Introduction
Autoimmune diseases are a clinically diverse group
of complex diseases that include rheumatoid arthritis (RA),
juvenile idiopathic arthritis, type 1 diabetes mellitus, multiple
sclerosis, systemic lupus erythematosus, psoriasis and psoriatic
arthritis. These diseases affect 4% of the global population
(1) and common treatments include
administration of non-steroidal anti-inflammatory drugs, such as
aspirin, corticosteroid drugs, methotrexate and anti-rheumatic
drugs, including gold compounds.
RA is a systemic chronic immuno-inflammatory disease
of unclear etiology involving progressive and destructive
polyarthritis in association with serological evidence of
auto-reactivity leading to persistent and progressive synovitis
(2). Although the exact etiology of RA
remains unknown, the interaction of immunological, genetic,
infectious agent, environmental and hormonal factors has been
demonstrated to contribute to its pathogenesis (3). Circulating antibodies, particularly the
rheumatoid factor and anti-citrullinated protein antibody (ACPA),
are important in the pathogenesis of RA (4). Synovial fibroblasts, macrophages, natural
killer cells, T CD8+ and T CD4+ lymphocytes,
B lymphocytes and plasma cells actively contribute to the inflamed
synovium (4).
However, genetic risk factors do not fully justify
the occurrence rate of RA; therefore, the role of environmental
factors must also be considered. Environmental factors, such as
weather, endemic microbes, lifestyle and diet are significantly
associated with RA. For example, smoking, by individuals who
express one of the predisposing alleles of RA, has been recognized
as a risk factor for RA and contributes to the generation of ACPA
(5).
Genetic studies are useful for determining
predictive factors for diagnosing patients, and numerous studies
regarding the genetics of RA have been performed (6–8). It is
assumed that genetic factors account for 60% of the cause of RA
susceptibility. The human leukocyte antigen (HLA)-DR4 allele
associated with major histocompatibility complex (MHC) class II and
other relevant alleles were recognized as the major genetic risk
factors in the occurrence of RA. Recent studies have revealed that
there is a very close association between MHC and the presence of
ACPA. In certain Korean populations, which are HLA-DRB1-0405 or
HLA-DRB1-0901 heterozygotes, the risk of RA susceptibility was
found to be particularly high (4).
However, HLA locus contributes to 30–50% of the genetic component
of RA susceptibility (4).
The lectin, galactoside-binding, soluble, 3 (LGALS3)
gene is located on chromosome 14 (14q22.3) and encodes galectin-3
protein. Galectins are lectins binding β-galactoside that have at
least one of the carbohydrate recognition domains (CRDs) in their
own structure. Approximately 15 members of this group have been
identified in mammals (9). Galectin-3,
a unique member of the galectins family, which has a CRD to
identify the carbohydrate in its C-terminal, and the N-terminal
domain, which is required for secretion (10). Galectin-3 has been identified in cells,
such as activated macrophages (11),
fibroblasts (12), dendritic cells
(13), eosinophils (14), mast cells (15) and osteoblasts (11,16).
Galectin-3 within the cell is important for regulation of post
transcription of mRNA in the nucleus (17), cell growth (18) and protecting cells from apoptosis via
Fas and tumor necrosis factor (TNF) (19,20).
Additionally, galectin-3 contributes to phagocytosis by macrophages
(21), and galectin-3 deficient
macrophages are incapable of phagocytosis of IgG-coated red blood
cells and apoptotic cells (21). In a
previous study, galectin-3 was introduced as an opsonin, which
facilitates the clearance of apoptotic neutrophils (21). In addition, the role of galectin-3 in
the morphogenesis and angiogenesis of endothelial cells has been
demonstrated (22).
There are various single nucleotide polymorphisms in
LGALS3 that affect its gene expression. For example, LGALS3 rs4644
C allele has recently been demonstrated to be associated with the
risk of RA in a population from Taiwan (23). Thus, due to the role of the LGALS3 gene
in the pathogenesis of various types of autoimmune disease,
including RA, it presents as a suitable candidate for genetic
studies. Therefore, the present study aimed to evaluate the
possible association between LGALS3 rs4652 A/C genetic variation
and the risk of RA in an Iranian population.
Materials and methods
Subjects
The present case-control study was performed on 120
patients (104 female, 16 male) with RA (mean age, 44.6±12.9 years)
who fulfilled the American College of Rheumatology (ACR) criteria
for RA (24). All the subjects were
patients of the Rheumatology Clinic at Zahedan University of
Medical Sciences (Zahedan, Iran) (25–27). The
control group consisted of 120 healthy individual (76 females, 44
male; mean age, 43.2±10.3 years) who were not related to the RA
patients. The Ethics Committee of Zahedan University of Medical
Sciences approved the project, and informed consent was obtained
from all patients and healthy individuals. Blood samples (3 ml)
from patients and healthy control subjects were collected in
EDTA-containing tubes. The characteristics of the subjects
participating in the study are presented in Table I.
 | Table I.Demographic and biochemical
characteristics of subjects from the current study. |
Table I.
Demographic and biochemical
characteristics of subjects from the current study.
| Characteristic | Rheumatoid arthritis
patients (n=120) | Control subjects
(n=120) |
|---|
| Gender
(female/male) | 104/16 | 76/44 |
| Age (years) | 44.6±12.9 | 43.2±10.3 |
| Disease duration
(years) | 6.05±5.85 | – |
| Rheumatoid factor
positivity (%) | 89.2 | – |
| Family history
(%) | 83.5 | – |
| CRP positivity
(%) | 91.7 | – |
| Anti-CCP positivity
(%) | 68.3 | – |
DNA extraction and polymerase chain
reaction (PCR)
Genomic DNA was extracted from the peripheral blood
samples that had been collected into tubes containing EDTA, as
described previously (25). The LGALS3
polymorphism was determined by a tetra-primer amplification
refractory mutation system-PCR (tetra-ARMS PCR). Two external
primers (forward outer, 5′-GGCTTATCCTGGACAGGCACCTC-3′ and reverse
outer, 5′-TTTTTGACTCTACCAACATACACCCAT-3′) as common primer for
control of PCR reaction and the two internal primers (forward
inner, 5′-CATCTTCTGGACAGCCAAGTGTCA-3′ specific for A allele and
reverse inner, 5′-AGTGGCAGGGTAGGCTCCAGG-3′ specific for C allele)
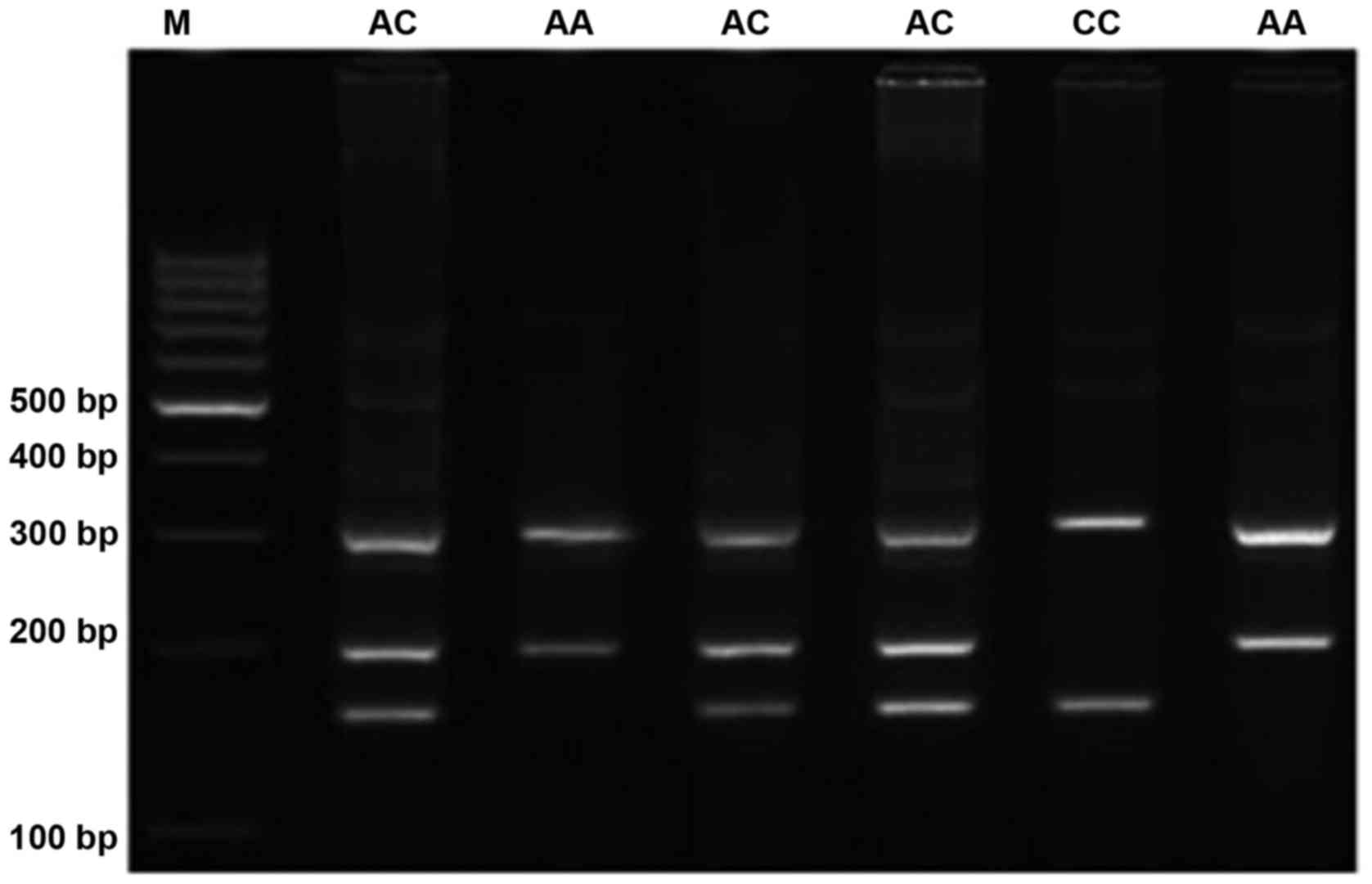
were designed and used. Product sizes were 203 bp for the A allele,
157 bp for the C allele and 314 bp for the control outer band. The
PCR was conducted using a commercially available PCR premix
(AccuPower PCR PreMix; Bioneer Corporation, Daejeon, Korea)
according to the manufacturer's instructions. Template DNA (1 µl;
~100 ng/µl), 0.6 µl of each inner forward and reverse primer (10
µM) and 1.5 µl of each outer forward and reverse primer (12 µM) and
14.8 µl DNase-free water were added into a 0.2-ml PCR tube
containing the AccuPower PCR PreMix. PCR cycling condition for
LGALS3 were as follows: 95°C for 5 min followed by 30 cycles of 30
sec at 95°C, 40 sec at 67°C, and 30 sec at 72°C, with a final step
at 72°C for 10 min. PCR products were visualized on 2% agarose gel
containing 0.5 µ/ml ethidium bromide, and images were obtained
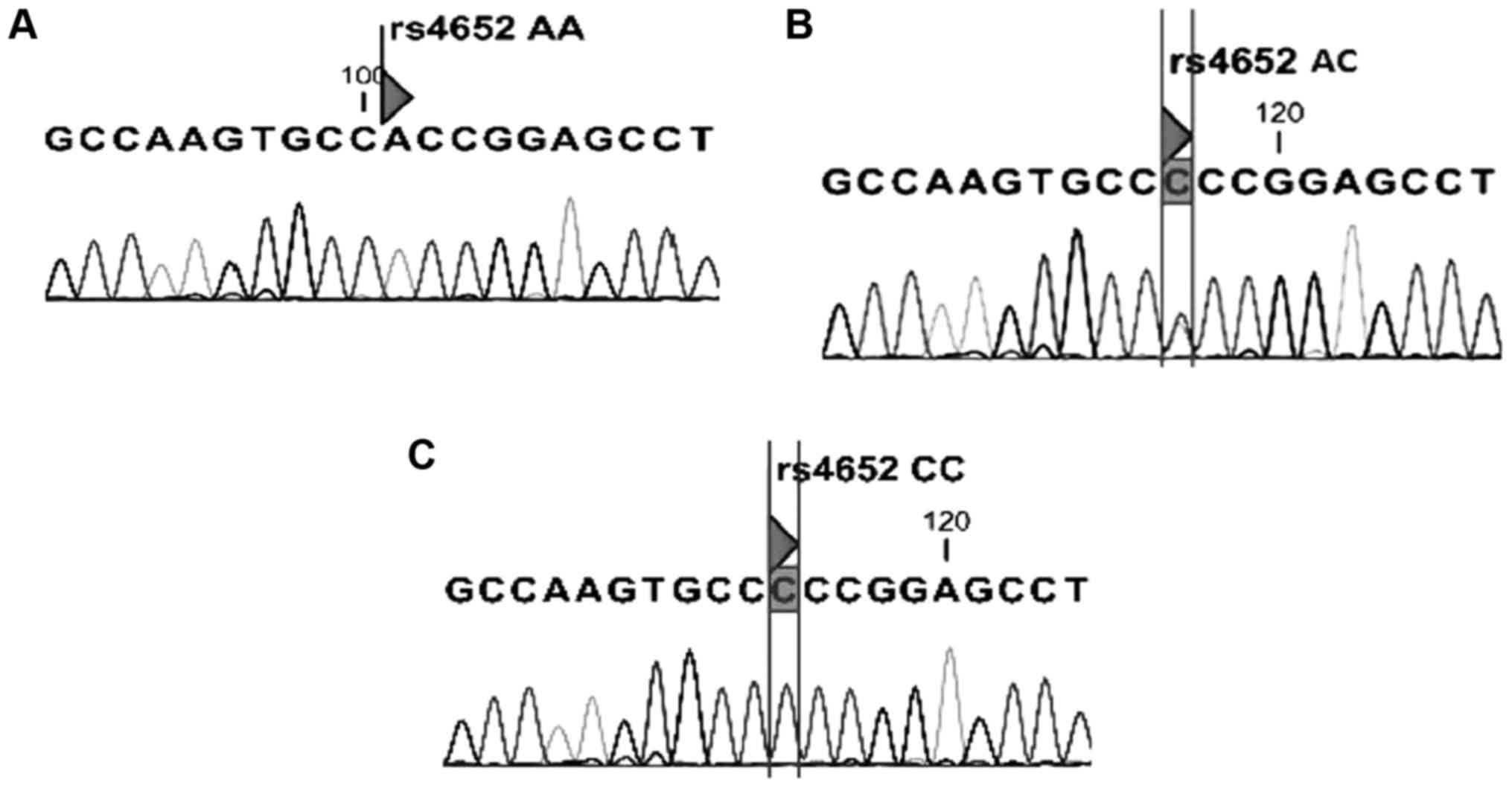
(Fig. 1). To ensure the tetra-ARMS-PCR
genotyping quality, random samples were sequenced. The genotypes
determined by PCR were concordant with those determined by
sequencing (Fig. 2).
Statistical analysis
Statistical analysis was performed using SPSS
version 18 software for windows (SPSS, Inc., Chicago, IL, USA). The
possible associations between the LGALS3 genotypes with RA were
assessed by computing the odds ratio (OR) and 95% confidence
intervals (95% CI) from logistic regression analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
LGALS3 rs4652 polymorphism data
The present study included 120 patients with RA and
120 healthy subjects. The genotypes and allele frequencies of
LGALS3 rs4652 polymorphism are presented in Table II. A significant difference was
identified between the groups regarding LGALS3 rs4652 polymorphism
(P=0.001), suggesting that the AC genotype was a risk factor for RA
(OR=11.622, 95% CI=4.473–28.656; P=0.001). However, the rs4652 C
allele was not a risk factor for RA (OR=0.653, 95% CI=0.069–6.198;
P=0.710). In addition, the possible association between LGALS3
rs4652 and gender and ethnicity was examined. The current data
indicated no significant association between rs4652 A/C
polymorphism with gender (male, 63.33% and female, 78.88%; P=0.50)
and ethnicity (Baluch, 73.38% and Fars, 76.72%; P =0.77).
 | Table II.Distribution of genotypes and allelic
frequencies of LGALS3 rs4652 A/C between patients with rheumatoid
arthritis (case; n=120) and healthy subjects (control; n=120). |
Table II.
Distribution of genotypes and allelic
frequencies of LGALS3 rs4652 A/C between patients with rheumatoid
arthritis (case; n=120) and healthy subjects (control; n=120).
| LGALS3 rs4652 | Case, n (%) | Control, n (%) | OR (95% CI) | P-value | OR (95%
CI)a | P-value |
|---|
| AA | 6 (5) | 42 (35) | Ref. | Ref. |
|
|
| AC | 113 (94.2) | 67 (55.8) | 11.80
(4.765–29.248) | 0.001 | 11.622
(4.473–28.656) | 0.001 |
| CC | 1 (0.8) | 11 (9.2) | 0.636
(0.069–5.851) | 0.690 | 0.653
(0.069–6.198) | 0.710 |
| CC+AC | 114 (95) | 78 (65) | 10.231
(4.149–25.229) | 0.001 | 9.840
(3.909–24.771) | 0.001 |
| Alleles |
|
|
|
|
|
|
| A
(wild) | 125 (52.1) | 151 (62.9) | Ref. |
|
|
|
| C
(mutant) | 115 (47.9) | 89 (37.1) | 1.560
(1.084–2.247) | 0.104 |
|
|
Discussion
In the present study the possible association
between LGALS3 rs4652 (+292 A/C) gene polymorphism and the risk of
RA was investigated. To the best of our knowledge this is the first
study to investigate this polymorphism (LGALS3 rs4652) in an
Iranian population. The results of the current study demonstrated
that rs4652 AC genotype significantly increased the risk of RA in
the Iranian population that was evaluated.
RA is a chronic systemic disease, which is
characterized by inflammation, alterations in the humoral and
cellular immune responses, and synovial hyperplasia. Galectins are
key in various pathologic conditions, including metastasis of tumor
cells, autoimmune diseases and inflammation. The galectin-3 gene is
composed of six exons and encodes a 32-kDa protein (28). Along with a number of surface
receptors, galectin-3 activates lymphoid and myeloid cells, which
results in interleukin (IL)-2 production in T lymphocytes (29) and IgE production in B lymphocytes
(30). Furthermore, galectin-3
increases the production and release of free superoxide radicals in
neutrophils and monocytes contributing to RA progression (31). Galectin-3 has been identified in the
synovial tissue of patients with RA, and it is considered to be a
positive regulator of proinflammatory chemokine and cytokine
production (32). Increased expression
levels of galectin-3 in the synovial tissue of RA patients promotes
the production of proinflammatory chemokines, such as IL-6,
granulocyte-macrophage colony-stimulating factor, TNF, C-X-C motif
chemokine ligand 8, C-C motif chemokine ligand 2, CCL3 and CCL5
(33–35), all of which contribute to the
inflammation, swelling, stiffness and joint destruction
characteristics of RA. Therefore, blocking of galectin-3 is
suggested as a potent strategy for the treatment of RA
patients.
Previous studies have shown that galectin-3 performs
different roles in various types of tissue and cell (11). In addition, galectin-3 has been
associated with certain extracellular functions (36,37).
Galectin-3 within the cell is important for mRNA splicing in the
nucleus (17), regulating cell growth
(18), and protecting cells from death
by Fas-induced apoptosis (19,20) and TNF (38). In addition, galectin-3 is known as an
opsonin in clearing apoptotic neutrophils by macrophages (21). Furthermore, in rats with arthritis
induced by collagen, increased expression levels of galectin-3 in
peripheral monocytes have been reported (39).
In the current study, the results demonstrated that
the frequencies of AA, AC, and CC were 5, 94.2 and 0.8% in RA
patients, and 35, 55.8 and 9.2% in the control subjects. Our
findings showed that the prevalence of the AC genotype in patients
with RA (94.2%) was significantly higher than in the control group
(55.8%), hence the rs4652 AC genotype was a risk factor for RA in
the population investigated in the current study (OR=11.7). In
contrast to our results, a study by Hu et al (23) indicated that the frequencies of LGALS3
rs4652 AA/AC/CC genotypes were 33.1, 47.7 and 19.2%, respectively
in RA patients, and 47.3, 38.5 and 14.3% in the control subjects.
The LGALS3 +292 C allele predisposed subjects to RA in a dominant
genotype. Subjects that carried the LGALS3 +292C allele (+292AC or
CC genotypes) were more susceptible to RA when compared with
subjects with the +292AA genotype (OR=1.8 and 95% confidence
interval=1.2–2.8; P=0.009). In addition, the frequency of the C
allele in the RA patients was identified to be 29 (19.2%) and 26
(14.3%) in the control group demonstrating a significant difference
between groups (P=0.043) (23).
Hu et al (23)
indicated that expression levels of cellular galectin-3 with the CC
genotype in macrophages tended to be higher than those of the other
genotypes. Furthermore, the authors showed that increased
galectin-3 expression is associated with defective monocyte
apoptosis in JIA. Such evidence highlights the important function
of intracellular galectin-3 in the pathogenesis of RA (23).
In conclusion, the current study demonstrated that
LGALS3 rs4652 AC was a risk factor for RA in a southeastern Iranian
population and patients with this polymorphism may be susceptible
to more severe clinical signs. Further studies with larger sample
sizes are required to confirm these results.
Acknowledgements
The authors would like to thank the subjects who
participated in the present study.
References
|
1
|
Hashemi M, Atabaki M, Daneshvar H, Zakeri
Z and Eskandari-Nasab E: Association of PTPN22 rs2476601 and EGFR
rs17337023 Gene polymorphisms and rheumatoid arthritis in Zahedan,
Southeast Iran. Int J Immunogenet. 40:299–305. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shalini P U, Debnath T, Jvs V, Kona LK,
Kamaraju SR, Kancherla R and Chelluri LK: A study on FoxP3 and
Tregs in paired samples of peripheral blood and synovium in
rheumatoid arthritis. Cent Eur J Immunol. 40:431–436. 2015.
|
|
3
|
Bowes J and Barton A: Recent advances in
the genetics of RA susceptibility. Rheumatology (Oxford).
47:399–402. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Perricone C, Ceccarelli F and Valesini G:
An overview on the genetic of rheumatoid arthritis: A never-ending
story. Autoimmun Rev. 10:599–608. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oliver JE and Silman AJ: Risk factors for
the development of rheumatoid arthritis. Scand J Rheumatol.
35:169–174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Amaya-Amaya J, Rojas-Villarraga A,
Molano-Gonzalez N, Montoya-Sánchez L, Nath SK and Anaya JM:
GDF15(MIC1) H6D Polymorphism Does Not Influence Cardiovascular
Disease in a Latin American Population with Rheumatoid Arthritis. J
Immunol Res. 2015:2707632015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang CM, Chen SY, Huang PH and Tsai FJ:
Effect of MPG gene rs2858056 polymorphism, copy number variation,
and level of serum MPG protein on the risk for rheumatoid
arthritis. PLoS One. 10:e01206992015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee YH, Bae SC and Song GG: Meta-analysis
of associations between functional prolactin −1149 G/T polymorphism
and susceptibility to rheumatoid arthritis and systemic lupus
erythematosus. Clin Rheumatol. 34:683–690. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sato S, St-Pierre C, Bhaumik P and
Nieminen J: Galectins in innate immunity: Dual functions of host
soluble beta-galactoside-binding lectins as damage-associated
molecular patterns (DAMPs) and as receptors for pathogen-associated
molecular patterns (PAMPs). Immunol Rev. 230:172–187. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Menon RP and Hughes RC: Determinants in
the N-terminal domains of galectin-3 for secretion by a novel
pathway circumventing the endoplasmic reticulum-Golgi complex. Eur
J Biochem. 264:569–576. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu FT, Patterson RJ and Wang JL:
Intracellular functions of galectins. Biochim Biophys Acta.
1572:263–273. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Moutsatsos IK, Wade M, Schindler M and
Wang JL: Endogenous lectins from cultured cells: Nuclear
localization of carbohydrate-binding protein 35 in proliferating
3T3 fibroblasts. Proc Natl Acad Sci USA. 84:6452–6456. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dietz AB, Bulur PA, Knutson GJ, Matasić R
and Vuk-Pavlović S: Maturation of human monocyte-derived dendritic
cells studied by microarray hybridization. Biochem Biophys Res
Commun. 275:731–738. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Truong MJ, Gruart V, Liu FT, Prin L,
Capron A and Capron M: IgE-binding molecules (Mac-2/epsilon BP)
expressed by human eosinophils. Implication in IgE-dependent
eosinophil cytotoxicity. Eur J Immunol. 23:3230–3235. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Craig SS, Krishnaswamy P, Irani AM, Kepley
CL, Liu FT and Schwartz LB: Immunoelectron microscopic localization
of galectin-3, an IgE binding protein, in human mast cells and
basophils. Anat Rec. 242:211–219. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Colnot C, Sidhu SS, Poirier F and Balmain
N: Cellular and subcellular distribution of galectin-3 in the
epiphyseal cartilage and bone of fetal and neonatal mice. Cell Mol
Biol (Noisy-le-grand). 45:1191–1202. 1999.PubMed/NCBI
|
|
17
|
Dagher SF, Wang JL and Patterson RJ:
Identification of galectin-3 as a factor in pre-mRNA splicing. Proc
Natl Acad Sci USA. 92:1213–1217. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sasaki S, Bao Q and Hughes RC: Galectin-3
modulates rat mesangial cell proliferation and matrix synthesis
during experimental glomerulonephritis induced by anti-Thy1.1
antibodies. J Pathol. 187:481–489. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fukumori T, Takenaka Y, Oka N, Yoshii T,
Hogan V, Inohara H, Kanayama HO, Kim HR and Raz A: Endogenous
galectin-3 determines the routing of CD95 apoptotic signaling
pathways. Cancer Res. 64:3376–3379. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hoyer KK, Pang M, Gui D, Shintaku IP,
Kuwabara I, Liu FT, Said JW, Baum LG and Teitell MA: An
anti-apoptotic role for galectin-3 in diffuse large B-cell
lymphomas. Am J Pathol. 164:893–902. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Karlsson A, Christenson K, Matlak M,
Björstad A, Brown KL, Telemo E, Salomonsson E, Leffler H and Bylund
J: Galectin-3 functions as an opsonin and enhances the macrophage
clearance of apoptotic neutrophils. Glycobiology. 19:16–20. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nangiamakker P, Thompson E, Hogan C,
Ochieng J and Raz A: Induction of tumorigenicity by galectin-3 in a
nontumorigenic human breast-carcinoma cell-line. Int J Oncol.
7:1079–1087. 1995.PubMed/NCBI
|
|
23
|
Hu CY, Chang SK, Wu CS, Tsai WI and Hsu
PN: Galectin-3 gene (LGALS3) +292C allele is a genetic
predisposition factor for rheumatoid arthritis in Taiwan. Clin
Rheumatol. 30:1227–1233. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arnett FC, Edworthy SM, Bloch DA, McShane
DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS,
et al: The American Rheumatism Association 1987 revised criteria
for the classification of rheumatoid arthritis. Arthritis Rheum.
31:315–324. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hashemi M, Moazeni-Roodi AK, Fazaeli A,
Sandoughi M, Bardestani GR, Kordi-Tamandani DM and Ghavami S: Lack
of association between paraoxonase-1 Q192R polymorphism and
rheumatoid arthritis in southeast Iran. Genet Mol Res. 9:333–339.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hashemi M, Moazeni-Roodi AK, Fazaeli A,
Sandoughi M, Taheri M, Bardestani GR, Zakeri Z, Kordi-Tamandani DM
and Ghavami S: The L55M polymorphism of paraoxonase-1 is a risk
factor for rheumatoid arthritis. Genet Mol Res. 9:1735–1741. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sandoughi M, Fazaeli A, Bardestani G and
Hashemi M: Frequency of HLA-DRB1 alleles in rheumatoid arthritis
patients in Zahedan, southeast Iran. Ann Saudi Med. 31:171–173.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dumic J, Dabelic S and Flögel M:
Galectin-3: An open-ended story. Biochim Biophys Acta.
1760:616–635. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hsu DK, Hammes SR, Kuwabara I, Greene WC
and Liu FT: Human T lymphotropic virus-I infection of human T
lymphocytes induces expression of the beta-galactoside-binding
lectin, galectin-3. Am J Pathol. 148:1661–1670. 1996.PubMed/NCBI
|
|
30
|
Kimata H: Enhancement of IgE production in
B cells by neutrophils via galectin-3 in IgE-associated atopic
eczema/dermatitis syndrome. Int Arch Allergy Immunol. 128:168–170.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yamaoka A, Kuwabara I, Frigeri LG and Liu
FT: A human lectin, galectin-3 (epsilon bp/Mac-2), stimulates
superoxide production by neutrophils. J Immunol. 154:3479–3487.
1995.PubMed/NCBI
|
|
32
|
Kuwabara I and Liu FT: Galectin-3 promotes
adhesion of human neutrophils to laminin. J Immunol. 156:3939–3944.
1996.PubMed/NCBI
|
|
33
|
Filer A, Bik M, Parsonage GN, Fitton J,
Trebilcock E, Howlett K, Cook M, Raza K, Simmons DL, Thomas AM, et
al: Galectin 3 induces a distinctive pattern of cytokine and
chemokine production in rheumatoid synovial fibroblasts via
selective signaling pathways. Arthritis Rheum. 60:1604–1614. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gao P, Simpson JL, Zhang J and Gibson PG:
Galectin-3: Its role in asthma and potential as an
anti-inflammatory target. Respir Res. 14:1362013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu Y, Yéléhé-Okouma M, Ea HK, Jouzeau JY
and Reboul P: Galectin-3: A key player in arthritis. Joint Bone
Spine. May 26–2016.(Epub ahead of print).
|
|
36
|
Krześlak A and Lipińska A: Galectin-3 as a
multifunctional protein. Cell Mol Biol Lett. 9:305–328. 2004.
|
|
37
|
Ochieng J, Furtak V and Lukyanov P:
Extracellular functions of galectin-3. Glycoconj J. 19:527–535.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Matarrese P, Tinari N, Semeraro ML, Natoli
C, Iacobelli S and Malorni W: Galectin-3 overexpression protects
from cell damage and death by influencing mitochondrial
homeostasis. FEBS Lett. 473:311–315. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shou J, Bull CM, Li L, Qian HR, Wei T, Luo
S, Perkins D, Solenberg PJ, Tan SL, Chen XY, et al: Identification
of blood biomarkers of rheumatoid arthritis by transcript profiling
of peripheral blood mononuclear cells from the rat collagen-induced
arthritis model. Arthritis Res Ther. 8:R282006. View Article : Google Scholar : PubMed/NCBI
|