1. Introduction
One of the most fundamental parts of a functioning
cell is the cellular membrane, where proteins closely related to
the membrane mediate important functions, such as selective
material mobility and information transmission between the cell and
its surroundings (1). Of note
>75% of the total membrane proteins are present in the
mitochondria and the remaining 25% are present in myelin membranes
(2). Membrane proteins can be
broadly classified into two types, extrinsic and intrinsic, based
on their function and are also classified based on their position
attached to the lipid membrane including integral, lipid anchored
and peripheral (3). Integral
proteins are intrinsic and have a cytoplasmic and extracellular
domain; they are transmembrane proteins found in an embedded form
with membrane-spanning domains, such as α-helices and multiple
β-strands (4). The integral
proteins can be otherwise termed as single-pass transmembrane and
multi-pass transmembrane proteins based on the polypeptide chain
crossing the lipid bilayer (5).
Bacteriorhodopsin (single-pass transmembrane protein) and
aquaporins (multi-pass transmembrane proteins) are examples of
integral proteins. Mostly they are glycosylated and located in the
extracellular space, and they are involved in endocytic and
secretory pathways (6,7). Peripheral membrane proteins are
extrinsic and have hydrophilic domains that transduce intracellular
signalling. G protein-coupled receptor (GPCR), receptor tyrosine
kinases, protein channels and transporters are some examples of
peripheral membrane proteins, and these are expressed in downstream
signalling pathways and are involved in cellular changes (8,9).
Lipid-anchored proteins are water soluble proteins attached
covalently and function either on one side of the cytoplasmic phase
or extracellular phase (10). One
group of proteins are anchored to the membrane by fatty acyl chain,
which are covalently linked to the N-terminal of glycine residue
(acylation), a second group of proteins are anchored to the
membrane by a hydrocarbon chain attached to the cysteine residue in
the C terminus (prenylation), and a third group
(glycophosphatidylinositol anchored proteins) bind to the cell
surface and are specialized proteins in the exoplasmic phase of
membranes (11).
When proteins are synthesized by pre-existing
membranes, they are inserted into the plasma membrane
asymmetrically and are distributed across the lipid bilayer. An
inside and outside asymmetry distribution of lipid bilayer occurs
by the external location of carbohydrates (oligosaccharide)
attached to the membrane proteins (12). Oligosaccharide chains are linked
via the H-bonds with the glycolipids attached on the extracellular
surface. Phospholipid-binding proteins recognize specific
phospholipids and transfer them from the plasma membrane to
mitochondria and peroxisomes (13). The present review discusses the
topics behind protein studies step by step in an aim to provide an
understanding of strategies used to perform experiments based on
the type of proteins and how they can be processed.
2. Protein separation and purification
Protein molecules can be separated based on the
charge, affinity, solubility, size and adsorption properties.
Electrophoresis and chromatography principles are the most popular
separation technique for protein molecules and some examples of
this include ion-exchange chromatography, affinity chromatography,
dialysis, ultrafiltration, size-exclusion chromatography (14) and electrophoresis [including
capillary electrophoresis, isoelectric focusing and sodium dodecyl
sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE)] utilized
to isolate protein samples (15).
During the biological experimentation process to
separate and purify protein samples, several detergent solutions
are used. The detergents utilized for protein separation can be
categorized into denaturing and non-denaturing detergents.
Denaturing detergents are polar molecules that contain charges
(anionic or cationic); one such widely used anionic detergent is
SDS and cationic detergents include cetyltrimethylammonium bromide.
Similarly, non-denaturing detergents were also utilized and are
classified based on the material and components used; these include
non-ionic detergents, zwitterionic detergents and bile salts. Some
popular non-denaturing detergents include Triton X-100, CHAPS and
cholate (16).
In a recent study, Zhou et al (17) established with a protocol for
isolating mitochondrial proteins. They utilized cultured 293T cells
for demonstration in which the mitochondrial fraction was isolated
from cultured 293T cells in the initial phase and membrane proteins
were localized by two methods, sonication and the sodium carbonate
method. The sodium carbonate method is utilized to extract integral
proteins and the sonication method is used to extract both integral
and peripheral proteins from the soluble protein. By contrast, the
non-ionizing detergent (Triton-X) and proteolytic enzyme
(proteinase K) are used to separate the outer membrane proteins
(17).
3. Protein identification and mapping
Protein appears in a 3-dimensional structure and to
determine its structure, mapping is performed to identify hot-spot
interactions and various other potential functions to recognize
functional binding sites and their roles. This can be performed
experimentally using various computer aided platforms, as well
bioinstrumentation. One such extraordinary analytical technique
invention for the identification of unknown proteins is
matrix-assisted laser desorption/ionization-time of flight mass
spectrometry (MS). It functions on the principle of peptide mass
fingerprinting (PMF) in which the unidentified protein molecules
are cleaved into small peptides and absolute masses are accurately
measured. Finally, the NIST mass spectral libraries is utilized
with the PMF data for the identification of anonymous proteins
(18). This technique can be
applied to various fields, such as microbiology, biotechnology,
food chemistry and environmental sciences to uncover the hidden
protein from the large population (19).
Nuclear magnetic resonance (NMR) spectroscopy is
another technique widely used to obtain structural information of
protein, nucleic acids and other biomolecules. It is performed in a
very lower energy frequency (radio frequency region of the
electromagnetic spectrum) and is based on nuclear spin interactions
with a magnetic field; the structure of a molecule is predicted. It
is a technique widely used by chemists to predict a chemical
structure of an unknown molecule. Chemical shifts and J-couplings
(internal nuclear spin interactions) are two key parameters
exclusively used by chemists to predict a chemical structure.
Processing samples in NMR should be diluted with deuterated
solvent, such as chloroform, deuterium oxide and other solvents
based on the nature of the material. Acetone, methanol,
trichloromethyl, nitrogen gas and a vacuum are some of the
substances used to cleanse the NMR tubes. This technique is widely
applied in pharmaceutical sciences, metabolomics research and food
chemistry (20-22).
In addition, reversed-phase-high pressurized liquid chromatography
(RP-HPLC) has been utilized in anti-platelet drug determination
(ticagrelor) in human blood plasma and in the determination of the
dosage form of chemical compounds in pharmaceuticals (23,24).
By contrast, for the diagnostic and monitoring parameters in
healthcare, enzyme-linked immunosorbent assay is utilized as a
significant approach to measure certain types of biomarkers
responsible for the cause of disease (25).
Additionally, some of the recent advancements in
protein identification include the identification of phosphoprotein
and signatures (cyclin D1, ERa and AR S650) using laser capture
microdissection-reverse phase protein microarrays by Gallagher
et al (26).
4. Evolution of sequencing approaches and
proteomic studies
The evolution of determining the protein sequence
began in the early 1950s with the sequencing of insulin using the
Edman degradation method. Edman degradation is the first used
method to sequence the amino acids in a peptide and is performed
without breaking the peptide bonds between other amino acid
residues, amino-terminal residue is labelled and separated from the
peptide. This method is also known as N-terminal sequencing, and is
utilized to identify unknown proteins, and the quality and identity
of proteins can also be determined. The main advantage of this
method is that it does not damage the protein entity and a
disadvantage is that it is not able to identify multiple proteins
simultaneously. Advancements to the Edman method led to the use of
nucleic acids and enzymes to sequence the protein, and sequencing
known as the Sanger method. The application of the Edman method is
widely applied to product development in chemical and
pharmaceutical industries by the development of enzyme/bio-catalyst
for large scale production, whereas the Sanger method is used in
biological molecules (DNA and RNA) for performing variant studies
(27).
A chemical reagent known as phenyl-isothiocyanate is
a widely used in HPLC, whereas in the Edman degradation method, the
same reagent is also called the Edman reagent (28). In the Edman method, only amino the
acid sequence in the peptide can be determined; hence, researchers
devised the addition of protein digestion and fractionization
protocols to determine the whole sequence of proteins. Protein
digestion can be performed with various enzymes, such as proteinase
K and fractionization is performed using HPLC, a chromatographic
method used to analyse and separate the digested protein samples in
liquid form. Later in the 1990s (29), following the discovery of MS,
researchers used MS combined with the HPLC technique as an
alternative to Edman degradation for the effective identification
and for the analysis of protein molecules (30).
The unidentified/unknown proteins from the large
sample population can be identified based on two approaches, the
bottom-up approach and top-down approach. In the bottom-up
approach, the protein is digested into peptides and peptides are
separated using MS techniques to determine the protein sequences.
The bottom-up approach is also known as shotgun proteomics and HPLC
combined with MS are utilized to determine the protein sequences.
In the top-down approach, protein is directly separated using MS
and other protocols remains the same, as in Shotgun proteomics to
determine protein sequences (31,32).
Moreover, recently, proteomic techniques have been
explored with several other potential proteins, including heat
shock proteins, metabolic enzymes, oxidative proteins, structural
proteins, and cell death and signalling regulators that can be
utilized in the application of drug or biomarker discovery
(33). The applications of
proteomics are widely employed by several industry sectors. These
include post-translational modification, targeted protein
quantification, protein-protein interaction analysis, chemical
proteomics, and protein expression profiling (34,35).
By contrast, recent developments in proteomics include the
monitoring of post-translational modifications by the capillary and
microchip electrophoresis techniques (36).
5. In silico analysis in
proteomics
Various computer-aided platforms have also made
analysis easier by performing the majority of the experiment in
silico. Diverse software and applications have been utilized
based on the objective of the study; these make the analysis
faster, accurate, as well as time and cost-effective, in comparison
to wet-lab experiments. Schrödinger (https://www.schrodinger.com/) is one such widely
utilized software for in silico experiments and the data can
be retrieved using a number of open-source platforms, such as
Ensembl (https://asia.ensembl.org/index.html), NCBI (https://www.ncbi.nlm.nih.gov/), EMBL (https://www.embl.org/), Expasy (https://www.expasy.org/resources/uniprotkb-swiss-prot)
and DDBJ (https://www.ddbj.nig.ac.jp/index-e.html). Some
experiments performed using in silico analysis involve
variant studies (37), molecular
docking (38), critical assessment
of structure prediction (CASP) experiments (39), homology modelling (40), pharmacophore modelling,
quantitative structure activity relationship (QSAR) modelling
(41), ab initio methods
(42), phylogenetic analysis,
sequence similarity searches, primer designing, and computational
fluid dynamics (43-45).
Over the past decade, bioinformatics has also played a crucial role
in computer programming approaches by algorithms (46). Hence as a state of the art, the
development of graph algorithms based on protein sequencing and
protein identification issues by dynamic programming would be a
future goal.
6. Focus on bacterial or fungal cell
membrane proteins
Cell membrane proteins and integral components can
be studied and are more extensively understood in prokaryotes
(bacteria and fungi) when compared to eukaryotic organisms. In
bacteria, cell division and synthesis occur with the influence of
cell membrane proteins. One such key protein includes, filamenting
temperature-sensitive mutant Z (FtsZ) and it is encoded by the FtsZ
gene (47). FtsZ is a complex
pinpoint in which all cytosolic and membrane proteins are detached.
Researchers have previously demonstrated that surfactant-free
membrane protein complex separation can be achieved by the presence
of FtsZ within penicillin-binding protein (PBP)2/2a nanoparticles
by utilizing anti-FtsZ antiserum for the purification of membrane
proteins by immuno-affinity chromatography. It was found that FtsZ,
PBP2 and PBP2a were captured by styrene-co-maleic acid-lipid
particles (SMALP) using an anti-FtsZ antibody, illustrating the
ability of the technique to remove significant protein complexes
(48).
RodA is a protein belonging to the shape,
elongation, division and sporulation (SEDS) family that plays a
crucial, yet ambiguous role in cell wall biosynthesis throughout
growth, division, and sporulation (49). RodA, a crucial core component of
the Rod complex that is highly conserved and serves as a dynamic
peptidoglycan-synthesizing tool that mediates the elongation of
rod-shaped bacteria. Meeske et al (49), performed several biotechnological
experiments to prove that SEDS proteins constitute a family of
peptidoglycan polymerases and the revelation that SEDS family
proteins are peptidoglycan glycosyltransferases (PGTs) with extra
cytoplasmic catalytic centres opens an alluring new option to
design antibiotics that specifically target this pathway.
Complex regulatory systems ensure that bacteria have
the required level of β-barrel outer membrane proteins (OMPs) to
facilitate habitat adaptation. The OMP islands are comprised of the
Bam complex, which catalyses the insertion of OMPs in the outer
membrane, and are distributed throughout the cell (50). The study by Rassam et al
(51) entrenched a mechanism of
binary OMP partitioning by utilizing fluorescent colicins as
OMP-specific probes, along with in vivo and in vitro
ensemble and single-molecule fluorescence microscopy, as well as
molecular dynamics simulations, to uncover the process underpinning
OMP turnover in Escherichia coli.
The presence of two adjacent folded subdomains with
an IgG-like structure distinguishes the family of proteins known as
the microbial surface components recognising adhesive matrix
molecules (MSCRAMMs). The ‘dock-lock-latch’ mechanism used to bind
fibrinogen or the ‘collagen hug’ mechanism used to bind collagen
are two examples of how these promote binding to ligands through
procedures that involve significant conformational changes.
MSCRAMMs function based on the clumping factors A and B, which play
a key role in the pathogenesis of Staphylococcus aureus
infections (52).
As the incidence of fungal diseases is increasing in
developed countries, it is critical to understand the pathological
mechanisms of fungi. The stimulation of host defences, including
phagocytosis and mediators of humoral immunity, as well as tissue
adherence, immune escape mechanisms and host defences are all
mediated by cell wall molecules. Endohydrolases, fucosyl
transferase, glucuronosyl transferase, chitinases, 1,3-β-glucan
synthase, chitin synthase and deacetylase, sialoglycoproteins and
uronic acid-containing glycoproteins are some of the enzymes
detached to the cell wall and need to be further investigated for
understanding the cell mechanisms and functions in fungi (53). To produce antifungal medications
against pathogenic fungus including Candida albicans,
Cryptococcus neoformans and Aspergillus fumigatus,
protein kinase-C (PKC) is a viable target. Thus, the growing body
of research on enzymes from various species has shown a keen
interest in fungal PKCs (54).
7. Proteins in structural biology
As numerous cell-membrane proteins are utilized in
therapeutics, it is important to understand the 3D structure of
proteins. X-ray crystallography is one such widely used method for
the characterization of the 3D structure of proteins. Effective
protein extraction, solubilization, stabilization and
crystallization are necessary for this method to be successful. For
those working to crystallize membrane proteins, each of these
processes may provide significant difficulties (55). Pre-crystallization screening needs
to be carried out after protein extraction, solubilization and
purification to evaluate the stability of proteins. The possibility
of crystallization and higher-resolution diffraction improves with
the discovery of factors that make pure proteins more stable. A
rapid, high-throughput technique utilized to assess the
thermostability of solubilized proteins is the thermal denaturation
experiment (56).
Dynamic light scattering (DLS) is a rapid and
sensitive alternative technique with proven effectiveness in
identifying membrane proteins in solution. DLS measures light
scattered by the macromolecules in solution, and depending on the
rate of light scattering fluctuations, it may assess the size,
homogeneity, and stability of samples (57). Based on the protein environment, a
precise method of crystallization is applied; this includes vapor
diffusion crystallization, in meso crystallization and
chaperone crystallization (58).
8. Challenges and future perspectives
The electrophoresis protocols are the most commonly
used assays for the separation of protein molecules. Among these,
SDS-PAGE is generally performed in all cases of separation of
biomolecules, whereas in multilevel proteomics, the determination
of the peptide, proteoform and protein complex is achieved by
capillary electrophoresis utilized with MS (59). In recombinant protein therapeutics
to perform quality assessment and unravel protein molecules,
capillary isoelectric focusing-MS can be utilized and charge
variant analysis can also be performed, demonstrated with
bispecific antibody (60). NMR and
MS are two techniques widely used to study and determine biological
molecules. However, in the field of metabolomics and flux-omics,
NMR has certain drawbacks as its resolution, sensitivity and
accessibility are low with multiple analytical technical approaches
(61). Other than NMR in
predicting structure, cryo-electron microscopy also plays a crucial
role in predicting the 3D structure of biomacromolecules and these
approaches will become advanced in future research applications in
analysing the peptide (62). In
determining the quality of food components, the majority of food
testing laboratories and industries use NMR and MS for quality
control; however, to date, there is no NMR database for foodstuffs
for comparative analysis and this has yet to be developed (63).
MS methods serve as effective tools for diagnosis
and prognosis by differentiating healthy vs. abnormal samples
(blood, urine or cerebrospinal fluid). Spatial metabolomics in
omics studies enable biomolecule localization, such that it can be
utilized with MS for generating imaging MS, and artificial
intelligence applications such as deep learning and machine
learning will also be utilized as a tool for image analysis
(64,65). In the development and advancement
of MS in multi-disciplinary studies, the study by Kuo et al
(66) described five future
directions that include constructing a public data repository,
creating a future automated platform for usage in a robotic
laboratory, moving towards on-site tests, broadening outreach and
blindly unravelling biomolecules in routine analysis. Various in
silico tools and projects based on web developments can also be
performed to advance the techniques bioanalytically.
However, beyond all the developed protein
technologies (for purification, detection, labelling and
fractionization), the major loophole that still exists is the
unknown complexity of protein structures in biological cells that
can used against the novel therapeutic technology and this can be
answered by the development of RNA-sequencing techniques and omics
approaches (67). In the recent
decade, numerous sequencing techniques for analysing RNA and
proteins brought advances to the therapeutic technology against
various diseases. Some of the recent ones include Illumina and
ion-torrent barcoding technologies (68).
Cell membrane proteins including the protein
kinases, GPCR, B-cell lymphoma and chaperones can be utilized in
the development of diagnostic and therapeutic products (69-71).
Likewise, cell membrane-coated nanoparticles have played a crucial
role in nanomedicine, vaccination and targeted drug delivery in
various diseases such as cancer, and metabolic disorders (72,73).
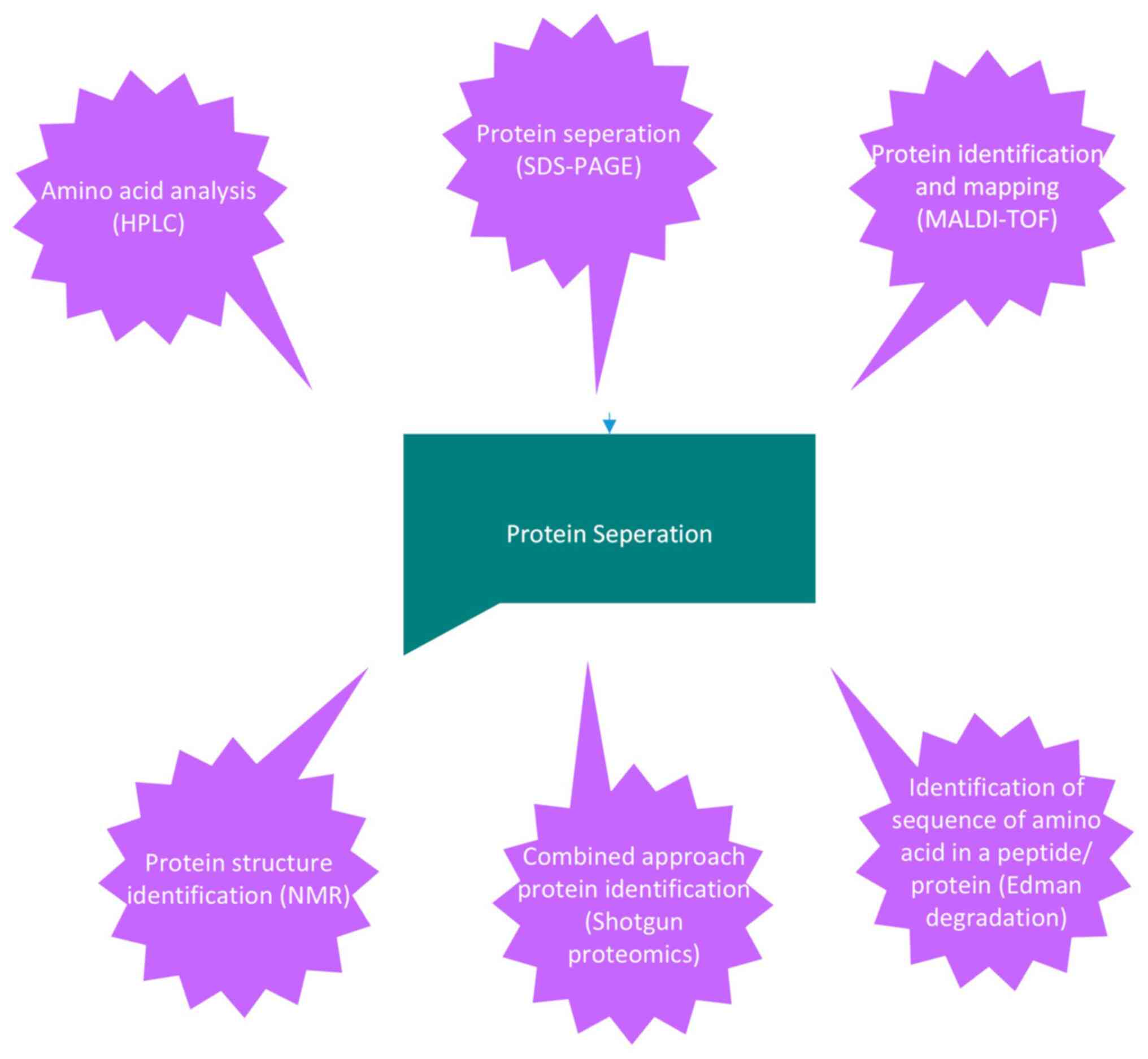
The various types of bioanalytical analyses used for protein
characterization are depicted in Fig.
1, along with a focus on prokaryotic organisms (Table I) for a better understanding of
cell membrane complexes and proteins.
 | Table IStudies on prokaryotic cell membranes
(published over the past 5 years). |
Table I
Studies on prokaryotic cell membranes
(published over the past 5 years).
| Prokaryotic cell
membrane proteins |
Authors/(Refs.) |
|---|
| Acinetobacter
baumannii | Nie et al
(74) |
| Pichia
pastoris | Chen et al
(75) |
| Beauveria
bassiana | Ding et al
(76) |
| Akkermansia
muciniphila | Wang et al
(77) |
| Aspergillus
flavus | Manju Devi et
al (78) |
| Lasiodiplodia
theobromae | Peng et al
(79) |
| Bacterial outer
membrane protein assembly | Nie et al
(74), Doyle and Bernstein
(80), Oluwole et al
(81), Peterson et al
(82), Sun et al (83), and Tomasek and Kahne (84) |
| Bacterial
respiratory membrane protein complexes | Muras et al
(85) |
| Bitopic membrane
proteins in bacterial cell division | den Blaauwen and
Luirink (86), and Nguyen et
al (87) |
| Bacteriophage
derived proteins | Grabowski et
al (88) and Sharma et
al (89) |
| Bacterial membrane
protein biogenesis | Hegde and Keenan
(6), Avila-Calderón et al
(90), and McDowell et al
(91) |
| Fungal membrane
protein biogenesis | Diederichs et
al (92), and Lübeck and
Lübeck (93) |
9. Conclusion
The present review mainly focused on bacterial and
fungal cell membrane proteins, which have been utilized in multiple
aspects of pharmaceuticals to produce therapeutic products.
Overall, the bioanalytical techniques, principles behind them, and
computer-aided platforms were also discussed in the present study.
For researchers who have a very keen interest in proteomics and
cell membrane proteins, these studies give the proposal and
clarification by helping in the development of problem statements,
novelty and applications.
Acknowledgements
Not applicable.
Funding
Funding: Not applicable.
Availability of data and materials
Not applicable.
Authors' contributions
MSAH and IRO were involved in the conceptualisation
of the study and in the editing of manuscript. MSAH performed the
literature search. KR and IRO reviewed the study. IRO was involved
in the submission of final manuscript and in correspondence. All
authors have read and approved the final manuscript. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
Not applicable.
References
|
1
|
Watson H: Biological membranes. Essays
Biochem. 59:43–69. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gopalakrishnan G, Awasthi A, Belkaid W, De
Faria O Jr, Liazoghli D, Colman DR and Dhaunchak AS: Lipidome and
proteome map of myelin membranes. J Neurosci Res. 91:321–334.
2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Muller MP, Jiang T, Sun C, Lihan M, Pant
S, Mahinthichaichan P, Trifan A and Tajkhorshid E: Characterization
of lipid-protein interactions and lipid-mediated modulation of
membrane protein function through molecular simulation. Chem Rev.
119:6086–6161. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wardhan R and Mudgal P: Membrane Proteins
in Textbook of Membrane Biology. Springer, Singapore, pp49-80,
2017.
|
|
5
|
Bai L and Li H: Structural insights into
the membrane chaperones for multi-pass membrane protein biogenesis.
Curr Opin Struct Biol. 79(102563)2023.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hegde RS and Keenan RJ: The mechanisms of
integral membrane protein biogenesis. Nat Rev Mol Cell Biol.
23:107–124. 2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Tzen JTC: Integral proteins in plant oil
bodies. ISRN Botany. 2012:1–16. 2012.
|
|
8
|
Che T: Advances in the treatment of
chronic pain by targeting GPCRs. Biochemistry. 60:1401–1412.
2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li Y, Zhang H, Kosturakis AK, Cassidy RM,
Zhang H, Kennamer-Chapman RM, Jawad AB, Colomand CM, Harrison DS
and Dougherty PM: MAPK signaling downstream to TLR4 contributes to
paclitaxel-induced peripheral neuropathy. Brain Behav Immun.
49:255–266. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Müller GA and Müller TD: (Patho)Physiology
of glycosylphosphatidylinositol-anchored proteins I: Localization
at plasma membranes and extracellular compartments. Biomolecules.
13(855)2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lee IH, Imanaka M, Modahl EH and
Torres-Ocampo AP: Lipid raft phase modulation by membrane-anchored
proteins with inherent phase separation properties. ACS Omega.
4:6551–6559. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Henderson JC, Zimmerman SM, Crofts AA,
Boll JM, Kuhns LG, Herrera CM and Trent MS: The power of asymmetry:
Architecture and assembly of the gram-negative outer membrane lipid
bilayer. Ann Rev Microbiol. 70:255–278. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lorent JH, Levental KR, Ganesan L,
Rivera-Longsworth G, Sezgin E, Doktorova M, Lyman E and levental I:
Plasma membranes are asymmetric in lipid unsaturation, packing and
protein shape. Nat Chem Biol. 16:642–652. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Coskun O: Separation techniques:
Chromatography. North Clin Istanb. 3:156–160. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Jorrin-Novo JV, Komatsu S, Sanchez-Lucas R
and de Francisco LE: Gel electrophoresis-based plant proteomics:
Past, present, and future. Happy 10th anniversary Journal of
Proteomics! J Proteomics. 198:1–10. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Orwick-Rydmark M, Arnold T and Linke D:
The use of detergents to purify membrane proteins. Curr Protoc
Protein Sci. 84:4.8.1–4.8.35. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhou D, Zhong S, Han X, Liu D, Fang H and
Wang Y: Protocol for mitochondrial isolation and sub-cellular
localization assay for mitochondrial proteins. STAR Protoc.
4(102088)2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Webster J and Oxley D: Protein
identification by MALDI-TOF mass spectrometry. Methods Mol Biol.
2012:227–240. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Santos IC, Hildenbrand ZL and Schug KA:
Applications of MALDI-TOF MS in environmental microbiology.
Analyst. 141:2827–2837. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Crook AA and Powers R: Quantitative
NMR-based biomedical metabolomics: Current status and applications.
Molecules. 25(5128)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hatzakis E: Nuclear magnetic resonance
(NMR) spectroscopy in food science: A comprehensive review. Compr
Rev Food Sci Food Saf. 18:189–220. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li M, Xu W and Su Y: Solid-state NMR
spectroscopy in pharmaceutical sciences. Trends Anal Chem.
135(116152)2021.
|
|
23
|
D'cruz D, Babu A and Joshy E:
Bioanalytical method development and validation of ticagrelor by
rp-Hplc. Int J App Pharm. 9(51)2017.
|
|
24
|
Aneesh TP, Radhakrishnan R, Sasidharan APM
and Choyal M: RP-HPLC method for simultaneous determination of
losartan and chlorthalidone in pharmaceutical dosage form. Int Res
J Pharm. 6:453–457. 2015.
|
|
25
|
Sobsey CA, Ibrahim S, Richard VR, Gaspar
V, Mitsa G, Lacasse V, Zahedi RP, Batist G and Borchers CH:
Targeted and untargeted proteomics approaches in biomarker
development. Proteomics. 20(1900029)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gallagher RI, Wulfkuhle J, Wolf DM,
Brown-Swigart L, Yau C, O'Grady N, Basu A, Lu R, Campbell MJ,
Magbanua MJ, et al: Protein signaling and drug target activation
signatures to guide therapy prioritization: Therapeutic resistance
and sensitivity in the I-SPY 2 trial. Cell Rep Med.
4(101312)2023.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Mann M: The rise of mass spectrometry and
the fall of edman degradation. Clin Chem. 62:293–294.
2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Matsudaira P: A Practical Guide to Protein
and Peptide Purification for Microsequencing. Matsudaira P (ed).
2nd edition. Academic Press, San Diego, CA, pp1-13, 1993.
|
|
29
|
Pomerantz SC and McCloskey JA: Analysis of
RNA hydrolyzates by liquid chromatography-mass spectrometry.
Methods Enzymol. 193:796–824. 1990.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Su D, Chan CT, Gu C, Lim KS, Chionh YH,
McBee ME, Russell BS, Babu IR, Begley TJ and Dedon PC: Quantitative
analysis of ribonucleoside modifications in tRNA by HPLC-coupled
mass spectrometry. Nat Protoc. 9:828–841. 2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Cupp-Sutton KA and Wu S: High-throughput
quantitative top-down proteomics. Mol Omics. 16:91–99.
2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Gao Y and Yates JR III: Protein analysis
by shotgun proteomics. In: Mass Spectrometry-Based Chemical
Proteomics. John Wiley & Sons, Ltd., Hoboken NJ, pp1-38,
2019.
|
|
33
|
Rozario LT, Sharker T and Nila TA: In
silico analysis of deleterious SNPs of human MTUS1 gene and their
impacts on subsequent protein structure and function. PLoS One.
16(e0252932)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Huang C, Hou C, Ijaz M, Yan T, Li X, Li Y
and Zhang D: Proteomics discovery of protein biomarkers linked to
meat quality traits in post-mortem muscles: Current trends and
future prospects: A review. Trends Food Sci Technol. 105:416–432.
2020.
|
|
35
|
Yokota H: Applications of proteomics in
pharmaceutical research and development. Biochim Biophys Acta
Proteins Proteom. 1867:17–21. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhao Y, Sun Q and Huo B: Focal adhesion
regulates osteogenic differentiation of mesenchymal stem cells and
osteoblasts. Biomater Transl. 2:312–322. 2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
de Oliveira Garcia FA, de Andrade ES and
Palmero EI: Insights on variant analysis in silico tools for
pathogenicity prediction. Front Genet. 13(1010327)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Deshpande RR, Tiwari AP, Nyayanit N and
Modak M: In silico molecular docking analysis for repurposing
therapeutics against multiple proteins from SARS-CoV-2. Eur J
Pharmacol. 886(173430)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Cramer P: AlphaFold2 and the future of
structural biology. Nat Struct Mol Biol. 28:704–705.
2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Cavasotto CN and Phatak SS: Homology
modeling in drug discovery: Current trends and applications. Drug
Discov Today. 14:676–683. 2009.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Taha MO, Atallah N, Al-Bakri AG,
Paradis-Bleau C, Zalloum H, Younis KS and Levesque RC: Discovery of
new MurF inhibitors via pharmacophore modeling and QSAR analysis
followed by in-silico screening. Bioorg Med Chem. 16:1218–1235.
2008.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Huang YZ, Chen SY and Deng F:
Well-characterized sequence features of eukaryote genomes and
implications for ab initio gene prediction. Comput Struct
Biotechnol J. 14:298–303. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Dereeper A, Audic S, Claverie JM and Blanc
G: BLAST-EXPLORER helps you building datasets for phylogenetic
analysis. BMC Evol Biol. 10(8)2010.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Meglécz E, Pech N, Gilles A, Dubut V,
Hingamp P, Trilles A, Grenier R and Martin J: QDD version 3.1: A
user-friendly computer program for microsatellite selection and
primer design revisited: Experimental validation of variables
determining genotyping success rate. Mol Ecol Resour. 14:1302–1313.
2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Jess R, Ling T, Xiong Y, Wright CJ and
Zhao F: Mechanical environment for in vitro cartilage tissue
engineering assisted by in silico models. Biomater Transl. 4:18–26.
2023.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wu YW, Simmons BA and Singer SW: MaxBin
2.0: An automated binning algorithm to recover genomes from
multiple metagenomic datasets. Bioinformatics. 32:605–607.
2016.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Xiao J and Goley ED: Redefining the roles
of the FtsZ-ring in bacterial cytokinesis. Curr Opin Microbiol.
34:90–96. 2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Paulin S, Jamshad M, Dafforn TR,
Garcia-Lara J, Foster SJ, Galley NF, Roper DI, Rosado H and Taylor
PW: Surfactant-free purification of membrane protein complexes from
bacteria: Application to the staphylococcal penicillin-binding
protein complex PBP2/PBP2a. Nanotechnology.
25(285101)2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Meeske AJ, Riley EP, Robins WP, Uehara T,
Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG and Rudner
DZ: SEDS proteins are a widespread family of bacterial cell wall
polymerases. Nature. 537:634–638. 2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Noinaj N, Kuszak AJ, Gumbart JC, Lukacik
P, Chang H, Easley NC, Lithgow T and Buchanan SK: Structural
insight into the biogenesis of β-barrel membrane proteins. Nature.
501:385–390. 2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Rassam P, Copeland NA, Birkholz O, Tóth C,
Chavent M, Duncan AL, Cross SJ, Housden NG, Kaminska R, Seger U, et
al: Supramolecular assemblies underpin turnover of outer membrane
proteins in bacteria. Nature. 523:333–336. 2015.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Foster TJ: The MSCRAMM family of
cell-wall-anchored surface proteins of gram-positive cocci. Trends
Microbiol. 27:927–941. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Lecointe K, Cornu M, Leroy J, Coulon P and
Sendid B: Polysaccharides cell wall architecture of mucorales.
Front Microbiol. 10(469)2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Heinisch JJ and Rodicio R: Protein kinase
C in fungi-more than just cell wall integrity. FEMS Microbiol Rev
42: 10.1093/femsre/fux051, 2018.
|
|
55
|
Kermani AA: A guide to membrane protein
X-ray crystallography. FEBS J. 288:5788–5804. 2021.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Alexandrov AI, Mileni M, Chien EYT, Hanson
MA and Stevens RC: Microscale fluorescent thermal stability assay
for membrane proteins. Structure. 16:351–359. 2008.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Kwan TOC, Reis R, Siligardi G, Hussain R,
Cheruvara H and Moraes I: Selection of biophysical methods for
characterisation of membrane proteins. Int J Mol Sci.
20(2605)2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Kermani AA, Aggarwal S and Ghanbarpour A:
Chapter 11 - Advances in X-ray crystallography methods to study
structural dynamics of macromolecules. Advanced Spectroscopic
Methods to Study Biomolecular Structure and Dynamics. 2023:309–355.
2023.
|
|
59
|
Chen D, McCool EN, Yang Z, Shen X,
Lubeckyj RA, Xu T, Wang Q and Sun L: Recent advances (2019-2021) of
capillary electrophoresis-mass spectrometry for multilevel
proteomics. Mass Spectrom Rev. 42:617–642. 2023.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Wu G, Yu C, Wang W, Du J, Xu G, Fu Z and
Wang L: Charge variants analysis of a bispecific antibody using a
fully automated one-step capillary isoelectric focusing-mass
spectrometry method. Curr Pharm Anal. 18:860–870. 2022.
|
|
61
|
Giraudeau P: NMR-based metabolomics and
fluxomics: Developments and future prospects. Analyst.
145:2457–2472. 2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Hiroaki H: Molecular mechanisms of
amyloid-β peptide fibril and oligomer formation: NMR-based
challenges. Biophys Physicobiol. 20(e200007)2023.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Sobolev AP, Thomas F, Donarskic J,
Ingallinad C, Circid S, Marincolae FC, Capitania D and Mannina L:
Use of NMR applications to tackle future food fraud issues. Trends
Food Sci Technol. 91:347–353. 2019.
|
|
64
|
Alexandrov T: Spatial metabolomics and
imaging mass spectrometry in the age of artificial intelligence.
Ann Rev Biomed Data Sci. 3:61–87. 2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Silantyev AS, Falzone L, Libra M, Gurina
OI, Kardashova KS, Nikolouzakis TK, Nosyrev AE, Sutton CW, Mitsias
PD and Tsatsakis A: Current and future trends on diagnosis and
prognosis of glioblastoma: From molecular biology to proteomics.
Cells. 8(863)2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Kuo TH, Dutkiewicz EP, Pei J and Hsu CC:
Ambient ionization mass spectrometry today and tomorrow: Embracing
challenges and opportunities. Anal Chem. 92:2353–2363.
2020.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Kroll AV, Fang RH and Zhang L:
Biointerfacing and applications of cell membrane-coated
nanoparticles. Bioconjug Chem. 28:23–32. 2017.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Ahmad R and Budnik B: A review of the
current state of single-cell proteomics and future perspective.
Anal Bioanal Chem. 415:6889–6899. 2023.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Khan WA: Chapter 3-Whole-exome and
whole-genome sequencing in the molecular diagnostic laboratory. In:
Diagnostic Molecular Pathology. Coleman WB and Tsongalis GJ (eds).
2nd edition. Academic Press, San Diego, CA pp27-38, 2024.
|
|
70
|
Wang F, Guo J, Wang S, Wang Y, Chen J, Hu
Y, Zhang H, Xu K, Wei Y, Cao L, et al: B-cell lymphoma-3 controls
mesenchymal stem cell commitment and senescence during skeletal
aging. Clin Transl Med. 12(e955)2022.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Al Huq Mohammed S and Rajamani K: Role of
chaperones and endoplasmic reticulum stress in protein complexity
associated with dyslipidemia: A future perspective to novel
therapeutics (Review). World Acad Sci J. 6:1–9. 2024.
|
|
72
|
Kingsak M, Maturavongsadit P, Jiang H and
Wang Q: Cellular responses to nanoscale substrate topography of
TiO2 nanotube arrays: Cell morphology and adhesion. Biomater
Transl. 3:221–233. 2022.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Xue X, Liu H, Wang S, Hu Y, Huang B, Li M,
Gao J, Wang X and Su J: Neutrophil-erythrocyte hybrid
membrane-coated hollow copper sulfide nanoparticles for targeted
and photothermal/ anti-inflammatory therapy of osteoarthritis.
Composites Part B: Engineering. 237(109855)2022.
|
|
74
|
Nie D, Hu Y, Chen Z, Li M, Hou Z, Luo X,
Mao X and Xue X: Outer membrane protein A (OmpA) as a potential
therapeutic target for Acinetobacter baumannii infection. J Biomed
Sci. 27(26)2020.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Chen Z, Wang Y, Cheng Y, Wang X, Tong S,
Yang H and Wang Z: Efficient biodegradation of highly crystallized
polyethylene terephthalate through cell surface display of
bacterial PETase. Sci Total Environ. 709(136138)2020.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Ding JL, Hou J, Feng MG and Ying SH:
Transcriptomic analyses reveal comprehensive responses of insect
hemocytes to mycopathogen Beauveria bassiana, and fungal
virulence-related cell wall protein assists pathogen to evade host
cellular defense. Virulence. 11:1352–1365. 2020.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Wang J, Xu W, Wang R, Cheng R, Tang Z and
Zhang M: The outer membrane protein Amuc_1100 of Akkermansia
muciniphila promotes intestinal 5-HT biosynthesis and extracellular
availability through TLR2 signalling. Food Funct. 12:3597–3610.
2021.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Devi SM, Raj N and Sashidhar RB: Efficacy
of short-synthetic antifungal peptides on pathogenic Aspergillus
flavus. Pestic Biochem Physiol. 174(104810)2021.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Peng J, Wu L, Zhang W, Zhang Q, Xing Q,
Wang X, Li X and Yan J: Systemic identification and functional
characterization of common in fungal extracellular membrane
proteins in lasiodiplodia theobromae. Front Plant Sci.
12(804696)2021.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Doyle MT and Bernstein HD: Bacterial outer
membrane proteins assemble via asymmetric interactions with the
BamA β-barrel. Nat Commun. 10(3358)2019.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Oluwole A, Shutin D and Bolla JR: Mass
spectrometry of intact membrane proteins: shifting towards a more
native-like context. Essays Biochem. 67:201–213. 2023.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Peterson JH, Doyle MT and Bernstein HD:
Small molecule antibiotics inhibit distinct stages of bacterial
outer membrane protein assembly. mBio. 13(e0228622)2022.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Sun J, Rutherford ST, Silhavy TJ and Huang
KC: Physical properties of the bacterial outer membrane. Nat Rev
Microbiol. 20:236–248. 2022.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Tomasek D and Kahne D: The assembly of
β-barrel outer membrane proteins. Curr Opin Microbiol. 60:16–23.
2021.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Muras V, Toulouse C, Fritz G and Steuber
J: Respiratory membrane protein complexes convert chemical energy.
Bacterial Cell Walls and Membranes. 2019:301–335. 2019.PubMed/NCBI View Article : Google Scholar
|
|
86
|
den Blaauwen T and Luirink J: Checks and
balances in bacterial cell division. MBio. 10:e00149–e00119.
2019.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Nguyen HTV, Chen X, Parada C, Luo AC, Shih
O, Jeng US, Huang CY, Shih YL and Ma C: Structure of the
heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the
bacterial divisome. Nat Commun. 14(1903)2023.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Grabowski L, Łepek K, Stasiłojć M,
Kosznik-Kwaśnicka K, Zdrojewska K, Maciąg-Dorszyńska M, Węgrzyn G
and Węgrzyn A: Bacteriophage-encoded enzymes destroying bacterial
cell membranes and walls, and their potential use as antimicrobial
agents. Microbiol Res. 248(126746)2021.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Sharma U, Vipra A and Channabasappa S:
Phage-derived lysins as potential agents for eradicating biofilms
and persisters. Drug Discovery Today. 23:848–856. 2018.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Avila-Calderón ED, Ruiz-Palma MDS,
Aguilera-Arreola MG, Velázquez-Guadarrama N, Ruiz EA, Gomez-Lunar
Z, Witonsky S and Contreras-Rodríguez A: Outer membrane vesicles of
gram-negative bacteria: An outlook on biogenesis. Front Microbiol.
12(557902)2021.PubMed/NCBI View Article : Google Scholar
|
|
91
|
McDowell MA, Heimes M and Sinning I:
Structural and molecular mechanisms for membrane protein biogenesis
by the Oxa1 superfamily. Nat Struct Mol Biol. 28:234–239.
2021.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Diederichs KA, Pitt AS, Varughese JT,
Hackel TN, Buchanan SK and Shaw PL: Mechanistic insights into
fungal mitochondrial outer membrane protein biogenesis. Curr Opin
Struct Biol. 74(102383)2022.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Lübeck M and Lübeck PS: Fungal cell
factories for efficient and sustainable production of proteins and
peptides. Microorganisms. 10(753)2022.PubMed/NCBI View Article : Google Scholar
|