Introduction
Environmental factors, such as temperature, salinity
and acidity (pH) can affect the growth of microorganisms and their
response to antibiotics. The spread of pathogens (vector-borne,
waterborne, airborne and foodborne) and the occurrence of disease
are sensitive to the effects of climate change. Fluctuations in
temperature, rainfall, acidity, salinity and El Niño Southern
Oscillation all affect the growth and spread of waterborne
pathogens, such as Vibrio spp. in aquatic environments.
Climate change is expected to increase the rate of antibiotic
resistance in human pathogens by increasing the lateral gene
transfer of mobile genetic resistance genes and increasing
bacterial growth rates, promoting environmental persistence,
carriage and transmission (1). The
study by Mora et al (2)
counted the number of microorganisms that have emerged and
disappeared as a result of climate change. They discovered that 85%
of the 357 infectious diseases controlled by humans have
proliferated, whereas only 16% have decreased due to climate change
(2). Climate change can cause
disease spread for a variety of reasons, including the geographic
expansion of vectors, such as mosquitoes, ticks, fleas, birds and
other mammals as a result of rain, floods, or drought; these
vectors transmit viruses, bacteria, fungi and parasites that have
caused several diseases and epidemics, including dengue fever,
plague, malaria, West Nile fever, cholera, trypanosomiasis,
echinococcosis, and Nipah and Ebola viruses. The change in weather
temperature has also placed individuals closer to pathogens and
their habitats, such as increased water activities, which has
increased the occurrence of waterborne illnesses such as cholera,
primary amoebic meningoencephalitis and gastroenteritis,
salmonellosis, shigellosis and others (2).
Another issue is Arctic ice melting due to global
warming, which has resulted in the spread of Bacillus anthracis due
to the reappearance of a previously frozen bacterial strain
(3). In addition, the increasing
rainfall has resulted in a decrease in water salinity, leading to
the appearance of cases of cholera due to the increased quantity of
Vibrio cholerae (V. cholerae) bacteria in the water
(4). In some situations, climate
change has negatively influenced particular types of diseases,
causing their decline or elimination, such as, SARS, COVID-19,
rotavirus and norovirus enteritis (5,6).
Generally, antimicrobial resistance and climate
change are two of the top health crises; they are connected
directly, and both affect human health by increasing the mortality
rates. On the other hand, indirectly, climate change causes
fluctuating temperatures, storms, forest fires and flooding. This
has produced shifts in the ecosystem, increasing the transmission
of vectors and infections and incorrect antibiotic usage, all of
which lead to increased antimicrobial resistance (7).
Numerous human pathogens, including V.
cholerae, are naturally found in aquatic environments in
rivers, coastal and estuarine habitats. V. cholerae is a key
pathogen that causes cholera and has played a crucial role in human
history. Its ability to survive in a variety of environments is
largely due to the development of a set of adaptation strategies
that enable these bacteria to survive under stresses such as
nutrient deficiency, variations in salinity and temperature, and
resistance to bacteriophage. Such a mechanism is the transformation
to a viable but nonculturable state and has the capacity to grow as
a biofilm on a variety of biotic (chitinous and gelatinous
zooplankton and phytoplankton) and abiotic surfaces, allowing V.
cholerae to disseminate to new watercourses through passive
transfer or mechanical transfer (8).
Similar to V. cholerae, Morganella
morganii (M. morganii) belongs to the
Gammaproteobacteria class of Gram-negative rod bacteria,
which are facultative anaerobic motile bacteria with flagella.
M. morganii is widely distributed in the natural environment
and as a normal flora in the intestinal tracts of humans and
different animals. It is an opportunistic bacterium that clinically
causes nosocomial infections in the urinary tract, hepatobiliary
tract, skin and soft tissue, wounds and blood (9). Previously, little consideration was
paid to these bacteria due to their rarity and low potential for
causing nosocomial epidemics. However, M. morganii was then
classified as a key pathogen due to its high mortality rate in some
infections because of its virulence and elevated antibiotic
resistance. This resistance is essentially acquired via
extragenetic and mobile elements (10).
The impact of climate change on fungal infections is
not yet fully known. Climate change is creating conditions that
encourage the introduction of novel fungal infections and preparing
fungi to adapt to previously hostile habitats. The present study
selected two types of fungi: Aspergillus niger (A.
niger) and Trichoderma harzianum (T. harzianum).
Aspergillus is a genus in the order Eurotiales that has a
global distribution and a diverse range of ecological habitats
(11). Fungi are easily influenced
by nutritional and physiological factors; therefore, even modest
environmental changes can alter their morphological traits
(12). In general, the dietary
requirements for fungal growth are uncomplicated; nonetheless,
various fungi require distinct physical, chemical, and nutritional
environments. As a result, understanding the fungal habitat is
contingent on knowing its temperature, hydrogen ion concentration
(pH) and nutritional requirements. These fungi usually thrive in a
laboratory at temperatures ranging from 0 to 40˚C. Additionally,
the maximum metabolic activity, cellular growth, conidial
generation and sporulation of Aspergillus spp. have been
found in liquid conditions with an optimum pH range of 5-7
(13-15).
Trichoderma, which belongs to the order Hypocreales, can be found
in all climate zones. These fungi are typically found in soil and
rotting wood (16). T.
harzianum has extensive temperature, salinity and pH growth
ranges, suggesting that it can adapt to changes in environmental
conditions (17). The present
study aimed to determine the effects of climate change on the
growth rate and antibiotic resistance of microorganisms living in
surface water.
Materials and methods
Sample collection and isolation of
bacteria
Water was collected from various locations along the
Tigris river in Baghdad, Iraq throughout the year. Water samples
(250 ml) were collected in sterile containers and filtered through
0.22-µm membrane filters. For the isolation of V. cholerae,
the membranes were then incubated in 100 ml alkaline peptone water
(HiMedia Laboratories, LLC) overnight at 37˚C. A loop full of the
overnight culture was streaked onto thiosulfate citrate bile
sucrose (TCBS) agar (HiMedia Laboratories, LLC) and incubated at
37˚C overnight, as previously described (18). Yellow colonies were examined under
a compound microscope (Olympus Corporation), and the comma-shaped
colonies were then diagnosed after 6-7 h using a Vitek 2 compact
system (Biomerieux). To isolate M. morganii, the membranes
were incubated for 24 h at 37˚C in 100 ml nutrient broth (Oxoid;
Thermo Fisher Scientific, Inc.). Subsequently, a loop full of
overnight culture was streaked on MacConkey agar (HiMedia
Laboratories, LLC), and the pale, non-lactose-fermented colonies
were diagnosed using a Vitek 2 Compact system after 4-5 h, as
previously descried (19).
Effects of different temperatures, pH
value and salinities on bacterial growth
To examine the effects of different environmental
conditions on bacterial growth in the laboratory, a 100-ml conical
flask was used with a volume of 50 ml nutrient broth (Oxoid; Thermo
Fisher Scientific, Inc.) and 1 ml inoculum at a concentration of
1.5x108 CFU/ml, depending on the 0.5 McFarland standard.
The flasks were incubated at different temperatures (17, 27 and
37˚C) for 24 h and aerated by shaking at 200 rpm to measure the
effects of temperature on V. cholerae and M. morganii
growth (20). The same conditions
were used to examine the effects of various pH values (5, 6, 7, 8,
9 and 10) and different percentages of salinity (0, 2, 4, 6, 8, 10
and 14%).
Antibiotic susceptibility
The antibiotic susceptibility of V. cholerae
and M. morganii was determined using the disk diffusion
method on Mueller-Hinton agar (Oxoid; Thermo Fisher Scientific,
Inc.), in accordance with the Clinical and Laboratory Standards
Institute (CLSI) guidelines (21).
The bacterial strains were examined under two sets of conditions as
follows: i) For V. cholerae, the normal growth conditions
were 37˚C, pH 8.6, and 2% salinity, whereas extreme conditions
included 27˚C, pH 10 and 4% salinity; ii) for M. morganii,
normal growth occurred at 37˚C, pH7, whereas extreme conditions
were set at 27˚C, pH 10, 6% salinity.
The following antibiotics from (Bioanalyse) were
used in the test: Tetracycline (30 µg), piperacillin/tazobactam (10
µg), ampicillin (25 µg), ampicillin (10 µg), colistin (10 µg),
amoxicillin/clavulanic acid (30 µg), amikacin (30 µg); amikacin (10
µg), cefotaxime (30 µg), and piperacillin (100 µg).
Sample collection and isolation of
fungi
Fungal isolates of A. niger and T.
harzianum were obtained from the aquatic environment in Iraq
(Tigris River). The isolation and cultivation were completed
depending on established protocols (22). A total of 1 ml of each sample was
aseptically inoculated onto sterile 9-cm glass Petri dishes, and
potato dextrose agar (PDA) medium (NEOGENLAB US) was then added
with chloramphenicol (250 mg dissolved in 250 ml distilled water)
to inhibit bacterial contamination. The plates were incubated at
25˚C for 48 h. The growing colonies were sub-cultured on PDA and
incubated for 7 days at 28˚C. Microscopic (Optika Microscopes)
examination and standard taxonomic keys were used to identify
fungal species (23).
Of note, two methods were employed for the
preparation of fungal inocula. First, spore suspensions were
generated by the addition of 10 ml sterile distilled water to
plates containing pure fungal colonies. A total of 5 ml of the
resulting suspension were transferred into sterile glass bottles
containing 95 ml sterile saline solution and were thoroughly mixed.
Spore concentrations were determined using a dilution technique to
achieve the desired inoculum density, as previously described
(23,24). The final spore suspension densities
were 1.7x10³ spores/ml for A. niger and 1.9x10³ spores/ml
for T. harzianum. Alternatively, 7-mm-diameter mycelial
discs were excised from actively growing fungal colonies and used
in downstream applications.
Analysis of the effects of salinity,
acidity and temperature
For the experiments, various concentrations of
sodium chloride were used, including 2, 4, 6, 8, 10, 12 and 14%,
which were added to the culture medium (PDA). Following
sterilization in an autoclave at 121˚C and 1.5 bar pressure, the
media were poured into sterile plastic petri dishes containing 1 ml
chloramphenicol (HiMedia Laboratories, LLC) (250 ppm) and left to
solidify. Subsequently, with a cork piercing a 7-mm size, each
fungal isolate was removed and placed in a dish with a control,
which was a Petri dish containing the antibiotic and culture media
and fungus only. The Petri dishes were incubated at 28˚C for 2-7
days, after which the diameters of the developing colonies and the
growth rates were measured. The previous steps used different pH
values ranging from 5-10 to measure the effect of acidity on fungal
growth. In the temperature experiment, three degrees were used: 17,
27 and 27˚C.
Antifungal resistance of A. niger and
T. harzianum under different growth conditions
In total, five types of antifungals were used from
(HiMedia Laboratories, LLC): Ketoconazole (KT), itraconazole (IT),
amphotericin B (AP), nystatin (NS 50) and miconazole (MIC 50), and
their effects on two fungal isolates (A. niger and T.
harzianum) were investigated. PDA was prepared at different pH
values and NaCl (HIMEDIA/India) concentrations (5 and 10%) and
autoclaved at 121˚C and 1.5 psi. After cooling, the media were
transferred to sterile plastic Petri dishes containing 1 ml spore
suspension, as previously described (25), for each fungal isolate and 1 ml of
the antibiotic, chloramphenicol (HiMedia Laboratories, LLC). They
were allowed to firm up before the antifungal tablets were applied
to each dish and incubated for 2-7 days at 28˚C.
Results
The growth and survival of microorganisms are
influenced by their strains and a range of environmental factors,
such as temperature and humidity, beneficial nutrients, pH, gas
conditions and osmotic pressure (26). In the present study, to determine
the effects of some of these parameters (TM, pH, and salinity) on
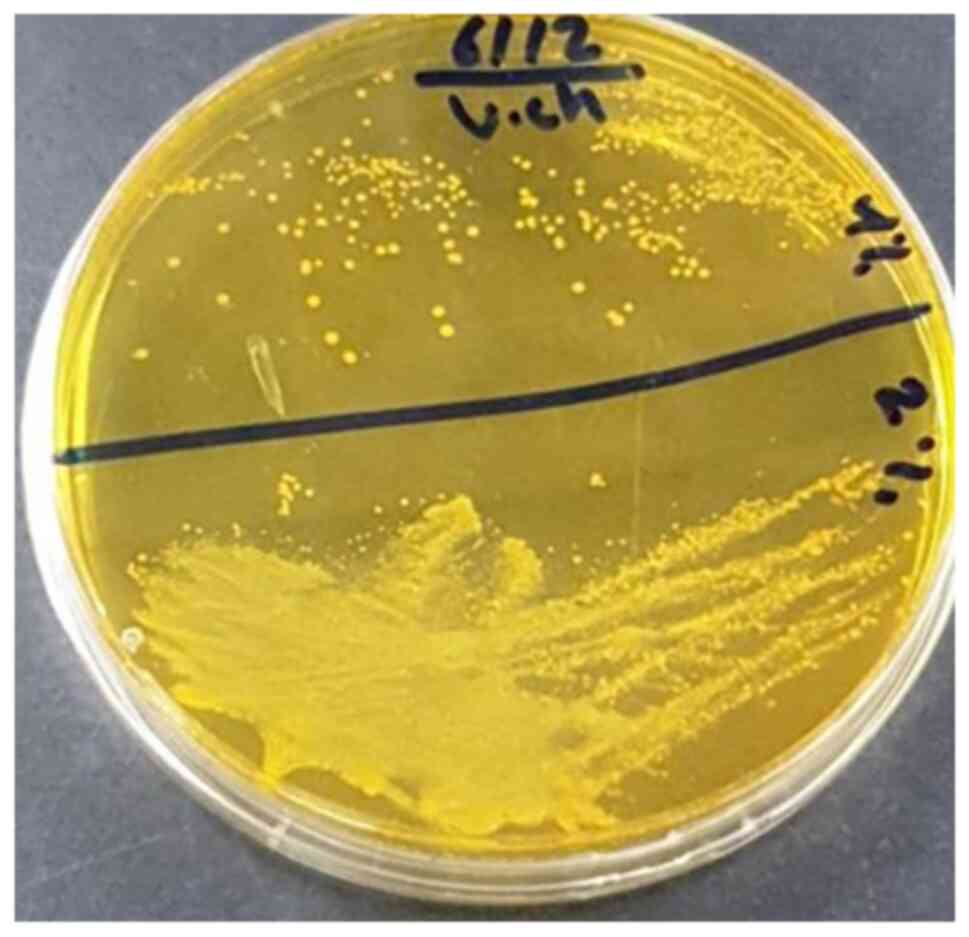
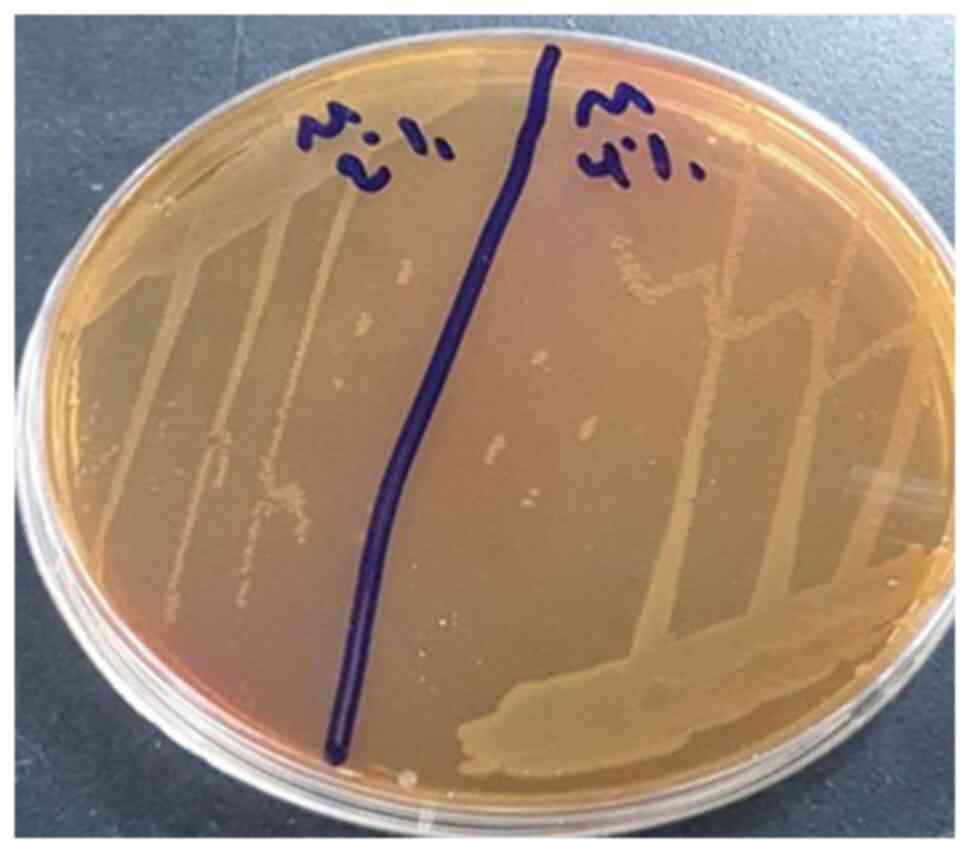
different microorganisms, V. cholerae (Fig. 1) and M. morganii (Fig. 2) were selected as bacteria, and
A. niger (Fig. 3) and T.
T. harzianum (Fig. 4) were
used as the fungi. All the microbes were isolated and diagnosed
locally. As shown in Table I, it
was found that V. cholerae could grow and survive at 4%
salinity and M. morganii could grow in 6% NaCl media at 37˚C
for 24 h of incubation; changing the acidity factor did not affect
bacterial growth under the same incubation conditions. It was found
that V. cholerae and M. morganii could survive
temperatures ranging from 17 to 37˚C. When incubated under extreme
growth conditions, V. cholerae thrived at 10 pH/4% NaCl/17˚C
and 10 pH/4% NaCl/27˚C, whereas M. morganii survived at 10
pH/6% NaCl/17˚C and 10 pH/6% NaCl/27˚C.
 | Table IImpact of environmental parameters on
the growth of microorganisms. |
Table I
Impact of environmental parameters on
the growth of microorganisms.
| | Growth of
bacteria | Growth of
fungi |
|---|
| Variables Salinity
(NaCl %) | Vibrio
cholerae | Morganella
morganii | Colony
diameter(mm) of Aspergillus niger | Colony diameter
(mm) of Trichoderma harzianum |
|---|
|
0 | + | + | 77.5 | 82.5 |
|
2 | + | + | 75 | 55 |
|
4 | + | + | 60 | 25 |
|
6 | - | + | 50 | 12.5 |
|
8 | - | - | 45 | - |
|
10 | - | - | 30 | - |
|
12 | - | - | - | - |
|
14 | - | - | - | - |
| Acidity (pH) | | | | |
|
5 | + | + | 75 | 78 |
|
6 | + | + | 55 | 78 |
|
7 | + | + | 60 | 78 |
|
8 | + | + | 60 | 79 |
|
9 | + | + | 65 | 79 |
|
10 | + | + | 65 | 79 |
| Temperature,˚C | | | | |
|
17 | + | + | - | - |
|
27 | + | + | 75 | 80 |
|
37 | + | + | - | - |
| pH/NaCl/TM | | | | |
| 10/4%/37˚C | + | + | - | - |
| 10/6%/37˚C | - | + | - | - |
| 10/4%/27˚C | + | + | - | - |
| 10/6%/27˚C | - | + | 30 | - |
| 10/10%/27˚C | - | - | - | - |
| 10/4%/17˚C | + | + | - | - |
| 10/6%/17˚C | - | + | - | - |
The growth diameter of the fungi varied depending on
the growth conditions. As demonstrated in Table I, it was found that A. niger
could live at 10% salinity with a colony diameter of 30 mm, whereas
T. T. harzianum could survive at 6% NaCl with a colony
diameter of 12.5 mm. Both fungi flourished at various acidity
levels, reaching a pH of 10. The effects of temperature on fungal
growth were evident, and the fungi could not tolerate temperatures
of 17 or 37˚C. When different variables were combined, only A.
niger produced a 30-mm colony at pH 10, 6% NaCl, and 27˚C.
Antimicrobial resistance is a critical aspect of the
recent century since the percentage of microorganisms that develop
antibiotic resistance has increased rapidly. The present study used
various types of antibacterial and antifungal agents under various
growth conditions for four types of microorganisms. The findings
presented in Table II
demonstrated how environmental growth conditions can affect
antibiotic resistance and antifungal resistance. V. cholerae
developed resistance to amikacin (10 µg), colistin (10 µg) and
tetracycline (100 µg) under extreme growth conditions, including pH
10, 4% NaCl and 27˚C, while it was intermediately resistant to
amikacin (10 µg), and sensitive to colistin (10 µg) and
tetracycline (100 µg) under normal growth conditions. Notably,
V. cholerae became sensitive to piperacillin (100 µg) after
growth under the same extreme conditions; however, its response to
the other tested antibiotics remained unaltered. The resistance of
M. morganii was largely unaffected by the altered growth
conditions, apart from ampicillin (10 µg), to which it developed
resistance under the same conditions.
 | Table IIInfluence of environmental growth
conditions on the antimicrobial resistance profile of selected
microorganisms. |
Table II
Influence of environmental growth
conditions on the antimicrobial resistance profile of selected
microorganisms.
| A, Antibiotics
(antibacterials) |
|---|
| | Vibrio
cholerae | Morganella
morganii |
|---|
| Medication | Concentration | Normal
conditions | Extreme
conditions | Normal
conditions | Extreme
conditions |
|---|
| Amikacin (AK) | 10 µg | I | R | I | I |
| Amikacin (AK) | 30 µg | S | S | S | S |
| Ampicillin
(AMP) | 10 µg | R | R | I | R |
| Ampicillin
(AMP) | 25 µg | R | R | R | R |
|
Amoxicillin/clavulanic acid (AMC) | 30 µg | R | R | R | R |
| Cefotaxime
(CTX) | 30 µg | R | R | R | R |
| Colistin (CLM) | 10 µg | S | R | R | R |
| Piperacillin
(PRL) | 100 µg | R | S | S | S |
|
Piperacillin/tazobactam (PIT) | 10 µg | R | R | R | R |
| Tetracycline | 100 µg | S | R | R | R |
| B, Antibiotics
(antifungals) |
| | Aspergillus
niger | Trichoderma
harzianum |
| Medication | Concentration | Normal
conditions | Extreme
conditions | Normal
conditions | Extreme
conditions |
| Ketoconazole
(KT) | 10 µg | S | R | R | R |
| Itraconazole
(IT) | 10 µg | S | I | R | R |
| Amphotericin B
(AP) | 100 µg | S | I | I | R |
| Nystatin (NS) | 50 µg | S | I | S | I |
| Miconozole
(Mic) | 50 µg | S | R | S | R |
As regards antifungal resistance, A. niger
exhibited resistance to ketoconazole (10 µg) and miconazole (50 µg)
under conditions of pH pH 10, 6% salinity and 27˚C; it also
exhibited intermediate resistance to itraconazole (IT) 10 µg,
amphotericin B (AP) 100 µg and nystatin (NS) 50 µg. Moreover, T.
T. harzianum was resistant to amphotericin B (100 µg) and
miconozole (50 µg) under the same conditions.
Discussion
Antibiotics and high or low temperatures are
examples of environmental stressors that can alter the selection
rules that exist in an ecosystem. These selection forces can have
an impact on organism population evolution, as well as their
ability to withstand environmental shocks (27,28).
The present study revealed that bacterial strains exhibited greater
resilience to diverse growth environments than fungal strains. The
observed outcome may be attributed to the ability of bacteria to
synthesize the sugar trehalose in response to different
environmental factors, including low temperatures, oxidative
stress, osmotic shock, acid stress and ethanol exposure (29).
Of note, environmental conditions can affect the
external appearance and internal composition of bacteria. Nhu et
al (30) found that variations
in the acidity of the surrounding environment affected colony
morphology, leading to the production of denser colonies with
irregular edges, their results illustrate how bacterial morphology
changes due to environmental stresses, which can affect their
pathogenicity and identification in changing environmental contexts
(30).
As regards the ability of V. cholerae to
adapt to pH and temperature, the growth characteristics observed in
the present study are consistent with those found in other
published research. According to previous research, the optimal pH
range for growth is between 6 and 9 (31-33),
and the optimal temperature range is between 30 and 37˚C, although
it can survive at 6˚C (34).
However, the results of the present study make a noteworthy
distinction by indicating salinity as a key determinant for the
growth and environmental persistence of V. cholerae. It is
important to note that at sodium chloride concentrations >4%, no
growth was observed. Furthermore, while this observation regarding
the effects of salinity contradicts the findings of Singleton et
al (35) and Huq et al
(31) regarding the growth of the
cholera vibrio strain at higher concentrations of sodium chloride,
it also suggests that strains may differ in their ability to
withstand osmotic pressure; these changes in environmental factors
can influence the conversion of environmental strains to pathogenic
strains and trigger anew outbreak at any time (36). As regards M. morganii, the
results were consistent with the findings of Frith et al
(37) that this bacterium can
tolerate various growth conditions of pH, salinity and temperature;
it was isolated from all sites investigated, despite the different
environmental circumstances.
The antimicrobial susceptibility of the
microorganisms under investigation varied depending on the growth
factors. Generally, at 27˚C, V. cholerae became resistant to
amikacin and tetracycline. These results are in agreement with
those in the study by Yuan et al (38), which reported that from 20 studies,
648 environmental V. cholerae isolates were investigated,
where the weighted pool resistance rate for tetracycline and
amikacin was 14 and 2%, respectively. Parvin et al (39) analyzed the data of tetracycline
resistance from 2000 to 2018 and demonstrated that 100% of V.
cholerae strains were susceptible to tetracycline between 2000
and 2004. Thereafter, a decline in susceptibility rate was
observed, decreasing to 6% between 2012 and 2017, then increasing
again to 76% in 2018(39).
These medications (amikacin and tetracycline), which
bind to ribosomes, have been demonstrated to either induce a
cold-shock-like pattern of protein production or to interact with
other stressors in a manner that is comparable to cold temperatures
in Escherichia coli <22-27˚C. Given that one of the main
effects of cold stress is translational block, the cellular
machinery involved in response to these antibiotics may overlap
with that involved in cold shock (40). When bacteria are repeatedly exposed
to various growth environments, cross-tolerance may arise. This
occurs when early exposure to a stressor leads to tolerance to a
different type of stress. A previous study suggested that the
mechanisms for tolerance may be shared among antibiotics that
target similar processes rather than through generalized cell
dormancy, creating multidrug-resistant bacteria (34). These shared tolerance mechanisms
can also adapt to various types of stressors (such as temperature,
pressure, or pH) (34).
After being subjected to a range of growth
conditions, V. cholerae becomes resistant to colistin.
Colistin is a last-resort antibiotic used to treat serious
infections caused by Gram-negative bacteria (41). According to the study by Sharif
et al (42), V.
cholerae is typically susceptible to this antibiotic, with 59%
of the 53 isolates being susceptible and 26% being resistant. As
the resistance gene (mcr) is carried on a plasmid that may be
transmitted from humans to animals, acquiring resistance to this
antibiotic poses a new threat to human life by decreasing the
effectiveness of eradicating harmful Gram-negative bacteria
(41).
Drug resistance in Enterobacteriaceae pathogens
(V. cholerae and M. morganii) is largely caused by
the frequent transfer of extrachromosomal mobile genetic elements
from nearby or distantly related bacterial species, even though
chromosomal alterations can also play a role in antimicrobial
resistance. The primary carriers of genetic characteristics
expressing antimicrobial resistance function found in the isolates
of enteric pathogens include plasmids, insertion sequences,
superintegrons, integrating conjugative elements and transposable
elements (43,44).
Antifungal resistance develops as a result of the
changes that affect how the antifungal interacts with its target
directly or indirectly. For example, mutations in genes encoding
lanosterol demethylase (ketoconazole and miconazole) can cause
changes in the binding site of the target that ultimately lead to
resistance. Increased drug efflux activity for intracellular
medications, such as azoles can increased target availability or
modifications to the effective drug concentration can also lead to
resistance (45,46). Polyene antifungals, such as
amphotericin B and nystatin target ergosterol, an essential
membrane component, rather than an essential cellular enzyme. This
distinct mode of action could potentially account for the
infrequent incidence of polyene resistance in fungi. However,
changing the sterol makeup of the plasma membrane is the most
well-known method of achieving polyene resistance (47). The findings of the present study
may represent a basis for future investigations into the effects of
climate factors on the phenotypic and genetic levels. Genetic
mutations are widely known as key drivers of novel epidemics and
diseases. Thence, it is recommended that following studies focus on
the elucidation of the genetic impacts of these factors on
microorganisms, which is essential for advancing the understanding
of pathogenic and disease evolution.
In conclusion, the present study demonstrates that
microbial growth and antimicrobial resistant are affected by
changes in climate and climate-related factors, such as
temperatures, salinity and environmental pH. These changes may
contribute to the emergence of new strains harboring multidrug
resistance genes.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
SAK was involved in the conception of the study, in
the bacteriological work, in the analysis of the results, and in
the writing of the manuscript. SHO was involved the fungal work,
and in the writing of the fungal-related sections of the
manuscript. Both authors have read and approved the final
manuscript. SAK and SHO confirm authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
SAK (PhD in Biotechnology; Masters degree in
Microbiology (Bacteriology) ORCID ID: https://orcid.org/0000-0002-4346-306X], resides and
works in Baghdad, Iraq under the scientific research commission as
a scientific researcher. SHO (Masters degree in Biology; ORCID ID:
https://orcid.org/0009-0000-2104-0990) resides and
works in Baghdad, Iraq under the scientific research
commission.
References
|
1
|
Cavicchioli R, Ripple WJ, Timmis KN, Azam
F, Bakken LR, Baylis M, Behrenfeld MJ, Boetius A, Boyd PW, Classen
AT, et al: 2019: Scientists' warning to humanity: microorganisms
and climate change. Nat Rev Microbiol. 17:569–586. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Mora C, McKenzie T, Gaw IM, Dean JM, von
Hammerstein H, Knudson TA, Setter RO, Smith CZ, Webster KM, Patz JA
and Franklin EC: Over half of known human pathogenic diseases can
be aggravated by climate change. Nat Clim Chang. 12:869–875.
2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Gross M: Permafrost thaw releases
problems. Curr Biol. 29:R39–R41. 2019.
|
|
4
|
Guzman Herrador BR, De Blasio BF,
MacDonald E, Nichols G, Sudre B, Vold L, Semenza JC and Nygård K:
Analytical studies assessing the association between extreme
precipitation or temperature and drinking water-related waterborne
infections: A review. Environ Health. 14(29)2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sobral MFF, Duarte GB, da Penha Sobral
AIG, Marinho MLM and de Souza Melo A: Association between climate
variables and global transmission oF SARS-CoV-2. Sci Total Environ.
729(138997)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chua PLC, Huber V, Ng CFS, Seposo XT,
Madaniyazi L, Hales S, Woodward A and Hashizume M: Global
projections of temperature-attributable mortality due to enteric
infections: a modelling study. Lancet Planet Health. 5:e436–e445.
2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Magnano San Lio R, Favara G, Maugeri A,
Barchitta M and Agodi A: how antimicrobial resistance is linked to
climate change: An global challenges overview of two intertwined.
Int J Environ Res Public Health. 20(1681)2023.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sampaio A, Silva V, Poeta P and
Aonofriesei F: Vibrio spp.: Life strategies, ecology, and risks in
a changing environment. Diversity. 14(97)2022.
|
|
9
|
Li H, Chen Z, Ning Q, Zong F and Wang H:
Isolation and identification of morganella morganii from rhesus
monkey (Macaca mulatta) in China. Vet Sci. 11(223)2024.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Liu H, Zhu J, Hu Q and Rao X: Morganella
morganii, a non-negligent opportunistic pathogen. Int J Infect Dis.
50:10–17. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wheeler KA, Hurdman BF and Pitt JI:
Influence of pH on the growth of some toxigenic species of
Aspergillus, Penicillium and Fusarium. Int J Food Microbiol.
12:141–149. 1991.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Rhouma A, Salem IB, M'hamdi M and
Boughalleb-M'Hamdi N: Relationship study among soils
physico-chemical properties and Monosporascus cannonballus
ascospores densities for cucurbit fields in Tunisia. Eur J Plant
Pathol. 153:65–78. 2019.
|
|
13
|
Abdel-Hadi A and Magan N: Influence of
physiological factors on growth, sporulation and ochratoxin A/B
production of the new Aspergillus ochraceus grouping. World
Mycotoxin J. 2:429–434. 2009.
|
|
14
|
Cao C, Li R, Wan Z, Liu W, Wang X, Qiao J,
Wang D, Bulmer G and Calderone R: The effects of temperature, pH,
and salinity on the growth and dimorphism of Penicillium marneffei.
Med Mycol. 45:401–407. 2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Abubakar A, Suberu HA, Bello IM,
Abdulkadir R, Daudu OA and Lateef AA: Effect of pH on mycelial
growth and sporulation of Aspergillus parasiticus. J Plant Sci.
1:64–67. 2013.
|
|
16
|
Druzhinina IS, Kopchinskiy AG, Komoń M,
Bissett J, Szakacs G and Kubicek CP: An oligonucleotide barcode for
species identification in Trichoderma and Hypocrea. Fungal Genet
Biol. 42:813–828. 2005.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Jones EG, Suetrong S, Sakayaroj J, Bahkali
AH, Abdel-Wahab MA, Boekhout T and Pang KL: Classification of
marine ascomycota, basidiomycota, blastocladiomycota and
chytridiomycota. In: Fungal Diversity. Vol 73. Springer, New York,
NY, pp1-72, 2015.
|
|
18
|
Daboul J, Weghorst L, DeAngelis C, Plecha
SC, Saul-McBeth J and Matson JS: Characterization of Vibrio
cholerae isolates from freshwater sources in northwest Ohio. PLoS
One. 15(e0238438)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Pincus DH: Microbial identification using
the bioMérieux Vitek® 2 system. In: Encyclopedia of Rapid
Microbiological Methods. Parenteral Drug Association, Bethesda, MD,
pp1-32, 2006.
|
|
20
|
Silva APRD, Longhi DA, Dalcanton F and
Aragão GMFD: Modelling the growth of lactic acid bacteria at
different temperatures. Braz arch biol Technol.
61(e18160159)2018.
|
|
21
|
CLSI: Methods for Antimicrobial Dilution
and Disk Susceptibility Testing of Infrequently Isolated or
Fastidious Bacteria. 3rd edition. Clinical and Laboratory Standards
Institute, Wayne, PA, 2015.
|
|
22
|
Romero MC, Hammer E, Hanschke R, Arambarri
AM and Schauer F: Biotransformation of biphenyl by filamentous
fungus Talaromyces helicus. World J Microbiol Biotechnol.
21:101–106. 2005.
|
|
23
|
Webster J (ed): Introduction to Fungi.
Cambridge University Press, New York, NY, p669, 1980.
|
|
24
|
Moor-Landecker E: Fundamentals of Fungi.
Prentice-Hall India, New Delhi, pp265-277, 1972.
|
|
25
|
CLSI: Method for Antifungal Disk Diffusion
Susceptibility Testing of Nondermatophyte Filamentous Fungi.
Clinical and Laboratory Standards Institute, Wayne, PA, 2010.
|
|
26
|
Qiu Y, Zhou Y, Chang Y, Liang X, Zhang H,
Lin X, Qing K, Zhou X and Luo Z: The effects of ventilation,
humidity, and temperature on bacterial growth and bacterial genera
distribution. Int J Environ Res Public Health.
19(15345)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Buckley LB and Huey RB: How extreme
temperatures impact organisms and the evolution of their thermal
tolerance. Integr Comp Biol. 56:98–109. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Donhauser J, Niklaus PA, Rousk J, Larose C
and Frey B: Temperatures beyond the community optimum promote the
dominance of heat-adapted, fast growing and stress resistant
bacteria in alpine soils. Soil Biol Biochem. 148(107873)2020.
|
|
29
|
Rodríguez-Verdugo A, Lozano-Huntelman N,
Cruz-Loya M, Savage V and Yeh P: Compounding effects of climate
warming and antibiotic resistance. IScience.
23(101024)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Nhu NT, Wang HJ and Dufour YS: Acidic pH
reduces Vibrio cholerae motility in mucus by weakening flagellar
motor torque. bioRxiv: Dec 10, 2019 (Epub ahead of print). doi:
https://doi.org/10.1101/871475.
|
|
31
|
Huq A, West PA, Small EB, Huq MI and
Colwell RR: Influence of water temperature, salinity, and pH on
survival and growth of toxigenic Vibrio cholerae serovar O1
associated with live copepods in laboratory microcosms. Appl
Environ Microbiol. 48:420–424. 1984.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Nhu NTQ, Lee JS, Wang HJ and Dufour YS:
Alkaline pH increases swimming speed and facilitates mucus
penetration for Vibrio cholerae. J Bacteriol. 203:e00607–20.
2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kostiuk B, Becker ME, Churaman CN, Black
JJ, Payne SM, Pukatzki S and Koestler BJ: Vibrio cholerae alkalizes
its environment via citrate metabolism to inhibit enteric growth in
vitro. Microbiol Spectr. 11(e0491722)2023.PubMed/NCBI View Article : Google Scholar
|
|
34
|
McCarthy SA: Effects of temperature and
salinity on survival of toxigenic Vibrio cholerae O1 in seawater.
Microb Ecol. 31:167–175. 1996.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Singleton FL, Attwell R, Jangi S and
Colwell RR: Effects of temperature and salinity on Vibrio cholerae
growth. Appl Environ Microbiol. 44:1047–1058. 1982.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Montilla R, Chowdhury MA, Huq A, Xu B and
Colwell RR: Serogroup conversion of Vibrio cholerae non-O1 to
Vibrio cholerae O1: Effect of growth state of cells, temperature,
and salinity. Can J Microbiol. 42:87–93. 1996.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Frith A, Hayes-Mims M, Carmichael R and
Björnsdóttir-Butler K: Effects of environmental and water quality
variables on histamine-producing bacteria concentration and species
in the northern Gulf of Mexico. Microbiol Spectr.
11(e0472022)2023.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Yuan XH, Li YM, Vaziri AZ, Kaviar VH, Jin
Y, Jin Y, Maleki A, Omidi N and Kouhsari E: Global status of
antimicrobial resistance among environmental isolates of Vibrio
cholerae O1/O139: A systematic review and meta-analysis. Antimicrob
Resist Infect Control. 11(62)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Parvin I, Shahunja KM, Khan SH, Alam T,
Shahrin L, Ackhter MM, Sarmin M, Dash S, Rahman MW, Shahid AS, et
al: Changing susceptibility pattern of Vibrio cholerae O1 isolates
to commonly used antibiotics in the largest diarrheal disease
hospital in Bangladesh during 2000-2018. Am J Trop Med Hyg.
103:652–658. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Cruz-Loya M, Kang TM, Lozano NA, Watanabe
R, Tekin E, Damoiseaux R, Savage VM and Yeh PJ: Stressor
interaction networks suggest antibiotic resistance co-opted from
stress responses to temperature. ISME J. 13:12–23. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Thaotumpitak V, Sripradite J, Atwill ER
and Jeamsripong S: Emergence of colistin resistance and
characterization of antimicrobial resistance and virulence factors
of Aeromonas hydrophila, Salmonella spp., and Vibrio cholerae
isolated from hybrid red tilapia cage culture. PeerJ.
11(e14896)2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Sharif N, Ahmed SN, Khandaker S, Monifa
NH, Abusharha A, Vargas DLR, Díez IT, Castilla AGK, Talukder AA,
Parvez AK and Dey SK: Multidrug resistance pattern and molecular
epidemiology of pathogens among children with diarrhea in
Bangladesh, 2019-2021. Sci Rep. 13(13975)2023.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Das B, Verma J, Kumar P, Ghosh A and
Ramamurthy T: Antibiotic resistance in Vibrio cholerae:
Understanding the ecology of resistance genes and mechanisms.
Vaccine. 38 (Suppl 1):A83–A92. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Luo XW, Liu PY, Miao QQ, Han RJ, Wu H, Liu
JH, He DD and Hu GZ: Multidrug resistance genes carried by a novel
transposon Tn 7376 and a genomic island named MMGI-4 in a
pathogenic morganella morganii isolate. Microbiology Spectrum.
10:e00265–22. 2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Robbins N, Caplan T and Cowen LE:
Molecular evolution of antifungal drug resistance. Annu Rev
Microbiol. 71:753–775. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Osset-Trénor P, Pascual-Ahuir A and Proft
M: Fungal drug response and antimicrobial resistance. J Fungi
(Basel). 9(565)2023.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Sanglard D, Ischer F, Parkinson T,
Falconer D and Bille J: Candida albicans mutations in the
ergosterol biosynthetic pathway and resistance to several
antifungal agents. Antimicrob Agents Chemother. 47:2404–2412.
2003.PubMed/NCBI View Article : Google Scholar
|