Introduction
Hepatitis B virus (HBV) infection is a major health
problem worldwide. Indonesia is an intermediate-to-high HBV endemic
region. Despite a decrease in the incidence of acute HBV infection
in Indonesia resulting from the universal newborn HBV immunization
program, the number of patients with advanced liver disease (ALD)
associated with chronic HBV infection is increasing (1,2). Chronic
HBV infection usually progresses to ALD, including liver cirrhosis
(LC) and hepatocellular carcinoma (HCC). Demographic,
environmental, viral and host-associated factors may contribute to
the risk of LC and HCC in chronic HBV-infected patients (3-5).
HBV is a double-stranded DNA virus that belongs to
the Hepadnaviridae family. The HBV genome contains 4 partially
overlapping open reading frames (ORFs) encoding the surface,
precore (PC)/core, polymerase and X proteins, which have diverse
mutation patterns (6). Mutations in
the 4 ORFs are associated with liver disease. The A1762T/G1764A,
T1753V, C1766T and T1768A mutations in the basal core promoter
(BCP), C1653T in the enhancer region II and G1896A in the PC region
are responsible for modulating the severity of the disease
presentation. Specific mutations in the HBV genome were identified
as predictors of LC and HCC in patients with chronic HBV infection
(7,8).
However, the specific mutations may differ depending on the HBV
genotype (3,9).
A total of 10 HBV (A-J) genotypes have been
identified worldwide (3,10). Genotypes B and C are predominant in
Asia. HBV genotypes are associated with the clinical
characteristics of patients (11).
Genotype B is associated with spontaneous hepatitis B e-antigen
(HBeAg) seroconversion at a younger age and a slower progression to
cirrhosis with active liver disease compared with genotype C. A
high viral load and genotype C are associated with HCC. HBeAg
positivity is more frequent in patients infected with HBV genotype
C compared with those infected with genotype B (12). Furthermore, subgenotypes have been
identified for certain HBV genotypes. HBV subgenotypes are
associated with geographical distribution and ethnic origin
(10).
In Indonesia, genotype B is the predominant HBV
genotype, particularly in the Western region, whereas genotype C is
predominant in the Eastern region (13). Samarinda is the capital city of the
East Kalimantan province. HBV infection is prevalent in the general
population in West Kalimantan province, and genotype B is
predominant among children in West Kotawaringin, in the Central
Kalimantan province (2,14). However, there are limited data on the
characteristics of HBV infection in Samarinda. The present study
investigated the distribution of HBV genotypes and mutations in
patients with ALD in Samarinda.
Materials and methods
Sample collection
Serum samples were collected from 41 patients with
ALD at Abdul Wahab Sjahranie Hospital (Samarinda, Indonesia) from
June 2012 to May 2013. A total of 23 patients with HCC (17 males
and 6 females), with a mean [± standard deviation (SD)] age of
52.8±9.3 years, and 18 LC patients (15 male and 3 female, mean
[±SD] age, 50.6±12.2 years) were included. Inclusion criteria were
as follows: A diagnosis of HCC or LC, and provision of informed
consent. LC was diagnosed by identification of abnormal liver
morphology using ultrasonography (USG). HCC was diagnosed by USG
indicating the presence of a tumor with α-fetoprotein (AFP) levels
≥200 ng/ml. The severity of the clinical manifestations was
determined by the clinicians. Alanine aminotransferase (ALT),
aspartate aminotransferase (AST) and AFP levels were obtained from
patient medical records. Patients were not diagnosed with LC or HCC
as part of the present study. Hepatitis B surface antigen
(HBsAg)-positive serum samples were obtained from blood donors
screened at the Red Cross Center, Samarinda from June to July 2012.
A total of 46 HBsAg-positive serum samples (male 44 and 2 female,
mean [±SD] age, 34.3±8.1 yeas) were randomly included in the
present study. The inclusion criteria for the blood donors were:
Positive for HBsAg and provision of informed consent. A total of
~1.5 ml of serum from each donor was provided at the Red Cross
Center (Samarinda, Indonesia). The serum samples collected and
stored at -20˚C in Abdul Wahab Sjahranie Hospital and the Red Cross
Center, and were then placed in styrofoam boxes with frozen ice
packs during the transportation to the Institute of Tropical
Diseases, Airlangga University (Surabaya, Indonesia). The samples
were stored at -80˚C until use in the Institute of Tropical
Diseases, Airlangga University. Ethical approval was obtained from
the Ethics Committees at Airlangga University, Mulawarman
University (Samarinda, Indonesia) and Kobe University (Kobe,
Japan). Informed consent for participation was obtained from each
individual.
Serological assay
All sera from patients with ALD were screened for
HBsAg by reverse passive hemagglutination (Mycell II HBsAg1AA1;
Institute of Immunology, Tokyo, Japan). Serum samples from blood
donors were also confirmed for HBsAg using the aforementioned kit.
HBeAg status was determined using an enzyme immunoassay method
(Immunis HBeAg EIA kit1A81; Institute of Immunology) for all serum
samples from patients with ALD and blood donors. In this assay,
absorbance of each microwell was measured at a wavelength of 450
nm. The controls for the HBsAg and HBeAg assays were included
according to the manufacturer's protocol.
Viral DNA extraction and PCR
amplification
Viral DNA was extracted from 200 µl serum using a
QIAamp DNA Blood Mini kit (Qiagen GmbH, Hilden, Germany) according
to the manufacturer's protocol. The presence of mutations within
the HBV genome was detected using polymerase chain reaction (PCR)
with Taq PCR master mix kit (Qiagen GmbH) targeting the ORFs of the
X gene (nt. 1374-1896) including the BCP, the PC (nt. 1814-1901),
and core (nt. 1901-2045). The primers used to determine the
mutations and genotypes were as follows: HB5F forward
(5'-CTCTGCCGATCCATACTGCGGAA-3'; nt. 1256-1278); HB5R reverse
(5'-TTAACCTAATCTCCT CCCCCA-3'; nt. 1761-1741); HB7F forward
(5'-GAGACCGTG AACGCCCA-3'; nt. 1611-1630); HB7R reverse (5'-CCTGAG
TGCTGTATGGTGAGG-3'; nt. 2072-2048); HB11F forward
(5'-GGGTCACCATATTCTTGGGAA-3'; nt. 2814-2834); HB11R reverse
(5'-GAACTGGAGCCAGCAGG-3'; nt. 75-56). The thermocycling conditions
for each primer pair were as follows: 94˚C for 5 min, followed by
40 cycles of 94˚C for 1 min, 55˚C for 1 min, and 72˚C for 1 min,
followed by a final extension of 72˚C for 5 min (15).
Direct sequencing and phylogenetic
analysis
Nucleotide sequences of the amplified fragments were
determined using the BigDye Deoxy Terminator v1.1 cycle sequencing
kit with an ABI Prism 310 Genetic Analyzer (Applied Biosystems;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). All sequence
data analyses were performed with Genetyx Windows v9 (Genetyx
Corporation, Tokyo, Japan). The sequences obtained in the present
study were aligned with those from the DNA Data Bank of Japan
(DDBJ) (16), European Molecular
Biology Laboratory (EMBL) (http://www.ebi.ac.uk/embl.html) and GenBank (17) DNA databases, and the variability of
the X, BCP, PC and core sequences was analyzed. HBV genotypes were
determined based on a percentage homology >96% between the pre-S
gene and HBV isolates from the DDBJ, EMBL and GenBank databases
(18) analyzed using the program
Genetyx Windows v9. A phylogenetic tree was constructed by the
Neighbor-Joining method to determine the HBV genotype/subgenotype,
and bootstrap resampling was performed 1,000 times using Molecular
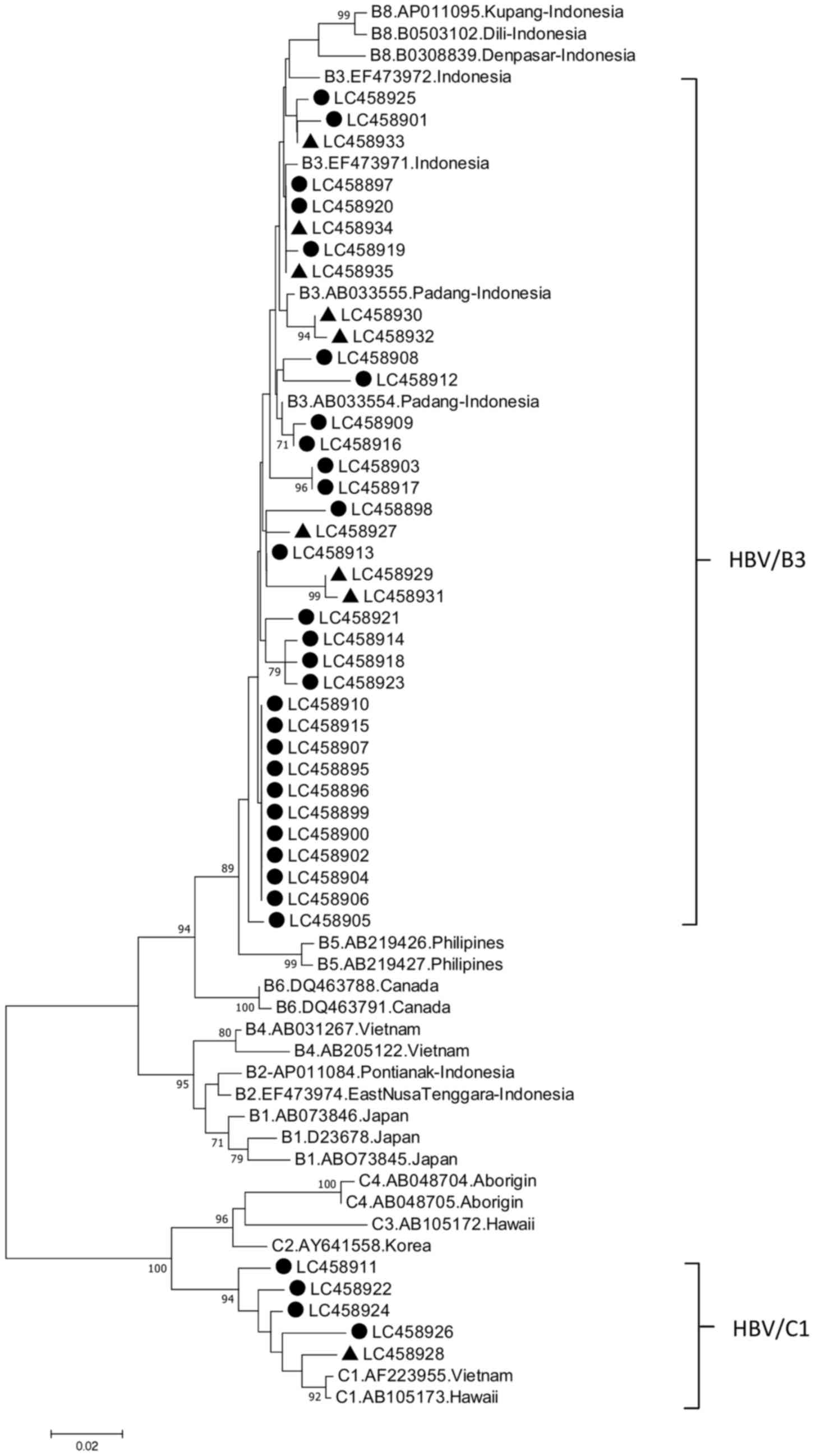
Evolutionary Genetic Analysis software v7(19). Phylogenetic tree analysis with the
representative samples from patients with ALD and HBV carriers was
performed. The representative 32 strains from patients with ALD and
9 from the HBV carriers were included (Fig. 1). The nucleotide sequences presented
in Fig. 1 have been deposited in the
GenBank database under accession numbers LC4558895-LC458926 and
LC458927-LC458935.
Quantification of HBV DNA
Viral load was assessed for all 41 samples from the
patients with ALD using a TaqMan quantitative (q)PCR method for
absolute quantification using an ABI 7300 real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Briefly, 2 µl
of HBV-DNA was amplified using a TaqMan Universal PCR master mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.), primer pairs
(SF1 forward, 5'-CACATCAGGATTCCTAGGACC-3'; nt. 166-186; SR1
reverse, 5'-GGTGAGTGATTGGAGGGTTG-3'; nt. 339-321) covering the
surfaceregion of HBV genome, and TaqMan probe
(HBSP1:FAM-5'-CAGAGTCTAGACTCGTGGTGGACT TC-3'-TAMRA; nt. 242-267).
The thermocycling conditions were as follows: Short incubation at
50˚C for 2 min, initial denaturation at 95˚C for 10 min, then 53
cycles of denaturation at 95˚C for 20 sec and annealing-extension
at 60˚C for 1 min (20).
Statistical analysis
Statistical analysis was performed using SPSS v22
(IBM Corp., Armonk, NY, USA) using the χ2 test or
Fisher's exact test for categorical variables. The t-test was used
for continuous variables. P<0.05 was considered to indicate a
statistically significant difference.
Results
Demographic and clinical
characteristics of patients with ALD
The study analyzed 41 patientswith ALD incomparison
with 46 HBV carriers as controls. Demographic and clinical
characteristics in patients with ALD (LC and HCC) are summarized in
Table I, and those of patients with
ALD and HBV carriers are described in Table II. The mean age was significantly
older in patients with ALD (51.9±10.6 years) compared with that in
HBV carriers (34.3±8.1 years). In the male patients and control
subjects, prevalence of HBV infection was significantly increased
in HBV carriers compared with that in the patients with ALD, and
HBV infection was more prevalent in males in the ALD and HBV
carrier control groups compared with females (Table II). The majority of the patients were
natives of Kalimantan (Dayak, Banjar and Kutai ethnic groups),
followed by Javanese and other ethnic groups (Buton, Sasak and
Bugis). AST levels in patients with HCC (243.7±237.0 U/l) were
significantly increased compared with those in the patients with LC
(105.0±55.7 U/l), while ALT levels in patients with HCC (104.8±99.8
U/l) were also increased compared with those in patients with LC
(73.7±75.2 U/l), but this difference was not significant (Table I).
 | Table IDemographic, virological and clinical
characteristics of HBV-infected patients with HCC and LC. |
Table I
Demographic, virological and clinical
characteristics of HBV-infected patients with HCC and LC.
| Characteristics | All | HCC | LC | P-value |
|---|
| Age, years | 51.9±10.6 | 52.8±9.3 | 50.6±12.2 | NS |
| Sex, male/female
(%) | 32/41 (78.0%) | 17/23 (73.9) | 15/18 (83.3) | NS |
| Ethnic group (%) | | | | NS |
|
Kalimantan
(Dayak, Banjar, Kutai) | 15.41 (36.6) | 9/23 (39.1) | 6/18 (33.3) | |
|
Java | 11/41 (26.8) | 6/23 (8.7) | 5/18 (27.8) | |
|
Others
(Buton, Sasak, Bugis) | 13/41 (31.7) | 6/23 (26.1) | 7/18 (38.9) | |
|
Unknown | 2/41 (4.87) | 2/23 (8.7) | 0/18 (0.0) | |
| Liver function | | | | |
|
ALT
(U/l) | 90.5±89.5 | 104.8±99.8 | 73.7±75.2 | NS |
|
AST
(U/l) | 178.0±189.6 | 243.7±237.0 | 105.0±55.7 | <0.05 |
| AFP, ng/ml (%) | | | | NS |
|
≥200 | 16/41 (39.0) | 11/23 (47.8) | 5/18 (27.8) | |
|
≤200 | 10/41 (24.4) | 3/23 (13.0) | 7/18 (38.9) | |
|
Unknown | 15/41 (36.6) | 9/23 (39.1) | 6/18 (33.3) | |
| HBsAg (R-PHA)
(%) | | | | NS |
|
Positive | 33/41 (80.5) | 18/23 (78.3) | 15/18 (83.3) | |
|
Negative | 5/41 (12.2) | 3/23 (13.0) | 2/18 (11.1) | |
|
Unknown | 3/41 (7.3) | 2/23 (8.7) | 1/18 (5.5) | |
| HBeAg (EIA)
(%) | | | | NS |
|
Positive | 3/41 (7.3) | 2/23 (8.7) | 1/18 (5.6) | |
|
Negative | 38/41 (92.7) | 21/23 (91.3) | 17/18 (94.4) | |
|
Quantitative
HBV DNA (log copies/ml) | 6.1±2.4 | 5.2±2.6 | 7.3±1.4 | NS |
| HBV DNA, positive
samples (%) | | | | |
|
Pre-S
region | 41/41 (100.0) | 23/23 (100.0) | 18/18 (100.0) | NS |
|
X
region | 38/41 (92.7) | 21/23 (91.3) | 17/18 (94.4) | NS |
|
Core
region | 40/41 (97.6) | 22/23 (95.6) | 18/18 (100.0) | NS |
| Subgenotype (Pre-S)
(%) | | | | NS |
|
B3 | 35/41 (85.4) | 21/23 (91.3) | 14/18 (77.8) | |
|
C1 | 6/41 (14.6) | 2/23 (8.7) | 4/18 (22.2) | |
 | Table IIDemographic and virological
characteristics of patients with ALD and HBV carriers. |
Table II
Demographic and virological
characteristics of patients with ALD and HBV carriers.
|
Characteristics | ALD | HBV carriers | P-value |
|---|
| Age, years | 52.8±9.3 | 50.6±12.2 | <0.05 |
| Sex, male/female
(%) | 17/23 (73.9) | 15/18 (83.3) | <0.05 |
| Ethnic group
(%) | | | NS |
|
Kalimantan
(Dayak, Banjar, Kutai) | 15/41 (36.6) | 18/46 (39.1) | |
|
Java | 11/41 (26.8) | 15/46 (32.6) | |
|
Others
(Buton, Sasak, Bugis) | 13/41 (31.7) | 11/46 (23.9) | |
|
Unknown | 2/41 (4.9) | 2/46 (4.3) | |
| HBeAg (EIA)
(%) | | | NS |
|
Positive | 3/41 (7.3) | 9/45 (5.6) | |
|
Negative | 38/41 (92.7) | 36/45 (94.4) | |
| HBV DNA, positive
samples (%) | | | NS |
|
Pre-S
region | 41/41 (100.0) | 46/46 (100.0) | |
|
X
region | 38/41 (91.3) | 46/46 (100.0) | |
|
Core
region | 40/41 (97.6) | 46/46 (100.0) | |
| Subgenotype (Pre-S)
(%) | | | <0.05 |
|
B3 | 35/41 (85.4) | 45/46 (97.8) | |
|
C1 | 6/41 (14.6) | 1/46 (2.2) | |
Phylogenetic analysis
Phylogenetic analysis in Pre-S1 gene of HBV strains
was performed. The HBV/B3 (85.4%) and HBV/C1 (14.6%) subgenotypes
were identified in patients with ALD, and HBV/B3 (97.8%) and HBV/C1
(2.2%) subgenotypes were identified in HBV carriers. The HBV/C1
subgenotype was significantly more prevalent in patients with ALD
than that in HBV carriers in East Kalimantan, Indonesia.
Mutations in the X, BCP, PC, and core
regions of patients with ALD
The following mutations were detected in the HBV
isolates from patients with ALD: C1505A, T1631C, C1638T and C1726A
in the X region; T1753V, A1762T/G1764A double mutation and T1768A
in the BCP; C1858T, G1896A and G1899A in the PC; and G1915T in the
core region. Compared with HBV carriers, patients with ALD
exhibited significantly increased incidence rates of HBV mutation
in the BCP region: double mutation A1762T/G1764A and T1753V, in PC
region: C1858T and in X region: C1505A. The G1896A mutation was
significantly more prevalent in HBV carriers compared with patients
with ALD (Table III).
 | Table IIIHBV mutations in patients with ALD
and HBV carriers. |
Table III
HBV mutations in patients with ALD
and HBV carriers.
| Mutations | ALD (%) | HBV carriers
(%) | P-value |
|---|
| X region | | | |
|
C1495T | 0/38 (0.0) | 0/46 (0.0) | NS |
|
C1505A | 11/38 (28.9) | 0/46 (0.0) | <0.05 |
|
T1631C | 21/38 (55.3) | 22/46 (47.8) | NS |
|
C1638T | 17/38 (44.7) | 13/46 (28.3) | NS |
|
C1726A | 5/36 (13.9) | 11/46 (23.9) | NS |
| BCP region | | | |
|
T1753V | 14/35(40) | 0/46 (0.0) | <0.05 |
|
A1762T/G1764A | 25/34 (73.5) | 0/46 (0.0) | <0.05 |
|
T1768A | 0/34 (0.0) | 5/46 (10.9) | NS |
| PC region | | | |
|
C1858T | 6/34 (17.6) | 0/46 (0.0) | <0.05 |
|
G1896A | 12/34 (35.3) | 29/46 (63.0) | <0.05 |
|
G1899A | 11/34 (32.3) | 6/46 (13.0) | NS |
| Core region | | | |
|
G1915T | 1/33 (3.0) | 0/46 (0.0) | NS |
Association between mutations and HBV
genotype in patients with ALD and HBV carriers
The association between mutations in the X, BCP, PC
and core regions and genotypes of patients with ALD was analyzed
(Table IV). Mutations in the X
region were detected only in HBV/B subgenotypes. In particular,
T1631C and C1638T mutations were significantly more prevalent in
the HBV/B genotypes compared with the HBV/C genotypes. The
prevalence of C1858T mutation was significantly increased in the
HBV/C genotypes compared with the HBV/B genotypes. A1762T/G1764A
mutations in the BCP region were frequently detected in HBV/B
(67.8%) and HBV/C (100%) genotypes.
 | Table IVHBV mutations in patients with
advanced liver disease according to genotype. |
Table IV
HBV mutations in patients with
advanced liver disease according to genotype.
| Mutations | Genotype B (%) | Genotype C (%) | P-value |
|---|
| X region | | | |
|
C1495T | 0/32 (0.0) | 0/6 (0.0) | NS |
|
C1505A | 11/32 (34.4) | 0/6 (0.0) | NS |
|
T1631C | 21/32 (65.6) | 0/6 (0.0) | <0.05 |
|
C1638T | 17/32 (53.1) | 0/6 (0.0) | <0.05 |
|
C1726A | 5/30 (16.7) | 0/6 (0.0) | NS |
| BCP region | | | |
|
T1753CV | 11/29 (37.9) | 3/6 (50.0) | NS |
|
A1762T/G1764A | 19/28 (67.8) | 6/6 (100.0) | NS |
|
T1768A | 0/28 (0.0) | 0/6 (0.0) | NS |
| PC region | | | |
|
C1858T | 0/28 (0.0) | 6/6 (100.0) | <0.05 |
|
G1896A | 10/28 (35.7) | 2/6 (33.3) | NS |
|
G1899A | 6/28 (21.4) | 5/6 (83.3) | NS |
| Core region | | | |
|
G1915T | 1/27 (3.7) | 0/6 (0.0) | NS |
Discussion
The present study enrolled 41 patients from East
Kalimantan, including patients with LC and HCC, and 46 HBV carriers
as controls. The transmigration problem in Indonesia, which
involves the migration of millions of people from the overcrowded
islands, primarily Java, to settlement areas (21), has resulted in a number of
transmigrants living in the outer islands, including Kalimantan.
Therefore, among the 41 patients with ALD and 46 HBV carriers, only
36.6 and 39.1%, respectively, belonged to the native population of
Kalimantan. The remaining patients belonged to other ethnic groups,
including Javanese, Buton, Sasak and Bugis. The average age among
patients with ALD was significantly older compared with the HBV
carriers. This observation is consistent with our previous study
conducted in Yogyakarta, which demonstrated that mean age among
patients with ALD was older (54 years) compared with patients with
chronic HBV infection (38 years) (7).
An additional study in Vietnam also revealed similar results, in
that the average age of patients with HCC (49.5 years) and LC (50.9
years) was older compared with HBV carriers (22.3 years). Older age
was one of independent risk factor for the development of HCC
(22).
Viral hepatitis infection may account for a marked
increase in aminotransferase levels. The increase in ALT associated
with hepatitis C infection is generally more marked compared with
that associated with hepatitis B (23). Despite the association between
elevated ALT levels and hepatocellular disease, the absolute peak
of the ALT increase is not correlated with the extent of liver cell
damage (24). In the present study,
AST levels were high in patients with LC and HCC, and no
significant difference between the two groups of patients was
observed, whereas AST levels were increased in patients with HCC
compared with patients with LC. AST and ALT are
hepatocyte-predominant enzymes (25);
it has been established that AST is more abundant in the liver and
that increases in AST levels are more sensitive to rates of
enzymatic clearance and mitochondrial injury compared with ALT
(26). A number of previous studies
revealed that the AST/ALT ratio is frequently used for the
assessment of hepatic fibrosis and the detection for HCC recurrence
(27-29),
which supports the results of the present study.
The present study analyzed patients with ALD
infected with HBV and HBV carriers to investigate HBV genotypes and
mutations associated the severity of the liver disease. Consistent
with previous studies (8,22,30,31),
mutations in the BCP region, particularly the double mutation
A1762T/G1764A and T1753V, were associated with the risk of ALD in
the present study. Similar results were described in a previous
study in Brazil, in which the 2 mutations in the BCP region
(combination double mutation A1762T/G1764A and T1753V) exhibited
higher frequencies in patients with HCC compared with patients
without HCC, even though difference was not significant (32). The mutation C1505A in the X region was
more frequently detected in ALD compared with chronic hepatitis B,
even though this comparison was not significantly different in our
study, and as previously demonstrated (7). In addition, the A1762T/G1764A double
mutation, T1753V, C1505A, and C1858T were not identified in HBV
carriers in the present study. The C1858T mutation is commonly
identified in Asia (9), and more
frequent in patients with HCC than in patients with chronic
hepatitis B in Vietnam (22).
However, a previous study conducted in China indicated that there
was no significant difference in the C1858T mutation between HBV
carriers and patients with HBV liver diseases (22,33). The
G1896A mutation was more frequent in HBV carriers compared with
patients with ALD in the present study, which is consistent with a
previous study conducted in Malaysia (9). A similar result was also revealed in a
study from Korea, in which the rates of G1896A mutation were not
significantly different between HCC and non-HCC groups (34). However, a Brazilian study suggested
that the rate of G1896A mutation was identified to be increased in
patients without HCC compared with patients with HCC, although this
difference was not significant (31).
By contrast, the G1896A mutation in the PC region is one of the
most commonly identified mutation in ALD and chronic HBV infection
(7,9,34).
The A1762T/G1764A double mutation in the BCP region,
which is responsible for decreased PC mRNA synthesis, results in
the decreased secretion of HBeAg and serves a significant role in
hepatocarcinogenesis, primarily in patients with HBeAg-negative
hepatitis (34). Previous studies
have demonstrated that A1762T/G1764A mutations decrease HBeAg
production to ~70% (30,31). The C1858T mutation decreases the
synthesis of HBeAg and is a precursor of the G1896A mutation. The
C1858T and G1896A mutations stabilize the structure of the epsilon
loop, which may result in increased viral replication (35). In the present study, the majority of
patients with ALD were HBeAg-negative, which may be associated with
the predominance of double mutations in the BCP region. However,
this was not confirmed as the majority of HBV carriers were also
HBeAg-negative in the study cohort. The G1896A mutation produces a
premature stop codon and decreases the synthesis of HBeAg. This
mutation serves a role in preventing the translation of HBeAg and
inhibits the production of HBeAg. The presence of a stop codon
mutation maybe a mechanism of immune evasion (9,23). The
predominance of the G1896A mutation in the PC region observed in
the present study may also be associated with the high prevalence
of patients who were HBeAg-negative.
In the present study, the two patient groups
demonstrated that the predominant HBV genotype/subgenotype was B3,
followed by C1, which was consistent with previous studies from
Indonesia revealing that the B and C genotypes are predominant in
this nation (13,18). The G1896A mutation was more frequent
in genotype B compared with genotype C among patients with
asymptomatic chronic HBV infection (9). A previous study from China indicated
that the double mutation in BCP region was detected with ALD in
genotypes B and C (30). The risk of
ALD is associated with certain HBV genotypes, and the prevalence of
HBV mutations in certain HBV genotypes (31).
In conclusion, the analysis of patients from East
Kalimantan demonstrated that the HBV genotype B was predominant,
followed by the genotype C. Several mutations were detected in
patients with ALD, including C1505A in the X region, T1753V and
A1762T/G1764A in the BCP region and C1858T in the PC region.
Mutations in the X gene were only detected in patients with
genotype B, and C1858T mutation in the PC was only detected in
patients with genotype C. A limitation of the present study was the
small number of samples analysed; additional studies with a larger
sample cohort are required to confirm the results suggesting that
viral factors affect the development of ALD.
Acknowledgements
Not applicable.
Funding
The present study was supported by a Grant-in-Aid
from the Ministry of Education, Culture, Sports, Science and
Technology, Japan (grant no. 16H05826) and a Grant-in-Aid from the
Japan Initiative for Global Research Network on Infectious Disease
supported by The Ministry of Education, Culture, Sports, Science
and Technology, Japan, and in part by a Grant-in-Aid from Professor
Dato' Sri Tahir through Tahir professorship, Indonesia.
Availability of data and materials
All data generated or analyzed in the present study
are included in this published article.
Authors' contributions
The study was conducted and designed by RMW and TU.
ISM, RMW, PBP, AI and MA performed the sample collection and
laboratory experiments. RMW, TU, J, and LNY were responsible for
the analysis and interpretation of the data. RMW and J wrote the
manuscript. MIL, S, YY, YH, TU, and J reviewed and edited the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from the Ethics
Committees at Airlangga University (Samarinda, Indonesia),
Mulawarman University (Samarinda, Indonesia) and Kobe University
(Kobe, Japan). Informed consent for participation was obtained from
each individual.
Patient consent for publication
All patients agreed to the publication of any
relevant data on the basis of anonymization of all personal
data.
Competing interest
The authors declare that they have no competing
interests.
References
|
1
|
Khan M, Dong JJ, Acharya SK, Dhagwahdorj
Y, Abbas Z, Jafri SMW, Mulyono DH, Tozun N and Sarin SK: Hepatology
issues in Asia: Perspective from regional leaders. J Gastroenterol
Hepatol. 19 (Suppl 7):S419–S430. 2004. View Article : Google Scholar
|
|
2
|
Purwono PB, Juniastuti Amin M, Bramanthi
R, Nursidah Resi EM, Wahyuni RM, Yano Y, Soetjipto Hotta H, et al:
Hepatitis B virus infection in Indonesia 15 years after adoption of
a universal infant vaccination program: Possible impacts of low
birth dose coverage and a vaccine-escape mutant. Am J Trop Med Hyg.
95:674–679. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Lindh M, Andersson AS and Gusdal A:
Genotypes, nt 1858 variants, and geographic origin of hepatitis B
virus - large-scale analysis using a new genotyping method. J
Infect Dis. 175:1285–1293. 1997.PubMed/NCBI
|
|
4
|
Fattovich G, Bortolotti F and Donato F:
Natural history of chronic hepatitis B: Special emphasis on disease
progression and prognostic factors. J Hepatol. 48:335–352.
2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Nishida N, Tokunaga K and Mizokami M:
Genome-Wide Association Study Reveals Host Genetic Factors for
Liver Diseases. J Clin Transl Hepatol. 1:45–50. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mizokami M, Orito E, Ohba K, Ikeo K, Lau
JY and Gojobori T: Constrained evolution with respect to gene
overlap of hepatitis B virus. J Mol Evol. 44 (Suppl 1):S83–S90.
1997.PubMed/NCBI
|
|
7
|
Heriyanto DS, Yano Y, Utsumi T,
Anggorowati N, Rinonce HT, Lusida MI, et al: Mutations Within
Enhancer II and BCP Regions of Hepatitis B Virus in Relation to
ALDs in Patients Infected With Subgenotype B3 in Indonesia. J Med
Virol. 84:44–51. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang X and Ding HG: Key role of hepatitis
B virus mutation in chronic hepatitis B development to
hepatocellular carcinoma. World J Hepatol. 7:1282–1286.
2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Suppiah J, Mohd Zain R, Bahari N, Haji
Nawi S and Saat Z: G1896A Precore mutation and association with
HBeAg status, genotype and clinical status in patients with chronic
hepatitis B. Hepat Mon. 15:e31490:2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Utsumi T, Yano Y and Hotta H: Molecular
epidemiology of hepatitis B virus in Asia. World J Med Genet.
4:19–26. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sunbul M: Hepatitis B virus genotypes:
Global distribution and clinical importance. World J Gastroenterol.
20:5427–5434. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Huang Y and Lok AS: Viral factors and
outcomes of chronic HBV infection. Am J Gastroenterol. 106:93–95.
2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Mulyanto Depamede SN, Surayah K, Tsuda F,
Ichiyama K, Takahashi M and Okamoto H: A nationwide molecular
epidemiological study on hepatitis B virus in Indonesia:
Identification of two novel subgenotypes, B8 and C7. Arch Virol.
154:1047–1059. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Darmawan E, Turyadi KE, El-Khobar KE,
Nursanty NK, Thedja MD and Muljono DH: Seroepidemiology and occult
hepatitis B virus infection in young adults in Banjarmasin,
Indonesia. J Med Virol. 87:199–207. 2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sugauchi F, Mizokami M, Orito E, Ohno T,
Kato H, Suzuki S, Kimura Y, Ueda R, Butterworth LA and Cooksley WG:
A novel variant genotype C of hepatitis B virus identified in
isolates from Australian Aborigines: Complete genome sequence and
phylogenetic relatedness. J Gen Virol. 82:883–892. 2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Tateno Y, Imanishi T, Miyazaki S,
Fukami-Kobayashi K, Saitou N, Sugawara H and Gojobori T: DNA Data
Bank of Japan (DDBJ) for genome scale research in life science.
Nucleic Acids Res. 30:27–30. 2002.PubMed/NCBI
|
|
17
|
Benson DA, Cavanaugh M, Clark K,
Karsch-Mizrachi I, Lipman DJ, Ostell J and Sayers EW: GenBank.
Nucleic Acids Res. 41 (D1):D36–D42. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lusida MI, Nugrahaputra VE, Soetjipto
Handajani R, Nagano-Fujii M, Sasayama M, Utsumi T and Hotta H:
Novel subgenotypes of hepatitis B virus genotypes C and D in Papua,
Indonesia. J Clin Microbiol. 46:2160–2166. 2008.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kumar S, Stecher G and Tamura K: MEGA7:
Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger
Datasets. Mol Biol Evol. 33:1870–1874. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Abe A, Inoue K, Tanaka T, Kato J, Kajiyama
N, Kawaguchi R, Tanaka S, Yoshiba M and Kohara M: Quantitation of
hepatitis B virus genomic DNA by real-time detection PCR. J Clin
Microbiol. 37:2899–2903. 1999.PubMed/NCBI
|
|
21
|
Fearnside PM: Transmigration in Indonesia:
Lessons from its environmental and social impacts. Environ Manage.
21:553–570. 1997. View Article : Google Scholar
|
|
22
|
Truong BX, Seo Y, Yano Y, Ho PTT, Phuong
TM, Long DV, Son NT, Long NC, Kato H, Hayashi Y, et al: Genotype
and variations in core promoter and pre-core regions are related to
progression of disease in HBV-infected patients from Northern
Vietnam. Int J Mol Med. 19:293–299. 2007.PubMed/NCBI
|
|
23
|
Marcellin P: Hepatitis C: The clinical
spectrum of the disease. J Hepatol. 31 (Suppl 1):9–16.
1999.PubMed/NCBI
|
|
24
|
Gowda S, Desai PB, Hull VV, Math AAK,
Vernekar SN and Kulkarni SS: A review on laboratory liver function
tests. Pan Afr Med J. 3(17)2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Giannini EG, Testa R and Savarino V: Liver
enzyme alteration: A guide for clinicians. CMAJ. 172:367–379.
2005.PubMed/NCBI
|
|
26
|
Okuda M, Li K, Beard MR, Showalter LA,
Scholle F, Lemon SM and Weinman SA: Mitochondrial injury, oxidative
stress, and antioxidant gene expression are induced by hepatitis C
virus core protein. Gastroenterology. 122:366–375. 2002.PubMed/NCBI
|
|
27
|
Giannini E, Botta F, Fasoli A, Ceppa P,
Risso D, Lantieri PB, Celle G and Testa R: Progressive liver
functional impairment is associated with an increase in AST/ALT
ratio. Dig Dis Sci. 44:1249–1253. 1999.PubMed/NCBI
|
|
28
|
Giannini E, Risso D, Botta F, Chiarbonello
B, Fasoli A, Malfatti F, Romagnoli P, Testa E, Ceppa P and Testa R:
Validity and clinical utility of the aspartate
aminotransferase-alanine aminotransferase ratio in assessing
disease severity and prognosis in patients with hepatitis C
virus-related chronic liver disease. Arch Intern Med. 163:218–224.
2003.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang ZX, Jiang CP, Cao Y, Zhang G, Chen WB
and Ding YT: Preoperative serum liver enzyme markers for predicting
early recurrence after curative resection of hepatocellular
carcinoma. Hepatobiliary Pancreat Dis Int. 14:178–185.
2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chen CH, Lee CM, Lu SN, Changchien CS, Eng
HL, Huang CM, Wang JH, Hung CH and Hu TH: Clinical significance of
hepatitis B virus (HBV) genotypes and precore and core promoter
mutations affecting HBV e antigen expression in Taiwan. J Clin
Microbiol. 43:6000–6006. 2005.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kim JK, Chang HY, Lee JM, Baatarkhuu O,
Yoon YJ, Park JY, Kim DY, Han KH, Chon CY and Ahn SH: Specific
mutations in the enhancer II/core promoter/precore regions of
hepatitis B virus subgenotype C2 in Korean patients with
hepatocellular carcinoma. J Med Virol. 81:1002–1008.
2009.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Araujo OC, Barros JJ, do Ó KM, Nabuco LC,
Luz CA, Perez RM, Niel C, Villela-Nogueira CA and Araujo NM:
Genetic variability of hepatitis B and C viruses in Brazilian
patients with and without hepatocelullar carcinoma. J Med Virol.
86:217–223. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yuan J, Zhou B, Tanaka Y, Kurbanov F,
Orito E, Gong Z, Xu L, Lu J, Jiang X, Lai W, et al: Hepatitis B
virus (HBV) genotypes/subgenotypes in China: Mutations in core
promoter and precore/core and their clinical implications. J Clin
Virol. 39:87–93. 2007.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Lee JH, Hong SP, Jang ES, Park SJ, Hwang
SG, Kang SK and Jeong SH: Analysis of HBV genotype, drug resistant
mutations, and pre-core/basal core promoter mutations in Korean
patients with acute hepatitis B. J Med Virol. 87:993–998.
2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lok AS, Akarca U and Greene S: Mutations
in the pre-core region of hepatitis B virus serve to enhance the
stability of the secondary structure of the pre-genome
encapsidation signal. Proc Natl Acad Sci USA. 91:4077–4081.
1994.PubMed/NCBI View Article : Google Scholar
|