Introduction
Diabetes mellitus (DM) is a chronic metabolic
disease that is characterized by a sustained high blood sugar level
due to a disruption in insulin secretion or/and action. The
prevalence of DM is continuously increasing at a rapid rate; thus,
this poses a critical public health concern. According to the World
Health Organization, there were >537 million adults with DM in
2021, a number which is prone to increase to 643 million by the
year 2030(1). Type 1 DM is caused
by autoimmune pancreatic β-cell destruction, whereas type 2 DM
results from insulin resistance, obesity and chronic low-grade
inflammation (2). Evidence
suggests a crucial role of immune and inflammatory signaling
pathways in the development of both types of DM, addressing the
relevance of cytokine regulation in the development and progression
of disease; this is a promising field which warrants investigation
(3).
Interleukin (IL)-10 is an anti-inflammatory cytokine
that is produced from different immune cells such as T-regulatory
cells, monocytes and macrophages. It plays an essential role in
immune regulation via attenuating pro-inflammatory actions and
maintaining immune homeostasis (4). As regards DM, IL-10 can potentially
protect against β-cell destruction in type 1 DM and modulate
systemic inflammation. Research has concluded that exogenous IL-10
can improve insulitis and delay the onset of type 1 DM in mice
(5). Additionally, a previous
study demonstrated an association between high IL-10 levels, and
improved insulin sensitivity and glycemic control in humans with
obesity-induced insulin resistance (6).
Previous studies have found a strong association
between the expression of specific genes and the prevalence of
certain diseases (7,8). The expression levels and functional
activity of IL-10 may be influenced by genetic variations in the
IL-10 gene, particularly single nucleotide polymorphisms
(SNPs) in the promoter region. Research has investigated
polymorphisms, including-1082A>G, -819C>T and -592C>A in
different populations and found that these variants can regulate
IL-10 transcription and, consequently, influence cytokine levels
and immune responses (9). Previous
studies have examined the association between IL-10 gene
polymorphisms and susceptibility to type 1 DM and type 2 DM, and
reported inconsistent results. Some reports have illustrated a
protective effect of high IL-10-producing genotypes
(10,11), whilst other studies have shown no
significant associations or even an elevated risk of disease with
certain alleles (12,13).
The gene-environment interaction is potentially
complex in the pathogenesis of diabetes as different contradictory
findings across different ethnic groups and geographic regions have
been reported. These discrepancies may be due to a variety of
factors, such as genetic background, lifestyle, comorbidities and
sample size variability (14,15).
In addition, the functional results of IL-10 polymorphisms
in vivo are not yet completely understood. In addition, the
mechanisms through which these genetic variations may affect other
pro-inflammatory and anti-inflammatory cytokines and consequently,
influence the immune milieu in patients with diabetes, remain to be
determined (16).
Due to the increasing evidence implicating the
association between inflammation and the onset and progression of
DM (17,18), studying the genetic basis of key
immunoregulatory markers, such as IL-10 could enhance the
understanding of the mechanisms of the disease, and could
potentially lead to the identification of novel biomarkers or
therapeutic targets. The present study aimed to investigate the
role of IL-10 gene variations in the development of DM and
to determine the nucleotide sequencing of this gene in Iraqi
women.
Subjects and methods
Study design
The present study was a case-control study that
included 45 women between the ages of 31 and 74 years. The
participants were divided into two groups as follows: 15 Healthy
women serving as the control group and 30 women diagnosed with DM.
The subjects of the study resided in Mosul, Iraq, and the study was
conducted during the period from December, 2024 to February, 2025.
The subjects in the control group were selected to match the cases
in terms of age (Table I). The
participants were further screened to exclude individuals with a
family history of diabetes, autoimmune diseases, or chronic
inflammatory conditions and those using medications to minimize
potential confounding bias. The present study was conducted with
the authorized approval from the Collegiate Committee for Medical
Research Ethics at the University of Mosul, Mosul, Iraq
(CCMRE-PHA-25-4). All participants provided written informed
consent.
 | Table IDemographic characteristics of the
study participants. |
Table I
Demographic characteristics of the
study participants.
| Variable | Diabetic group
(n=30) | Control group
(n=15) |
|---|
| Age, years (mean ±
SD) | 54.2±9.1 years | 52.7±8.4 years |
| BMI (mean ± SD) | 31.4±4.6 kg/m² | 28.2±3.9 kg/m² |
| Family history of DM
(%) | 20 (66.7%) | 3 (20.0%) |
| Hypertension (%) | 16 (53.3%) | 4 (26.7%) |
Collection of blood samples and DNA
extraction
Blood specimens were obtained from all subjects
using EDTA tubes. DNA was isolated from whole blood samples using a
Whole Blood Genomic DNA Extraction kit (AddBio Inc.) following the
manufacturer's instructions. The extracted DNA quality was assessed
by measuring the absorbance at 260 and 280 nm using a nanodrop
spectrophotometer (Thermo Fisher Scientific, Inc.). DNA was
obtained from the blood of all 45 samples and thereafter preserved
at -20˚C for use in further experiments.
Tetra-primer amplification refractory
mutation system-polymerase chain reaction (tetra-ARMS-PCR)
The DNA concentration in each sample was modified
using TE buffer solution (10 mM Tris-HCl with 1m M
EDTA•Na2) (Bioneer Corporation) to achieve 25 ng/µl for
PCR amplification. In total, four primers were used for primer
reactions, including F-outer and R-outer throughout the gene. The
forward outer-reverse inner primers are used for the mutant allele,
in place of those for the normal allele. Nucleic acid from each
sample was combined with suitable primers for the specific
mutations and master mix components to create the PCR reaction
mixture in 0.2-ml PCR tubes. The mixture was rapidly centrifuged at
3,000 x g for 1 min at room temperature to yield the ideal
components. The PCR tubes were subjected to cycling in a
thermocycler using tailored protocols for specific mutations. The
reaction product (at 2% concentration) was introduced into the
wells of a prepared agarose gel following the injection of a DNA
ladder from Bio-Lab Ltd. into designated wells. Following 40 min of
electrophoresis for sample migration, the bands were visualized
using a horizontal gel electrophoresis system (BiocomDirect).
Tetra-ARMS-PCR was used to evaluate the genetic variation of the
IL-10 gene at the locus (rs1800896).
Determination of genetic variation of
the IL-10 gene in situ (rs1800896) using tetra-ARMS-PCR
The detection of the A à G mutation in the
IL-10 gene at the rs1800896 site was performed by the
addition of 4 µl (100 nanograms) of template DNA to the contents of
the master mix (Macrogen) which contains 1 µl (10 picomol) of each
mutation-specific primer for the IL-10 gene (Table II) (19). Subsequently, a multiplication
reaction was performed on the reaction mix using the thermocycler
and according to the special program for the reaction presented in
Table III. Using the gradient
program in the thermocycler device (GeneAmp PCR System 9700,
Applied Biosystems, Thermo Fisher Scientific, Inc.), the optimal
temperature for the primer bonding was deduced and the gradient was
±5. Following this, 2% agarose gel (AddBio) was used to separate
the PCR reaction.
 | Table IIPrimers used for detection of the
genetic variation at rs1800896. |
Table II
Primers used for detection of the
genetic variation at rs1800896.
| Locus | Primer | Sequence | Band size | Annealing |
|---|
| rs1800896 | F-outer |
5'-GAATTTGGTTTCCTCACCCTACTG-3' | 390 bp | 61˚C |
| for IL-10 | R-outer |
5'-CTGAAGAAGTCCTGATGTCACTGC-3' | | |
| | F-inner |
5'-TTTCCTCTTACCTATCCCTACTTCCACT-3' | 190 bp | |
| | R-inner |
5'-AAGACAACACTACTAAGGCTTCTTTGGTAG-3' | 250 bp | |
 | Table IIIThermocycling conditions used in
ARMS-PCR technique for the detection of the mutation at
rs1800896. |
Table III
Thermocycling conditions used in
ARMS-PCR technique for the detection of the mutation at
rs1800896.
| No. | Stage | locus | Temperature | Time | Cycle number |
|---|
| 1 | Initial
denaturation | For all sites | 95˚C | 6 min. | 1 |
| 2 | Denaturation | For all sites | 95˚C | 45 sec. | 35 |
| 3 | Annealing | (rs1800896) | 61˚C | 1 min. | |
| 4 | Extension | For all sites | 72˚C | 1 min. | |
| 5 | Final
extension | For all sites | 72˚C | 5 min. | 1 |
| 6 | Stop reaction | For all sites | 4˚C | 5 min. | 1 |
DNA Sanger sequencing technology of
the IL-10 gene
The sequence of the nitrogenous bases of the gene
under investigation was ascertained, and the results of the PCR
reaction were directed to the aforementioned gene using the primers
of the resultant bands. The 3130 Genetic Analyzer (supplied by
Applied Biosystems, Hitachi, Ltd.) was used to determine the gene
sequences in the 30 DNA samples. The gene sequences were compared
with those recorded in the National Centre for Biotechnology
Information (NCBI).
Statistical analysis
Statistical analysis was conducted by calculating
confidence intervals (CIs) value, P-values and odds ratios (ORs)
using MedCalc statistical software, version 20.009 https://www.medcalc.org. The association between
genotypes or alleles and DM was evaluated using Fisher's exact
test. The sample size for genotype and allele frequency comparisons
was n=30 patients and n=15 controls. A value of P<0.05 was
considered to indicate a statistically significant difference and
95% CI values were calculated for all ORs. The following
frequencies were calculated based on the following dependency:
Allelic frequency of the normal allele=2 (number of homozygous
individuals) + (number of heterozygous individuals)/2 (total
number), e.g., frequency of G=(2xGG + AG)/(2 x total sample size).
The allelic frequency of the mutant allele=2 (number of
heterozygotes) + (number of heterozygotes)/2(total number).
Results
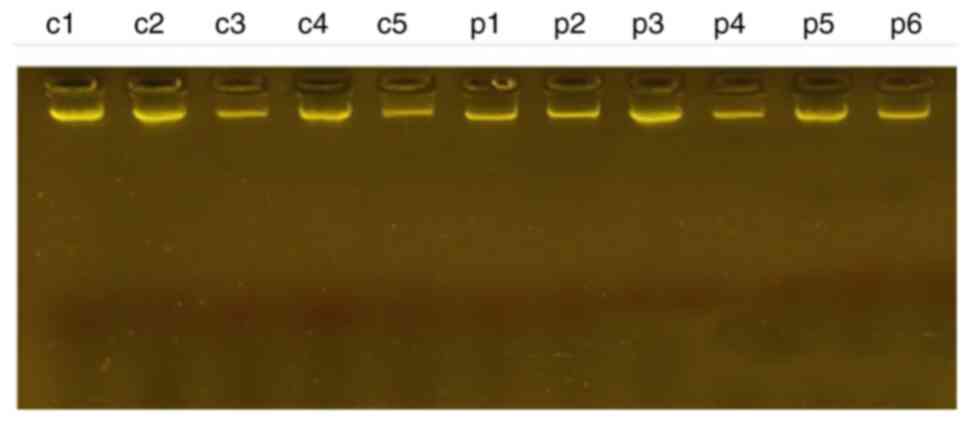
DNA extraction from blood samples
The genome packages were extracted from blood
samples and illustrated in Fig. 1.
The concentration ranged from (50-125 ng/µl) and the purity of the
DNA sample ranged between (1.5-1.7).
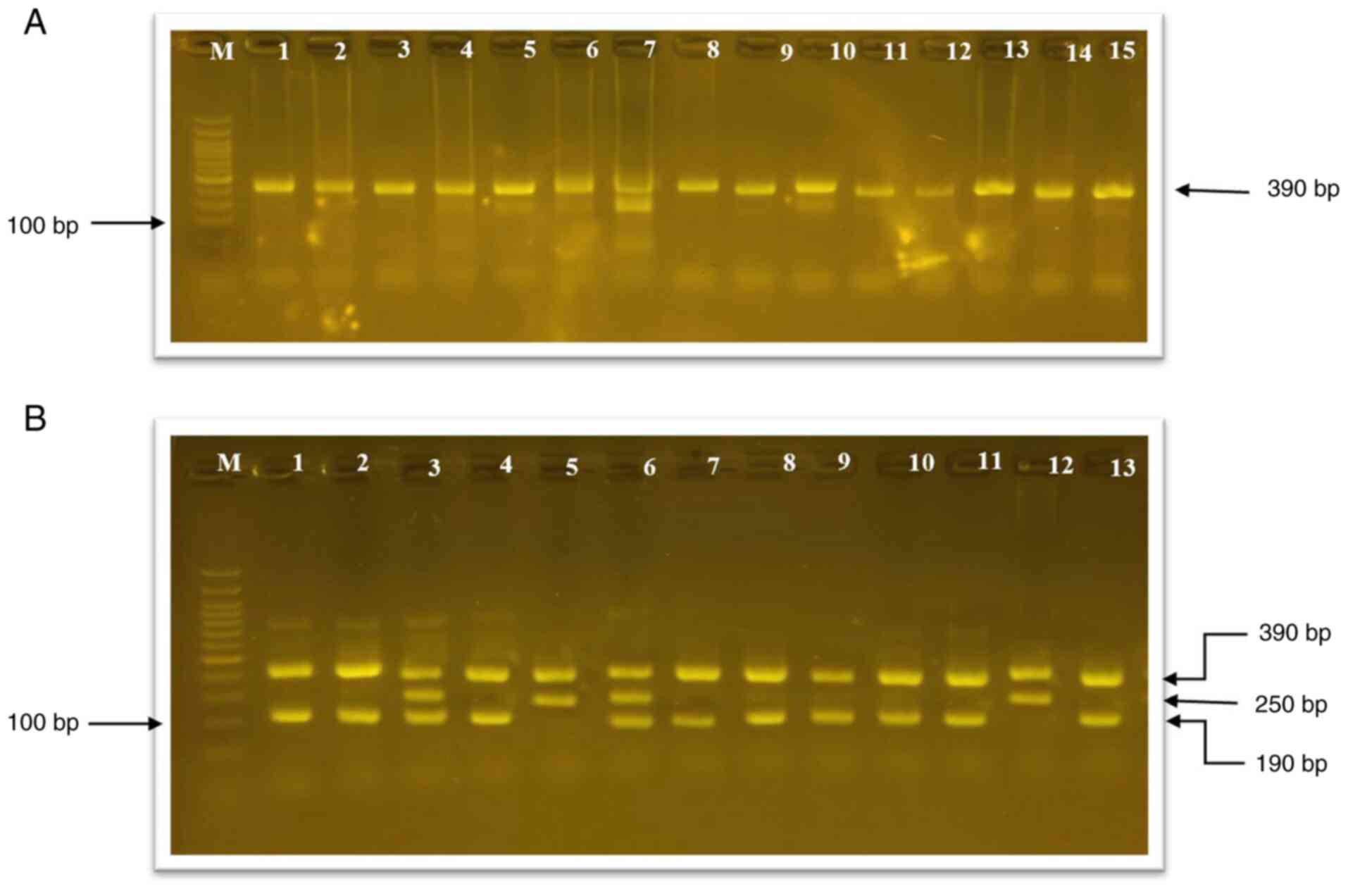
Determination of genetic variations of
the IL-10 gene in situ (rs1800896) using tetra-ARMS-PCR
The expression of the IL-10 gene (rs1800896)
on chromosome 1 and its variations in both healthy women and in
those with diabetes were investigated using tetra-ARMS-PCR. The PCR
reaction revealed that there was a genetic variation of the
IL-10 gene in women with diabetes, which appeared as three
genotypes AA, AG and GG; however, in the control samples, there was
only one genotype (Fig. 2).
The percentage of allelic observations and the
frequency of the different genotypes of the IL-10 gene at
rs1800896 are presented in Table
IV. The results for women with diabetes revealed that the
frequency of the mutant genotype GG was the highest in women with
diabetes at 13.3% compared with the mutant genotype in the control
group at 0%, while the percentage of the normal genotype AA was the
lowest in women with diabetes, at 80%, compared to 100% in the
control group. The percentage of the variant genotype AG was 6% in
the patient group in comparison to 0% in the control group. The
percentage of the allelic frequency observed for the mutant G
allele was high in the patient group (16.7%) compared to the
control group (0%). By contrast, the percentage of the normal
allele in patients was 83%, in comparison with that of the control
group, which was 100%.
 | Table IVDistribution of allelic viewing and
genotype of the IL-10 gene at the locus (rs1800896) among
the group of controls and women with diabetes. |
Table IV
Distribution of allelic viewing and
genotype of the IL-10 gene at the locus (rs1800896) among
the group of controls and women with diabetes.
| Comparison | Patients (n=30), n
(%) | Controls (n=15), n
(%) | OR | 95% CI |
P-valuea |
|---|
| AA vs. (AG +
GG) | 24 (80.0) | 15 (100.0) | 0.00 | Not estimable | 0.1575 |
| AG vs. (AA +
GG) | 2 (6.7) | 0 (0.0) | | Not estimable | 0.5455 |
| GG vs. (AA +
AG) | 4 (13.3) | 0 (0.0) | | Not estimable | 0.2847 |
| A vs. G | A=50 (83.3) | A=30 (100.0) | 0.00 | Not estimable | 0.0273 |
| | G=10 (16.7) | G=0 (0.0) | | | |
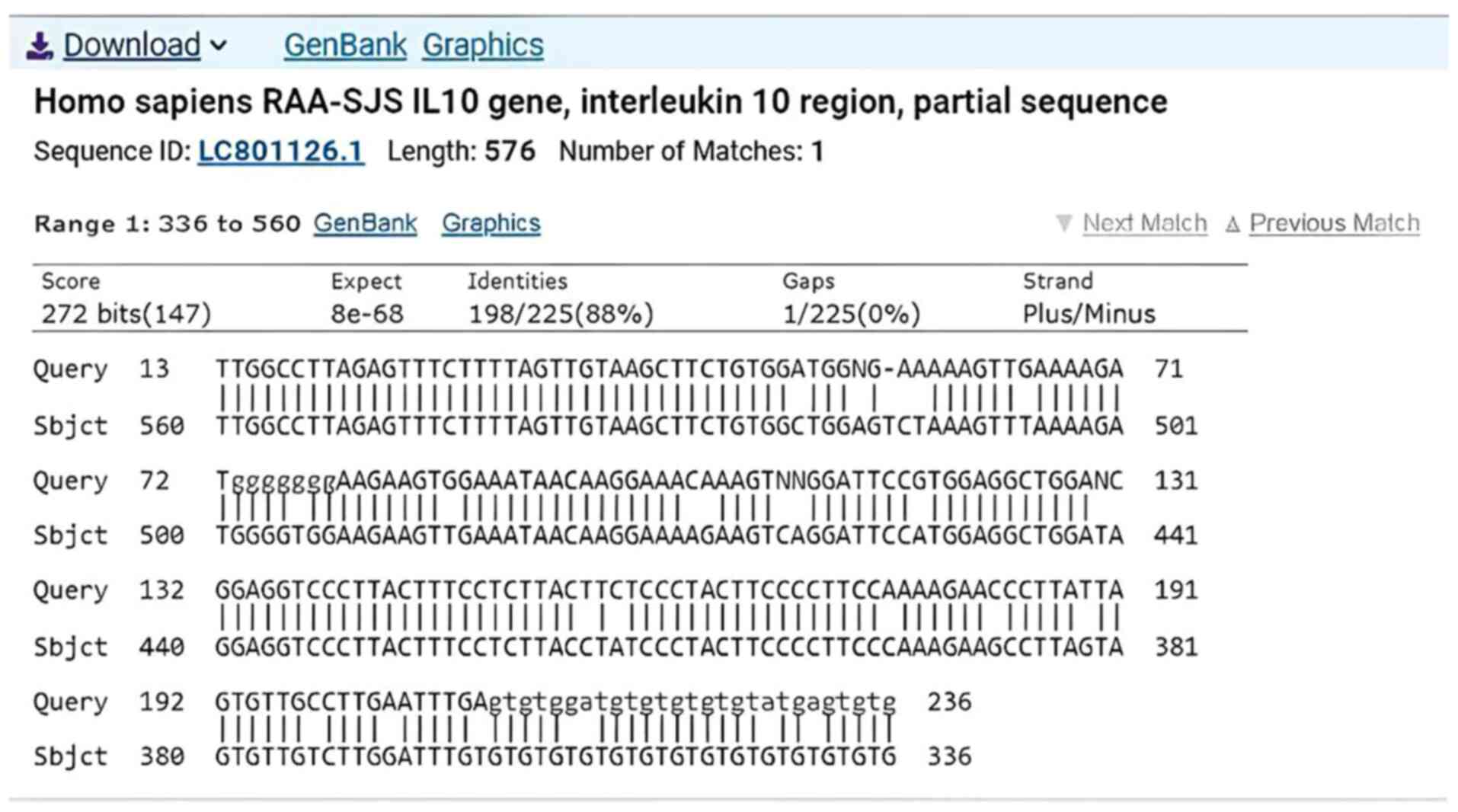
DNA sequencing technology for IL-10
gene
The results of DNA sequencing technology for the
amplified IL-10 gene revealed that there were differences in
the number of nucleotides, as illustrated in Fig. 3. DNA sequencing confirmed the
presence of the rs1800896 (A>G) polymorphism in the promoter
region of the IL-10 gene. The variation was identified in
yhr AA, AG and GG genotypes. The SNP is located at position
chr1:206946930 (GRCh38) and may influence IL-10
transcriptional activity due to its regulatory location.
In addition, the Sanger sequencing analysis revealed
varied types of genetic mutations and their locations on the
IL-10 gene after comparing them with the gene sequences at
the NCBI site (Table V). In
addition, a representative sequencing chromatogram illustrated in
Fig. S1.
 | Table VTypes and locations of mutations in
the IL-10 gene in patients with diabetes. |
Table V
Types and locations of mutations in
the IL-10 gene in patients with diabetes.
| ID sequence | Nucleotide | Location | Mutation type | Identity (%) | Gaps (%) |
|---|
| LC801126.1 | T→G | 507 | Transition | 88 | 0 |
| LC801126.1 | T→A | 514 | Trans version | 88 | 0 |
| LC801126.1 | C→A | 515 | Transition | 88 | 0 |
| LC801126.1 | T→- | 516 | Deletion | 88 | 0 |
| LC801126.1 | A→N | 518 | | 88 | 0 |
Discussion
The present study examined the association between
IL-10 gene polymorphism at rs1800896 and the incidence of DM
in women, particularly genotype and allele frequencies in diabetic
vs. healthy individuals. The present study identified a clear
distinction in the distribution of IL-10 genotypes and
alleles between diabetic and non-diabetic women by using
tetra-ARMS-PCR and Sanger sequencing, which suggests a potential
genetic predisposition influenced by IL-10
polymorphisms.
The present study found that three genotypes AA
(wild-type), AG (heterozygous) and GG (homozygous mutant) are
present among women with diabetes, whereas only the AA genotype was
observed in the control group, underscoring a significant genetic
variation in the patient population. Notably, the GG genotype was
limited to the diabetic patients (13.3%), and the AG genotype was
also detected only in the diabetic group (6.7%), whereas this was
absent in the control group. These findings are in accordance with
those in the study by Zietz et al (20), which linked IL-10 gene
polymorphisms with an altered immune regulation and an elevated
risk of metabolic disorders, including type 2 DM. The rs1800896
polymorphism, which occurs in the promoter region of the
IL-10 gene on chromosome 1q31-32, can affect transcriptional
activity and cytokine expression levels (21). Notably, chronic low-grade
inflammation is associated with the incidence of type 2 DM, whereas
IL-10 plays a crucial role as an anti-inflammatory mediator.
The findings of the present study about allele
frequency support the hypothesis of a genetic predisposition. The
mutant G allele was absent in the healthy individuals, while it was
noted in 16.7% of the patients with diabetes. The absence of this
genotype in healthy individuals highlights its possible
contribution to disease vulnerability, particularly as regards the
allele-level data. The findings of the present study are consistent
with those of the study by Van Exel et al (6), which demonstrated a positive
association between IL-10 gene variants and the incidence of
metabolic syndrome and type II DM, and a negative association
between IL-10 expression and insulin resistance and systemic
inflammation. In addition, Pociot and Lernmark (22) examined the factors affecting the
genetic architecture of diabetes and reported a significant role in
inflammatory gene polymorphisms, including IL-10. Notably, a
substantial link has been reported between DM and inflammation, and
a number of anti-diabetic drugs have anti-inflammatory effects that
aid in ameliorating diabetes-related complications (23).
As regards sequencing analysis, the present study
revealed nucleotide differences and different types of genetic
variations, such as transversions and deletions in comparison with
the reference IL-10 sequence provided by NCBI. These
multiple point mutations and structural variability in the
IL-10 gene could influence transcription factor binding or
mRNA stability, ultimately affecting cytokine production. Due to
these probabilities, further functional studies are required to
investigate the precise impact of the mutations observed.
Previous studies have reported conflicting results
about the role of the G allele at rs1800896. For instance, studies
revealed that an elevated IL-10 secretion may play a
protective role in autoimmune and inflammatory conditions (24,25).
However, the present study demonstrated that the high frequency of
the G allele in patients with diabetes does not play a protective
role; instead, it reflects an adaptive or dysregulated immune
response in attempting to counterbalance systemic inflammation.
This dual effect of IL-10 has been noted in
other chronic diseases, in which an increased IL-10
production may inhibit essential immune activation, leading to
metabolic dysfunction (26). The
discrepancies among different studies may be potentially due to
that various factors can modulate the effect of IL-10
polymorphisms, such as gene-environment interactions, ethnic
backgrounds of the populations studied, dietary habits and
lifestyle factors.
The present study has certain limitations which
should be mentioned. The present study had a relatively small
sample size and only focused on a single SNP. This was due to the
costly materials and techniques used and that the study was
self-funded. Increasing the sample size, adjusting for
environmental, clinical and metabolic variables, and including
additional IL-10 polymorphisms or other cytokine genes could
provide a more comprehensive understanding of genetic vulnerability
to DM. The functional consequences of these genetic variations are
recommended to be investigated via correlating genotypic data with
IL-10 serum levels and some clinical indicators (e.g., HbA1c and
HOMA-IR). Inflammatory status activation and estimation can differ
between different genders (27,28).
Although the present study focused exclusively on women to control
for the impact of sex hormones on immune regulation and cytokine
gene expression, future studies including both sexes are
recommended for the generalizability and expansion of these
results.
In conclusion, the present study revealed a
potential link between the IL-10 gene polymorphism
(rs1800896) and the disruption in glucose homeostasis (DM) in women
and suggested that the presence of the G allele and GG genotype may
be regarded as a risk factor. However, functional validation and
clinical correlation analyses are required to confirm these
findings. In addition, these results highlight the significance of
inflammation and immune regulation in the pathogenesis of DM and
support the inclusion of immunogenetic profiling in future risk
assessment and therapeutic strategies.
Supplementary Material
Representative DNA sequencing
chromatogram of the IL-10 gene promoter region.
Acknowledgements
The authors would like to extend their sincere and
warm thanks to the College of Pharmacy at the University of Mosul
and the College of Dentistry at the Alnoor University (Mosul, Iraq)
for their support via access to institutional infrastructure,
administrative/logistical help in organizing the workflow and
ethical approval procedures.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
AAA and SSIB designed the study. SJS performed the
experiments. AAA and SSIB were involved in the writing of the draft
of the manuscript. SSIB and SJS confirm the authenticity of all the
raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Ethics approval was obtained from the Collegiate
Committee for Medical Research Ethics (CCMRE-PHA-25-4). All
participants provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
WHO: World Diabetes Day 2024, 2024.
Available from: https://www.emro.who.int/media/news/world-diabetes-day-2024.html.
|
|
2
|
Popoviciu MS, Kaka N, Sethi Y, Patel N,
Chopra H and Cavalu S: Type 1 diabetes mellitus and autoimmune
diseases: A critical review of the association and the application
of personalized medicine. J Pers Med. 13(422)2023.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen L, Deng H, Cui H, Fang J, Zuo Z, Deng
J, Li Y, Wang X and Zhao L: Inflammatory responses and
inflammation-associated diseases in organs. Oncotarget.
9:7204–7218. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Carlini V, Noonan DM, Abdalalem E, Goletti
D, Sansone C, Calabrone L and Albini A: The multifaceted nature of
IL-10: Regulation, role in immunological homeostasis and its
relevance to cancer, COVID-19 and post-COVID conditions. Front
Immunol. 14(1161067)2023.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Rabinovitch A: An update on cytokines in
the pathogenesis of insulin-dependent diabetes mellitus. Diabetes
Metab Rev. 14:129–151. 1998.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Van Exel E, Gussekloo J, de Craen AJM,
Frölich M, Bootsma-Van Der Wiel A and Westendorp RGJ: Leiden 85
Plus Study. Low production capacity of interleukin-10 associates
with the metabolic syndrome and type 2 diabetes: The Leiden 85-plus
study. Diabetes. 51:1088–1092. 2002.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Younis MA, Ibrahim AM and Ahmad AA:
Association between blood group and TAS2R gene with COVID-19
infection. Ann Coll Med Mosul. 46:63–68. 2024.
|
|
8
|
Ismael SS and Al-Shamaa SD: Mutation in
microtubule-associated protein tau MAPT coding gene and its
correlation with Alzheimer's disease. Int J Res Pharm Sci.
11:5150–5157. 2020.
|
|
9
|
Vakili M, Shirinzadeh-Dastgiri A, Ershadi
R, Dastgheib SA, Shiri A, Aghasipour M, Barahman M, Manzourolhojeh
M, Aghili K, Neamatzadeh H and Akbarian E: Correlation between
rs1800871, rs1800872 and rs1800896 polymorphisms at IL-10 gene and
lung cancer risk. Asian Pac J Cancer Prev. 25:287–298.
2024.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Abbood SJA, Anvari E and Fateh A:
Association between interleukin-10 gene polymorphisms (rs1800871,
rs1800872, and rs1800896) and severity of infection in different
SARS-CoV-2 variants. Hum Genomics. 17(19)2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Naing C, Htet NH, Basavaraj AK and Nalliah
S: An association between IL-10 promoter polymorphisms and diabetic
nephropathy: A meta-analysis of case-control studies. J Diabetes
Metab Disord. 17:333–343. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Peng X, Xu J, Wang P, Zhou J and Guo H:
Interleukin-10-1082A/G polymorphism and diabetic nephropathy: A
meta-analysis. Med Sci Monit. 21:890–894. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hua Y, Shen J, Song Y, Xing Y and Ye X:
Interleukin-10-592C/A, -819C/T and -1082A/G polymorphisms with risk
of type 2 diabetes mellitus: A HuGE review and meta-analysis. PLoS
One. 8(e66568)2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mittal R, Camick N, Lemos JRN and Hirani
K: Gene-environment interaction in the pathophysiology of type 1
diabetes. Front Endocrinol (Lausanne). 15(1335435)2024.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Geng T and Huang T: Gene-environment
interactions and type 2 diabetes. Asia Pac J Clin Nutr. 29:220–226.
2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rodrigues KF, Pietrani NT, Sandrim VC,
Vieira CMAF, Fernandes AP, Bosco AA and Gomes KB: Association of a
large panel of cytokine gene polymorphisms with complications and
comorbidities in type 2 diabetes patients. J Diabetes Res.
2015(605965)2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Grossmannm V, Schmitt VH, Zeller T,
Panova-Noeva M, Schulz A, Laubert-Reh D, Juenger C, Schnabel RB,
Abt TGJ, Laskowski R, et al: Profile of the immune and inflammatory
response in individuals with prediabetes and type 2 diabetes.
Diabetes Care. 38:1356–1364. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
van Greevenbroek MMJ, Schalkwijk CG and
Stehouwer CDA: Obesity-associated low-grade inflammation in type 2
diabetes mellitus: Causes and consequences. Neth J Med. 71:174–187.
2013.PubMed/NCBI
|
|
19
|
Al-Mawlah1 YH, Alasadi YF, Hadi AM and
Abdul-Abbas HS: Association between some candidate gene variants
and the development of osteoporosis. Nat Volatiles Essent Oils.
8:1125–1142. 2021.
|
|
20
|
Zietz B, Watzlawek E, Palitzsch KD,
Schölmerich J and Schäffler A: GG-genotype in the promotor region
of uncoupling-protein-1 gene is associated with lower level of
dehydroepiandrosterone in type 2 diabetes. Exp Clin Endocrinol
Diabetes. 109:102–106. 2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Turner DM, Williams DM, Sankaran D,
Lazarus M, Sinnott PJ and Hutchinson IV: An investigation of
polymorphism in the interleukin-10 gene promoter. Eur J
Immunogenet. 24:1–8. 1997.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pociot F and Lernmark Å: Genetic risk
factors for type 1 diabetes. Lancet. 387:2331–2339. 2016.
|
|
23
|
Hadid KA, Alassaf FA and Abed MN: Beyond
blood sugar: Exploring the anti-inflammatory frontier of
antidiabetic medications to alleviate diabetic complications. Rom J
Med Pract. 19:92–99. 2024.
|
|
24
|
Iyer SS and Cheng G: Role of interleukin
10 transcriptional regulation in inflammation and autoimmune
disease. Crit Rev Immunol. 32:23–63. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Guo B: IL-10 modulates Th17 pathogenicity
during autoimmune diseases. J Clin Cell Immunol.
7(400)2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Couper KN, Blount DG and Riley EM: IL-10:
The master regulator of immunity to infection. J Immunol.
180:5771–5777. 2008.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hammo AA, Ahmad AA and Althanoon ZA: Role
of gender in the protection against doxorubicin-induced oxidative
stress. Pharmacogn J. 14:782–788. 2022.
|
|
28
|
Mohammed AA, Abdulla AA and Karam AA:
Measurement of inflammation-related biomarkers in different chronic
kidney diseases in humans: Role of aging and gender? IIUM Med J
Malaysia. 20:37–43. 2021.
|