1. Introduction
Rabies is an acute, often fatal viral disease that
primarily affects the central nervous system. It is typically
transmitted through bites from infected domestic dogs and wild
carnivorous animals. All warm-blooded animals, including humans,
are susceptible to rabies infection. The rabies virus (RABV), a
member of the Lyssavirus genus, is commonly found in the
salivary glands of rabid animals and is excreted in their saliva.
When an infected animal bites, the virus enters the blood through a
fresh wound. Once introduced, RABV propagates along nerve tissue to
the brain, establishing itself within the central nervous system.
Subsequently, it spreads via nerves to the salivary glands, often
resulting in symptoms such as excessive salivation. The disease
initially manifests with excitation of the central nervous system,
characterized by irritability and aggression. Infected animals may
be highly contagious even before symptoms manifest (1). Despite effective vaccines for pre-
and post-exposure prophylaxis (PEP), rabies remains a significant
public health challenge, particularly in developing countries where
access to PEP is limited. Once clinical symptoms appear, rabies is
almost always fatal, underscoring the urgent need for effective
therapeutic interventions. Rabies is prevalent in >150
countries, mainly in Asia and Africa, where exposure without timely
PEP results in a mortality rate >99%. The RABV genome encodes
five essential proteins: Nucleoprotein (N), phosphoprotein (P),
matrix protein (M), glycoprotein (G) and polymerase (L). These
proteins play critical roles in the viral life cycle, facilitating
attachment to host cells, entry via endocytosis, uncoating and
replication within the host cytoplasm.
Rabies poses a severe threat to human health and
economic stability, particularly in developing countries. According
to the World Health Organization (WHO) 2023 report, rabies causes
~59,000 human deaths annually, with 99% of cases resulting from dog
bites (2). The highest mortality
rates are reported in Asia and Africa, where access to PEP is often
limited. India alone accounts for almost 36% of global
rabies-related deaths, translating to an estimated 20,000
fatalities per year (3). By
contrast, developed nations have largely controlled rabies through
extensive vaccination programs and improved surveillance
measures.
The economic impact of rabies is profound, resulting
in over $8.6 billion in global annual losses. These losses stem
primarily from the costs associated with PEP, the loss of
livestock, and reduced productivity due to illness and death. The
financial burden of rabies is particularly acute for low-income
populations, where the average estimated cost of PEP can be
prohibitive (4). This situation
emphasizes the urgent need for novel therapeutic interventions, as
there are currently no effective antiviral treatments available
once clinical symptoms appear.
Recent epidemiological studies highlight the ongoing
challenges in controlling rabies in specific regions. For instance,
Malaysia has faced recurrent outbreaks of dog-mediated human
rabies, with a significant association between low vaccination
rates and increased incidence of cases (5). Another study conducted from 2015 to
2023 revealed that dogs accounted for 89.35% of confirmed rabies
cases in animals, emphasizing the importance of vaccination
campaigns (6). Therefore, rabies
continues to pose a significant public health threat globally,
particularly in developing regions where access to preventive
measures is limited. Enhanced vaccination efforts and improved
public health strategies are essential to combat this preventable
disease effectively. The urgent need for therapeutic interventions
for symptomatic patients further highlights the complexities
involved in managing rabies as a public health concern.
Effective vaccines have been developed for PEP;
timely administration can prevent rabies development following
exposure. PEP includes wound cleansing, vaccine administration and
equine or human rabies immune globulins (ERIG or HRIG). However,
PEP is ineffective once neurological signs appear. Although there
are rare reports of human rabies survivors, no established
therapies exist. Advances in treatment often stem from key studies
on rabies pathogenesis in animal models (7).
While vaccines have proven effective in preventing
rabies transmission, there is a pressing need for therapeutic
strategies to combat the disease in symptomatic patients. This need
is particularly critical in resource-limited settings where
vaccination coverage remains insufficient. The WHO has initiated
efforts to improve access to rabies vaccines through partnerships
with organizations such as Gavi, focusing on strengthening
surveillance and reporting systems while encouraging multisectoral
collaborations (8).
Conventional treatment methods include
pharmacological therapies, surgical intervention and physical
therapy. These methods have been the cornerstone of medical
practice for decades, as they are often backed by extensive
clinical research, providing established protocols for healthcare
providers. In addition, this approach allows for direct patient
assessment, enabling healthcare providers to tailor treatments
based on individual patient needs (9). In a number of cases, conventional
treatments can yield immediate results, such as pain relief or
symptom management. On the contrary, these approaches can be
resource-intensive, requiring significant time and personnel for
administration and monitoring, and can sometimes be costly in
resource-limited settings (10).
Therefore, treatment strategies have incorporated in silico
approaches, as they provide a better understanding of the treatment
design, disease modelling and drug discovery, which include
molecular docking, virtual screening and machine learning (ML)
algorithms that analyze large datasets to predict treatment
outcomes.
In silico methods can process vast amounts of
data quickly, significantly reducing the time required for drug
discovery and development compared to traditional laboratory
methods. These approaches can easily scale to accommodate larger
datasets without a corresponding increase in resource requirements.
This scalability is particularly beneficial in addressing complex
diseases with multifactorial causes in a cost-effective manner,
while minimizing laboratory work and animal testing. However,
computational predictions need to be validated through experimental
studies, as there is a risk of false positives or negatives. Their
effectiveness relies heavily on the availability and quality of
data. They may not always accurately reflect the complexities of
human biology or disease progression (11).
In recent years, computational methods have emerged
as powerful tools in drug discovery and vaccine design for viral
infections such as rabies. In silico approaches, such as
molecular docking, homology modelling, virtual screening, molecular
dynamics simulations and quantitative structure-activity
relationship (QSAR) models, offer cost-effective and time-efficient
means of identifying potential antiviral compounds and
understanding their mechanisms of action. These methods can predict
interactions between viral proteins and potential inhibitors,
facilitating targeted therapy development (12). However, systematic clinical
research into combination therapies faces challenges due to the
sporadic occurrence of rabies cases. There is a pressing need for
medical approaches that accelerate effective therapy development
through veterinary care and investigational treatments of naturally
infected dogs when appropriate. Understanding the pathogenesis of
RABV in both humans and dogs has advanced significantly, providing
critical insight into severe neurological dysfunction associated
with the virus. Of note, four disease processes need to be managed:
Viral propagation, neuronal degeneration, inflammation and systemic
compromise (13).
The present review aimed to provide a comprehensive
overview of recent advancements in in silico research
focused on RABV and focusses on novel ligand identification,
structural elucidation of key viral proteins, drug repurposing
strategies as potential treatments for rabies, and explores the
design of multi-epitope peptide vaccines. By emphasizing these
developments, we aim to illustrate the potential of computational
approaches in combating rabies and inspire further research in this
vital area. While the biological mechanisms of rabies are
well-documented, addressing its therapeutic challenges necessitates
innovative approaches. Computational methods, with their
cost-effectiveness and predictive capabilities, have emerged as a
powerful solution in the quest for novel treatments.
2. Overview of structure-based drug
discovery methods
Given the lack of effective treatments
post-infection, structure-based drug discovery offers a promising
avenue by exploiting detailed molecular insights into the key
proteins of the virus. In silico methods, which encompass
computer-based techniques, have become essential in drug discovery.
These methods utilize computational tools and algorithms to
predict, analyze and optimize interactions between ligands
(potential drug molecules) and biological targets, such as proteins
or nucleic acids. By simulating molecular interactions and
screening extensive libraries of compounds, in silico
approaches significantly expedite the identification of promising
drug candidates, while reducing the costs and time associated with
traditional experimental methods. Structure-based drug discovery
has emerged as a promising and efficient strategy for identifying
novel and potent drug candidates in a more efficient and
cost-effective manner than conventional methods (14). This approach relies on the
three-dimensional structures of biological protein targets to
elucidate the molecular basis of diseases. To virtually identify
drug candidates, several preparatory steps for proteins and
compounds are essential for obtaining accurate results. Key steps
include homology modelling, virtual screening, QSAR models and
pharmacophore modelling, molecular docking and molecular dynamics
simulations (12,15). Molecular docking predicts the
preferred orientation of one molecule when bound to another to form
a stable complex. By contrast, molecular dynamics simulations study
the physical movements of atoms and molecules, providing a dynamic
view of molecular interactions. These techniques are cost-effective
and precise enough to predict mechanical characteristics, optimize
structures, and simulate the natural motion of biological
macromolecules (16).
Structure-based virtual screening involves several computational
phases, including target and database preparation, docking,
post-docking analysis, and prioritization of compounds for testing.
Homology modelling predicts protein structures from amino acid
sequences by aligning them with known templates, constructing
models and refining them through various steps (14). QSAR models establish associations
between the properties of chemical substances and their biological
activities to predict the activities of new chemical entities.
Moreover, pharmacophore modelling identifies essential structural
features necessary for a molecule to interact with a specific
biological target, aiding in virtual screening and lead compound
optimization (15). An overview of
the in silico methods used in the drug discovery process,
with their advantages and disadvantages is provided in Table I.
 | Table IIn silico methods,
applications and tools. |
Table I
In silico methods,
applications and tools.
| Method | Description | Application in
rabies drug discovery | Commonly used
tools | Advantages | Limitations | (Refs.) |
|---|
| Molecular
docking | Predicts the
preferred binding orientation of a ligand (drug candidate) with a
target protein. | Identifies
potential inhibitors for rabies nucleoprotein (N), glycoprotein
(G), and RNA-dependent RNA polymerase (RdRp). | AutoDock, AutoDock
Vina, SwissDock, Glide (Schrödinger), GOLD | Cost-effective,
high-throughput screening. | May not account for
protein flexibility and solvent effects. | (23,58) |
| Molecular dynamics
(MD) simulations | Simulates the
physical movements of atoms and molecules over time. | Evaluates the
stability and dynamic interactions of drug candidates with rabies
virus proteins. | GROMACS, AMBER,
CHARMM, NAMD | Provides insight
into binding stability and conformational changes. | Computationally
intensive, requires high-performance computing. | (12) |
| Virtual
screening | Screens large
compound libraries for potential drug candidates based on binding
affinity. | Identifies small
molecules with potential antiviral activity against rabies virus
proteins. | PyRx, Schrodinger's
Glide, MOE, OpenEye FRED, LigandScout | Rapidly analyzes
thousands of compounds. | Requires accurate
target structures for reliable predictions. | (12,15) |
| Homology
modeling | Predicts the 3D
structure of proteins when an experimentally determined structure
is unavailable. | Used to model the
structure of rabies virus RdRp, nucleoprotein, and glycoprotein for
drug discovery. | SWISS-MODEL,
MODELLER, Phyre2, I-TASSER | Provides structural
insights when crystal structures are unavailable. | Accuracy depends on
template selection and alignment quality. | (26) |
| Quantitative
structure- activity relationship (QSAR) models | Establishes
mathematical relationships between chemical properties and
biological activity. | Predicts the
effectiveness of new rabies antiviral compounds before experimental
testing. | KNIME, DeepChem,
PaDEL- Descriptor, ChemOffice, QSARINS | Enables
optimization of drug candidates. | Requires extensive
datasets for training reliable models. | (12,15) |
| Pharmacophore
modeling | Identifies
essential molecular features required for ligand- receptor
interactions. | Helps in the design
of novel inhibitors targeting rabies virus proteins. | LigandScout,
PharmaGist, Discovery Studio, MOE | Highlights key
functional groups for drug interaction. | May miss non-
obvious interactions not included in the model. | (12,58) |
3. Structural and functional insights into
rabies virus proteins
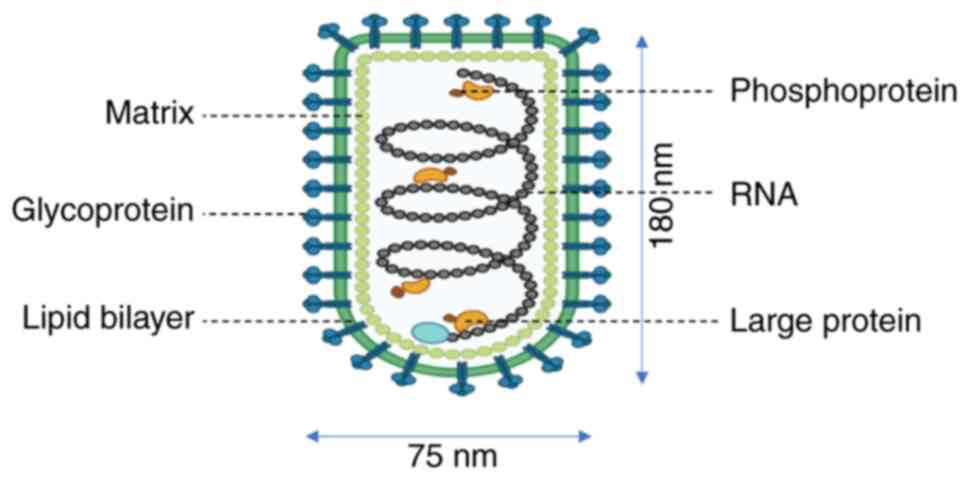
RABV, a member of the Rhabdoviridae family,
encodes five structural proteins: Nucleoprotein (N), phosphoprotein
(P), matrix protein (M), glycoprotein (G) and the large
RNA-dependent RNA polymerase (L or RdRp) (Fig. 1). Each protein plays a critical
role in viral replication, immune evasion and pathogenesis.
Understanding their structural and functional properties provides
essential insight into antiviral drug targeting. The N protein
encapsulates the viral RNA genome, forming the ribonucleoprotein
complex crucial for viral transcription and replication, shielding
the viral genome from host immune responses (17). The G protein mediates viral entry
by interacting with host cell receptors, such as the nicotinic
acetylcholine receptor and p75 neurotrophin receptor. It is also
the primary target for neutralizing antibodies, rendering it a key
focus for vaccine and antiviral drug development (18). The L protein, in association with
the P protein, carries out viral genome replication and
transcription, rendering it a promising target for broad-spectrum
antivirals (19).
Computational approaches in targeting
RABV proteins
Modern computational techniques have significantly
advanced rabies antiviral research. Structure-based drug design,
homology modelling via SWISS-MODEL (https://swissmodel.expasy.org/), molecular docking
using AutoDock Vina (https://vina.scripps.edu/) and GOLD (https://www.ccdc.cam.ac.uk/discover/blog/gold-the-all-in-one-molecular-docking-package/),
and molecular dynamics simulations have provided valuable insight
into protein-ligand interactions. Additionally, artificial
intelligence (AI) and ML are enhancing ligand identification and
optimization, expediting therapeutic discoveries. However, in
vitro experiments to validate the in silico results are
mandatory for robust drug discovery.
Key viral targets for drug
discovery
Viral proteins serve as crucial targets in antiviral
drug discovery due to their vital roles in the viral life cycle,
including replication, transcription and host cell entry. Targeting
these proteins can disrupt viral propagation and provide effective
therapeutic strategies (19).
The nucleoprotein is critical for encapsulating
viral RNA and forming a ribonucleoprotein complex essential for
replication and transcription. Structural analysis (PDB ID: 2GTT)
has revealed its strong RNA-binding affinity, rendering it a viable
target for small-molecule inhibitors. Molecular docking studies
identified ZINC01530604 and ZINC01530605 as promising candidates
targeting phosphorylation sites essential for nucleoprotein
function. Additionally, QSAR modelling analyzed >450
physicochemical parameters, leading to the identification of
peptide inhibitors, such as P16b6, that can effectively bind to the
phosphoprotein and suppress viral replication (20). While molecular docking is a
powerful tool for narrowing down candidates, further validation
steps are essential for confirming antiviral activity.
The RABV virus glycoprotein (RVG) facilitates host
cell entry and neurotropism, making it a main antiviral target.
Docking studies have suggested polyethylene glycol 4000 as an
inhibitor, effectively blocking receptor binding sites to prevent
viral spread (21). Structural
studies have also highlighted conserved tyrosine and threonine
residues contributing to glycoprotein stability. Furthermore,
computational techniques have facilitated the identification of
neutralizing monoclonal antibodies, such as 523-11, which bind to
RVG and prevent membrane fusion (22).
RABV RdRp is essential for viral RNA synthesis.
Homology models based on vesicular stomatitis virus (PDB ID: 5A22)
have provided structural insight into its catalytic mechanism.
Molecular docking analyses identified compounds binding to key
catalytic residues (Met585, Glu620, Lys621, Trp622 and Glu696),
inhibiting polymerase activity. A virtual screening of 2,045
compounds from the National Cancer Institute (NCI) identified
several promising inhibitors with strong binding affinity to RdRp
active sites, supporting further experimental validation (23).
Thus, these viral proteins, nucleoprotein,
glycoprotein and RdRp, are promising targets for antiviral drug
discovery due to their critical functions in the viral lifecycle
and their amenability to targeted inhibition. Progress with
small-molecule inhibitors, peptides, and neutralizing antibodies
against these targets may lead to the development of effective
antiviral therapies. Hence, focusing drug discovery efforts on
critical viral proteins with validated roles in replication and
infection, supported by structural and computational studies, are
fundamental for developing potent antiviral agents that can
effectively disrupt viral propagation and combat viral
diseases.
Across the literature, there is a general consensus
that structure-based computational methods accelerate the
identification of antiviral candidates for RABV, particularly for
targets, such as nucleoprotein, glycoprotein and RdRp. However,
discrepancies remain as regards the translation of in silico
findings to in vitro and in vivo efficacy. This
emphasizes the necessity for integrated pipelines that combine
computational screening with experimental validation, a direction
increasingly advocated in recent reviews and original research
articles.
4. AI and ML in rabies treatment
The advent of ML and AI has markedly facilitated the
understanding of complex biological systems and accelerated the
drug discovery process. One area where ML/AI has played a vital
role is protein structure prediction, which is essential to
understanding the biological mechanisms involved in the disease.
DeepMind's AlphaFold is the game changer in this scenario,
surpassing all other existing tools and recognized as the recipient
of the 2024 Nobel Prize in Chemistry (24). Secondly, the role of AI in drug
discovery has been monumental. In the field of RABV drug discovery,
these technologies facilitate the analysis of genetic,
epidemiological and pharmacological datasets to predict treatment
efficiency, optimize antiviral drugs and identify novel therapeutic
targets by significantly reducing the cost and time involved in
developing new rabies treatments. AI-based computational models are
speeding up the discovery of host factors and RABV proteins that
can serve as therapeutic targets by analyzing viral genomes and
host-pathogen interactions to detect the critical molecular
pathways essential for RABV replication (25). In addition, predicting the
conserved viral epitopes could enable the development of vaccines
or anti-viral drugs (26).
Likewise, AI also supports the identification of host proteins
involved in immune evasion or viral invasion that could open new
avenues in therapeutics (27).
Another area where AI is playing a transformative
role is in the development of antiviral drugs for rabies. ML
approaches enable rapid screening of millions of compounds to
identify leads as potential inhibitors of viral replication by
analyzing extensive molecular datasets and predicting drug-receptor
interactions, followed by lead optimization to enhance their
efficacy (28). Drug repurposing
is another interesting area of research where AI can expedite the
identification of FDA-approved drugs with activity against the RABV
(29). AI also predicts drug-virus
interactions, identifying promising antiviral candidates inhibiting
replication. Additionally, the amalgamation of AI and CRISPR
genome-editing technology is advancing research into RABV
evolution, diagnosis, and treatment (30). Together, these efforts are paving
the way for safer, more effective therapeutic options for rabies
prevention and treatment.
5. Drug repurposing
Research into the molecular basis of rabies, which
affects the central nervous system, has demonstrated that the virus
targets neurons, leading to severe neurological symptoms (31). Currently, the only available
treatment option is post-exposure prophylaxis, which comes with
important drawbacks, including high costs and complicated
administration. These limitations necessitate alternative
strategies where drug repurposing could benefit rabies treatment.
By re-examining existing drugs for novel therapeutic applications,
the drug development process can be accelerated with reduced cost
and potentially reveal new treatment pathways. Repurposed
medications may provide a more rapid route to clinical application,
addressing the urgent need for more accessible and effective
therapies for rabies (10).
In a previous study, notable candidates, such as
cidofovir, emtricitabine, famciclovir, acyclovir, and stavudine
were shown to exhibit potential efficacy against the RABV
glycoprotein (32). Advanced
docking analyses were conducted using the CB-Dock2 program to
assess these candidates. This sophisticated approach searched the
database of the server for template ligands with strong topological
similarity (FP2 ≥0.4) through an integrated homologous
template-based blind docking mechanism (32). Following this, cavity detection was
performed using an in-house technique known as FitDock to aid
molecular docking with specific templates. The docking analyses
identified five potential binding cavities for Emtricitabine within
the target protein, with volumes ranging from 7209 ų to 1703 Å
with significant binding affinities, varying from -5.0 to -6.1;
lower scores suggest stronger binding affinity. The root means
square deviation values were observed around 10 Å, and the hydrogen
bond count fluctuated around 25, indicating stable interactions
with limited conformational deviation and persistent hydrogen
bonding (32).
In addition to computational methods for drug
repurposing, the validation of these compounds is crucial for
establishing their efficacy against rabies. Repositioning already
approved medications can significantly lower drug development costs
and timelines, while reducing the likelihood of unexpected
side-effects. Computational repositioning allows for the rapid
screening of candidates in silico, which renders it an
attractive option for prioritizing potential treatments. In a
previous study, the ChEMBL library was filtered from 2,354,965
compounds down to 6,554 that had passed through any phase of
clinical trials to avoid cytotoxic inhibitors (33). The cryo-electron microscopy
(cryo-EM) structure of the respiratory syncytial virus (RSV) strain
A2 polymerase (PDB: 6UEN) was prepared for AutoDock Vina docking
studies. Compounds with flexible ligand structures or poor binding
affinities were eliminated, narrowing the selection to 4,919
candidates. Protein-Ligand Interaction Profiler was used for
interaction analyses with micafungin, trombopag, trovafloxacin,
azlocillin Na, trospium, amiodarone HCl, perflubron, mephenesin and
methadone. Among these candidates, micafungin demonstrated
exceptional promise with a binding affinity of -14.32 kcal/mol and
an inhibition constant of 0.0317 nM, outperforming existing
inhibitors, such as AZ-27 (0.47997 nM), ALS-8112 (2.56 nM) and
ribavirin (6.98 nM) (33). In
vitro assays further confirmed the efficacy of micafungin by
reducing RSV polymerase activity to only 29.6% of the control
levels; by contrast, verubecestat (97.4%), ribavirin (98.4%) and
ALS-8112 (89.5%) retained markedly higher levels of activity
compared with the control. These findings collectively demonstrate
that computational approaches can effectively identify promising
repurposed compounds for rabies treatment while validating their
efficacy through rigorous screening processes (33).
Phthalazine derivatives have gained attention in
recent times due to their pharmacological properties (34). Their unique structural
characteristics make them promising candidates in combating viral
infections. High-throughput screening methods have played a key
role in discovering compounds that inhibit RABV replication, while
maintaining low cytotoxicity. Comprehending the mechanisms of
action of these compounds is vital for augmenting their therapeutic
potential. Phthalazine and its derivatives, nitrogen-containing
heterocyclic compounds, have gathered attention in medicinal
chemistry due to its versatile pharmacological properties. Of note,
one phthalazine derivative compound, 7671954, was found to be
particularly effective against RABV infections across different
Lyssavirus species with low cytotoxicity (IC50 values <30
µM) (34). Experiments established
that these compounds inhibited viral replication by targeting the
replication complex (34).
Furthermore, these derivatives interact with host receptor, such as
the p75 neurotrophin receptor and nicotinic acetylcholine receptor,
both of which play critical roles in viral entrance, potentially
increasing immune responses while limiting viral interactions with
host cells (18).
In summary, drug repurposing provides a
cost-effective and accelerated path for rabies treatment,
identifying promising candidates such as emtricitabine,
famciclovir, acyclovir, stavudine and cidofovir. Computational
screening and high-throughput assays have enabled the rapid
identification of drugs with potent antiviral potential against
rabies. These approaches address the urgent need for affordable and
accessible therapies beyond current post-exposure prophylaxis,
which remains costly and limited in availability. The continued
research and validation of these repurposed drugs could transform
rabies management and save countless lives globally.
6. Multi-epitope peptide vaccines
Generally, drug discovery focuses on treatment
post-infection, whereas vaccines provide proactive protection.
Computational methods have facilitated vaccine design through
precise epitope mapping. Since viral entry is the key to infection,
the RABV glycoprotein (RABV-G) is mainly targeted for vaccine
development. Several immunoinformatic tools are utilized to
identify the antigenic sites and further design multiepitope
peptide vaccines (35).
The Immune Epitope Database (IEDB) is employed to
predict epitopes based on the Bepipred Linear Epitope Prediction
2.0 method. Initial sequence analysis that includes antigenicity,
allergenicity and virulence assessments is carried out using
AllerTOP v2.0, DeepTMHMM, VirulentPred and VaxiJen. This is
followed by BLASTP analysis to ascertain no cross reactivity with
human proteins. Physico-chemical properties and structure
prediction studies are performed using Protparam, PSIPRED and
GalaxyRefine followed by the final prediction of immune response
prediction using C-ImmSim server. Studies were carried out using
this approach for RABV proteins (36,37).
In a previous study, AR16 and hPAB, two conserved
RABV peptides, were combined with Gp96 adjuvant to create an
oil-in-water emulsion. By day 14, the vaccine candidates produced
virus-neutralizing antibodies and were safe. By day 28, they peaked
at 5-6 IU/ml in mice and 7-9 IU/ml in beagles. The promise of
multi-epitope peptide vaccines for rabies prevention was
demonstrated by the 70-80% protective efficacy against a virulent
rabies strain in mice (36). In
another study, using computational immunoinformatic methods, B-cell
epitopes from the RABV glycoprotein and nucleoprotein were found
(37). The eukaryotic vector
pcDNA3.1(-) was used to express the highly antigenic areas after
they were put together into a multi-epitope construct. Oral
delivery was facilitated by adopting food-grade Escherichia
coli as a carrier. Experiments conducted both in vitro
and in vivo demonstrated that the vaccination could produce
a robust immune response in mice, markedly increasing survival
following viral challenge without causing any negative
side-effects. This method demonstrates a viable, scalable approach
to rabies prevention, especially in populations of wild animals
(37).
7. Phytochemical-based compound
candidates
In recent times, natural forms of therapy have
gained importance based on popular beliefs, skills, and indigenous
practices owing to their no-side-effect advantage. The role of
traditional medicine has taken a center stage, primarily including
plants as the source of innumerable compounds with enormous
therapeutic benefits (38).
Although this approach is used for rabies management, there are
limited data available to support the efficacy of these methods.
Phytochemicals provide an alternative approach for antiviral
treatment as they are known to inhibit viral replication (39).
Compounds such as coumarins, lignans, alkaloids,
flavonoids and terpenoids exhibit antioxidant activity and may
suppress viral genome activity. A previous study identified
anti-rabies phytochemicals from Salix subserrata and onion,
screened for toxicity using SwissADME and Protox-II servers. Among
the candidates, (S+)-catechin and kaempferol exhibited promising
binding interactions with RABV glycoprotein based on docking
results. (+)-Catechin displayed an improved binding affinity, full
fitness and estimated ΔG values than kaempferol with the formation
of conventional hydrogen bonds with the target protein, rendering
it a potential natural inhibitor against rabies; this demonstrates
the role of natural product-based drugs (40).
Likewise, dodecandrin from Phytolacca
dodecandra L'Hérit., a ribosomal inactivating protein, is
expected to have an antiviral effect by depurinating the
α-sarcin/ricin loop of the ribosomal RNA, thereby halting protein
synthesis and inhibiting viral replication (41). Datura metel Linn. contains
anticholinergic agents such as atropine, which can affect nervous
system function by acting as competitive antagonists of muscarinic
acetylcholine receptors. This blockade can regulate nervous system
activity, perhaps relieving neurological signs of rabies (42). Salix subserrata and
Silene macroselen were also previously investigated for the
ability of their extracts to extend the survival rates of mice
infected with rabies, although no specific bioactive compounds were
reported (40). Although these
plants have exhibited some potential in laboratory settings, they
have not been proven effective for human rabies treatment, setting
the stage for additional pharmacological and clinical studies
(9). Additionally, salviifoside A
from Alangium salviifolium, was previously demonstrated to
exhibit potential in selectively inhibiting RVG interactions with
promising binding scores derived from docking analyses against RVG
protein structures obtained from UniProt (Accession no. A3RM22)
(43). That study utilized
homology modelling with SWISS-MODEL to create potential structures
for docking analysis.
In summary, phytochemical-derived compounds present
an encouraging and diverse pool of natural antiviral substances
with possible activity against RABV targets; however, their
therapeutic promise has yet to be definitively confirmed by
thorough pharmacological and clinical trials to convert these
initial results into efficient human therapies. An overview of the
type of ligands, their targets and the mechanism of action is
presented in Table II.
 | Table IIIdentified ligands, drug repurposing
and vaccine design. |
Table II
Identified ligands, drug repurposing
and vaccine design.
| Compound/drug | Type | Target protein | Binding affinity
(Kcal/mol) | Predicted effect/
mechanism of action | In silico
method used | (Refs.) |
|---|
| ZINC01530604 | Ligand | Nucleoprotein
(N) | -7.5 | Inhibits viral
replication | Molecular
docking | (21) |
| ZINC01530605 | Ligand | Nucleoprotein
(N) | -7.2 | Inhibits viral
replication | Molecular
docking | (21) |
| (+)-Catechin | Phytochemical | Nucleoprotein
(N) | -8.1 | Natural
inhibitor | Molecular
docking | (40) |
| Kaempferol | Phytochemical | Nucleoprotein
(N) | -7.9 | Natural
inhibitor | Molecular
docking | (40) |
| Emtricitabine | Drug
repurposing | Polymerase (L) | -7.8 | Potential
inhibitor | Molecular
docking | (32) |
| Famciclovir | Drug
repurposing | Polymerase (L) | -7.5 | Potential
inhibitor | Molecular
docking | (32) |
| Multi-epitope
peptide 1 | Vaccine design | Various | Varies | Induces immune
response | IEDB, Bepipred | (36,37) |
| Multi- epitope
peptide 2 | Vaccine design | Various | Varies | Induces immune
response | IEDB, Bepipred | (36,37) |
8. RNA-based therapeutics
RNA-based therapeutics have emerged as a promising
strategy for the prevention and treatment of rabies, leveraging
advanced vaccine platforms such as mRNA and self-replicating RNA
(srRNA) technologies.
RNA interference (RNAi)
Initial studies investigated RNAi as a
post-transcriptional gene silencing mechanism against rabies. A
previous study demonstrated that there was a 5-fold decrease in the
RABV titer in BHK-21 cells by using short-interfering RNAs (siRNAs)
against the mRNA of the N protein (44). Research has highlighted that
microRNAs can significantly reduce genomic RNA and mRNA of the
proteins in replication and pathogenesis, indicating the potential
of RNAi as an antiviral agent (44).
Animal studies have supported these findings. Gupta
et al (14) documented
extensive protection in mice intracerebrally infected with an
adenovirus vector expressing siRNA coding-sequences against the N
and L proteins, and subsequent challenge with fixed RABV. In
addition, another study found decreased N protein synthesis, fewer
clinical signs and reduced morbidity in mice intracerebrally
inoculated with plasmids encoding for siRNAs against the N protein
prior to challenge with the CVS fixed strain of RABV (45). Aptamers and siRNAs were tested by
coupling them to each other and administering them with
Lipofectamine. One of the siRNAs (N53) produced an 80.13% decrease
in viral RNA, while aptamer UPRET 2.03 led to a 61.3% decrease when
applied alone at 2 h post-infection (p.i). Chimera UPRET 2.03-N8
(aptamer-siRNA) at 24 h p.i. resulted in 36.5% inhibition of viral
replication (46).
mRNA vaccines
mRNA technology has emerged as a rapid and scalable
platform for the development of a rabies vaccine. In a previous
study, an optimized mRNA vaccine construct (LVRNA001) encoding the
RABV glycoprotein (RABV-G) induced neutralizing antibody titers and
a robust Th1 cell-mediated immune response in mice (47). Protection against lethal challenge
with RABV was demonstrated in mice and dogs. The post-exposure
immunization of dogs with LVRNA001 yielded a 100% survival rate,
compared to 33.33% for the inactivated vaccine (47).
The thermostability of mRNA rabies vaccines was also
explored in another study. It was established that extended storage
at temperatures between -80 and +70˚C, or exposure to alternating
temperatures, did not impair the protective potential of the
vaccine in mice, suggesting that a rigorous cold chain may not be
necessary (48). Another study
focused on nucleoside-modified mRNA-lipid nanoparticle (LNP)
vaccines. A single immunization of RABV-G mRNA-LNP in mice induced
stronger humoral and T-cell immune responses than three doses of an
inactivated vaccine. This single-dose mRNA vaccine provided
complete protection against rabies, with the immune response
persisting for at least 25 weeks, and possibly longer with a
two-dose regimen (49).
CureVac AG engineered a RABV (RABV) mRNA vaccine,
CV7201, using the cationic protein protamine to encapsulate mRNA
that encodes the RABV glycoprotein. CV7201 was thermostable and
effectively elicited a WHO-defined antibody response in 70.3% of
subjects through three intradermal immunizations (Clinical trial
no. NCT02241135). Optimizing the LNP formulation from CV7201,
CureVac AG engineered CV7202, which employs the identical mRNA
antigen as CV7201. The optimized LNP contains an ionizable amino
lipid, a PEG-lipid-modified one, phospholipid and cholesterol
(50). CV7202 exhibited acceptable
tolerance in a clinical trial (Clinical trial no. NCT03713086)
(50). Stokes et al
(51) used a cationic nanoemulsion
to encapsulate saRNA coding for alphavirus RNA-dependent RNA
polymerase and the rabies glycoprotein G.
RNA-based adjuvants
The application of RNA adjuvants to boost the
immunogenicity of rabies vaccines has also been investigated
(52). A first-in-concept human
study evaluated the safety, tolerability and immunogenicity of the
TLR 7/8/RIG-I agonist RNA adjuvant CV8102, used alone or blended
with a commercial rabies vaccine (Rabipur®). The results
revealed that CV8102 was safe and well-tolerated, with doses of 25
and 50 µg markedly increasing the immunogenicity of reduced doses
of the rabies vaccine (52).
Consequently, RNA-based therapeutics, including RNAi, mRNA vaccines
and RNA-based adjuvants, exhibit tremendous potential in developing
rabies prevention and treatment methods, with further research
aimed at maximizing their efficacy, safety and accessibility for
global use.
9. Regional variations and implications for
treatment
Understanding the genetic diversity of the RABV
(Table III) is crucial for
designing effective vaccines. A previous study analyzing 50
dog-brain samples from suspected rabies cases in India provided
insight into regional viral variations (53). PCR screening targeting the
nucleoprotein (N) and glycoprotein (G) genes, followed by
sequencing, revealed that six isolates from Mumbai belonged to a
single Arctic lineage which traced back to an Indian lineage I from
2006. Bayesian coalescent analysis estimated the time to the most
recent common ancestor (TMRCA) for these sequences to be 1993,
indicating long-standing geographic clustering. These phylogenetic
data underscore the importance of tracking viral evolution for
epidemiological studies and vaccine development (53). Sequence analysis further revealed
amino acid polymorphisms in G-protein epitopes, influencing
antigenicity and pathogenicity. These findings highlight the
necessity for region-specific vaccine strategies to account for
genetic variations in circulating RABV strains (53).
 | Table IIIRabies virus strains and
therapeutics |
Table III
Rabies virus strains and
therapeutics
| Strain | Geographical
location | Host | Genetic
variations | Impact on
pathogenicity | Target for
therapeutics | Therapeutic
approach (Refs.) |
|---|
| RABV1 | Africa | Domestic
animals | Single nucleotide
polymorphisms | Increased virulence
in certain strains | Nucleoprotein
(N) | Drug/ligand
discovery (64-66) |
| RABV2 | Asia | Bats | Insertions and
deletions | Variable
pathogenicity | Glycoprotein
(G) | Vaccine design
(64-66) |
| RABV3 | Americas | Wild animals | Amino acid
substitutions | Reduced efficacy of
some vaccines | Polymerase (L) | Drug repurposing
(64-66) |
10. Bioinformatics and integrative
strategies to fight rabies virus
Computational studies play a critical role in
decoding the intricacies of RABV pathogenesis. Research into
intrinsically disordered protein regions of the RABV proteome has
yielded key insights (51). High
intrinsic disorder levels in the phosphoprotein (P-protein) and
nucleoprotein (N-protein) are linked to Negri body formation and
host immune suppression. Proteomics analysis of highly purified
RABV (RABV) has revealed 47 host proteins that become entrapped
inside viral particles upon assembly. Of these, 11 are highly
disordered. A comprehensive study on five of these highly
disordered mouse proteins, neuromodulin, Chmp4b, DnaJB6, Vps37B and
Wasl, utilized bioinformatics tools such as FuzDrop, D2P2, UniProt,
RIDAO, STRING, AlphaFold and ELM to investigate their intrinsic
disorder propensity (54). These
disordered host proteins appear to play a major role in RABV
pathogenicity, immune evasion, and possibly resistance to antiviral
drugs. That study highlights the complex interplay between virus
and host, in which intrinsic disorder plays a key role in viral
pathogenic processes. This suggests that these intrinsically
disordered proteins and their corresponding host interactions might
be good candidates for therapeutic targets (54).
In addition, dysregulation in the matrix (M) protein
could play a role in viral budding and transmission, affecting the
capacity of the virus to spread. Phylogenetic analysis, comparing
RABV isolates from different host species and geographic regions,
has revealed clear patterns of clustering consistent with
host-specific adaptation of the virus (55). These patterns highlight the
ecological niche of Lyssavirus, which is strongly shaped by their
mammalian hosts, with implications for transmission dynamics and
vaccine development.
Overcoming limitations of in silico
approaches
While in silico approaches have
revolutionized vaccine and drug discovery, it is important to
recognize their limitations. One of the main challenges is the
predictive reliability and accuracy of computational models, which
are highly dependent on structural information. Flaws in this
information, due to low-resolution inputs or errors in homology
modelling, can undermine the efficacy of the models. Moreover,
molecular docking and dynamics simulation tend to over-simplify
biological interactions with the result of predictions being often
at variance from actual practice. Incorporating high-resolution
experimental information, e.g., cryo-electron microscopy, can
effectively improve the fidelity of such models.
A key aspect of enhancing in silico
approaches is strengthening the connection between computational
methods and in vitro and in vivo confirmations. The
inability of most predicted candidates to succeed experimentally
demonstrates the need to improve the practical applicability of
predictions. Additionally, present computational models lack
consideration of host-pathogen interactions, immune systems, or
PK/PD profiles, which are critical aspects. Using systems biology
methods and AI-based simulations can make predictions more
biologically relevant. Computational complexity and accessibility
are also major challenges. Sophisticated approaches, including
molecular dynamics simulations and quantum chemistry calculations,
are computationally costly and hence scale poorly. Optimizing
parallel processing algorithms and utilizing cloud-based platforms
can help overcome these limitations. Furthermore, most
sophisticated tools need specialized skills and hence access is
restricted in low-resource environments where rabies is endemic.
Providing improved user interfaces and training programs can
facilitate access.
The use of combined in silico and in
vitro approaches is crucial in the design of novel rabies
therapeutics. Computational methods, such as molecular dynamics
simulation and QSAR models, enable the prediction of the behavior
of putative drug candidates before they are synthesized (56-60).
These models can screen large compound libraries, thus accelerating
the identification of lead candidates while reducing drug
development time and costs. In addition, in silico
approaches can model RABV interactions with host cells, providing
insight into the life cycle of the virus and identifying novel
therapeutic targets. The synergy of computational prediction and
experimental confirmation is central to expediting the development
of useful rabies vaccines and treatments.
Translation to practical
application
Even with significant success in the use of
computational resources towards therapeutic interventions for
rabies, the translation of these in silico discoveries into
viable treatments is a challenge. To address this issue, tackling
the underreporting and misdiagnosis of cases, particularly in rural
locations in endemic nations, calls for the creation of affordable,
simple, and usable point-of-care diagnostic devices. Strengthening
surveillance systems with a view to collecting accurate data in a
timely manner is indispensable for successful control of rabies
(61). Likewise, a One Health
approach can be adopted which was proven successful in several
countries such as Bhutan, Bangladesh and Sri Lanka (62). Further improvements in the
prediction accuracy of epidemiological models, by incorporating
real-time data and environmental factors, can result in more
efficient resource deployment and enhanced predictive capabilities.
Public education programs for raising awareness and adherence to
rabies prevention strategies, like dog vaccination and prompt
post-exposure prophylaxis, are also essential (63).
Finally, bioinformatics and computational approaches
are crucial for progressing the knowledge on the pathogenesis of
RABV and establishing efficient strategies for prevention and
treatment. Overcoming the disconnect between computational
prediction and experimental investigations is necessary for
bridging in silico discovery and practical implementation
and, eventually mitigating the global disease burden of rabies.
11. Conclusion and future perspectives
Addressing rabies at the therapeutic level remains a
major scientific and public health challenge, particularly in
regions with limited access to post-exposure interventions. While
vaccines have substantially reduced transmission, effective
treatments for symptomatic cases are still lacking. The present
review highlighted how computational tools are transforming the
drug discovery landscape for rabies by enabling the exploration of
novel targets and mechanisms. Notably, in silico platforms
have facilitated the rational design of peptide vaccines, the
identification of antiviral leads, and re-evaluation of existing
drug libraries with improved precision and efficiency. However, the
success of these approaches centers on their integration with
experimental validation pipelines. Future efforts are required to
prioritize translational studies, interdisciplinary collaborations
and capacity building in endemic regions to harness the full
potential of bioinformatics-driven research. Ultimately, a
data-driven, systems-level approach may provide the most promising
route toward effective and equitable rabies control.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
RR drafted the manuscript. SS contributed to the
conceptualization of the study, assisted in drafting the manuscript
and supervised the study. Both authors were involved in the
literature search. Both authors have read and approved the final
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nayak JB, Chaudhary JH, Bhavsar PP,
Anjaria PA, Brahmbhatt MN and Mistry UP: Rabies: Incurable
biological threat. In: Zoonosis of Public Health Interest.
IntechOpen. London, 2022.
|
|
2
|
World Health organization (WHO). In: WHO
Expert Consultation on Rabies: Third Report. Abela-Ridder B (ed).
WHO, Geneva, 2018.
|
|
3
|
World Health Organization (WHO). Rabies -
India. WHO, Geneva. https://www.who.int/india/health-topics/rabies.
|
|
4
|
World Health Organization (WHO). Rabies.
WHO, Geneva, 2023. https://www.who.int/news-room/fact-sheets/detail/rabies.
|
|
5
|
Wada YA, Mazlan M, Noordin MM, Mohd-Lila
MA, Fong LS, Ramanoon SZ and Zahli NIU: Rabies epidemiology in
Malaysia (2015-2023): A cross-sectional insights and strategies for
control. Vaccine. 42(126371)2024.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yu Q, Liu J, Zhao H, Chen H, Xiang Y, Liu
Q, Mei L, Zhang W, Cheng M, Li Z, et al: Canine rabies vaccination,
surveillance and public awareness programme in Beijing, China,
2014-2024. Bull World Health Organ. 103:247–254. 2025.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Gnanadurai CW, Huang CT, Kumar D and Fu
ZF: Novel approaches to the prevention and treatment of rabies. Int
J Virol Stud Res. 3:8–16. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
WHO Rabies Modelling Consortium. The
potential effect of improved provision of rabies post-exposure
prophylaxis in Gavi-eligible countries: A modelling study. Lancet
Infect Dis. 19:102–111. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Beasley EA, Wallace RM, Coetzer A, Nel LH
and Pieracci EG: Roles of traditional medicine and traditional
healers for rabies prevention and potential impacts on
post-exposure prophylaxis: A literature review. PLoS Negl Trop Dis.
16(e0010087)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sreenivasan N, Li A, Shiferaw M, Tran CH,
Wallace R, Blanton J, Knopf L, Abela-Ridder B and Hyde T: Working
group on Rabies PEP logistics. Overview of rabies post-exposure
prophylaxis access, procurement and distribution in selected
countries in Asia and Africa, 2017-2018. Vaccine. 37 (Suppl
1):A6–A13. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Uzundurukan A, Nelson M, Teske C, Islam
MS, Mohamed E, Christy JV, Martin HJ, Muratov E, Glover S and Fuoco
D: Meta-analysis and review of in silico methods in drug discovery
- part 1: Technological evolution and trends from big data to
chemical space. Pharmacogenomics J. 25(8)2025.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Alazman I, Mishra MN, Alkahtani BS and
Goswami P: Computational analysis of rabies and its solution by
applying a fractional operator. Appl Math Sci Eng.
32(2340607)2024.
|
|
13
|
Knobel DL, Jackson AC, Bingham J, Ertl
HCJ, Gibson AD, Hughes D, Joubert K, Mani RS, Mohr BJ, Moore SM, et
al: A one medicine mission for an effective rabies therapy. Front
Vet Sci. 9(867382)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Gupta Y, Savytskyi OV, Coban M, Venugopal
A, Pleqi V, Weber CA, Chitale R, Durvasula R, Hopkins C, Kempaiah P
and Caulfield TR: Protein structure-based in-silico approaches to
drug discovery: Guide to COVID-19 therapeutics. Mol Aspects Med.
91(101151)2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Prinsa Saha S, Bulbul MZH, Ozeki Y, Alamri
MA and Kawsar SMA: Flavonoids as potential KRAS inhibitors: DFT,
molecular docking, molecular dynamics simulation and ADMET
analyses. J Asian Nat Prod Res. 26:955–992. 2024.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Alamri MA, Prinsa Kawsar SMA and Saha S:
Exploring marine-derived bioactive compounds for dual inhibition of
Pseudomonas aeruginosa LpxA and LpxD: Integrated bioinformatics and
cheminformatics approaches. Mol Divers. 29:1033–1047.
2025.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang PH and Xing L: The roles of rabies
virus structural proteins in immune evasion and implications for
vaccine development. Can J Microbiol. 70:461–469. 2024.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lian M, Hueffer K and Weltzin MM:
Interactions between the rabies virus and nicotinic acetylcholine
receptors: A potential role in rabies virus induced behavior
modifications. Heliyon. 8(e10434)2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Nakagawa K, Kobayashi Y, Ito N, Suzuki Y,
Okada K, Makino M, Goto H, Takahashi T and Sugiyama M: Molecular
function analysis of rabies virus RNA polymerase L protein by using
an L gene-deficient virus. J Virol. 91:e00826–17. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lu Y, Cheng L and Liu J: Optimisation of
inhibitory peptides targeting phosphoprotein of rabies virus. Int J
Pept Res Ther. 26:1043–1049. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tomar NR, Singh V, Marla SS, Chandra R,
Kumar R and Kumar A: Molecular docking studies with rabies virus
glycoprotein to design viral therapeutics Indian J Pharm. Sci.
72:486–490. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yang F, Lin S, Ye F, Yang J, Qi J, Chen Z,
Lin X, Wang J, Yue D, Cheng Y, et al: Structural analysis of rabies
virus glycoprotein reveals pH-dependent conformational changes and
interactions with a neutralizing antibody. Cell Host Microbe.
27:441–453.e7. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kiriwan D and Choowongkomon K: In silico
structural elucidation of the rabies RNA-dependent RNA polymerase
(RdRp) toward the identification of potential rabies virus
inhibitors. J Mol Model. 27(183)2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Jumper J, Evans R, Pritzel A, Green T,
Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A,
Potapenko A, et al: Highly accurate protein structure prediction
with AlphaFold. Nature. 596:583–589. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sahoo D, Katkar GD, Khandelwal S,
Behroozikhah M, Claire A, Castillo V, Tindle C, Fuller M, Taheri S,
Rogers TF, et al: AI-guided discovery of the invariant host
response to viral pandemics. EbioMedicine.
68(103390)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yurina V and Adianingsih OR: Predicting
epitopes for vaccine development using bioinformatics tools. Ther
Adv Vaccines Immunother. 10(25151355221100218)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Elste J, Saini A, Mejia-Alvarez R, Mejía
A, Millán-Pacheco C, Swanson-Mungerson M and Tiwari V: Significance
of artificial intelligence in the study of virus-host cell
interactions. Biomolecules. 14(911)2024.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tarasova O and Poroikov V: Machine
learning in discovery of new antivirals and optimization of viral
infections therapy. Curr Med Chem. 28:7840–7861. 2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Almulhim M, Ghasemian A, Memariani M,
Karami F, Yassen ASA, Alexiou A, Papadakis M and Batiha GE: Drug
repositioning as a promising approach for the eradication of
emerging and re-emerging viral agents. Mol Divers: Mar 18, 2025
(Epub ahead of print).
|
|
30
|
Koujah L, Shukla D and Naqvi AR:
CRISPR-Cas based targeting of host and viral genes as an antiviral
strategy. Semin Cell Dev Biol. 96:53–64. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fooks A, Cliquet F, Finke S, Freuling C,
Hemachudha T, Mani RS, Müller T, Nadin-Davis S, Picard-Meyer E,
Wilde H and Banyard AC: Rabies. Nat Rev Dis Primers.
3(17091)2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hameed A: Drug repurposing for rabies
treatment: A computational analysis of 14 agents. ChemRxiv,
2024.
|
|
33
|
Xu E, Park S, Calderon J, Cao D and Liang
B: In silico identification and in vitro validation of repurposed
compounds targeting the RSV polymerase. Microorganisms.
11(1608)2023.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Perraud V, Vanderhoydonck B, Bouvier G,
Dias de Melo G, Kilonda A, Koukni M, Jochmans D, Rogée S, Ben
Khalifa Y, Kergoat L, et al: Mechanism of action of phthalazinone
derivatives against rabies virus. Antiviral Res.
224(105838)2024.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Abdulhameed Odhar H, Hashim AF, Humadi SS
and Ahjel SW: Design and construction of multi epitope-peptide
vaccine candidate for rabies virus. Bioinformation. 19:167–177.
2023.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Niu Y, Liu Y, Yang L, Qu H, Zhao J, Hu R,
Li J and Liu W: Immunogenicity of multi-epitope-based vaccine
candidates administered with the adjuvant Gp96 against rabies.
Virol Sin. 31:168–175. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ding Y, Gao Y, Chen R, Zhang Z, Li Q, Jia
T, Zhang T, Xu R, Shi W, Chen L, et al: Development of a novel
multi-epitope oral DNA vaccine for rabies based on a food-borne
microbial vector. Int J Biol Macromol. 255(128085)2024.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Rizvi SAA, Einstein GP, Tulp OL, Sainvil F
and Branly R: Introduction to traditional medicine and their role
in prevention and treatment of emerging and re-emerging diseases.
Biomolecules. 12(1442)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Behl T, Rocchetti G, Chadha S, Zengin G,
Bungau S, Kumar A, Mehta V, Uddin MS, Khullar G, Setia D, et al:
Phytochemicals from plant foods as potential source of antiviral
agents: An overview. Pharmaceuticals (Basel).
14(381)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Deressa A, Hussen K, Abebe D and Gera D:
Evaluation of the efficacy of crude extracts of Salix subserrata
and Silene macroselen for the treatment of rabies in Ethiopia.
Ethiop Vet J. 14:1–16. 2011.
|
|
41
|
Admasu P, Deressa A, Mengistu A, Gebrewold
G and Feyera T: In vivo antirabies activity evaluation of
hydroethanolic extract of roots and leaves of Phytolacca
dodecandra. Glob Vet. 12:12–18. 2014.
|
|
42
|
Roy S, Samant L, Ganjhu R, Mukherjee S and
Chowdhary A: Assessment of in vivo antiviral potential of Datura
metel Linn. extracts against rabies virus. Pharmacogn Res.
10:109–112. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Maheswari UM, Ebenezer NS and Priyakumari
JC: An In silico approach: Homology modelling and docking studies
of rabies virus glycoprotein with salviifoside a of alangium
salviifolium. Int J Sci Res. 6:531–534. 2017.
|
|
44
|
Yang YJ, Zhao PS, Zhang T, Wang HL, Liang
HR, Zhao LL, Wu HX, Wang TC, Yang ST and Xia XZ: Small interfering
RNAs targeting the rabies virus nucleoprotein gene. Virus Res.
169:169–174. 2012.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Trobaugh DW and Klimstra WB: MicroRNA
regulation of RNA virus replication and pathogenesis. Trends Mol
Med. 23:80–93. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Scott TP and Nel LH: Rabies prophylactic
and treatment options: An in vitro study of siRNA- and
aptamer-based therapeutics. Viruses. 13(881)2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Li J, Liu Q, Liu J, Wu X, Lei Y, Li S,
Zhao D, Li Z, Luo L, Peng S, et al: An mRNA-based rabies vaccine
induces strong protective immune responses in mice and dogs. Virol
J. 19(184)2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Stitz L, Vogel A, Schnee M, Voss D, Rauch
S, Mutzke T, Ketterer T, Kramps T and Petsch B: A thermostable
messenger RNA based vaccine against rabies. PLoS Negl Trop Dis.
11(e0006108)2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Bai S, Yang T, Zhu C, Feng M, Zhang L,
Zhang Z, Wang X, Yu R, Pan X, Zhao C, et al: Corrigendum: A single
vaccination of nucleoside-modified rabies mRNA vaccine induces
prolonged highly protective immune responses in mice. Front
Immunol. 14(1146466)2023.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Aldrich C, Leroux-Roels I, Huang KB, Bica
MA, Loeliger E, Schoenborn-Kellenberger O, Walz L, Leroux-Roels G,
von Sonnenburg F and Oostvogels L: Proof-of-concept of a low-dose
unmodified mRNA-based rabies vaccine formulated with lipid
nanoparticles in human volunteers: A phase 1 trial. Vaccine.
39:1310–1318. 2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Stokes A, Pion J, Binazon O, Laffont B,
Bigras M, Dubois G, Blouin K, Young JK, Ringenberg MA, Ben
Abdeljelil N, et al: Nonclinical safety assessment of repeated
administration and biodistribution of a novel rabies
self-amplifying mRNA vaccine in rats. Regul Toxicol Pharmacol.
113(104648)2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Doener F, Hong HS, Meyer I, Tadjalli-Mehr
K, Daehling A, Heidenreich R, Koch SD, Fotin-Mleczek M and
Gnad-Vogt U: RNA-based adjuvant CV8102 enhances the immunogenicity
of a licensed rabies vaccine in a first-in-human trial. Vaccine.
37:1819–1826. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Pharande RR, Majee SB, Gaikwad SS,
Moregoankar SD, Bannalikar AK, Doiphode A, Gandge R, Dighe D, Ingle
S and Mukherjee S: Evolutionary analysis of rabies virus using the
partial nucleoprotein and glycoprotein gene in Mumbai region of
India. J Gen Virol. 102:2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Ashraf HN and Uversky VN: Intrinsic
disorder in the host proteins entrapped in rabies virus particles.
Viruses. 16(916)2024.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Cai M, Liu H, Jiang F, Sun Y, Wang W, An
Y, Zhang M, Li X, Liu D, Li Y, et al: Analysis of the evolution
infectivity and antigenicity of circulating rabies virus strains.
Emerg Microbes Infect. 11:1474–1484. 2022.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Quayum ST, Esha NJI, Siraji S, Abbad SSA,
Alsunaidi ZHA, Almatarneh MH, Rahman S, Alodhayb AN, Alibrahim KA,
Kawsar SMA and Uddin KM: Exploring the effectiveness of flavone
derivatives for treating liver diseases: Utilizing DFT, molecular
docking, and molecular dynamics techniques. MethodsX.
12(102537)2024.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Saha S, Gupta V, Hossain A, Prinsa P,
Ferdous J, Lokhande KB, Jakhmol V and Kawsar Sarkar MA:
Computational Investigation of the Unveils NSD2 Inhibition
Potential of Berberis vulgaris, Sambucus nigra, and Morus alba
through Virtual Screening, Molecular Docking, MD Simulation, and
DFT Analyses, Karbala International J. Modern Sci. 11:129–143.
2025.
|
|
58
|
M A Kawsar S, Hosen MA, Ahmad S, El Bakri
Y, Laaroussi H, Ben Hadda T, Almalki FA, Ozeki Y and Goumri-Said S:
Potential SARS-CoV-2 RdRp inhibitors of cytidine derivatives:
Molecular docking, molecular dynamic simulations, ADMET, and POM
analyses for the identification of pharmacophore sites. PLoS One.
17(e0273256)2022.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Saha S, Prinsa and Sarkar MAK:
Natural Flavonoids as Primary Amoebic Meningoencephalitis
Inhibitor: Virtual Screening, Molecular Docking, MD Simulation,
MMPBSA, Density Functional Theory, Principal Component, and Gibbs
Free Energy Landscape Analyses. Chem Biodiv.
22(e202402521)2025.
|
|
60
|
Karim T, Almatarneh MH, Rahman S, Alodhayb
AN, Albrithen H, Md. Hossain MM, Sarkar M. A. Kawsar, Poirier RA
and Uddin KM: In silico prediction of antibacterial activity of
quinolone derivatives. ChemistrySelect. 9(e202402780)2024.
|
|
61
|
Sudarshan MK and Ashwath Narayana DH:
Appraisal of surveillance of human rabies and animal bites in seven
states of India. Indian J Public Health. 63 (Supplement):S3–S8.
2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Acharya KP, Acharya N, Phuyal S, Upadhyaya
M and Lasee S: One-health approach: A best possible way to control
rabies. One Health. 10(100161)2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Changalucha J, Steenson R, Grieve E,
Cleaveland S, Lembo T, Lushasi K, Mchau G, Mtema Z, Sambo M, Nanai
A, et al: The need to improve access to rabies post-exposure
vaccines: Lessons from Tanzania. Vaccine. 37 (Suppl 1):A45–A53.
2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Bourhy H, Reynes JM, Dunham EJ, Dacheux L,
Larrous F, Huong VTQ, Xu G, Yan J, Miranda MEG and Holmes EC: The
origin and phylogeography of dog rabies virus. J Gen Virol. 89 (Pt
11):2673–2681. 2008.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Rupprecht CE, Hanlon CA and Hemachudha T:
Rabies re-examined. Lancet Infect Dis. 2:327–343. 2002.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Banyard AC, Evans JS, Luo TR and Fooks AR:
Lyssaviruses and bats: Emergence and zoonotic threat. Viruses.
6:2974–2990. 2014.PubMed/NCBI View Article : Google Scholar
|