Introduction
Oral squamous cell carcinoma (OSCC) is an aggressive
malignancy with high global incidence, particularly in Southeast
Asia (1). Its tendency for
late-stage diagnosis and early lymphatic spread contributes to poor
outcomes, particularly in technically unresectable cases (2). While early-stage OSCC is associated
with favorable survival rates, prognosis significantly exacerbates
in advanced-stage disease (3,4).
Identifying reliable molecular markers is crucial for early
detection, prognostication and personalized treatment strategies.
Recent advances in genomic and transcriptomic technologies have
enabled the identification of differentially expressed genes (DEGs)
associated with tumor growth, immune regulation and patient
survival in OSCC (5). These
molecular signatures provide valuable insight for early diagnosis,
treatment stratification and monitoring therapeutic outcomes
(6). In parallel, epigenetic
alterations, such as DNA methylation and copy number variations
(CNVs) are known to influence gene expression by silencing tumor
suppressor genes or activating oncogenes, contributing to disease
progression (7,8). The tumor microenvironment,
particularly immune cell infiltration, also plays a critical role
in shaping the clinical behavior of OSCC and influencing treatment
response (9).
Transposable elements, such as Alu sequences
contribute to genomic instability and have been implicated in
cancer development through mechanisms such as insertional
mutagenesis and RNA editing (10,11).
Although typically found in introns, some Alu insertions can
disrupt gene function by terminating transcription early (12,13).
Furthermore, Alu sequences harbor multiple transcription factor
binding sites, which can influence gene expression and potentially
activate oncogenic pathways. Elevated fragmented Alu DNA in
circulating DNA has been linked to a high tumor burden, suggesting
their potential as non-invasive cancer biomarkers (14). Beyond standard treatment
approaches, there is growing interest in exploring phytochemicals
and repurposed drugs as alternative means to interfere with
oncogenic signaling in OSCC. Naturally occurring compounds derived
from plants have demonstrated anticancer activity and are valued
for their potential to reduce toxicity, while inhibiting tumor
progression (15). At the same
time, established chemotherapeutic agents, such as vinorelbine are
being reconsidered for new applications, particularly through the
analysis of their binding affinity to cancer-related molecular
targets. In silico techniques, such as molecular docking and
molecular dynamics simulations currently play a key role in
predicting and validating the therapeutic relevance of these
candidate compounds (16).
The present study focused on combining genomic,
epigenetic, immunological and transposable element analyses to
uncover key biomarkers and therapeutic targets associated with
OSCC. Gaining insight into the molecular events that drive OSCC
progression can support the advancement of precision medicine
strategies and enable the development of biomarker-guided
interventions, with the goal of enhancing early diagnosis,
prognostic accuracy and treatment outcomes.
Materials and methods
Analysis of DEGs
The analysis was performed using RNA sequencing data
from the Gene Expression Omnibus (GEO) database for oral cancer
(GSE246050). DEGs were identified using GEO2R, an interactive tool
for analyzing high-throughput gene expression data. The DEGs were
identified based on the criteria of a log fold change (LogFC) of ≥1
for upregulated genes and <-1 for downregulated genes. Genes
meeting these thresholds and exhibiting a false discovery rate
(FDR)-adjusted P-value ≤0.05 were deemed statistically significant
(17).
Interaction network construction
The protein-protein interaction (PPI) network of
DEGs was constructed using the STRING database. To further analyze
the network, the list of genes was imported into Cytoscape (version
3.9.1) for the visualization and topological analysis of the
structural properties of the network. A high-confidence interaction
score (≥0.7) was applied to assess the degree of connectivity
within the network. Key hub genes were identified based on a degree
threshold of ≥10, indicating their central role in the interaction
network.
Survival analysis and heatmap
visualization of the survival associated genes
The overall survival (OS) and disease-free survival
(DFS) analyses of genes from the GEO dataset were performed using
the Gene Expression Profiling Interactive Analysis (GEPIA) online
tool (18), which has been cited
in >400 studies (19). GEPIA
performs a survival analysis on the basis of the gene expression
level, and a hypothesis is evaluated using the log-rank test
(Mantel-Cox test). Hazard ratios (HR) were calculated using the Cox
proportional hazards model with 95% confidence intervals (CIs).
Genes that had a log-rank P-value of ≤0.05 were regarded as
significant. To visualize the expression pattern of these important
genes heatmap was created using the Complex Heatmap package in R
studio (20).
cBioPortal analysis
To assess the impact of survival-related DEGs on
patient outcomes, the expression data of head and neck squamous
cell carcinoma (HNSCC) were downloaded from the Firehose Legacy
dataset of cBioPortal (https://www.cbioportal.org/datasets). The dataset
contained the American Joint Committee on Cancer (AJCC)
pathological staging, survival data, patient IDs and gene
expression values. Gene expression values were categorized as high
or low based on the median ± 2SEM cut-off. The receiver operating
characteristic (ROC) curve analysis was conducted to identify the
prognostic value of the selected genes in relation to OS using
survival duration in months and dichotomized levels of gene
expression (high and low). Genes with a greater area under the
curve (AUC) were regarded as having a greater predictive potential
and were selected to be further analyzed. Moreover, the ROC
analysis was also utilized to examine the association between the
patterns of gene expression and the advanced T4 tumor stage
classification, which also provides information about their
association with the severity of the disease. OriginPro (version
2019b; https://www.originlab.com/) was used all
statistical analysis since it offers advanced statistical analysis
tools to enable comprehensive analysis of data. ROC curves were
generated based on a classification model at all classification
thresholds, incorporating true positive rate (sensitivity) and
false positive rate (1-specificity) as key parameters. A P-value
<0.05 was considered to indicate a statistically significant
difference.
Gene Ontology (GO) and pathway
analysis
Functional enrichment analysis was performed using
the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt; https://www.webgestalt.org/), which includes GO
annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analysis (21).
Association of selected genes with
methylation probes
The methylation profiling of selected genes was
conducted using the MethSurv database, which enables CpG
site-specific survival analysis based on methylation level
(22). The platform performs
univariable and multivariable survival analyses for CpG sites
located within or near the query gene using Cox proportional
hazards models. Differences in survival between highly methylated
and lowly methylated patient groups are visualized using
Kaplan-Meier (KM) plots.
CNV and gene expression correlation
analysis
To validate the correlation between CNVs and
survival-associated DEGs in an independent cohort, RNA sequencing
data from The Cancer Genome Atlas (TCGA) were accessed via
cBioPortal (http://www.cbioportal.org). CNV data
were pre-processed using Genomic Identification of Significant
Targets in Cancer (GISTIC), which estimates somatic copy number
alterations for each annotated gene. The CNV values were
categorized as follows: -2 (homozygous deletion), -1 (heterozygous
loss), 0 (diploid), 1 (single-copy gain), and 2 (high-level
amplification or multiple-copy gain). A Spearman's correlation
analysis was conducted to determine the correlation between CNVs
and the level of gene expression using a linear regression model.
The Rho values of Spearman's correlation were used to evaluate the
correlation.
Correlation between amphiregulin
(AREG) and C-X3-C motif chemokine receptor 1 (CX3CR1) and immune
infiltration
The association between AREG and
CX3CR1 expression and tumor-infiltrating immune cells was
analyzed using the TIMER 2.0 database (http://timer.comp-genomics.org/) (23). The infiltration scores were
calculated using the single-sample Gene Set Enrichment Analysis
(ssGSEA) method. Additionally, heatmaps were generated to visualize
the correlation between the expression of genes and the immune cell
infiltration based on Spearman's Rho values. The analysis provides
the understanding of the possible role of the survival-related DEGs
in immune modulation in the tumor microenvironment.
Patients and ethics statement
The present study enrolled 18 patients from Parul
Sevashram Hospital, Vadodara, India, including 9 patients with oral
cancer and 9 patients with technically unresectable oral cancer
(TUOC). The study protocol was reviewed and approved by the Ethics
Committee of Parul University (Approval no.
PUIECHR/PIMSR/00/081734/5307) and was conducted in accordance with
institutional ethical standards and regulatory guidelines. All
patients signed an informed consent form. In addition, blood
samples were collected from 6 healthy individuals to serve as
controls for the DNA-based assays (Table I). All procedures performed in the
present study were in accordance with the principles for medical
research of the 1964 Declaration of Helsinki and its later
amendments or comparable ethical standards.
 | Table IClinical and pathological
characteristics of the patients and controls included in the
present study. |
Table I
Clinical and pathological
characteristics of the patients and controls included in the
present study.
| Group | Age (years) | Sex | Stage/substage | Sample type | HIV status | HCV status |
|---|
| OSCC | 65 | Female | CT2aN1M0 | Blood | Positive | Non-reactive |
| OSCC | 58 | Male | CT4aN1M0 | Blood | Positive | Non-reactive |
| OSCC | 72 | Male | CT1aN1M0 | Blood | Information not
available | Information not
available |
| OSCC | 62 | Male | CT4aN1M0 | Blood | Positive | Non-reactive |
| OSCC | 62 | Male | CT2aN1 M0 | Blood | Positive | Non-reactive |
| OSCC | 32 | Male | CT2aN1 M0 | Blood | Information not
available | Information not
available |
| OSCC | 49 | Female | CT2aM0 | Blood | Information not
available | Information not
available |
| OSCC | 42 | Male | CT4aM0 | Blood | Information not
available | Information not
available |
| OSCC | 52 | Female | CT2aN1 | Blood | Positive | Non-reactive |
| TUOC | 62 | Male | CT1aN1M0 | Blood | Information not
available | Information not
available |
| TUOC | 39 | Female | CT4aN1M0 | Blood | Information not
available | Information not
available |
| TUOC | 53 | Male | CT2aN2M0 | Blood | Information not
available | Information not
available |
| TUOC | 42 | Female | CT4aN2M0 | Blood | Positive | Information not
available |
| TUOC | 45 | Male | CT4aN2M0 | Blood | Positive | Non-reactive |
| TUOC | 44 | Male | CT4aN2M0 | Blood | Positive | Non-reactive |
| TUOC | 43 | Female | CT2aN2M0 | Blood | Positive | Non-reactive |
| TUOC | 67 | Male | CT1aM0 | Blood | Positive | Non-reactive |
| TUOC | 57 | Female | CT2aN2 | Blood | Positive | Non-reactive |
| Control | 24 | Female | - | - | - | - |
| Control | 27 | Female | - | - | - | - |
| Control | 23 | Female | - | - | - | - |
| Control | 24 | Female | - | - | - | - |
| Control | 23 | Male | - | - | - | - |
| Control | 23 | Male | - | - | - | - |
Patient cohort
The clinical and pathological details of all
patients included in the study are summarized in Table I. The table provides information on
age, sex, tumor stage, grade, tumor size, lymph node status and HIV
results. In the OSCC group, the age of the patients ranged from 32
to 72 years, with a median age of 58±4.18 years. In the TUOC group,
the age ranged from 39 to 67 years, with a median age of 45±3.30
years. Data on HCV status were not available for all cases, as this
information was not routinely recorded in patient files. Since the
study focused primarily on gene expression profiling rather than
viral-associated carcinogenesis, the absence of HCV data does not
affect the interpretation of the findings (24).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from fresh blood using the
QIAamp® RNA Blood Mini kit (cat. no. 52304; Qiagen,
Inc.) and RNA purity was confirmed by spectrophotometric analysis
(A260/A280 ratio between 1.8-2.0). cDNA was synthesized from 20 ng
RNA using the G-Biosciences cDNA Synthesis kit (cat. no. 786-5020;
G-Biosciences) and used for qPCR on the Rotor-Gene Q system with
SYBR-Green Master mix (cat. no. 786-5062; G-Biosciences). qPCR was
run in triplicate under standard cycling conditions as follows:
95˚C for 3 min, followed by 40 cycles of 95˚C for 15 sec and 60˚C
for 60 sec. The relative expression levels of the target gene mRNAs
were calculated using the comparative Cq method (relative
expression, 2-ΔΔCq) (25), using GAPDH as an internal control.
The primer sequences were as follows: AREG forward,
5'-GTGGTGCTGTCGCTCTTGATACTC-3' and reverse,
5'-TCAAATCCATCAGCACTGTGGTC-3'; CX3CR1 forward,
5'-GGGACTGTGTTCCTGTCCAT-3' and reverse, 5'-GACACTCTTGGGCTTCTTGC-3';
and GAPDH forward, 5'-CCCACTCCTCCACCTTTGAC-3' and reverse,
5'-TTCCTCTTGTGCTCTTGCTG-3'.
Alu DNA quantification
Blood DNA was extracted from patients with 18 oral
cancers using the QIAamp DNA Blood Mini kit (cat. no. 51104;
Qiagen, Inc.) and stored at -80˚C. AluY sequences and subtypes were
obtained from the Dfam database (26), and qPCR was conducted in triplicate
using primers for AluY, (forward, 5'-GTAATCCCAGCACTTTGGG-3' and
reverse, 5'-TAGTAGAGACGGGGTTTCAC-3'), AluY-K12 (forward,
5'-TACAAAAAATTAGCCGGGCG-3' and reverse, 5'-CATGCCATTCTCCTGCCTC-3'
and AluY-I6 (forward, 5'-GTAATCCCAGCACTTTGGG-3' and reverse,
5'-GTTCACGCCATTCTCCTGC-3'). Each 10 µl PCR reaction consisted of 5
µl SYBR-Green mix (G-Biosciences), 1 µl forward/reverse primer (10
µM), and 100 pg DNA. The cycling conditions were 95˚C for 10 min,
followed by 45 cycles at 95˚C for 15 sec and 60˚C for 60 sec, and a
hold at 60˚C for for ALUY and ALUY-K12. For ALUY-I6, the conditions
were 95˚C for 10 min, 30 repeats at 95˚C for 15 sec, and 65˚C for
60 sec. The specificity of the PCR products was confirmed by
melting curve analysis. The specificity of the PCR products was
confirmed by melting curve analysis. Quantification of total blood
Alu DNA content was based on a standard curve prepared from serial
dilutions of genomic DNA (10 ng to 0.01 pg) from healthy donors.
Each qPCR run included a no-template control, and the results are
reported as the mean ± SEM.
Molecular docking analysis of AREG
inhibitors
A library of 1,525 plant-derived bioactive compounds
was initially screened using SWISS-ADME to evaluate their
physicochemical properties and drug-likeness, resulting in the
selection of 174 potential candidates. In parallel, 50 clinically
reported anticancer drugs were shortlisted based on literature
evidence for their relevance in cancer therapy. Molecular docking
was performed between these compounds and the AREG protein using
PyRx, with the AREG structure obtained from PDB (2RNL) and compound
structures retrieved from PubChem (27). Among the screened molecules, the
top two compounds exhibiting the highest binding affinity and key
hydrogen bonding interactions with AREG were selected for molecular
dynamics simulations to further assess their stability and
inhibitory potential.
Molecular dynamics simulations
Molecular dynamics simulations were carried out for
cabraleadiol (CID-21625899) and vinorelbine in complex with AREG
using the NAMD3 simulation package with the CHARMM force field. Key
structural parameters, including root mean square deviation (RMSD),
root mean square fluctuation (RMSF) and radius of gyration (Rg),
were calculated using NAMD3 system-building commands and visualized
with the Bio3D v2.3-0 package (http://thegrantlab.org/bio3d_v2/download). The
simulation was run for a total of 200 nsec, and the binding free
energy (ΔG) of each drug-protein complex was estimated using the
NAMD Energy plugin.
Statistical analysis
The expression of DEGs in GEO2R was assessed using
moderated t-tests with the Benjamini-Hochberg FDR correction
(|LogFC| ≥1). Survival analyses were conducted in GEPIA using Cox
proportional hazards models and log-rank tests, with HRs and 95%
CIs reported. ROC curves (OriginPro 2019b) were generated using
survival duration and dichotomized gene expression, and AUC values
assessed prognostic performance. Spearman's correlation analysis
was used to analyze the correlations between CNV and gene
expression and immune infiltration scores in TIMER 2.0. MethSurv
applied Cox regression to compare survival between high- and
low-methylation groups. qPCR data from the three study groups
(resectable oral cancer, n=9; technically unresectable oral cancer,
n=9; and healthy controls, n=6) were analyzed using the
2-ΔΔCq method and are presented as
the mean ± SEM. Comparisons among the three groups were performed
using one-way ANOVA, followed by Tukey's post hoc test to identify
pairwise differences. Alu DNA quantification was performed on two
groups (resectable oral cancer vs. technically unresectable oral
cancer; n=9 per group) and compared using an unpaired two-tailed
Student's t- test. For all analyses, a P-value <0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were performed using OriginPro 2019b
(https://www.originlab.com/).
Results
Identification of prognostic
biomarkers in OSCC using integrated bioinformatics analysis of GEO
datasets
A total of 20,030 genes were identified from the
GSE246050 dataset using GEO2R analysis. Among these, 794 genes were
significantly upregulated and 1,331 were downregulated in the OSCC
samples compared to the healthy controls. To focus on the most
relevant molecular interactions, only genes with an interaction
degree of ≥10 in Cytoscape were retained for further analysis. This
filtering step yielded 318 upregulated and 221 downregulated genes
(Table SI), which were then
subjected to survival analysis using GEPIA. Among the upregulated
genes, 46 exhibited a significant association with OS, 28 with DFS,
and six genes were linked to both OS and DFS. Similarly, among the
downregulated genes, 41 genes were associated with OS, 24 with DFS,
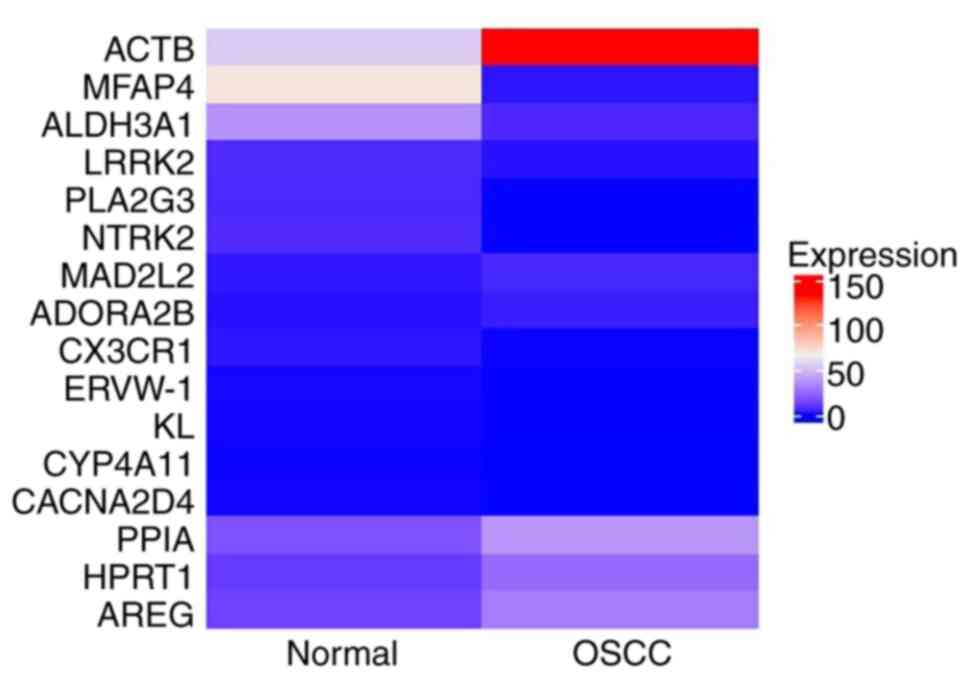
and ten genes displayed significance in both categories (Table SII). Heatmap analysis of genes
affecting both OS and DFS revealed six upregulated genes (ACTB,
HPRT1, PPIA, MAD2L2, AREG, ADORA2B) and ten downregulated genes
(MFAP4, ALDH3A1, CX3CR1, ERVW-1, CYP4A11, PLA2G3, LRRK2, NTRK2,
CACNA2D4, KL), as shown in Fig.
1. These differentially expressed genes, which affect both OS
and DFS, were identified as potential survival-associated
biomarkers for further investigation.
GO and KEGG pathway enrichment
analysis of 16 survival-associated genes
GO functional enrichment and KEGG pathway analyses
were used to examine the biological functions and molecular
pathways of genes related to both OS and DFS. Enriched GO terms
were categorized into three groups, including biological process
(BP), cellular component (CC) and molecular function (MF) (Table SIII). Under the BP category, key
terms such as phosphorylation, cell activation and
leukocyte-mediated immunity were identified, which suggests their
involvement in immune response and cellular signaling. The CC
category featured the enrichment of presynapse, axon terminus,
neuron projection terminus and elastic fibers, suggesting that they
are involved in neural signaling and extracellular matrix
organization. Among the enriched terms in the MF category,
G-protein coupled receptor activity, chemokine binding, growth
factor binding, and fatty acid hydroxylase activity were
identified, indicating the variety of functions in the process of
signal transduction and metabolism. In addition, KEGG pathway
enrichment analysis revealed several signaling pathways related to
the target genes (Table SIV).
These were the MAPK signaling pathway, which is crucial in cell
proliferation and survival, the oxytocin signaling pathway, which
is involved in cellular communication and immune regulation, the
Hippo signaling pathway, which controls the size of the organs and
cell growth, and the pathways of bacterial invasion of epithelial
cells, which suggests its potential implication in
infection-associated oncogenesis. These results provide information
on the functional role of the identified genes and their role in
the key oncogenic and immune control pathways.
Validation of selected genes in the
cBioPortal database
The Head and Neck Squamous Cell Carcinoma (TCGA,
Firehose Legacy) dataset was obtained from cBioPortal, and RSEM
expression data for genes associated with both OS and RFS were
extracted. Among the ten identified downregulated genes, only seven
(excluding ERVW-1, LRRK2 and KL) were available in
the cBioPortal dataset. All gene expression data were analyzed to
determine RSEM cut-off values for classifying high and low
expression levels for each gene. Patients were stratified into
high- and low-risk groups based on the cut-off value, determined as
the median ± 2SEM of gene expression level and on the basis of
their impact on overall survival. Out of the 13 genes initially
identified through survival analysis using cBioPortal data, seven
genes (ACTB, AREG, HPRT1, MFAP4, ALDH3A1, CYP4A11 and
CACNA2D4) demonstrated statistically significant
associations with either OS, RFS, or both, indicating their
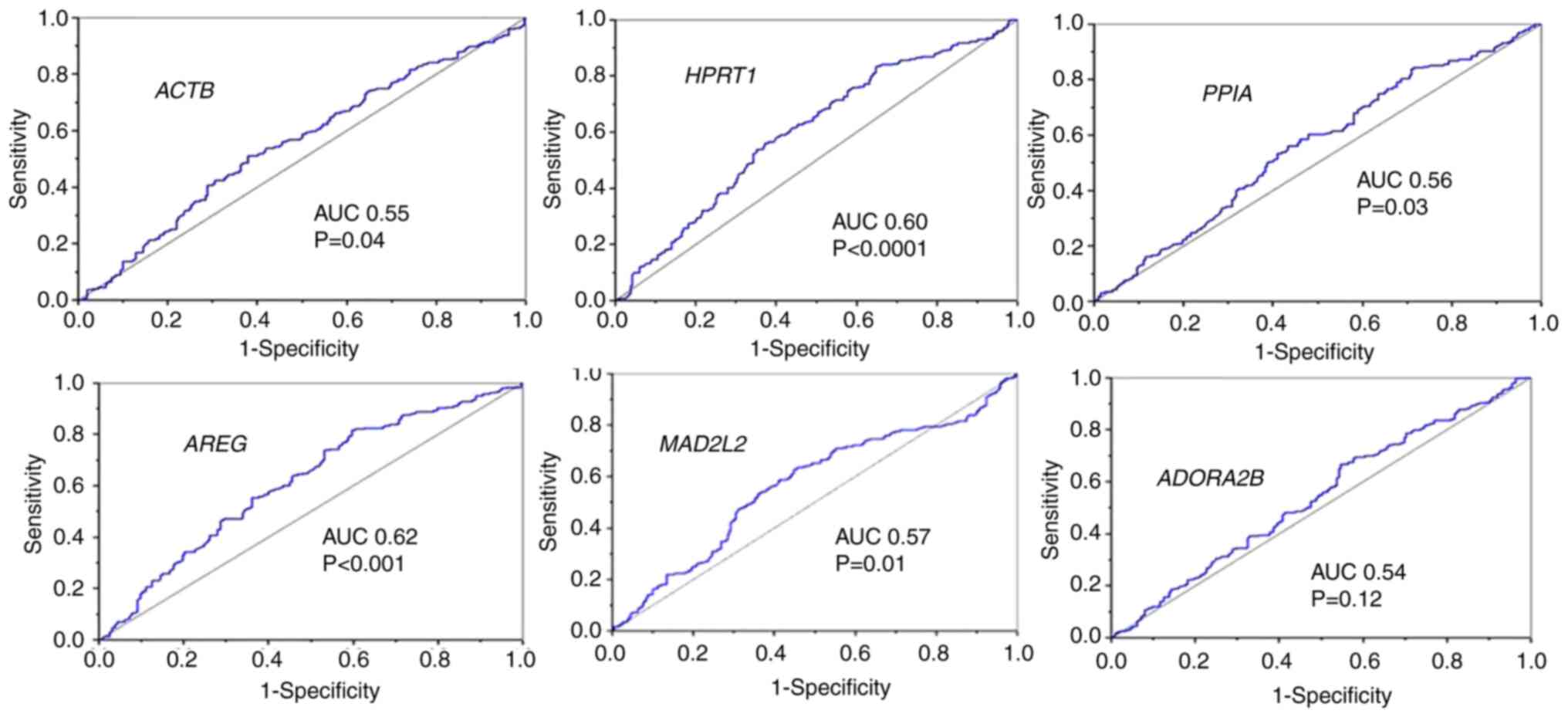
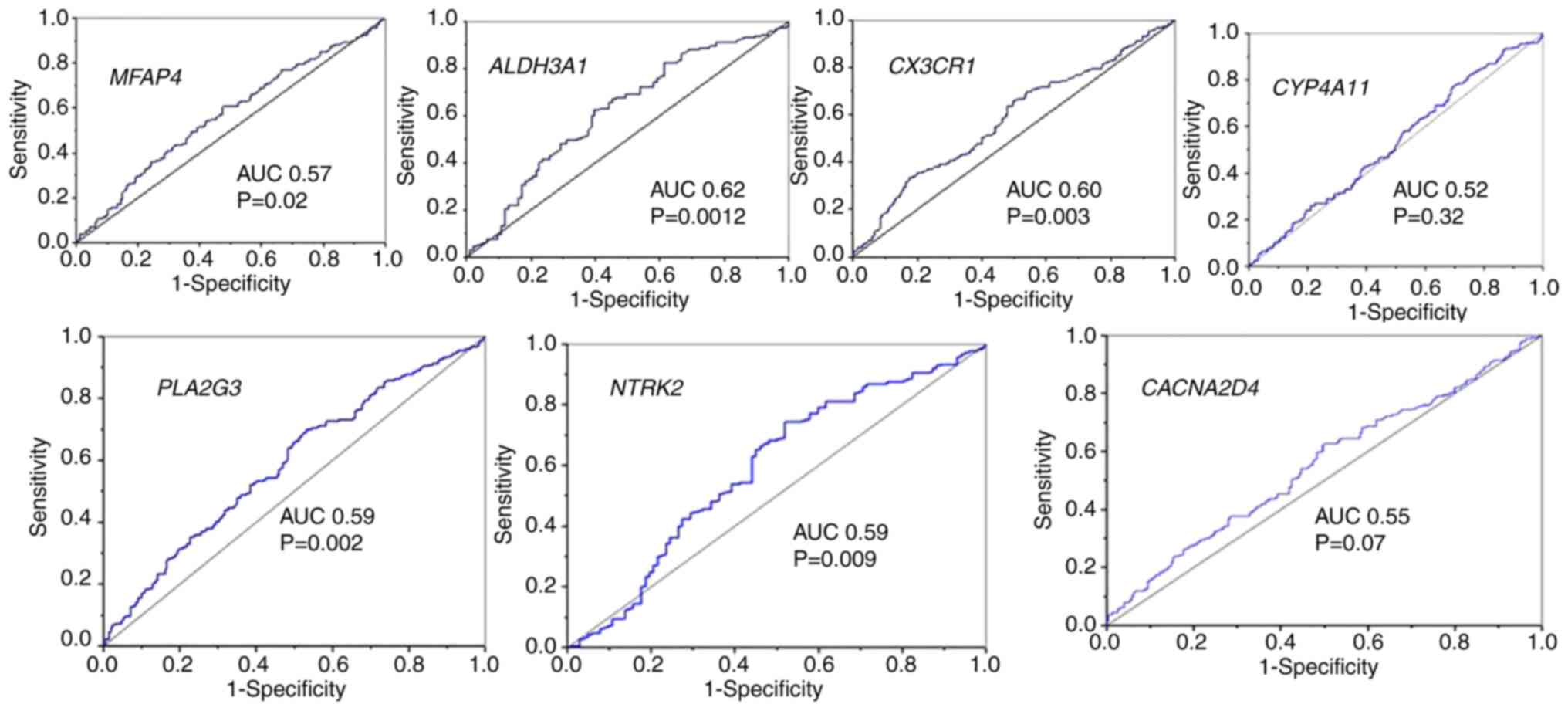
potential as prognostic markers in OSCC (Table II). Further evaluation through ROC
curve analysis revealed that only AREG, HPRT1,
ALDH3A1 and CX3CR1 had an AUC ≥0.60, suggesting their
moderate diagnostic value (Figs. 2
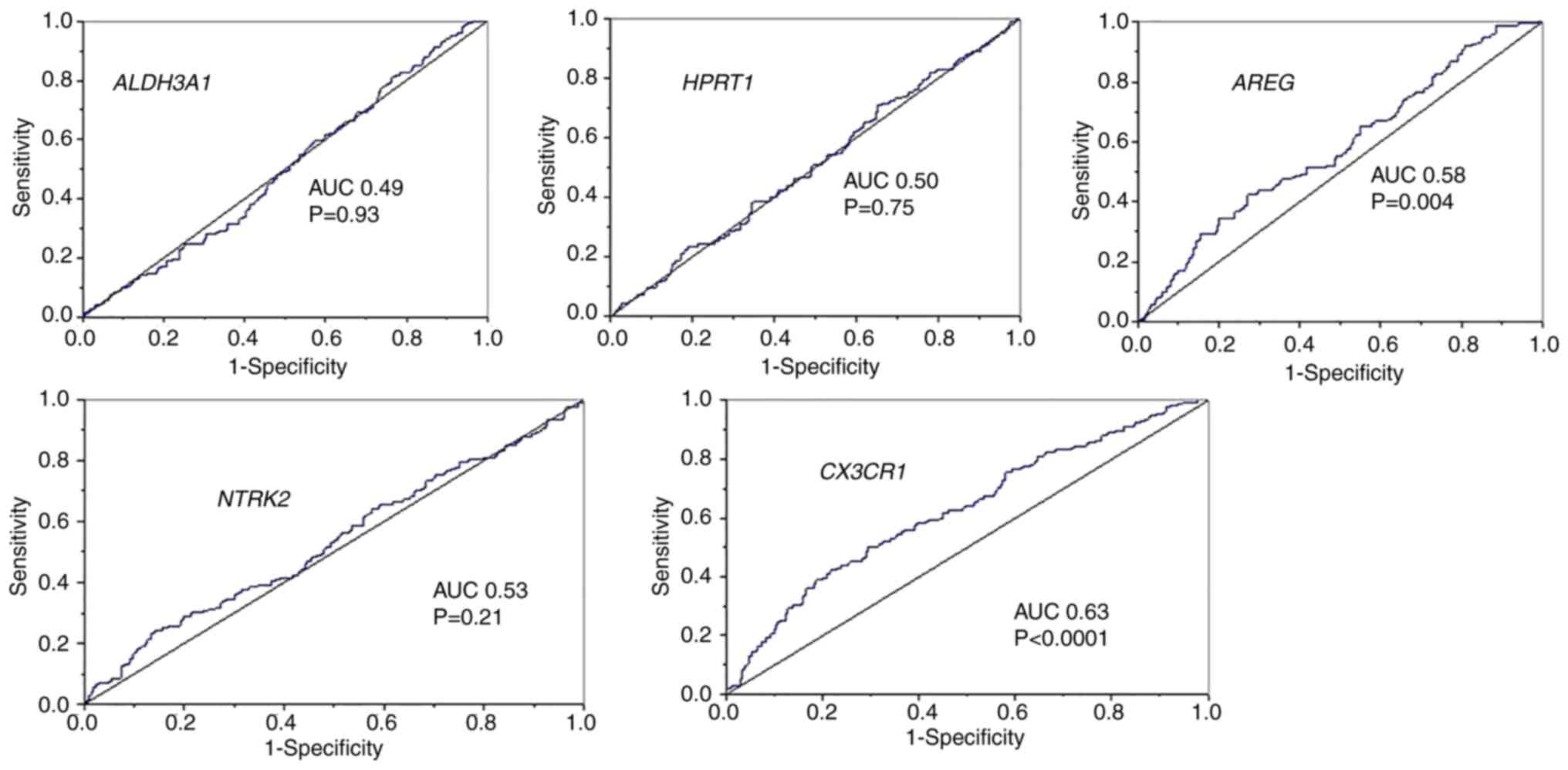
and 3). To assess the relevance of
these four genes in advanced disease, ROC analysis was performed
using T4-stage classification. Among the candidate genes,
AREG (upregulated) and CX3CR1 (downregulated)
exhibited the highest AUC values (~0.6), indicating relatively
better discrimination of advanced-stage OSCC compared to the others
(Fig. 4). Although their overall
diagnostic performance was moderate, their stage-specific
expression trends and biological links to tumor progression and
immune regulation supported their selection for detailed downstream
analysis (28-30).
 | Table IISurvival analysis of the
survival-associated DEGs identified in oral cancer from the
cBioPortal database. |
Table II
Survival analysis of the
survival-associated DEGs identified in oral cancer from the
cBioPortal database.
| Gene symbol | Risk status (no. of
patients) | Survival/deceased
status (no. of patients) | P-value | DFS/RFS (no. of
patients) | P-value |
|---|
| Upregulated
genes |
|---|
| ACTB | High risk
(245) | OS (129); Deceased
(116) | 0.0062a | DFS (108); Recurred
(73) | 2.9662 |
|
CO:139086.35±130633.33 | Low risk (242) | OS (157); Deceased
(85) | | DFS (131); Recurred
(61) | |
| AREG | High risk
(234) | OS (121); Deceased
(113) | 0.0319a | DFS (97); Recurred
(76) | 0.0067a |
|
CO:789.911±514.86 | Low risk (234) | OS (144); Deceased
(90) | | DFS (124); Recurred
(53) | |
| HPRT1 | High risk
(244) | OS (121); Deceased
(123) | 0.0001a | DFS (97); Recurred
(74) | 0.0071a |
|
CO:964.36±884.77 | Low risk (238) | OS (159); Deceased
(79) | | DFS (135); Recurred
(57) | |
| PPIA | High risk
(235) | OS (132); Deceased
(103) | 0.3980 | DFS (109); Recurred
(73) | 0.2849 |
| | Low risk (218) | OS (131); Deceased
(87) | | DFS (108); Recurred
(57) | |
| MAD2L2 | High risk
(234) | OS (127); Deceased
(107) | 0.2077 | DFS (104); Recurred
(74) | 2.1575 |
|
CO:743.67±656.76 | Low risk (240) | OS (144); Deceased
(96) | | DFS (120); Recurred
(62) | |
| ADORA2B | High risk
(243) | OS (132); Deceased
(111) | 0.1296 | DFS (107); Recurred
(76) | 0.7371 |
|
CO:429.28±377.05 | Low risk (227) | OS (139); Deceased
(88) | | DFS (117); Recurred
(56) | |
| Downregulated
genes |
| MFAP4
CO:373.16±166.62 | High risk
(211) | OS (131); Deceased
(80) | 0.0363a | DFS (112); Recurred
(49) | 0.0062a |
| | Low risk (195) | OS (101); Deceased
(94) | | DFS (80); Recurred
(67) | |
| ALDH3A1 | High risk
(197) | OS (118); Deceased
(79) | 0.0054a | DFS (99); Recurred
(46) | 0.0312a |
|
CO:2152.11±184.34 | Low risk (94) | OS (40); Deceased
(54) | | DFS (32); Recurred
(29) | |
| CX3CR1 | High risk
(232) | OS (138); Deceased
(94) | 0.4080 | DFS (119); Recurred
(58) | 2.9959 |
| CO:25.29±16.56 | Low risk (214) | OS (119); Deceased
(95) | | DFS (96); Recurred
(69) | |
| CYP4A11,
CO:0.55±0.20 | High risk
(201) | OS (135); Deceased
(66) | 0.0026a | DFS (116); Recurred
(47) | 0.0074a |
| | Low risk (227) | OS (120); Deceased
(107) | | DFS (97); Recurred
(73) | |
| PLA2G3, | High risk
(220) | OS (133); Deceased
(87) | 0.1919 | DFS (116); Recurred
(51) | 3.4236 |
|
CO:140.43±66.95 | Low risk (212) | OS (115); Deceased
(97) | | DFS (93); Recurred
(63) | |
| NTRK2 | High risk
(210) | OS (126); Deceased
(84) | 0.1103 | DFS (106); Recurred
(50) | 0.0378a |
|
CO:1166.69±98.61 | Low risk (103) | OS (52); Deceased
(51) | | DFS (41); Recurred
(35) | |
|
CACNA2D4 | High risk
(233) | OS (145); Deceased
(88) | 0.0398a | DFS (123); Recurred
(62) | 0.1449 |
| CO:24.26±19.58 | Low risk (222) | OS (117); Deceased
(105) | | DFS (93); Recurred
(65) | |
Association of AREG and CX3CR1 with
methylation probes
According to methylation profile data from the
MethSurv database, AREG is associated with five specific
methylation probes: cg02009651, cg03244277, cg02334660, cg26611070
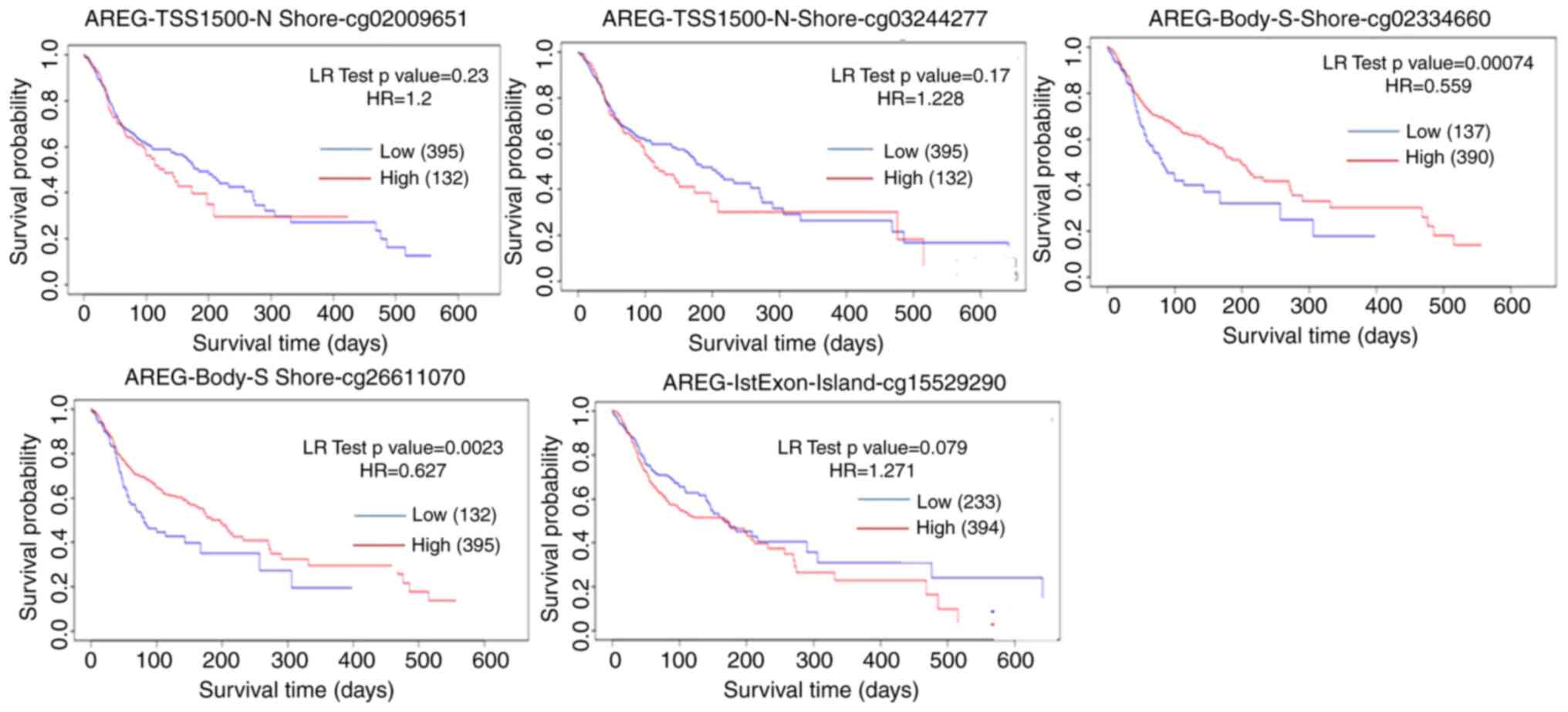
and cg15529290 (Fig. 5). Among
these, methylation levels at CpG sites cg02009651, cg03244277 and
cg15529290 were negatively associated, whereas methylation at
cg02334660 and cg26611070 was associated with a favorable prognosis
correlation. Additionally, the analysis of the association between
these CpG sites and demographic/clinical factors including
ethnicity, race, body mass index (BMI), age and clinical events
revealed that cg02334660 and cg26611070 were positively association
with these variables, while cg15529290 (at 1st exon), cg02009651
and cg03244277 exhibited a negative association.
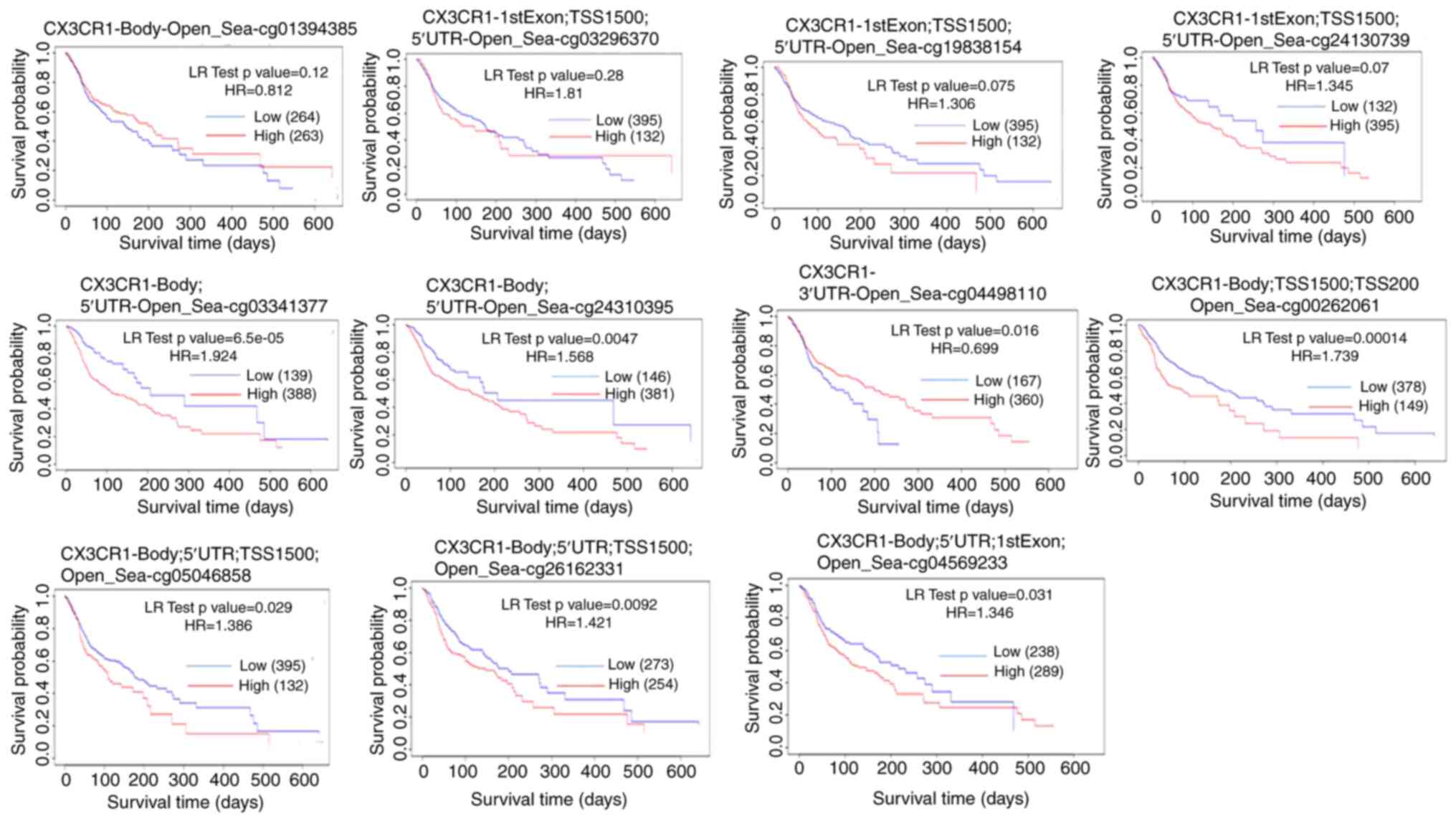
From the CX3CR1 methylation profile data from
the MethSurv database, CX3CR1 was shown to be associated
with 11 specific methylation probes: cg24130739, cg03296370,
cg19838154, cg01394385, cg26162331, cg00262061, cg05046858,
cg04498110, cg04569233, cg03341377 and cg24310395 (Fig. 6). Methylation-survival analyses
were performed using the MethSurv web tool, which fits Cox
proportional hazards models and reports hazard ratios, log-rank
test p-values, and a proportional hazards assumption test for each
CpG site (22). The log-rank test
has optimum power under the assumption of proportional hazard
rates. However, this assumption is often violated, particularly
when two survival curves cross each other. In Fig. 6, the late crossing of the survival
curve can be observed in the last plot. The authors were not able
to restrict the time range in the software to remove the late-stage
crossover. In addition, survival analyses for methylation were
obtained from the MethSurv platform using aggregate output; thus,
the application of alternative weighted tests was not possible.
Therefore, this may be a limitation of the present study. Among
these, methylation levels at cg24130739, cg03296370, cg19838154,
cg03341377, CpG sites were negatively associated, whereas
methylation at cg00262061, cg04498110, cg03341377, cg24310395,
cg04569233, cg05046858 and cg26162331 was associated with a
favorable prognosis. Additionally, the analysis of the association
between these CpG sites and demographic/clinical factors, including
ethnicity, race, BMI, age and clinical events revealed that
methylation at cg01394385, cg26162331, cg00262061 and cg05046858
were positively associated with these variables, while methylation
at cg24130739, cg03296370, cg19838154, cg04498110, cg04569233,
cg03341377 and cg24310395 exhibited a negative association
(Fig. 6).
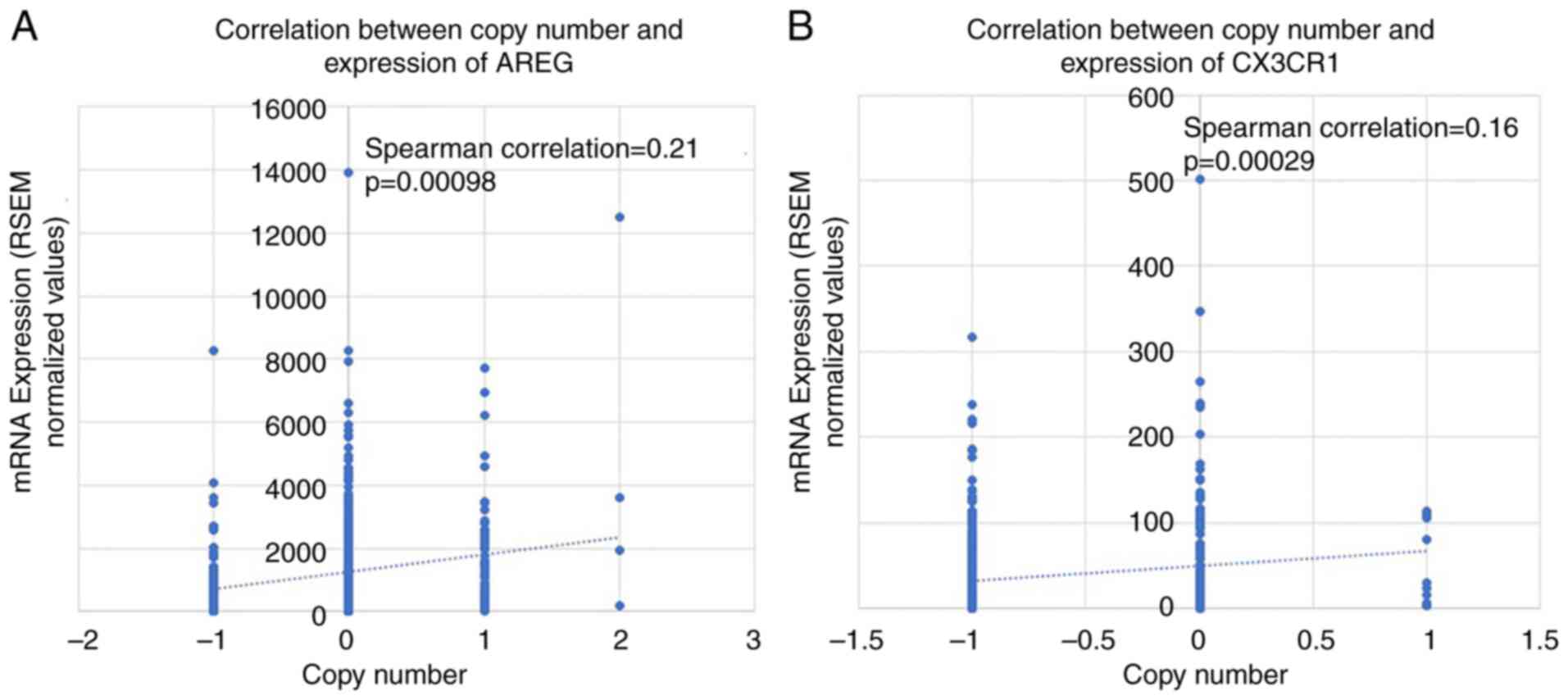
Copy number analysis
CNV is known to influence gene expression; however,
the association between CNVs of AREG and CX3CR1 and
their impact on gene expression in oral cancer remain to be fully
elucidated. Correlation analysis was performed using the HNSCC
dataset hosted on cBioPortal, based on patients with paired
copy-number and expression data (AREG, n=234; CX3CR1, n=524). In
the present study, the analysis revealed a significant effect of
CNV on gene expression (P<0.001; (Fig. 7). From a tumor biology perspective,
GISTIC2 analysis identified significant CNV events, which may
contribute to the observed differences in gene expression. To
further investigate this association, patients were stratified into
high- and low-risk groups based on a cut-off value determined as
the median ± SEM of gene expression levels. CNV analysis revealed
that copy number deletions were significantly associated with a
decreased gene expression of both AREG (high, 35/230; low,
71/232; P=0.000137) and CX3CR1 (high, 142/227; low, 163/213;
P=0.0058) in the cBioPortal dataset for HNSCC. However, no
significant differences were observed for copy number
amplifications (Table III).
 | Table IIIDistribution of AREG and
CXC3CR1 copy number alterations in the high- and
low-expression groups. |
Table III
Distribution of AREG and
CXC3CR1 copy number alterations in the high- and
low-expression groups.
| | Deletion | No change | Amplification | Total |
|---|
| AREG | | | | |
|
High | 35 | 155 | 40 | 230 |
|
Low | 71 | 138 | 23 | 232 |
| CX3CR1 | | | | |
|
High | 142 | 80 | 5 | 227 |
|
Low | 163 | 46 | 4 | 213 |
To further validate the correlations between CNV and
gene expression, a linear regression model was applied using HNSCC
data from cBioPortal. The analysis revealed a weak correlation
between gene expression and copy number alterations for both AREG
(ρ=0.213, P=0.00098) and CX3CR1 (ρ=0.159, P=0.00029), as shown in
Fig. 7A and B. Although the correlation coefficients
indicate weak associations, they are statistically significant due
to the large sample size.
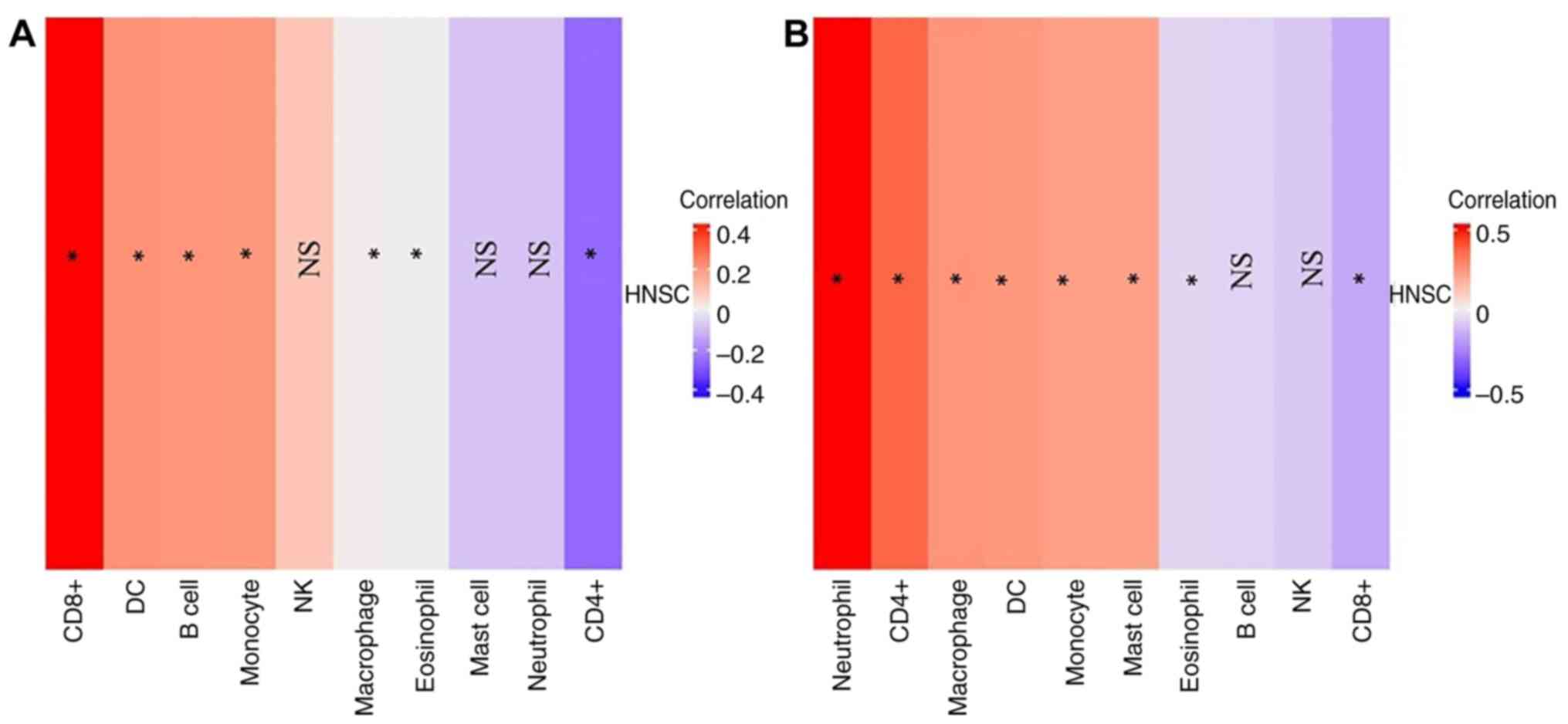
AREG and CX3CR1 expression are
associated with immune cell infiltration
The results of TIMER analysis suggested that
AREG and CX3CR1 exhibit differential immune
infiltration patterns in HNSCC. Notably, CX3CR1 appears to
be more strongly associated with neutrophil and CD4+
T-cell infiltration, while AREG is associated more with
CD8+ T-cells (Fig. 8).
These findings may provide insight into the immunomodulatory roles
of these genes in the HNSCC tumor microenvironment.
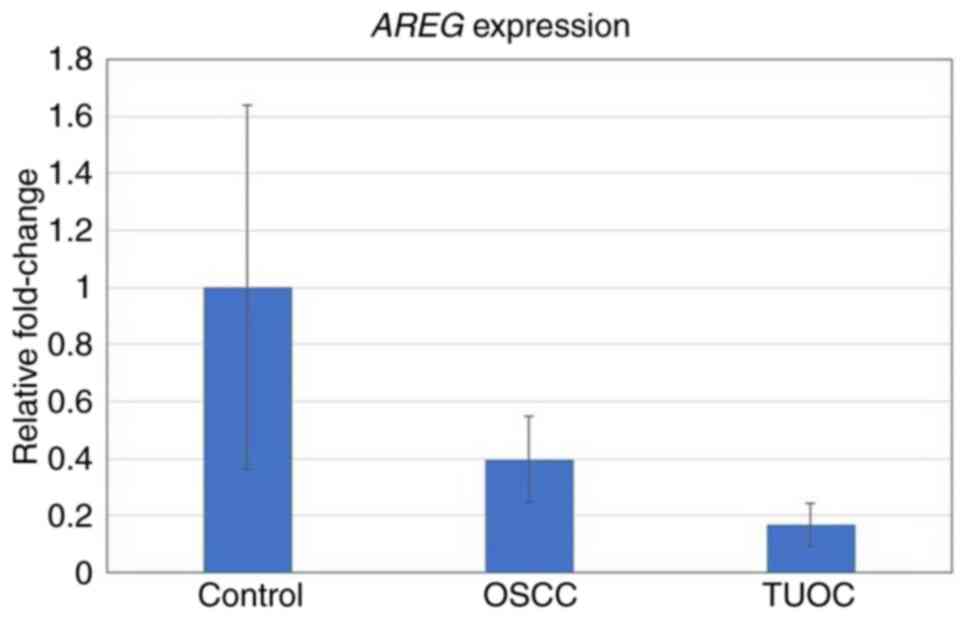
RT-qPCR analysis of AREG and CX3CR1 in
oral cancer
Subsequently, the expression levels of AREG
and CX3CR1 were evaluated in the blood samples of patients
with oral cancer. AREG expression was found to be reduced in
technically unresectable cases (TUOC, 0.16-fold change ± 0.08)
compared to patients with resectable cancer (0.39-fold change ±
0.15) (Fig. 9). By contrast,
CX3CR1 expression could not be reliably detected, as its Cq
value was >42, indicating levels below the measurable limit.
Phytochemicals as AREG potential
inhibitors
Of the 1,525 phytochemicals screened, 274 displayed
drug-like properties, indicating potential as drug candidates.
Similarly, Gene Set Cancer Analysis (GSCA) identified 84 inhibitors
for AREG by co-relating drug sensitivity and gene expression.
Binding affinity scores were calculated for both 274 phytochemicals
and 84 AREG inhibitors. The (25s)-neospirost-4-en-3-one for
phytochemical and vinorelbine for inhibitors demonstrate highest
binding affinity to AREG and therefore, were prioritized for
binding energy calculation. The list of top 20 phytochemicals and
inhibitors is presented in Tables
IV and V, respectively.
 | Table IVList of top 20 phytochemical
inhibitors demonstrating binding affinity scores and hydrogen bond
interactions. |
Table IV
List of top 20 phytochemical
inhibitors demonstrating binding affinity scores and hydrogen bond
interactions.
| Name | Binding
affinity | Hydrogen bonds |
|---|
|
(25s)-neospirost-4-en-3-one | -7.5 | TYR-27 |
| Limonin | -7.1 | ASN-10, LYS-26 |
| 2-(4-hydroxyphenyl)
naphthalic anhydride | -6.7 | TYR-27 |
|
Cucurbita-5_23(e)-diene-3-beta_7-beta_25-triol | -6.7 | HIS-30, LYS-9 |
| Betulinic_acid | -6.7 | LYS-9 |
| Caloxanthone_a | -6.7 | LYS-9 |
|
Garcimangosxanthone_b | -6.7 | GLN-17 |
|
2_3-dihydrowithaferin_a | -6.6 | CYS-38, LYS-37 |
|
Garcidepsidone_b | -6.6 | TYR-27, GLU-15 |
|
3-beta-acetoxy-8-beta-isobutyryloxyreynosin | -6.5 | TYR-27 |
|
8-prenyl_erythrinin_c | -6.5 | TYR-42, GLN-39,
CYS-38, LYS-37 |
| Caloxanthone_a | -6.5 | LYS- 9, GLU-15 |
|
Eichlerialactone | -6.5 | LYS-26 |
|
Garcimangosxanthone_c | -6.5 | GLY-17, CYS-25,
GLY-23 |
|
3-beta__12-dihydroxy-13-methyl-6_8_11_13-podocarpatetraen | -6.4 | TYR-27, ASN-10
GLY-7, LYS-26 |
|
Metrhylmitrekaurenate | -6.3 | TYR-27 |
| Xanthone V1 | -6.3 | LYS-9, GLU-32
TYR-27, GLU-29 |
| Escobarine A | -6.2 | GLY-7, LYS-9
TYR-27 |
| Garcidepsidone
A | -6.2 | TYR-27 |
|
2r_3r)-3_5-dihydroxy-7-methoxy_avanone | -6.1 | TYR-27 |
 | Table VList of top 20 AREG inhibitors
demonstrating binding affinity scores and hydrogen bond
interactions. |
Table V
List of top 20 AREG inhibitors
demonstrating binding affinity scores and hydrogen bond
interactions.
| Ligand | Binding
affinity | Hydrogen bonds |
|---|
| Vinorelbine | -9.8 | LYS-9 |
| Docetaxel | -8.6 | GLU-29, TYR-27,
GLY-7, SER-6 |
| Tipifarnib | -8.1 | TYR-27 |
| Tanespimycin | -8.1 | GLU-32, TYR-27 |
| AKT inhibitor
VIII | -7.9 | SER-5, GLU-15 |
| AZ628 | -7.8 | GLN-39, GLN-40,
CYS-38, TYR-42, GLY-44 |
| A-770041 | -7.7 | HIS-22, LYS-26,
ASN-10 |
|
S-Trityl-L-cysteine | -7.6 | TYR-27 |
| OSU-03012 | -7.6 | LYS-9, GLU-29 |
| Nilotinib | -7.5 | LYS-9, TYR-27 |
| LY317615 | -7.5 | GLU-15, TYR-27 |
| Etoposide | -7.5 | ASN-10, ASN-13,
CYS-25 |
| GSK1904529A | -7.3 | LYS-26 |
| BMS-509744 | -7.2 | LYS-9, VAL-34 |
| ERK5-IN-1 | -7.2 | GLN-17, CYS-25 |
| PHA-665752 | -7.2 | ASN-13, CYS-12,
CYS-25 |
| Doramapimod | -7.1 | SER-5, SER-6 |
| FR-180204 | -7.0 | GLY-44, CYS-38,
GLN-39 |
| Cyclopamine | -7.0 | TYR-27 |
| BMS-708163 | -7.0 | GLU-32 |
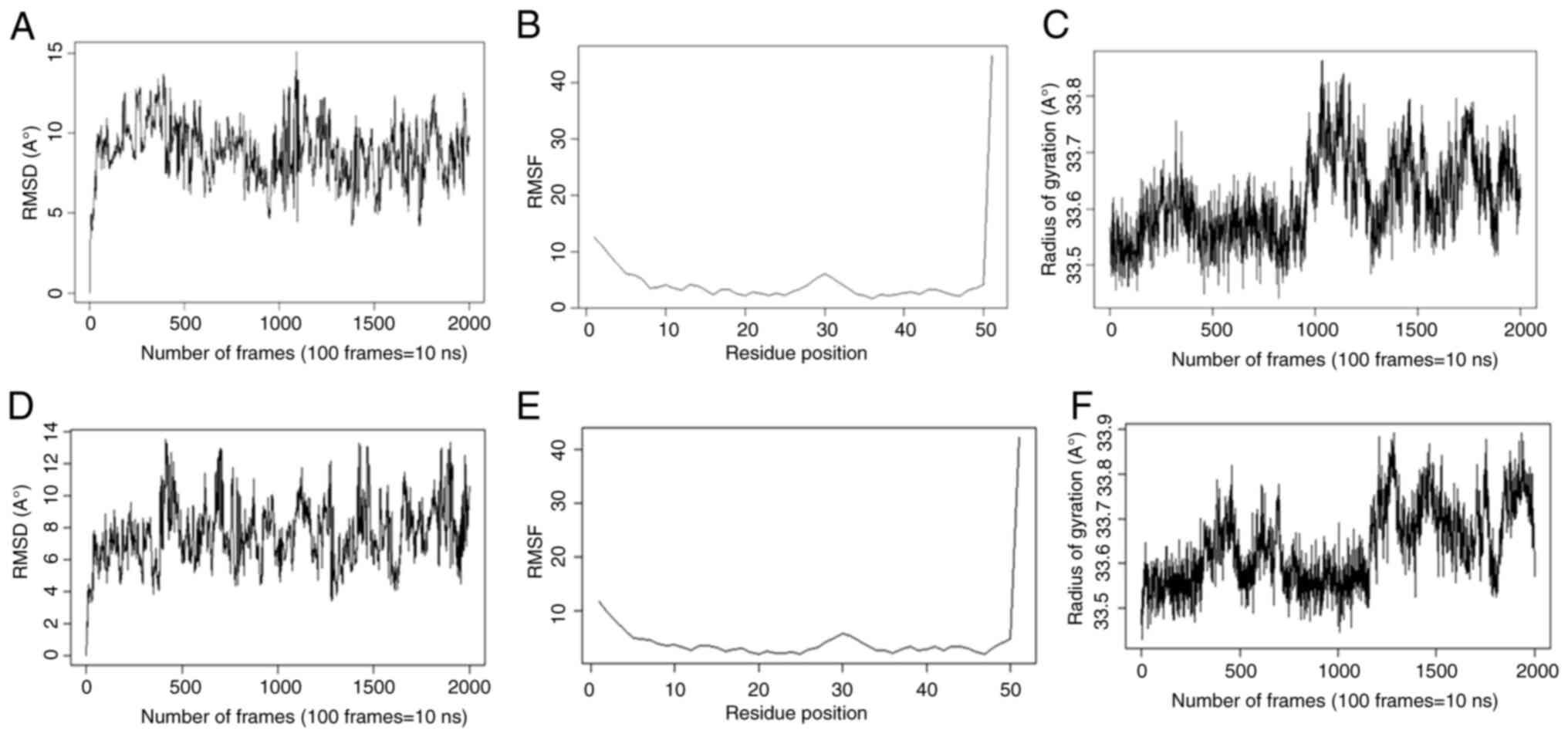
Molecular docking and molecular
dynamics simulation
Docking analysis revealed the highest binding
affinity between AREG- (25s)-neospirost-4-en-3-one and
AREG-vinorelbine (Tables IV and
V).
The stability and compactness of these complexes
were assessed through RMSD and radius of gyration (Rg), as
demonstrated in Table VI and
Fig. 10. The RMSD and Rg did not
fluctuate for the 200 nsec, confirming the stability of the 3D
protein structure during molecular dynamics simulations.
Additionally, the RMSF was stable over 200 ns of simulation,
suggesting least residue-level fluctuations. These findings
strongly support the stable interaction of the complexes during the
200 nsec simulation, confirming their structural stability and
potential effectiveness.
 | Table VIComparative analysis of AREG-
(25s)-neospirost-4-en-3-one and AREG-vinorelbine. |
Table VI
Comparative analysis of AREG-
(25s)-neospirost-4-en-3-one and AREG-vinorelbine.
| SN | RMSD (A˚) | Rg (A˚) | RMSF (nm) |
|---|
| AREG | 5.33±0.03 | 33.59±0.002 | 3.15±0.29 |
|
(25s)-neospirost-4-en-3-one | 1.17±0.001 | 4.79±0.001 | 0.41±0.04 |
| Vinorelbine | 3.40±0.01 | 5.35±0.001 | 2.27±0.10 |
| AREG-
(25s)-neospirost-4-en-3-one | 7.61±0.04 | 33.64±0.002 | 3.76±0.27 |
|
AREG-vinorelbine | 8.85±0.04 | 33.61±0.002 | 3.93±0.31 |
The results also revealed that the binding energy
between AREG-vinorelbine was -15.26 kcal/mol, while the binding
energy between AREG- (25s)-neospirost-4-en-3-one was +3.6 kcal/mol
(Table VII). These results
suggest a strong binding interaction of vinorelbine with AREG in
comparison to (25s)-neospirost-4-en-3-one.
 | Table VIIFree binding energy of
(25s)-neospirost-4-en-3-one and vinorelbine with AREG. |
Table VII
Free binding energy of
(25s)-neospirost-4-en-3-one and vinorelbine with AREG.
| SN | Electrostatic
energy (kcal/mol) | Vander Waals energy
(kcal/mol) | Free binding energy
of (ΔG) AREG-drug complex (kcal/mol) |
|---|
|
(25s)-neospirost-4-en-3-one | -47.24±0.01
SEM | 5.69±0.02 SEM | NA |
| Vinorelbine | 105.55±0.03
SEM | 33.40±0.03 SEM | NA |
| AREG | -351.46±1.59
SEM | -116.96±0.24
SEM | NA |
|
AREG-(25s)-neospirost-4-en-3-one | -326.85±1.80
SEM | -115.10±0.25
SEM | +3.6 |
|
AREG-Vinorelbine | -400.60±2.26
SEM | -116.802±0.29
SEM | -15.26 |
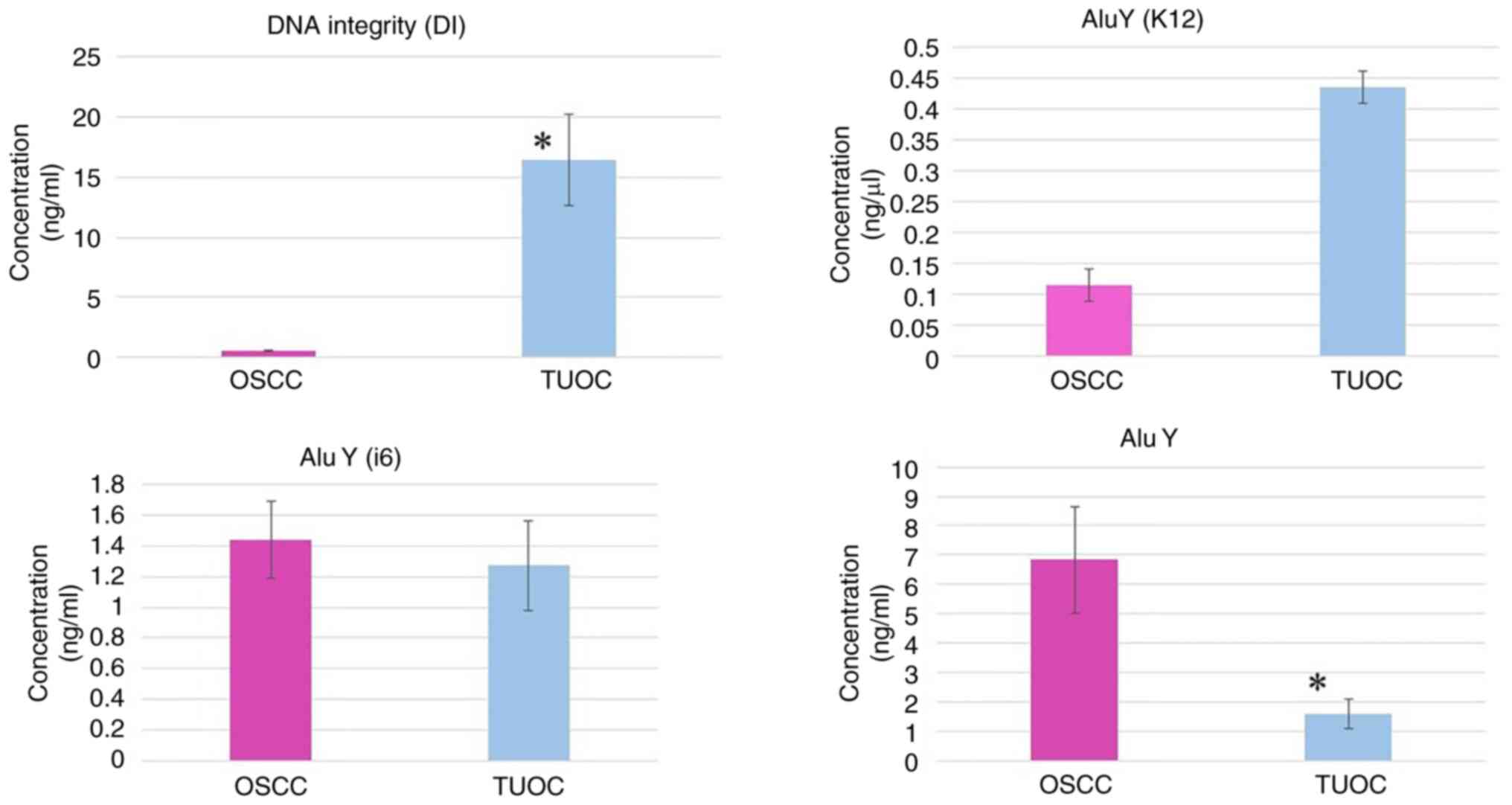
Analysis of Alu family expression in
patients with oral cancer and TUOC
In the present study, Alu DNA levels were quantified
in serum samples from patients with resectable oral cancer and TUOC
to evaluate their potential as biomarkers. The DNA integrity (DI)
ratio, measured using Alu115/Alu247, was significantly elevated in
patients with TUOC (16.46±3.82) compared to patients with
resectable OSCC (0.55±0.11), with a P-value of 0.0007, indicating
increased genomic instability in advanced-stage disease (Fig. 11). The analysis of AluY
subfamilies revealed a non-significant increase in AluY (K12)
expression in patients with TUOC (0.43±0.22) vs. patients with
resectable OSCC (0.11±0.03; P=0.1715), and a similar trend was
observed for AluY (i6) (TUOC, 1.44±0.25; resectable OSCC,
1.27±0.29; P=0.6589) (Fig. 11).
Of note, total AluY family expression, encompassing all subtypes,
was significantly higher in patients with resectable OSCC
(6.83±1.83) than in those with TUOC (1.60±0.51; P=0.0143) (Fig. 11). These results suggest that
while individual AluY subfamilies may not distinctly reflect
disease stage, elevated DI ratios and changes in total AluY
expression may serve as potential indicators of disease progression
and early-stage tumor activity in OSCC.
Discussion
OSCC is an aggressive malignancy and has low chances
of survival because of late diagnosis, resistance to therapy and
lack of effective molecular biomarkers. Although previous research
has examined the significance of genomic modifications, epigenetic
modifications, and immune responses to define possible diagnostic
and therapeutic targets, there is limited clinical translation of
these results (31). The present
study used an integrative strategy that consisted of
bioinformatics, transcriptomic profiling, epigenetic analysis,
immune infiltration evaluation, and molecular docking to provide
further information on the molecular landscape of OSCC.
Previous studies have reported frequently
dysregulated genes in oral cancer, such as TP53, EGFR,
NOTCH1 and CCND1 that are associated with proliferation,
apoptosis and resistance to treatment (32,33).
The present study demonstrated that AREG and CX3CR1
were the most relevant genes associated with progressive OSCC,
which are associated with immune infiltration and poor prognosis
indicators. The clinical importance of AREG is further supported by
a study conducted by Bourova-Flin et al (28), which reported that high expression
of AREG, along with Cyclin A1 and
DDX20/Gemin3, was indicative of aggressive OSCC and
correlated with unfavorable clinical outcomes. This reinforces the
observation that AREG may contribute to tumor progression
and immune-related changes in advanced disease. Equally, CX3CL1 has
also been demonstrated to facilitate the migration and invasion of
OSCC cells by increasing ICAM-1 expression via CX3CR1 receptor,
which underscores its contribution to tumor invasiveness (34). These reports are consistent with
the findings of the present study, in which CX3CR1
expression was modulated in terms of immune cell invasion and
disease severity. Collectively, these data support the notion that
AREG and CX3CR1 are potentially useful biomarkers to
detect and describe OSCC cases. Nonetheless, the diagnostic and
prognostic ability of both genes seems to be limited as both of
them showed only moderate AUC values (~0.6) in the ROC analyses.
Thus, the results are to be considered as preliminary, and further
research is required to confirm their applicability to clinical
settings in larger patient groups.
The epigenetic modifications including CNVs and DNA
methylation are the focus of OSCC development. In line with the
earlier reports that reported promoter hypermethylation-induced
silencing of tumor suppressors, such as CDKN2A, DAPK1 and
MGMT (35,36), the present study reported
hypermethylation of AREG and CX3CR1, with certain CpG
sites being associated with poor survival. CNV analysis identified
deletions of CX3CR1 that were associated with decreased
expression, indicating its tumor-suppressive activity, whereas
AREG was not affected by deletion of CNV, indicating further
epigenetic or post-transcriptional regulation. On the immunological
front, previous research has linked CD8+ T-cell
infiltration with improved prognosis, and increased Tregs or MDSCs
with poorer outcomes (37,38). The present study demonstrated that
AREG and CX3CR1 were associated with CD8+
T-cells and neutrophils and CD4+ T-cells respectively,
which indicated differential roles in immune modulation. Taken
together, these results support previous data of chemokine and
growth factor implication by modulating tumor microenvironment in
OSCC and emphasize their role in the regulation of the epigenome,
as well as tumor-immune interactions (39,40).
In a previous study on non-small cell lung cancer
and HNSCC, changes in serum levels of EGFR and its ligands, such as
AREG, were described, and lower levels of AREG
expression were found in serum samples of patients (41). These findings are in line with the
findings of the present study, which also revealed the reduced
expression of AREG in the blood of patients with TUOC.
Although the present study was not able to do tissue-level
validation, several previous studies have established that
AREG is overexpressed in OSCC tissues (28,29,42,43).
This, along with the observation of lower AREG levels in
circulation in the present study, confirms the hypothesis of the
‘dual faces’, in which local tumor upregulation is accompanied by
lower systemic levels. These findings also indicate that local
tumor expression can be more useful in prognosis compared to
circulating levels. CX3CR1 was, however, not detectable in
serum samples, presumably due to very low levels of expression or
degradation, suggesting that tissue-based analysis can be more
informative on this marker.
Transposable elements, particularly Alu elements,
are critical in the progression of cancer (44). Consistent with previous studies,
the present study also discovered that the DNA integrity ratio
(Alu115/Alu247) was significantly higher in patients with TUOC and
indicated more genomic instability (45,46).
Whereas AluY (K12) expression was more elevated in TUOC, the total
expression of the AluY family was more elevated in resectable OSCC,
indicating stage-specific roles of Alu subfamilies. The results are
in line with previous research demonstrating that some Alu
subfamilies differ in expression across cancer types, which
supports their possible use as biomarkers of disease staging and
monitoring (45).
Due to the drawbacks of traditional OSCC therapy,
natural compounds have become the focus of interest as possible
anticancer agents as they can regulate oxidative stress,
inflammation and apoptosis pathways (47,48).
The present study identified 274 drug-like compounds; however, only
vinorelbine, an approved drug, exhibited a strong and stable
binding with AREG in molecular docking and molecular dynamics
simulations. These results suggest that vinorelbine has a higher
therapeutic potential compared to the identified natural compounds.
Although limited by a small validation cohort and reliance on
bioinformatics data, the results are exploratory and indicate that
AREG, CX3CR1 and Alu elements can be considered as a
potential therapeutic targets and biomarkers in OSCC. To validate
these findings and enhance biomarker-based treatment strategies,
larger, multi-center and functional studies are required.
Overall, the analyses of genomic, epigenetic,
immunological and transposable elements were combined in the
present study to identify novel biomarkers and therapeutic targets
in OSCC. The results enhance the current understanding of the
molecular mechanisms of OSCC and may be applied to designing the
individual treatment plans in accordance with the profile of
biomarkers.
Supplementary Material
List of upregulated and downregulated
genes.
DEGs associated with OS and RFS in
oral cancer.
GO function and pathway enrichment
analysis of common genes in OSCC.
KEGG pathways analysis of DEGs genes
in OSCC.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the intramural
funding from Parul University, Vadodara, India (grant no.
RDC/IMSL/213).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
All authors (NK, CJ, MP and RG) contributed to the
conception and design of the study. Material preparation, data
collection and analysis were performed by NK and CJ. Molecular
dynamics simulation and qPCR were performed by MP. NK and RG
confirm the authenticity of all the raw data. The first draft of
the manuscript, editing and supervision was performed by RG, and
all authors commented on previous versions of the manuscript. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was reviewed and approved by the
Ethics Committee of Parul University (Approval no.
PUIECHR/PIMSR/00/081734/5307) and was conducted in accordance with
institutional ethical standards and regulatory guidelines. All
patients signed an informed consent form, and all procedures
performed in this study were in accordance with the principles for
medical research of the 1964 Declaration of Helsinki and its later
amendments or comparable ethical standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Shimojukkoku Y, Nguyen PT, Ishihata K,
Ishida T, Kajiya Y, Oku Y, Kawaguchi K, Tsuchiyama T, Saijo H,
Shima K and Sasahira T: Role of early growth response-1 as a tumor
suppressor in oral squamous cell carcinoma. Discov Oncol.
15(714)2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kijowska J, Grzegorczyk J, Gliwa K, Jędras
A and Sitarz M: Epidemiology, diagnostics, and therapy of oral
cancer-update review. Cancers (Basel). 16(3156)2024.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Yosefof E, Edri N, Kurman N, Bachar G,
Shpitzer T, Mizrachi A and Popovtzer A: The role of adjuvant
radiotherapy for early-stage oral cavity cancer with minor adverse
features; A single institute experience. Head Neck. 47:1839–1847.
2025.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Caputo WL, de Souza MC, Basso CR, Pedrosa
VA and Seiva FRF: Comprehensive profiling and therapeutic insights
into differentially expressed genes in hepatocellular carcinoma.
Cancers (Basel). 15(5653)2023.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zakari S, Niels NK, Olagunju GV, Nnaji PC,
Ogunniyi O, Tebamifor M, Israel EN, Atawodi SE and Ogunlana OO:
Emerging biomarkers for non-invasive diagnosis and treatment of
cancer: A systematic review. Front Oncol. 4(1405267)2024.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ye M, Huang X, Wu Q and Liu F: Senescent
stromal cells in the tumor microenvironment: Victims or
accomplices? Cancers (Basel). 15(1927)2023.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lakshminarasimhan R and Liang G: The role
of DNA methylation in cancer. Adv Exp Med Biol. 945:151–172.
2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yi L, Wu G, Guo L, Zou X and Huang P:
Comprehensive analysis of the PD-L1 and immune infiltrates of m6A
RNA methylation regulators in head and neck squamous cell
carcinoma. Mol Ther Nucleic Acids. 21:299–314. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang Y, Zong W, Ju S, Jing R and Cui M:
Promising member of the short interspersed nuclear elements (Alu
elements): Mechanisms and clinical applications in human cancers. J
Med Genet. 56:639–645. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen LL and Yang L: ALUternative
regulation for gene expression. Trends Cell Biol. 27:480–490.
2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Masson E, Maestri S, Bordeau V, Cooper DN,
Férec C and Chen JM: Alu insertion-mediated dsRNA structure
formation with pre-existing Alu elements as a disease-causing
mechanism. Am J Hum Genet. 111:2176–2189. 2024.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang ZY, Ge LP, Ouyang Y, Jin X and Jiang
YZ: Targeting transposable elements in cancer: Developments and
opportunities. Biochim Biophys Acta Rev Cancer.
1879(189143)2024.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lehle S, Emons J, Hack CC, Heindl F, Hein
A, Preuß C, Seitz K, Zahn AL, Beckmann MW, Fasching PA, et al:
Evaluation of automated techniques for extraction of circulating
cell-free DNA for implementation in standardized high-throughput
workflows. Sci Rep. 13(373)2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Choudhari AS, Mandave PC, Deshpande M,
Ranjekar P and Prakash O: Phytochemicals in cancer treatment: From
preclinical studies to clinical practice. Front Pharmacol.
10(1614)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Othman B, Beigh S, Albanghali MA, Sindi
AAA, Shanawaz MA, Ibahim MAEM, Marghani D, Kofiah Y, Iqbal N and
Rashid H: Comprehensive pharmacokinetic profiling and molecular
docking analysis of natural bioactive compounds targeting oncogenic
biomarkers in breast cancer. Sci Rep. 15(5426)2025.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Patel MS, Sinngh P, Gandupalli L and Gupta
R: Identification and evaluation of survival-associated common
chemoresistant genes in cancer. Biomed Biotechnol Res J. 8:320–327.
2024.
|
|
18
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res.
45(W1):W98–W102. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47(W1):W556–W560.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Gu Z, Eils R and Schlesner M: Complex
heatmaps reveal patterns and correlations in multidimensional
genomic data. Bioinformatics. 32:2847–2849. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liao Y, Wang J, Jaehnig EJ, Shi Z and
Zhang B: WebGestalt 2019: Gene set analysis toolkit with revamped
UIs and APIs. Nucleic Acids Res. 47(W1):W199–W205. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Modhukur V, Iljasenko T, Metsalu T, Lokk
K, Laisk-Podar T and Vilo J: MethSurv: A web tool to perform
multivariable survival analysis using DNA methylation data.
Epigenomics. 10:277–288. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48(W1):W509–W514. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kingsley JP, Pranay G, Priya A and
Chandrakumar MJ: Human papilloma virus testing in oral squamous
cell carcinoma in Southern India: A case-control study. Current
Medical Issues. 19:12–18. 2021.
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hubley R, Finn RD, Clements J, Eddy SR,
Jones TA, Bao W, Smit AF and Wheeler TJ: The Dfam database of
repetitive DNA families. Nucleic Acids Res. 44(D1):D81–D89.
2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Dallakyan S and Olson AJ: Small-molecule
library screening by docking with PyRx. Methods Mol Biol.
1263:243–250. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Bourova-Flin E, Derakhshan S, Goudarzi A,
Wang T, Vitte AL, Chuffart F, Khochbin S, Rousseaux S and
Aminishakib P: The combined detection of Amphiregulin, Cyclin A1
and DDX20/Gemin3 expression predicts aggressive forms of oral
squamous cell carcinoma. Br J Cancer. 125:1122–1134.
2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gao J, Ulekleiv CH and Halstensen TS:
Epidermal growth factor (EGF) receptor-ligand based molecular
staging predicts prognosis in head and neck squamous cell carcinoma
partly due to deregulated EGF-induced amphiregulin expression. J
Exp Clin Cancer Res. 35(151)2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
He C, Wu Y, Nan X, Zhang W, Luo Y, Wang H,
Li M, Liu C, Liu J, Mou X and Liu Y: Induction of CX3CL1 expression
by LPS and its impact on invasion and migration in oral squamous
cell carcinoma. Front Cell Dev Biol. 12(1371323)2024.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ravindran S, Ranganathan S, R K, J N, A S,
Kannan SK, Prasad K D, Marri J and K R: The role of molecular
biomarkers in the diagnosis, prognosis, and treatment
stratification of oral squamous cell carcinoma: A comprehensive
review. J Liq Biopsy. 7(100285)2025.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kunesch K: GATA3-expressing regulatory T
cells accumulate in response to early cancerous changes via
TCR-dependent local proliferation: Dissertation, RWTH Aachen
University, 2024.
|
|
33
|
Gintoni I, Vassiliou S, Chrousos GP and
Yapijakis C: Review of disease-specific microRNAs by strategically
bridging genetics and epigenetics in oral squamous cell carcinoma.
Genes (Basel). 14(1578)2023.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wu CY, Peng PW, Renn TY, Lee CJ, Chang TM,
Wei AI and Liu JF: CX3CL1 induces cell migration and invasion
through ICAM-1 expression in oral squamous cell carcinoma cells. J
Cell Mol Med. 27:1509–1522. 2023.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Izumi T, Rychahou P, Chen L, Smith MH and
Valentino J: Copy number variation that influences the ionizing
radiation sensitivity of oral squamous cell carcinoma. Cells.
12(2425)2023.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Agarwal N and Jha AK: DNA hypermethylation
of tumor suppressor genes among oral squamous cell carcinoma
patients: A prominent diagnostic biomarker. Mol Biol Rep.
52(44)2024.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kondoh N and Mizuno-Kamiya M: The role of
immune modulatory cytokines in the tumor microenvironments of head
and neck squamous cell carcinomas. Cancers (Basel).
14(2884)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Fu C and Jiang A: Dendritic cells and CD8
T cell immunity in tumor microenvironment. Front Immunol.
9(3059)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Sahingur SE and Yeudall WA: Chemokine
function in periodontal disease and oral cavity cancer. Front
Immunol. 6(214)2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Mesgari H, Esmaelian S, Nasiri K,
Ghasemzadeh S, Doroudgar P and Payandeh Z: Epigenetic regulation in
oral squamous cell carcinoma microenvironment: A comprehensive
review. Cancers (Basel). 15(5600)2023.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Lemos-Gonzalez Y, Rodríguez-Berrocal FJ,
Cordero OJ, Gómez C and Páez de la Cadena M: Alteration of the
serum levels of the epidermal growth factor receptor and its
ligands in patients with non-small cell lung cancer and head and
neck carcinoma. Br J Cancer. 96:1569–1578. 2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li M, Wei Y, Huang W, Wang C, He S, Bi S,
Hu S, You L and Huang X: Identifying prognostic biomarkers in oral
squamous cell carcinoma: An integrated single-cell and bulk RNA
sequencing study on mitophagy-related genes. Sci Rep.
14(19992)2024.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhou J, Xu Y, Li Y, Zhang Q, Zhong L, Pan
W, Ji K, Zhang S, Chen Z, Liu Y, et al: Cancer-associated
fibroblasts derived amphiregulin promotes HNSCC progression and
drug resistance of EGFR inhibitor. Cancer Lett.
622(217710)2025.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Park MK, Lee JC, Lee JW and Hwang SJ: Alu
cell-free DNA concentration, Alu index, and LINE-1 hypomethylation
as a cancer predictor. Clin Biochem. 94:67–73. 2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Nair MG, Ramesh RS, Naidu CM, Mavatkar AD,
V P S, Ramamurthy V, Somashekaraiah VM, C E A, Raghunathan K,
Panigrahi A, et al: Estimation of ALU repetitive elements in plasma
as a cost-effective liquid biopsy tool for disease prognosis in
breast cancer. Cancers (Basel). 15(1054)2023.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Lehner J, Stötzer OJ, Fersching D, Nagel D
and Holdenrieder S: Circulating plasma DNA and DNA integrity in
breast cancer patients undergoing neoadjuvant chemotherapy. Clin
Chim Acta. 425:206–211. 2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Farhan M: Revisiting the
antioxidant-prooxidant conundrum in cancer research. Med Oncol.
41(179)2024.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Huang J, Xu Y, Wang Y, Su Z, Li T, Wu S,
Mao Y, Zhang S, Weng X and Yuan Y: Advances in the study of
exosomes as drug delivery systems for bone-related diseases.
Pharmaceutics. 15(220)2023.PubMed/NCBI View Article : Google Scholar
|