing Tax
protein (106). By triggering the
E2F/RB pathway, TAX enhances the expression of PIN1. Furthermore,
PIN1 binds to Tax when phosphorylated in the p-Ser160-Pro motif in
the presence of mitotic kinases (106). Due to PIN1 regulation,
phosphorylated Tax interacts with IKKγ to enhance NF-κB activation,
promoting cancer progression (107). Furthermore, PIN1 can determine
Tax stability by regulating its ubiquitination and lysosomal
degradation (106).
High-risk human papillomavirus
(HR-HPV)
HR-HPV can cause cancers of the cervix, vagina,
vulva, penis, anus and the back of the throat, including the base
of the tongue and tonsils (oropharynx), in both males and females
(108). The most frequent type of
cancer caused by HR-HPV is cervical cancer, with an annual
incidence of >500,000 new cases worldwide (109). Of note, ~14 HR-HPV types are
responsible for the majority of cervical cancers, mainly HPV16 and
HPV18. The viral protein E2 is a determinant factor that manages
viral replication and transcription, and it is employed as an early
indicator of HPV infection (110). Notably, HPV-infected cervical
lesions exhibit an elevated level of PIN1(44). The increased expression of PIN1 in
cervical cancer can enhance the nuclear sequestration of NF-κB
(46) and stimulate the
transactivation of STAT3, thereby advancing the onset of cancer
(48,111). Even a slight, yet significant
increase in PIN1 expression has been observed in E2-transfected 293
cells; E2 has been reported to amplify the functionality of PIN1.
This evidence suggests that PIN1 activity is addressed by the E2
modulation of the transcription factors, NF-κB and STAT3, driving
thus cancer progression (112).
Human immunodeficiency virus
(HIV)
HIV is a lentivirus that targets predominantly
CD4+ T-lymphocytes and macrophages. Since these T-cells
are the regulators of the adaptive immune system, their depletion
effectively weakens the immune system, leading to acquired immune
deficiency syndrome (AIDS) (113). Several host factors have been
shown to play an essential role in the HIV life cycle (114). PIN1 is one of these factors,
enhancing HIV infection by being involved in three vital stages of
the HIV replication cycle: Cell entry, reverse transcription and
host genome integration (115,116). The HIV core depends on PIN1 to
discard capsid proteins. PIN1 links to the p-Ser16-Pro17 motif of
the HIV capsid protein, reorganizing its structure and removing the
capsid from the HIV core (117,118). Furthermore, PIN1 eases the
reverse transcription of the HIV genome. The host protein A3G can
be packaged into viral particles and induces alterations in DNA
during HIV genome reverse transcription. HIV-1 expresses Vif
protein to resist the activity of A3G by mediating A3G degradation.
Regarding this, PIN1 diminishes A3G expression and prevents A3G
from entering HIV particles (87,119). Finally, Saleh et al
(116) reported that PIN1 bound
to the HIV integrase through its pSer57-Pro motif, thus stabilizing
and promoting integrase activity. Consequently, PIN1 improved the
insertion of the HIV cDNA into the host genome (116).
Severe acute respiratory syndrome
coronavirus 2 (SARS-CoV-2)
The unprecedented outbreak of novel coronavirus
disease 2019 (COVID-19) emerged as a worldwide pandemic, causing
profound effects on societies across the globe. SARS-CoV-2 is a
highly transmissible and pathogenic enveloped, plus-stranded RNA
virus with a single-stranded RNA genome (120). Recently, it was established that
PIN1 plays a crucial role as a cellular component essential for the
propagation of SARS-CoV-2. Yamamotoya et al (121), by using the siRNA-mediated
silencing of PIN1 expression, significantly reduced the
proliferation of SARS-CoV-2 in VeroE6/TMPRSS2 cells. Several
recently developed PIN1 inhibitors exhibited potent suppressive
effects on SARS-CoV-2 proliferation, as evidenced by reductions in
both viral mRNA and protein synthesis, leading to the mitigation of
the cytopathic effect on VeroE6/TMPRSS2 cells (121). Furthermore, Ino et al
(122) reported that PIN1 bound
precisely to the p-Ser79-Pro motif of the SARS-CoV-2 N protein.
However, further in-depth investigations are warranted to elucidate
the effects of this interaction on the viral progression. One
particular compound, H-77, has demonstrated the ability to block
SARS-CoV-2 proliferation with an EC50 <5 µM, regardless of
whether it is administered to the culture medium before or
following SARS-CoV-2 infection (121). The inhibition of viral N protein
mRNA synthesis by H-77 suggests that the underlying molecular
mechanism responsible for SARS-CoV-2 suppression likely involves
viral gene transcription or earlier stages of infection (121). Another PIN1 inhibitor, all-trans
retinoic acid, known to activate the retinoic acid receptor while
inhibiting PIN1 activity, also reduced SARS-CoV-2 proliferation.
These findings collectively suggest that PIN1 inhibitors are
promising therapeutic agents for combatting COVID-19(121).
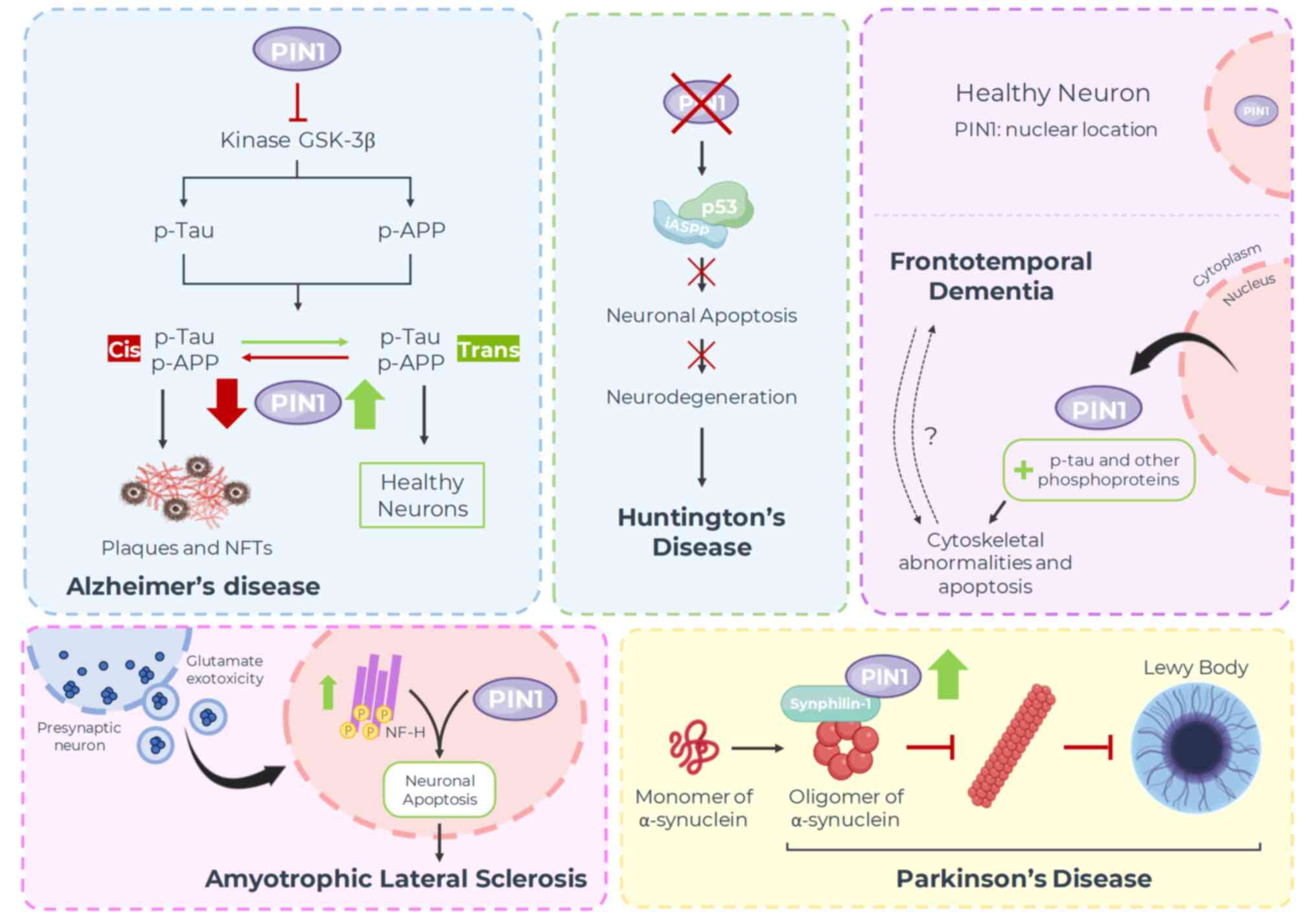
8. Neurodegenerative diseases
Apart from its role in bodily functions, an
increasing amount of data point to the vital participation of PIN1
in neurodegenerative disorders, such as Alzheimer's disease (AD),
Parkinson's disease (PD), frontotemporal dementia (FTD),
Huntington's disease (HD) and amyotrophic lateral sclerosis (ALS).
It exhibits a broad spectrum of impacts in these conditions, from
safeguarding nerve cells to causing them harm (Fig. 5).
PD
PD is recognized as the most common
neurodegenerative movement disorder. It affects multiple systems,
entailing a variety of motor and non-motor symptoms.
Pathologically, Lewy bodies (LBs) are the characteristic protein
conglomerates in tissues of PD, spanning from the gastrointestinal
tract to the neocortex (123).
LBs are primarily composed of α-synuclein (124), which is an unstructured protein
in its natural state, but can be induced to form an indissoluble
α-synuclein cluster in the pathological condition (125). Synphilin-1 is a protein that can
interact with α-synuclein, leading to the formation of debris
inclusion bodies in the cytoplasm; this interaction plays a central
role in creating LBs (126). Due
to the absence of a p-Ser/Thr-Pro pattern in α-synuclein, PIN1 does
not bind directly to free molecules, but influences α-synuclein
through indirect mechanisms (127). When synphilin-1 is
phosphorylated, PIN1 can bind through Ser211-Pro and Ser215-Pro
patterns, thus indirectly interacting with α-synuclein. The
increased expression of PIN1 can impede the degradation of
α-synuclein, prolong its half-life, promote its insolubility and
contribute to the formation of debris inclusion bodies (128). Hence, it can be speculated that
inhibitors targeting PIN1 might alleviate the progression of
PD.
AD
AD is a progressive neurodegenerative disorder
primarily affecting cognitive functions, particularly memory,
thinking and behavior. It is the most common cause of dementia
among older adults (129). The
disease is characterized by the abnormal accumulation of protein
aggregates, including plaques and intracellular neurofibrillary
tangles (NFTs). NFTs are clumps of microtubules formed due to the
excessive phosphorylation of Tau protein (130). Extracellular plaques mainly
consist of aggregates of amyloid-β-peptides resulting from the
increased processing of amyloid precursor protein (APP) (131). In the neuronal cells of patients
with AD, PIN1 expression is typically downregulated and
demonstrates an inverse association with the degeneration of
neuronal fibers. In addition, PIN1 facilitates the conformational
conversion of GSK-3β-mediated phosphorylated Tau proteins from the
dysfunctional cis form to the functional trans form,
thus promoting the degradation of Tau proteins (132). Furthermore, PIN1 induces the
phosphorylation of APP Thr668-Pro, transitioning it to trans
isomer and redirecting APP processing towards non-amyloidogenic
pathways (1). PIN1 can directly
impede the activation of GSK-3β by binding to the phosphorylated
Thr330-Pro motif and catalyzing its isomerization (133). There is evidence to suggest that
a decrease in PIN1 expression in the brain generally increases
susceptibility to AD and, on the other hand, its overexpression in
mature neurons can protect against neurodegeneration caused by Tau
hyperphosphorylation (134).
HD
HD is a progressive neurodegenerative disorder
characterized by abnormal and repetitive expansions within exon 1
of the gene encoding CAG in the huntingtin protein (HTT),
responsible for the degeneration of striatal neurons in the brain
(135). The mutated huntingtin
protein (mHTT) forms an intranuclear inclusion through misfolding
and aggregation (136), which is
harmful and it leads to glial proliferation of astrocytes and
selective loss of striatal neurons (137). Additionally, it can induce the
DNA damage response (DDR) in neurons (138), a significant pathological
characteristic of HD. Research has discovered that p53 mediates
this cytotoxicity in HD cells and transgenic animal models, and its
inhibitors hinder this process (138). Conversely, when PIN1 is silenced,
p53 binds to iASPP (a critical inhibitor oncoprotein of p53)
regardless of mHTT expression, resulting in the failure to induce
apoptosis and consequently preventing mHTT-related
neurodegeneration (139).
Furthermore, PIN1 is also implicated in DDR and the regulation of
DNA double-strand break repair (140).
FTD
FTD is a clinical disorder associated with the
neurodegeneration of the cortex of the frontal and temporal lobes,
often in conjunction with the degeneration of subcortical brain
areas. In consequence, behavior, executive function, or language
are compromised. FTD is caused by genetic mutations, environmental
factors and protein abnormalities (141). Recently, researchers have
observed decreases in PIN1 levels across various forms of FTD.
These observations have not only been observed in FTD cases with
Tau pathology, such as FTD with Tau mutation, Pick disease and
cortico-basal degeneration, but also in cases without Tau
pathology, such as FTD with motor neuron-type inclusions and
neuronal intermediate filament inclusion disease (134).
Additionally, in neurons sourced from the middle
frontal gyrus, a shift of PIN1 from the nucleus to the cytoplasm
has been identified in all FTD cases, as opposed to typical brains,
which conversely exhibit a primary nuclear localization of
PIN1(142). Research has
demonstrated that the anomalous detected translocation of the
mitotic regulator PIN1 depends on the presence of p-Tau, along with
the elevated quantity of other target phosphoproteins in neuronal
cytoplasm, such as mitotic phospho-epitopes and cell
cycle–associated proteins (143).
A buildup of these proteins has been noted in diverse pathological
contexts (e.g., AD, FTD, parkinsonism linked to chromosome 17,
progressive supranuclear palsy and cortico-basal degeneration) and
described as signs of the interrupted mitotic process causing
cytoskeletal abnormalities and neuronal apoptosis (144). However, additional studies are
required to determine whether PIN1 translocation to the cytoplasm
represents an early event driving the neurodegenerative processes
or if it emerges due to FTD.
ALS
ALS is a nerve-deteriorating disease affecting the
upper and lower motor neurons, resulting in the paralysis of
voluntary muscles, swallowing difficulties, speech impairment and
respiratory collapse (145).
Recently, Iridoy et al (146) demonstrated a notable
downregulation in PIN1 expression in the spinal cord and non-motor
cortex of a select group of patients with ALS, signifying PIN1
expression as a potential neurodegeneration indicator. Nonetheless,
the existing understanding of the expression profile of PIN1 in ALS
is exceedingly limited, as is its role in the pathophysiology of
the disease. The literature suggests it may encourage the abnormal
piling up of phosphorylated neurofilament proteins in the
perikaryon, a significant characteristic of ALS and other
nerve-deteriorating diseases. Consistently, PIN1 has been reported
to link with phosphorylated neurofilament heavy chain (NF-H) in
neurons and to co-locate in ALS-impacted spinal cord neuronal
inclusions (147). In rat dorsal
root ganglion cultures, exposed to excitotoxic stress to provoke
the accumulation of phosphorylated NF-H within the cell body to
imitate neurodegeneration, glutamate-induced harm has been shown to
heighten phosphorylated NF-H in perikaryal accumulations that
co-localized with PIN1 and induce neuronal apoptosis. These effects
are mitigated through pharmacological intervention or
siRNA-mediated reduction of PIN1 expression, implying that in the
face of neurotoxic stress, PIN1 can potentially promote cell death
by triggering the assembly of phosphorylated NF-H in the perikaryal
(148).
9. PIN1 inhibitors
Given the involvement of PIN1 in a wide range of
pathological conditions, pharmaceutical firms and several academic
research teams have directed their efforts toward formulating novel
strategies for PIN1 inhibition, aiming to exploit the therapeutic
promise offered by these inhibitors.
Some natural compounds that have exhibited the
ability to bind to PIN1 were identified in the past. Juglone, or
5-hydroxy-1,4-naphthoquinone, was the first of these to be
described (149). This molecule,
produced by the walnut tree, functions as an inactivator (not an
inhibitor), forming Michael adducts with Cys113 of PIN1. The
PIN1-juglone complex has a reduced stability within cells, and is
ubiquitinated and rapidly degraded via the proteasome pathway,
rendering it inappropriate for commercialization (149). Another natural compound that can
bind to PIN1 is epigallocatechin-3-gallate (EGCG) (150). This compound is the main
flavonoid in green tea and has been shown to exhibit antitumor
activity. X-ray information obtained from the co-structures has
indicted that EGCG can bind to both the WW and PPIase domains.
However, subsequent analyses have indicated that its PIN1
inhibitory activity is due to binding to the WW domain (150).
The study of the complexes that these inhibitors
form with PIN1 has provided critical information about the
structural biology of this protein, its interactions and the
identification of possible key amino acids for its function. The
first developments of specific PIN1 inhibitors were based on the
information provided by these natural compounds. However, a number
of molecules with different chemical compositions have been
developed over the past decades. These compounds can be broadly
divided according to whether they bind to the PPIase or WW
domains.
PPIase inhibitors
Since the PPI domain is the catalytic domain that
provides PIN1 with its isomerase activity, it is not surprising
that the vast majority of efforts to find a specific inhibitor of
PIN1 have focused on this region of the protein. Below, the
developments that were consider most relevant, grouped according to
the chemical nature of the inhibitors are described.
Peptides
The primary strategy used to develop PIN1
inhibitors was based on peptide molecules. In 2005, Bayer et
al (151) developed a library
of synthetic peptides inhibiting PIN1 using the natural peptide
pepticinamin E as the base structure. Thereafter, various
developments of PIN1 peptide inhibitors emerged with different
rational modifications to improve the binding capacity to this
protein and, at the same time, problems caused by the nature of
this type of molecule, such as its low-cell permeability and high
degradation, highlighting the development of modified cyclic
peptides that substantially increase their stability and ability to
cross cell membranes. These peptides were shown to exert an
anti-proliferative effect in vitro on HeLa cells (152). It should be noted that although
the translation of these peptides to PIN1 inhibitor drugs is
extremely complex, all these efforts and research provided a large
volume of information that allowed for the characterization of
different mechanisms to inhibit the PPI domain, which were
elegantly reviewed by He et al (153).
Small molecules
As regards the evaluation of small molecules
derived from drug design based on the structure of PPI domain
inhibitors, in 2009, Pfizer searched PIN1 inhibitors with a massive
analysis of more than one million compounds. Although several
candidate compounds were obtained, orthogonal assays for biological
activity or co-crystallization were not performed (154).
In a recent study, Russo Spena et al
(155) searched for PIN1
inhibitors using a library of 35,000 low-molecular-weight compounds
through virtual screening based on docking, targeting the PPI
domain. This strategy was complemented with molecular dynamics
simulations to reduce the number of candidates. Among these, they
identified a compound that demonstrated binding to PIN1, and
simultaneously, this compound exhibited antiproliferative effects
in four different ovarian cancer cell models. It is worth noting
that treatments with the candidate compound also reduced signaling
pathways associated with PIN1(155).
Covalent inhibitors
Covalent inhibitors are a group of chemical
compounds that establish permanent bonds with specific proteins,
thereby modifying their function and regulation. These inhibitors
are highly valued in research and drug development due to their
exceptional selectivity and potency. However, they also present
challenges in terms of design and the potential for unintended
side-effects due to off target nonspecific irreversible
binding.
The first PIN1 covalent inhibitor described after
the natural compound Juglone was KPT-6566. This inhibitor obtained
by mechanism-based screening can selectively inhibit PIN1 and
target it for degradation by binding to the PPI domain. This
interaction induces cell death in cancer cells in vitro and
reduces the growth of lung metastasis in vivo (156).
In a recent study, Liu et al (157) screened using an in-house library
of 2,000 electrophilic compounds. Following a series of structural
optimizations, they successfully obtained a more potent compound
named ZL-Pin13. The covalent binding of this compound with PIN1 was
confirmed through crystallography. This compound exerted
anti-proliferative effects in breast cancer cell models with IC50
values <3 µM. It also reduced the levels of proteins associated
with PIN1, such as Cmyc, Mcl-1 and cyclin D1(157).
WW inhibitors
The WW domain is essential, granting PIN1 the
unique ability to specifically recognize and bind to pSer/Thr-Pro
motifs of its target proteins (158). It is important to note that
recent research has highlighted the critical importance of the WW
site for the proper functioning of the PIN1 protein:
The PIN1 WW domain is phosphorylated on Ser16 both
in vitro and in vivo. This phosphorylation regulates
the ability of the WW domain to mediate Pin1 substrate interaction.
Thus, Ser16 may be critical for regulating WW domain binding
activity and Pin1 function (159).
On the other hand, it has been demonstrated that a
mutant with the Trp34Ala mutation, which is a highly conserved
amino acid in the WW domain of PIN1, experiences a 20-fold
reduction in its binding affinity for substrates due to sustained
inter-domain contact (160).
Consequently, the catalytic pocket remains in a suboptimal
conformation and exhibits diminished enzymatic activity (161).
Despite the crucial role of this domain, efforts to
develop specific inhibitors targeting WW have been notably limited
compared to those directed toward the catalytic PPI domain of PIN1.
Nonetheless, it is considered that the flat and shallow interface
of the WW domain does not provide a favorable starting point for
the rational design of potent PIN1 inhibitors (153).
The most prominent example of an inhibitor designed
for the WW domain of PIN1 is a semi-synthetic compound derived from
the natural compound, acetyl-11-keto-β-boswellic acid. This
inhibitor been shown to exert anti-proliferative and pro-apoptotic
effects in prostate cancer cells, underscoring the therapeutic
potential of targeting this domain (162).
In this regard, the authors' research team has
achieved significant progress by embarking on the first de
novo design of inhibitors explicitly tailored to bind to the
PIN1 WW domain. The authors previously conducted structural
analyses of Trp34 and its vicinity within the WW domain. Through
this series of analyses, a novel allosteric region was identified
within this domain, constituting a novel pharmacological pocket. By
employing a combination of in silico virtual screening based
on docking and in vitro biophysical assessments, four small
molecules out of 450,000 compounds were successfully identified,
that exhibited a high affinity for PIN1(163).
Their distinctiveness compared to other PIN1
inhibitors lies in their exclusive and specific targeting of the WW
region, in contrast to other inhibitors that interact with the
conserved catalytic PPI domain of PIN1, a feature shared by other
peptidyl prolyl isomerases. These efforts represent an essential
step in expanding therapeutic options for addressing pathologies
associated with the PIN1 protein and highlight the
as-yet-unexplored potential of this domain as a therapeutic
target.
10. Conclusions and future perspectives
In conclusion, the present review has contributed
to a better understanding of the implications and potential
functions of PIN1 in multiple ailments. The majority of
pharmaceutical products developed to target PIN1 are focused on
cancer therapy, considering its upregulation in cancerous tissues
and its role in tumor progression. Conversely, there is a notable
lack of research and application of PIN1 inhibitors and agonists in
other pathologies, indicating the need for more comprehensive and
more profound studies to unveil the therapeutic potential of PIN1
modulators. Several investigations strongly suggest that upstream
regulatory signals and downstream targets of PIN1 establish an
attractive field that has not yet been fully explored, which may
provide new insight into the treatment of the multiple pathologies
reviewed herein.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Agencia Argentina
para la Promoción Científica y Tecnológica (ANPCyT) (grant/award
nos. PICT-2018 N° 2372; PICT-START UP 2020 N°0001; PICT-INVI-2021
N°00036; and PICT-GRFI-2021 N°00029) and by Universidad Nacional de
Quilmes (grant/award no. PUNQ N°2283/22).
Availability of data and materials
Not applicable.
Authors' contributions
JM wrote the ‘Introduction’, ‘Cancer’ and ‘PIN1
inhibitors’ Sections. RGA wrote the ‘Cardiovascular diseases’,
‘Metabolic diseases’ and ‘Osteoporosis’ sections. LB wrote the
‘Autoimmune diseases’ section. RNV wrote the ‘Neurodegenerative
diseases’ section. MDPC and DLMG wrote the ‘Viral infections’
section. RGA and LB prepared the figures and tables. DEG and DLMG
functioned as co-supervisors and were also involved in the writing
of the sections. DEG conceived the study and, compiled and
corrected the final version of the manuscript. All authors have
read and approved the final manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
Not applicable.
References
|
1
|
Liou YC, Zhou XZ and Lu KP: Prolyl
isomerase Pin1 as a molecular switch to determine the fate of
phosphoproteins. Trends Biochem Sci. 36:501–514. 2011.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Alvarez E, Northwood IC, Gonzalez FA,
Latour DA, Seth A, Abate C, Curran T and Davis RJ:
Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate
protein phosphorylation. Characterization of the phosphorylation of
c-myc and c-jun proteins by an epidermal growth factor receptor
threonine 669 protein kinase. J Biol Chem. 266:15277–15285.
1991.PubMed/NCBI
|
|
3
|
Gurung D, Danielson JA, Tasnim A, Zhang
JT, Zou Y and Liu JY: Proline Isomerization: From the chemistry and
biology to therapeutic opportunities. Biology (Basel).
12(1008)2023.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lu KP, Hanes SD and Hunter T: A human
peptidyl-prolyl isomerase essential for regulation of mitosis.
Nature. 380:544–547. 1996.PubMed/NCBI View Article : Google Scholar
|
|
5
|
El Boustani M, De Stefano L, Caligiuri I,
Mouawad N, Granchi C, Canzonieri V, Tuccinardi T, Giordano A and
Rizzolio F: A Guide to PIN1 function and mutations across cancers.
Front Pharmacol. 9(1477)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Rustighi A, Tiberi L, Soldano A, Napoli M,
Nuciforo P, Rosato A, Kaplan F, Capobianco A, Pece S, Di Fiore PP
and Del Sal G: The prolyl-isomerase Pin1 is a Notch1 target that
enhances Notch1 activation in cancer. Nat Cell Biol. 11:133–142.
2009.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ryo A, Liou YC, Wulf G, Nakamura M, Lee SW
and Lu KP: PIN1 is an E2F target gene essential for Neu/Ras-induced
transformation of mammary epithelial cells. Mol Cell Biol.
22:5281–5295. 2002.PubMed/NCBI View Article : Google Scholar
|
|
8
|
MacLachlan TK, Somasundaram K, Sgagias M,
Shifman Y, Muschel RJ, Cowan KH and El-Deiry WS: BRCA1 effects on
the cell cycle and the DNA damage response are linked to altered
gene expression. J Biol Chem. 275:2777–2785. 2000.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Luo ML, Gong C, Chen CH, Lee DY, Hu H,
Huang P, Yao Y, Guo W, Reinhardt F, Wulf G, et al: Prolyl isomerase
Pin1 acts downstream of miR200c to promote cancer stem-like cell
traits in breast cancer. Cancer Res. 74:3603–3616. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Eckerdt F, Yuan J, Saxena K, Martin B,
Kappel S, Lindenau C, Kramer A, Naumann S, Daum S, Fischer G, et
al: Polo-like kinase 1-mediated phosphorylation stabilizes Pin1 by
inhibiting its ubiquitination in human cells. J Biol Chem.
280:36575–36583. 2005.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen CH, Chang CC, Lee TH, Luo M, Huang P,
Liao PH, Wei S, Li FA, Chen RH, Zhou XZ, et al: SENP1 deSUMOylates
and regulates Pin1 protein activity and cellular function. Cancer
Res. 73:3951–3962. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hamdane M, Dourlen P, Bretteville A, Sambo
AV, Ferreira S, Ando K, Kerdraon O, Bégard S, Geay L, Lippens G, et
al: Pin1 allows for differential Tau dephosphorylation in neuronal
cells. Mol Cell Neurosci. 32:155–160. 2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liou YC, Sun A, Ryo A, Zhou XZ, Yu ZX,
Huang HK, Uchida T, Bronson R, Bing G, Li X, et al: Role of the
prolyl isomerase Pin1 in protecting against age-dependent
neurodegeneration. Nature. 424:556–561. 2003.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fahed G, Aoun L, Bou Zerdan M, Allam S,
Bou Zerdan M, Bouferraa Y and Assi HI: Metabolic Syndrome: Updates
on pathophysiology and management in 2021. Int J Mol Sci.
23(786)2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Saltiel AR: Insulin signaling in health
and disease. J Clin Invest. 131(e142241)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Nakatsu Y, Mori K, Matsunaga Y, Yamamotoya
T, Ueda K, Inoue Y, Mitsuzaki-Miyoshi K, Sakoda H, Fujishiro M,
Yamaguchi S, et al: The prolyl isomerase Pin1 increases β-cell
proliferation and enhances insulin secretion. J Biol Chem.
292:11886–11895. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Inoue MK, Nakatsu Y, Yamamotoya T, Hasei
S, Kanamoto M, Naitou M, Matsunaga Y, Sakoda H, Fujishiro M, Ono H,
et al: Pin1 plays essential roles in NASH development by modulating
multiple target proteins. Cells. 8(1545)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Han Y, Lee SH, Bahn M, Yeo CY and Lee KY:
Pin1 enhances adipocyte differentiation by positively regulating
the transcriptional activity of PPARγ. Mol Cell Endocrinol.
436:150–158. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Nakatsu Y and Asano T: Prolyl isomerase
Pin1 impacts on metabolism in muscle and adipocytes. Yakugaku
Zasshi. 142:449–456. 2022.PubMed/NCBI View Article : Google Scholar : (In Japanese).
|
|
20
|
Kanna M, Nakatsu Y, Yamamotoya T,
Kushiyama A, Fujishiro M, Sakoda H, Ono H, Arihiro K and Asano T:
Hepatic Pin1 expression, particularly in nuclei, is increased in
NASH patients in accordance with evidence of the role of Pin1 in
lipid accumulation shown in hepatoma cell lines. Int J Mol Sci.
24(8847)2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Azeez TA: Osteoporosis and cardiovascular
disease: A review. Mol Biol Rep. 50:1753–1763. 2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Xu J, Yu L, Liu F, Wan L and Deng Z: The
effect of cytokines on osteoblasts and osteoclasts in bone
remodeling in osteoporosis: A review. Front Immunol.
14(1222129)2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Islam R, Yoon WJ and Ryoo HM: Pin1, the
master orchestrator of bone cell differentiation. J Cell Physiol.
232:2339–2347. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Park KR, Kim S, Cho M, Kang SW and Yun HM:
Effects of PIN on osteoblast differentiation and matrix
mineralization through runt-related transcription factor. Int J Mol
Sci. 21(9579)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kurakula K, Hagdorn QAJ, van der Feen DE,
Vonk Noordegraaf A, Ten Dijke P, de Boer RA, Bogaard HJ, Goumans MJ
and Berger RMF: Inhibition of the prolyl isomerase Pin1 improves
endothelial function and attenuates vascular remodelling in
pulmonary hypertension by inhibiting TGF-β signalling.
Angiogenesis. 25:99–112. 2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Shin HR, Bae HS, Kim BS, Yoon HI, Cho YD,
Kim WJ, Choi KY, Lee YS, Woo KM, Baek JH and Ryoo HM: PIN1 is a new
therapeutic target of craniosynostosis. Hum Mol Genet.
27:3827–3839. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Lee SH, Jeong HM, Han Y, Cheong H, Kang BY
and Lee KY: Prolyl isomerase Pin1 regulates the osteogenic activity
of Osterix. Mol Cell Endocrinol. 400:32–40. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Cho E, Lee JK, Lee JY, Chen Z, Ahn SH, Kim
ND, Kook MS, Min SH, Park BJ and Lee TH: BCPA {N,
N'-1,4-Butanediylbis[3-(2-chlorophenyl)acrylamide]} Inhibits
Osteoclast Differentiation through Increased Retention of
Peptidyl-Prolyl cis-trans Isomerase Never in Mitosis A-Interacting
1. Int J Mol Sci. 19(3436)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gao Y, Chen N, Fu Z and Zhang Q: Progress
of wnt signaling pathway in osteoporosis. Biomolecules.
13(483)2023.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li S, Cui Y, Li M, Zhang W, Sun X, Xin Z
and Li J: Acteoside derived from cistanche improves
glucocorticoid-induced osteoporosis by activating PI3K/AKT/mTOR
pathway. J Invest Surg. 36(2154578)2023.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhao P, Xiao L, Peng J, Qian YQ and Huang
CC: Exosomes derived from bone marrow mesenchymal stem cells
improve osteoporosis through promoting osteoblast proliferation via
MAPK pathway. Eur Rev Med Pharmacol Sci. 22:3962–3970.
2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yoshida G, Kawabata T, Takamatsu H, Saita
S, Nakamura S, Nishikawa K, Fujiwara M, Enokidani Y, Yamamuro T,
Tabata K, et al: Degradation of the NOTCH intracellular domain by
elevated autophagy in osteoblasts promotes osteoblast
differentiation and alleviates osteoporosis. Autophagy.
18:2323–2332. 2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Vazgiourakis VM, Zervou MI, Papageorgiou
L, Chaniotis D, Spandidos DA, Vlachakis D, Eliopoulos E and
Goulielmos GN: Association of endometriosis with cardiovascular
disease: Genetic aspects (Review). Int J Mol Med.
51(29)2023.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sarmah N, Nauli AM, Ally A and Nauli SM:
Interactions among endothelial nitric oxide synthase,
cardiovascular system, and nociception during physiological and
pathophysiological states. Molecules. 27(2835)2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Fagiani F, Vlachou M, Di Marino D,
Canobbio I, Romagnoli A, Racchi M, Govoni S and Lanni C: Pin1 as
molecular switch in vascular endothelium: Notes on its putative
role in age-associated vascular diseases. Cells.
10(3287)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Kennard S, Ruan L, Buffett RJ, Fulton D
and Venema RC: TNFα reduces eNOS activity in endothelial cells
through serine 116 phosphorylation and Pin1 binding: Confirmation
of a direct, inhibitory interaction of Pin1 with eNOS. Vascul
Pharmacol. 81:61–68. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liu M, Yu P, Jiang H, Yang X, Zhao J, Zou
Y and Ge J: TThe essential role of Pin1 via NF-κB signaling in
vascular inflammation and atherosclerosis in ApoE-/-Mice. Int J Mol
Sci. 18(644)2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Liang G, Wang S, Shao J, Jin YJ, Xu L, Yan
Y, Günther S, Wang L and Offermanns S: Tenascin-X mediates
Flow-induced suppression of EndMT and atherosclerosis. Circ Res.
130:1647–1659. 2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Huminiecki L, Goldovsky L, Freilich S,
Moustakas A, Ouzounis C and Heldin CH: Emergence, development and
diversification of the TGF-beta signalling pathway within the
animal kingdom. BMC Evol Biol. 9(28)2009.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Gentile C, Muise-Helmericks RC and Drake
CJ: VEGF-mediated phosphorylation of eNOS regulates angioblast and
embryonic endothelial cell proliferation. Dev Biol. 373:163–175.
2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Rai N, Sydykov A, Kojonazarov B, Wilhelm
J, Manaud G, Veeroju S, Ruppert C, Perros F, Ghofrani HA, Weissmann
N, et al: Targeting peptidyl-prolyl isomerase 1 in experimental
pulmonary arterial hypertension. Eur Respir J.
60(2101698)2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kim MR, Choi HS, Heo TH, Hwang SW and Kang
KW: Induction of vascular endothelial growth factor by
peptidyl-prolyl isomerase Pin1 in breast cancer cells. Biochem
Biophys Res Commun. 369:547–553. 2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Toko H, Konstandin MH, Doroudgar S,
Ormachea L, Joyo E, Joyo AY, Din S, Gude NA, Collins B, Völkers M,
et al: Regulation of cardiac hypertrophic signaling by prolyl
isomerase Pin1. Circ Res. 112:1244–1252. 2013.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Chen Y, Wu YR, Yang HY, Li XZ, Jie MM, Hu
CJ, Wu YY, Yang SM and Yang YB: Prolyl isomerase Pin1: A promoter
of cancer and a target for therapy. Cell Death Dis.
9(883)2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chuang HH, Zhen YY, Tsai YC, Chuang CH,
Huang MS, Hsiao M and Yang CJ: Targeting Pin1 for modulation of
cell motility and cancer therapy. Biomedicines.
9(359)2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Wu W, Xue X, Chen Y, Zheng N and Wang J:
Targeting prolyl isomerase Pin1 as a promising strategy to overcome
resistance to cancer therapies. Pharmacol Res.
184(106456)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Cohn GM, Liefwalker DF, Langer EM and
Sears RC: PIN1 provides dynamic control of MYC in response to
extrinsic signals. Front Cell Dev Biol. 8(224)2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Nashaat S, Henen MA, El-Messery SM and
Eisa H: New benzimidazoles targeting breast cancer: Synthesis, Pin1
inhibition, 2D NMR binding, and computational studies. Molecules.
27(5245)2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Ueda K, Nakatsu Y, Yamamotoya T, Ono H,
Inoue Y, Inoue MK, Mizuno Y, Matsunaga Y, Kushiyama A, Sakoda H, et
al: Prolyl isomerase Pin1 binds to and stabilizes acetyl CoA
carboxylase 1 protein, thereby supporting cancer cell
proliferation. Oncotarget. 10:1637–1648. 2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Tan X, Zhou F, Wan J, Hang J, Chen Z, Li
B, Zhang C, Shao K, Jiang P, Shi S, et al: Pin1 expression
contributes to lung cancer: Prognosis and carcinogenesis. Cancer
Biol Ther. 9:111–119. 2010.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Kim G, Bhattarai PY and Choi HS:
Peptidyl-prolyl cis/trans isomerase NIMA-interacting 1 as a
molecular target in breast cancer: A therapeutic perspective of
gynecological cancer. Arch Pharm Res. 42:128–139. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Chen Y, Wu Y, Yu S, Yang H, Wang X, Zhang
Y, Zhu S, Jie M, Liu C, Li X, et al: Deficiency of microRNA-628-5p
promotes the progression of gastric cancer by upregulating PIN1.
Cell Death Dis. 11(559)2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chen M, Xia Y, Tan Y, Jiang G, Jin H and
Chen Y: Downregulation of microRNA-370 in esophageal squamous-cell
carcinoma is associated with cancer progression and promotes cancer
cell proliferation via upregulating PIN1. Gene. 661:68–77.
2018.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Kuramochi J, Arai T, Ikeda S, Kumagai J,
Uetake H and Sugihara K: High Pin1 expression is associated with
tumor progression in colorectal cancer. J Surg Oncol. 94:155–160.
2006.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Wang T, Liu Z, Shi F and Wang J: Pin1
modulates chemo-resistance by up-regulating FoxM1 and the
involvements of Wnt/β-catenin signaling pathway in cervical cancer.
Mol Cell Biochem. 413:179–187. 2016.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Kim G, Bhattarai PY, Lim SC, Kim JY and
Choi HS: PIN1 facilitates ubiquitin-mediated degradation of
serine/threonine kinase 3 and promotes melanoma development via TAZ
activation. Cancer Lett. 499:164–174. 2021.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Maggio J, Cardama GA, Armando RG, Balcone
L, Sobol NT, Gomez DE and Mengual Gómez DL: Key role of PIN1 in
telomere maintenance and oncogenic behavior in a human glioblastoma
model. Oncol Rep. 49(91)2023.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Li Q, Dong Z, Lin Y, Jia X, Li Q, Jiang H,
Wang L and Gao Y: The rs2233678 polymorphism in PIN1 promoter
region reduced cancer risk: A meta-analysis. PLoS One.
8(e68148)2013.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Wulf G, Garg P, Liou YC, Iglehart D and Lu
KP: Modeling breast cancer in vivo and ex vivo reveals an essential
role of Pin1 in tumorigenesis. EMBO J. 23:3397–3407.
2004.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Takahashi K, Akiyama H, Shimazaki K,
Uchida C, Akiyama-Okunuki H, Tomita M, Fukumoto M and Uchida T:
Ablation of a peptidyl prolyl isomerase Pin1 from p53-null mice
accelerated thymic hyperplasia by increasing the level of the
intracellular form of Notch1. Oncogene. 26:3835–3845.
2007.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Girardini JE, Napoli M, Piazza S, Rustighi
A, Marotta C, Radaelli E, Capaci V, Jordan L, Quinlan P, Thompson
A, et al: A Pin1/mutant p53 axis promotes aggressiveness in breast
cancer. Cancer Cell. 20:79–91. 2011.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Zucchi D, Silvagni E, Elefante E,
Signorini V, Cardelli C, Trentin F, Schilirò D, Cascarano G,
Valevich A, Bortoluzzi A and Tani C: Systemic lupus erythematosus:
One year in review 2023. Clin Exp Rheumatol. 41:997–1008.
2023.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Baek WY, Choi YS, Lee SW, Son IO, Jeon KW,
Choi SD and Suh CH: Toll-like receptor signaling inhibitory peptide
improves inflammation in animal model and human systemic lupus
erythematosus. Int J Mol Sci. 22(12764)2021.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Khoryati L, Augusto JF, Shipley E,
Contin-Bordes C, Douchet I, Mitrovic S, Truchetet ME, Lazaro E,
Duffau P, Couzi L, et al: IgE inhibits Toll-like receptor 7- and
Toll-like receptor 9-mediated expression of interferon-α by
plasmacytoid dendritic cells in patients with systemic lupus
erythematosus. Arthritis Rheumatol. 68:2221–2231. 2016.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Salloum R and Niewold TB: Interferon
regulatory factors in human lupus pathogenesis. Transl Res.
157:326–331. 2011.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Tun-Kyi A, Finn G, Greenwood A, Nowak M,
Lee TH, Asara JM, Tsokos GC, Fitzgerald K, Israel E, Li X, et al:
Essential role for the prolyl isomerase Pin1 in Toll-like receptor
signaling and type I interferon-mediated immunity. Nat Immunol.
12:733–741. 2011.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Wei S, Yoshida N, Finn G, Kozono S,
Nechama M, Kyttaris VC, Zhen Zhou X, Tsokos GC and Ping Lu K:
Pin1-Targeted therapy for systemic lupus erythematosus. Arthritis
Rheumatol. 68:2503–2513. 2016.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Jacob N and Stohl W: Cytokine disturbances
in systemic lupus erythematosus. Arthritis Res Ther.
13(228)2011.PubMed/NCBI View
Article : Google Scholar
|
|
71
|
Ding J, Su S, You T, Xia T, Lin X, Chen Z
and Zhang L: Serum interleukin-6 level is correlated with the
disease activity of systemic lupus erythematosus: A meta-analysis.
Clinics (Sao Paulo). 75(e1801)2020.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Tackey E, Lipsky PE and Illei GG:
Rationale for interleukin-6 blockade in systemic lupus
erythematosus. Lupus. 13:339–343. 2004.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Takeno M, Gunn J, Suzuki JT, Kim NP, Kang
J, Finn TB, Vazirpour M, Martin W and Leung CJ: A novel role of
peptidyl-prolyl isomerase-1 as inducer of IL-6 expression in
systemic lupus erythematosus. Am J BioMedicine. 3:439–450.
2015.
|
|
74
|
Jang S, Kwon EJ and Lee JJ: Rheumatoid
arthritis: Pathogenic roles of diverse immune cells. Int J Mol Sci.
23(905)2022.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Nagaoka A, Takizawa N, Takeuchi R, Inaba
Y, Saito I, Nagashima Y, Saito T and Aoki I: Possible involvement
of peptidylprolyl isomerase Pin1 in rheumatoid arthritis. Pathol
Int. 61:59–66. 2011.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Makarov SS: NF-kappa B in rheumatoid
arthritis: A pivotal regulator of inflammation, hyperplasia, and
tissue destruction. Arthritis Res. 3:200–206. 2001.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Araki Y and Mimura T: Matrix
metalloproteinase gene activation resulting from disordred
epigenetic mechanisms in rheumatoid arthritis. Int J Mol Sci.
18(905)2017.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Li X and Makarov SS: An essential role of
NF-kappaB in the ‘tumor-like’ phenotype of arthritic synoviocytes.
Proc Natl Acad Sci USA. 103:17432–17437. 2006.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Ma Y, Hong FF and Yang SL: Role of
prostaglandins in rheumatoid arthritis. Clin Exp Rheumatol.
39:162–172. 2021.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Kondo N, Kuroda T and Kobayashi D:
Cytokine networks in the pathogenesis of rheumatoid arthritis. Int
J Mol Sci. 22(10922)2021.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Jeong HG, Pokharel YR, Lim SC, Hwang YP,
Han EH, Yoon JH, Ahn SG, Lee KY and Kang KW: Novel role of Pin1
induction in type II collagen-mediated rheumatoid arthritis. J
Immunol. 183:6689–6697. 2009.PubMed/NCBI View Article : Google Scholar
|
|
82
|
M'Koma AE: Inflammatory bowel disease:
Clinical diagnosis and pharmaceutical management. Med Res Arch.
11(10)2023.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Matsunaga Y, Hasei S, Yamamotoya T, Honda
H, Kushiyama A, Sakoda H, Fujishiro M, Ono H, Ito H, Okabe T, et
al: Pathological role of Pin1 in the development of DSS-Induced
colitis. Cells. 10(1230)2021.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Shao BZ, Wang SL, Pan P, Yao J, Wu K, Li
ZS, Bai Y and Linghu EQ: Targeting NLRP3 inflammasome in
inflammatory bowel disease: Putting out the fire of inflammation.
Inflammation. 42:1147–1159. 2019.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Dong R, Xue Z, Fan G, Zhang N, Wang C, Li
G and Da Y: Pin1 promotes NLRP3 inflammasome activation by
phosphorylation of p38 MAPK pathway in septic shock. Front Immunol.
12(620238)2021.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Dagenais A, Villalba-Guerrero C and
Olivier M: Trained immunity: A ‘new’ weapon in the fight against
infectious diseases. Front Immunol. 14(1147476)2023.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Kanna M, Nakatsu Y, Yamamotoya T, Encinas
J, Ito H, Okabe T, Asano T and Sakaguchi T: Roles of peptidyl
prolyl isomerase Pin1 in viral propagation. Front Cell Dev Biol.
10(1005325)2022.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Stroh LJ and Krey T: Structural insights
into hepatitis C virus neutralization. Curr Opin Virol.
60(101316)2023.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Lim YS, Tran HT, Park SJ, Yim SA and Hwang
SB: Peptidyl-prolyl isomerase Pin1 is a cellular factor required
for hepatitis C virus propagation. J Virol. 85:8777–8788.
2011.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Jeng WJ, Papatheodoridis GV and Lok ASF:
Hepatitis B. Lancet. 401:1039–1052. 2023.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Kojima Y and Ryo A: Pinning down viral
proteins: A new prototype for virus-host cell interaction. Front
Microbiol. 1(107)2010.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Pang R, Lee TK, Poon RT, Fan ST, Wong KB,
Kwong YL and Tse E: Pin1 interacts with a specific serine-proline
motif of hepatitis B virus X-protein to enhance
hepatocarcinogenesis. Gastroenterology. 132:1088–1103.
2007.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Shang S, Hua F and Hu ZW: The regulation
of β-catenin activity and function in cancer: Therapeutic
opportunities. Oncotarget. 8:33972–33989. 2017.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Ao R, Zhang DR, Du YQ and Wang Y:
Expression and significance of Pin1, β-catenin and cyclin D1 in
hepatocellular carcinoma. Mol Med Rep. 10:1893–1898.
2014.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Nishi M, Miyakawa K, Matsunaga S, Khatun
H, Yamaoka Y, Watashi K, Sugiyama M, Kimura H, Wakita T and Ryo A:
Prolyl Isomerase Pin1 regulates the stability of hepatitis B virus
core protein. Front Cell Dev Biol. 8(26)2020.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Kwon H, Kim J, Song C, Sajjad MA, Ha J,
Jung J, Park S, Shin HJ and Kim K: Peptidyl-prolyl cis/trans
isomerase Pin1 interacts with hepatitis B virus core particle, but
not with HBc protein, to promote HBV replication. Front Cell Infect
Microbiol. 13(1195063)2023.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Womack J and Jimenez M: Common questions
about infectious mononucleosis. Am Fam Physician. 91:372–376.
2015.PubMed/NCBI
|
|
98
|
Hutcheson RL, Chakravorty A and Sugden B:
Burkitt lymphomas evolve to escape dependencies on Epstein-Barr
virus. Front Cell Infect Microbiol. 10(606412)2020.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Narita Y, Murata T, Ryo A, Kawashima D,
Sugimoto A, Kanda T, Kimura H and Tsurumi T: Pin1 interacts with
the Epstein-Barr virus DNA polymerase catalytic subunit and
regulates viral DNA replication. J Virol. 87:2120–2127.
2013.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Chang ET, Ye W, Zeng YX and Adami HO: The
evolving epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol
Biomarkers Prev. 30:1035–1047. 2021.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Xu M, Cheung CC, Chow C, Lun SW, Cheung ST
and Lo KW: Overexpression of PIN1 enhances cancer growth and
aggressiveness with cyclin D1 induction in EBV-Associated
nasopharyngeal carcinoma. PLoS One. 11(e0156833)2016.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Yu JH, Im CY and Min SH: Function of PIN1
in cancer development and its inhibitors as cancer therapeutics.
Front Cell Dev Biol. 8(120)2020.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Letafati A, Soheili R, Norouzi M,
Soleimani P and Mozhgani SH: Therapeutic approaches for
HTLV-1-associated adult T-cell leukemia/lymphoma: A comprehensive
review. Med Oncol. 40(295)2023.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Ernzen KJ and Panfil AR: Regulation of
HTLV-1 transformation. Biosci Rep. 42(BSR20211921)2022.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Peloponese JM Jr, Yasunaga J, Kinjo T,
Watashi K and Jeang KT: Peptidylproline cis-trans-isomerase Pin1
interacts with human T-cell leukemia virus type 1 tax and modulates
its activation of NF-kappaB. J Virol. 83:3238–3248. 2009.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Jeong SJ, Ryo A and Yamamoto N: The prolyl
isomerase Pin1 stabilizes the human T-cell leukemia virus type 1
(HTLV-1) Tax oncoprotein and promotes malignant transformation.
Biochem Biophys Res Commun. 381:294–299. 2009.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Mesnard JM, Barbeau B, Cesaire R and
Peloponese JM: Roles of HTLV-1 basic zip factor (HBZ) in viral
chronicity and leukemic transformation. Potential new therapeutic
approaches to prevent and treat HTLV-1-related diseases. Viruses.
7:6490–6505. 2015.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Szymonowicz KA and Chen J: Biological and
clinical aspects of HPV-related cancers. Cancer Biol Med.
17:864–878. 2020.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Fowler JR, Maani EV, Dunton CJ and Jack
BW: Cervical Cancer. In: StatPearls. Treasure Island (FL)
ineligible companies. Disclosure: Elizabeth Maani declares no
relevant financial relationships with ineligible companies.
Disclosure: Charles Dunton declares no relevant financial
relationships with ineligible companies. Disclosure: Brian Jack
declares no relevant financial relationships with ineligible
companies, 2023.
|
|
110
|
McBride AA: The papillomavirus E2
proteins. Virology. 445:57–79. 2013.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Hareza DA, Wilczynski JR and Paradowska E:
Human papillomaviruses as infectious agents in gynecological
cancers. Oncogenic properties of viral proteins. Int J Mol Sci.
23(1818)2022.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Prabhavathy D, Vijayalakshmi R, Kanchana
MP and Karunagaran D: HHPV16 E2 enhances the expression of NF-κB
and STAT3 target genes and potentiates NF-κB activation by
inflammatory mediators. Cell Immunol. 292:70–77. 2014.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Masenga SK, Mweene BC, Luwaya E, Muchaili
L, Chona M and Kirabo A: HIV-Host Cell Interactions. Cells.
12(1351)2023.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Sarkar S, Balakrishnan K, Chintala K,
Chintala K, Mohareer K, Luedde T, Vasudevan AAJ, Münk C and
Banerjee S: Tough Way In, Tough Way Out: The complex interplay of
host and viral factors in nucleocytoplasmic trafficking during
HIV-1 infection. Viruses. 14(2503)2022.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Watashi K, Khan M, Yedavalli VR, Yeung ML,
Strebel K and Jeang KT: Human immunodeficiency virus type 1
replication and regulation of APOBEC3G by peptidyl prolyl isomerase
Pin1. J Virol. 82:9928–9936. 2008.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Saleh S, Lu HK, Evans V, Harisson D, Zhou
J, Jaworowski A, Sallmann G, Cheong KY, Mota TM, Tennakoon S, et
al: HIV integration and the establishment of latency in
CCL19-treated resting CD4(+) T cells require activation of NF-κB.
Retrovirology. 13(49)2016.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Dochi T, Nakano T, Inoue M, Takamune N,
Shoji S, Sano K and Misumi S: Phosphorylation of human
immunodeficiency virus type 1 capsid protein at serine 16, required
for peptidyl-prolyl isomerase-dependent uncoating, is mediated by
virion-incorporated extracellular signal-regulated kinase 2. J Gen
Virol. 95:1156–1166. 2014.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Rossi E, Meuser ME, Cunanan CJ and Cocklin
S: Structure, function, and interactions of the HIV-1 Capsid
Protein. Life (Basel). 11(100)2021.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Bao Q and Zhou J: Various strategies for
developing APOBEC3G protectors to circumvent human immunodeficiency
virus type 1. Eur J Med Chem. 250(115188)2023.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Hu B, Guo H, Zhou P and Shi ZL:
Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol.
19:141–154. 2021.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Yamamotoya T, Nakatsu Y, Kanna M, Hasei S,
Ohata Y, Encinas J, Ito H, Okabe T, Asano T and Sakaguchi T: Prolyl
isomerase Pin1 plays an essential role in SARS-CoV-2 proliferation,
indicating its possibility as a novel therapeutic target. Sci Rep.
11(18581)2021.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Ino Y, Nishi M, Yamaoka Y, Miyakawa K,
Jeremiah SS, Osada M, Kimura Y and Ryo A: Phosphopeptide enrichment
using Phos-tag technology reveals functional phosphorylation of the
nucleocapsid protein of SARS-CoV-2. J Proteomics.
255(104501)2022.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Ye H, Robak LA, Yu M, Cykowski M and
Shulman JM: Genetics and pathogenesis of Parkinson's syndrome. Annu
Rev Pathol. 18:95–121. 2023.PubMed/NCBI View Article : Google Scholar
|
|
124
|
Hallacli E, Kayatekin C, Nazeen S, Wang
XH, Sheinkopf Z, Sathyakumar S, Sarkar S, Jiang X, Dong X, Di Maio
R, et al: The Parkinson's disease protein alpha-synuclein is a
modulator of processing bodies and mRNA stability. Cell.
185:2035–2056 e2033. 2022.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Gao V, Briano JA, Komer LE and Burre J:
Functional and Pathological Effects of α-Synuclein on Synaptic
SNARE Complexes. J Mol Biol. 435(167714)2023.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Carvajal-Oliveros A, Dominguez-Baleon C,
Sanchez-Diaz I, Zambrano-Tipan D, Hernández-Vargas R, Campusano JM,
Narváez-Padilla V and Reynaud E: Parkinsonian phenotypes induced by
Synphilin-1 expression are differentially contributed by
serotonergic and dopaminergic circuits and suppressed by nicotine
treatment. PLoS One. 18(e0282348)2023.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Ghosh A, Saminathan H, Kanthasamy A,
Anantharam V, Jin H, Sondarva G, Harischandra DS, Qian Z, Rana A
and Kanthasamy AG: The peptidyl-prolyl isomerase Pin1 up-regulation
and proapoptotic function in dopaminergic neurons: Relevance to the
pathogenesis of Parkinson disease. J Biol Chem. 288:21955–21971.
2013.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Ryo A, Togo T, Nakai T, Hirai A, Nishi M,
Yamaguchi A, Suzuki K, Hirayasu Y, Kobayashi H, Perrem K, et al:
Prolyl-isomerase Pin1 accumulates in lewy bodies of parkinson
disease and facilitates formation of alpha-synuclein inclusions. J
Biol Chem. 281:4117–4125. 2006.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Weller J and Budson A: Current
understanding of Alzheimer's disease diagnosis and treatment.
F1000Res 7: Faculty Rev-1161, 2018.
|
|
130
|
Karran E and De Strooper B: The amyloid
hypothesis in Alzheimer disease: New insights from new
therapeutics. Nat Rev Drug Discov. 21:306–318. 2022.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Ma C, Hong F and Yang S: Amyloidosis in
Alzheimer's Disease: Pathogeny, etiology, and related therapeutic
directions. Molecules. 27(1210)2022.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Bianchi M and Manco M: Pin1 modulation in
physiological status and neurodegeneration. Any contribution to the
pathogenesis of type 3 diabetes? Int J Mol Sci.
19(2319)2018.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Wang SC, Hu XM and Xiong K: The regulatory
role of Pin1 in neuronal death. Neural Regen Res. 18:74–80.
2023.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Fagiani F, Govoni S, Racchi M and Lanni C:
The Peptidyl-prolyl Isomerase Pin1 in neuronal Signaling: From
neurodevelopment to neurodegeneration. Mol Neurobiol. 58:1062–1073.
2021.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Palaiogeorgou AM, Papakonstantinou E,
Golfinopoulou R, Sigala M, Mitsis T, Papageorgiou L, Diakou I,
Pierouli K, Dragoumani K, Spandidos DA, et al: Recent approaches on
Huntington's disease (Review). Biomed Rep. 18(5)2023.PubMed/NCBI View Article : Google Scholar
|
|
136
|
D'Egidio F, Castelli V, Cimini A and
d'Angelo M: Cell rearrangement and oxidant/antioxidant imbalance in
huntington's disease. Antioxidants (Basel). 12(571)2023.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Pereira CAS, Medaglia NC, Ureshino RP,
Bincoletto C, Antonioli M, Fimia GM, Piacentini M, Pereira GJDS,
Erustes AG and Smaili SS: NAADP-Evoked Ca2+ signaling
leads to mutant huntingtin aggregation and autophagy impairment in
murine astrocytes. Int J Mol Sci. 24(5593)2023.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Sap KA, Geijtenbeek KW, Schipper-Krom S,
Guler AT and Reits EA: Ubiquitin-modifying enzymes in Huntington's
disease. Front Mol Biosci. 10(1107323)2023.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Napoli M, Girardini JE, Piazza S and Del
Sal G: Wiring the oncogenic circuitry: Pin1 unleashes mutant p53.
Oncotarget. 2:654–656. 2011.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Steger M, Murina O, Hühn D, Ferretti LP,
Walser R, Hänggi K, Lafranchi L, Neugebauer C, Paliwal S, Janscak
P, et al: Prolyl isomerase PIN1 regulates DNA double-strand break
repair by counteracting DNA end resection. Mol Cell. 50:333–343.
2013.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Ulugut H and Pijnenburg YAL:
Frontotemporal dementia: Past, present, and future. Alzheimers
Dement. 19:5253–5263. 2023.PubMed/NCBI View Article : Google Scholar
|
|
142
|
Thorpe JR, Mosaheb S, Hashemzadeh-Bonehi
L, Cairns NJ, Kay JE, Morley SJ and Rulten SL: Shortfalls in the
peptidyl-prolyl cis-trans isomerase protein Pin1 in neurons are
associated with frontotemporal dementias. Neurobiol Dis.
17:237–249. 2004.PubMed/NCBI View Article : Google Scholar
|
|
143
|
Cataldo AM, Peterhoff CM, Troncoso JC,
Gomez-Isla T, Hyman BT and Nixon RA: Endocytic pathway
abnormalities precede amyloid beta deposition in sporadic
Alzheimer's disease and Down syndrome: Differential effects of APOE
genotype and presenilin mutations. Am J Pathol. 157:277–286.
2000.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Husseman JW, Nochlin D and Vincent I:
Mitotic activation: A convergent mechanism for a cohort of
neurodegenerative diseases. Neurobiol Aging. 21:815–828.
2000.PubMed/NCBI View Article : Google Scholar
|
|
145
|
Mead RJ, Shan N, Reiser HJ, Marshall F and
Shaw PJ: Amyotrophic lateral sclerosis: A neurodegenerative
disorder poised for successful therapeutic translation. Nat Rev
Drug Discov. 22:185–212. 2023.PubMed/NCBI View Article : Google Scholar
|
|
146
|
Iridoy MO, Zubiri I, Zelaya MV, Martinez
L, Ausín K, Lachen-Montes M, Santamaría E, Fernandez-Irigoyen J and
Jericó I: Neuroanatomical quantitative proteomics reveals common
pathogenic biological routes between amyotrophic lateral sclerosis
(ALS) and frontotemporal dementia (FTD). Int J Mol Sci.
20(4)2018.PubMed/NCBI View Article : Google Scholar
|
|
147
|
Perrot R and Eyer J: Neuronal intermediate
filaments and neurodegenerative disorders. Brain Res Bull.
80:282–295. 2009.PubMed/NCBI View Article : Google Scholar
|
|
148
|
Kesavapany S, Patel V, Zheng YL, Pareek
TK, Bjelogrlic M, Albers W, Amin N, Jaffe H, Gutkind JS, Strong MJ,
et al: Inhibition of Pin1 reduces glutamate-induced perikaryal
accumulation of phosphorylated neurofilament-H in neurons. Mol Biol
Cell. 18:3645–3655. 2007.PubMed/NCBI View Article : Google Scholar
|
|
149
|
Chao SH, Greenleaf AL and Price DH:
Juglone, an inhibitor of the peptidyl-prolyl isomerase Pin1, also
directly blocks transcription. Nucleic Acids Res. 29:767–773.
2001.PubMed/NCBI View Article : Google Scholar
|
|
150
|
Xi L, Wang Y, He Q, Zhang Q and Du L:
Interaction between Pin1 and its natural product inhibitor
epigallocatechin-3-gallate by spectroscopy and molecular dynamics
simulations. Spectrochim Acta A Mol Biomol Spectrosc. 169:134–143.
2016.PubMed/NCBI View Article : Google Scholar
|
|
151
|
Bayer E, Thutewohl M, Christner C, Tradler
T, Osterkamp F, Waldmann H and Bayer P: Identification of hPin1
inhibitors that induce apoptosis in a mammalian Ras transformed
cell line. Chem Commun (Camb). 516–518. 2005.PubMed/NCBI View Article : Google Scholar
|
|
152
|
Jiang B and Pei D: A selective,
cell-permeable nonphosphorylated bicyclic peptidyl inhibitor
against Peptidyl-Prolyl isomerase Pin1. J Med Chem. 58:6306–6312.
2015.PubMed/NCBI View Article : Google Scholar
|
|
153
|
He S, Li L, Jin R and Lu X: Biological
function of Pin1 in vivo and its inhibitors for preclinical study:
Early development, current strategies, and future directions. J Med
Chem. 66:9251–9277. 2023.PubMed/NCBI View Article : Google Scholar
|
|
154
|
Guo C, Hou X, Dong L, Dagostino E,
Greasley S, Ferre R, Marakovits J, Johnson MC, Matthews D,
Mroczkowski B, et al: Structure-based design of novel human Pin1
inhibitors (I). Bioorg Med Chem Lett. 19:5613–5616. 2009.PubMed/NCBI View Article : Google Scholar
|
|
155
|
Russo Spena C, De Stefano L, Poli G,
Granchi C, El Boustani M, Ecca F, Grassi G, Grassi M, Canzonieri V,
Giordano A, et al: Virtual screening identifies a PIN1 inhibitor
with possible antiovarian cancer effects. J Cell Physiol.
234:15708–15716. 2019.PubMed/NCBI View Article : Google Scholar
|
|
156
|
Campaner E, Rustighi A, Zannini A,
Cristiani A, Piazza S, Ciani Y, Kalid O, Golan G, Baloglu E,
Shacham S, et al: A covalent PIN1 inhibitor selectively targets
cancer cells by a dual mechanism of action. Nat Commun.
8(15772)2017.PubMed/NCBI View Article : Google Scholar
|
|
157
|
Liu L, Zhu R, Li J, Pei Y, Wang S, Xu P,
Wang M, Wen Y, Zhang H, Du D, et al: Computational and
structure-based development of high potent cell-active covalent
inhibitor targeting the Peptidyl-Prolyl isomerase
NIMA-Interacting-1 (Pin1). J Med Chem. 65:2174–2190.
2022.PubMed/NCBI View Article : Google Scholar
|
|
158
|
Born A, Henen MA and Vogeli B: Activity
and Affinity of Pin1 Variants. Molecules. 25:2019.PubMed/NCBI View Article : Google Scholar
|
|
159
|
Lu PJ, Zhou XZ, Liou YC, Noel JP and Lu
KP: Critical role of WW domain phosphorylation in regulating
phosphoserine binding activity and Pin1 function. J Biol Chem.
277:2381–2384. 2002.PubMed/NCBI View Article : Google Scholar
|
|
160
|
Verdecia MA, Bowman ME, Lu KP, Hunter T
and Noel JP: Structural basis for phosphoserine-proline recognition
by group IV WW domains. Nat Struct Biol. 7:639–643. 2000.PubMed/NCBI View Article : Google Scholar
|
|
161
|
Wang JZ, Xi L, Lin T, Wang Y, Zhu GF and
Du LF: The structural and functional role of the three tryptophan
residues in Pin1. J Photochem Photobiol B. 146:58–67.
2015.PubMed/NCBI View Article : Google Scholar
|
|
162
|
Li K, Li L, Wang S, Li X, Ma T, Liu D,
Jing Y and Zhao L: Design and synthesis of novel 2-substituted
11-keto-boswellic acid heterocyclic derivatives as anti-prostate
cancer agents with Pin1 inhibition ability. Eur J Med Chem.
126:910–919. 2017.PubMed/NCBI View Article : Google Scholar
|
|
163
|
Maggio J, Cabrera M, Armando R, Chinestrad
P, Pifano M, Menna PL, Gomez DE and Gómez DLM: Rational design of
PIN1 inhibitors for cancer treatment based on conformational
diversity analysis and docking based virtual screening. J Biomol
Struct Dyn. 40:5858–5867. 2022.PubMed/NCBI View Article : Google Scholar
|