Introduction
Structural chromosome aberrations that result in the
production of fusion oncogenes are one of the most common causes of
oncogenesis. In the past they have been reported in many classes of
hematological malignancies and mesenchymal tumors (1,2), and
recently in a few types of epithelial carcinomas (3–5). A
fusion gene comprising portions of the EML4 gene and the
ALK gene that resulted from a small inversion in chromosome
2p was recently discovered in a subset of non-small cell lung
carcinomas (NSCLCs) (4). The fused
mRNA based on the gene fusion encodes the N-terminal portion of
EML4 ligated to the intracellular region of the receptor-type
protein tyrosine kinase ALK. EML4-ALK oligomerizes constitutively
in cells through the coiled-coil domain within the EML4 region and
becomes activated to exert oncogenicity both in vitro and
in vivo (4,6). Several types of EML4-ALK variants
have been found in NSCLCs (4,6–18),
and although one NSCLC containing KIF5B-ALK and another NSCLC
containing TFG-ALK have been found (13,15),
all of the other ALK fusions detected in NSCLCs have been EML4-ALK
fusions.
Notably, recent studies have shown that ALK
inhibitors have potential therapeutic efficacy for NSCLCs that are
positive for ALK fusion proteins (4,6,16,19).
Thus, the development of a diagnostic system for NSCLCs expressing
ALK fusion proteins will be essential to identifying subgroups of
NSCLC patients for treatment with ALK inhibitors.
Immunohistochemical analysis of paraffin-embedded sections during
routine pathologic diagnosis is a convenient means of examining the
level of protein expression when the analytical condition is
determined. Takeuchi et al recently reported an effective
means of immunohistochemical detection of EML4-ALK by the
intercalated antibody-enhanced polymer (iAEP) method (13). However, another group reported
difficulty detecting EML4-ALK immunohistochemically (14), and it is speculated that the low
expression level of EML4-ALK protein may be attributable to a low
level of EML4 transcriptional activity or to instability of
EML4-ALK in cells (13). Moreover,
based on the results of a fluorescence in situ hybridization
(FISH) analysis, Perner et al reported finding that only a
subset of tumor cells contains the 2p rearrangement that leads to
the formation of EML4-ALK (10).
Thus, a system for immunohistochemical detection of ALK in NSCLCs
would need to be established in order to diagnose tumors containing
ALK fusions and elucidate the expression status of ALK fusion
proteins. We also believe that immunohistochemical screening for
ALK fusions may lead to the identification of novel EML4-ALK
variants or novel fusions with ALK in addition to known EML4-ALK
variants. Moreover, although the only carcinomas in which ALK
fusions have been found thus far are NSCLCs, ALK fusions may be
present in other types of carcinomas. However, no studies using the
iAEP method, except a study by Takeuchi et al (13), have been published. Therefore, in
the present study, we immunohistochemically evaluated a total of
302 NSCLCs and 291 gastric carcinomas for ALK expression using the
iAEP method and then investigated RNA-available,
immunohistochemically ALK-positive tumors for expression of EML4-,
KIF5B- and TFG-ALK fusions.
Materials and methods
Surgical specimens
Samples of surgical specimens from 302 NSCLC and 291
gastric carcinoma patients who underwent surgery for their cancer
at Hamamatsu University School of Medicine, University Hospital or
Mikatahara Seirei General Hospital were obtained. The mean age of
the 302 NSCLC patients was 63.9 years [standard deviation (SD)
10.7], and they consisted of 168 men and 134 women. The NSCLC
tumors were histologically classified as adenocarcinoma in 184
cases, squamous cell carcinoma in 98 cases, large-cell carcinoma in
9 cases and adenosquamous carcinoma in 11 cases. The mean age of
the 291 gastric carcinoma patients was 65.4 years (SD 11.8), and
they consisted of 206 men and 85 women. The gastric tumors were
histologically classified as intestinal-type adenocarcinoma in 151
cases, diffuse-type adenocarcinoma in 138 cases and adenosquamous
carcinoma in 2 cases. This study was approved by the Institutional
Review Board (IRB) of Hamamatsu University School of Medicine and
the IRB of Mikatahara Seirei General Hospital.
Immunohistochemical staining
Immunostaining for ALK using the iAEP method was
performed as described previously (13) with slight modifications. In brief,
paraffin-embedded tissue sections were deparaffinized, rehydrated
and boiled at 96°C for 40 min in Target Retrieval Solution (pH 9.0)
(Dako, Kyoto, Japan) for antigen retrieval. Endogenous peroxidase
activity was blocked by incubation for 5 min in a 3% hydrogen
peroxide solution. Next, the sections were incubated with a Protein
Block, Serum-free (Dako) for 10 min at room temperature (RT) and
then with a mouse anti-ALK monoclonal antibody (clone 5A4; Abcam,
Cambridge, UK) at a dilution of 1:50 for 30 min at RT. To increase
the sensitivity of detection, the sections were incubated with
polyclonal rabbit anti-mouse immunoglobulin at a dilution of 1:500
for 15 min at RT. After washing, the sections were incubated for 30
min at RT with an amino acid polymer conjugated with goat
anti-rabbit IgG and horseradish peroxidase (Histofine Simple Stain
MAX-PO Kit; Nichirei, Tokyo, Japan). The antigen-antibody complex
was visualized with 3,3′-diaminobenzidine tetrahydrochloride, and
the sections were counterstained with hematoxylin. The staining was
performed with a Dako autostainer (Dako) (20).
Reverse transcription (RT)-polymerase
chain reaction (PCR)
Total RNA was extracted from lung tissue samples
with an RNeasy Kit (Qiagen, Valencia, CA, USA) and converted to
first-strand cDNA with a SuperScript First-Strand Synthesis System
for RT-PCR (Invitrogen, Carlsbad, CA, USA) by following the
supplier’s protocol. PCR was performed in 20-μl reaction mixtures
containing HotStarTaq DNA polymerase (Qiagen) under the following
conditions: 30 sec at 94°C, 30 sec at 61°C and 90 sec at 72°C for
45 cycles. A total of five different PCR primer pairs for EML4-ALK,
three PCR primer pairs for KIF5B-ALK and one PCR primer pair for
TFG-ALK were used for the RT-PCR. The forward PCR primers were:
5′-GCC TCA GTG AAA AAA TCA GTC TCA AG-3′ for the sequence on exon 2
of EML4, 5′-ACA AAT TCG AGC ATC ACC TTC TCC-3′ for the sequence on
exon 4 of EML4, 5′-GTG CAG TGT TTA GCA TTC TTG GGG-3′ for the
sequence on exon 13 of EML4, 5′-CTG TGG GAT CAT GAT CTG AAT CCT
G-3′ for the sequence on exon 14 of EML4, 5′-CTT CCT GGC TGT AGG
ATC TCA TGA C-3′ for the sequence on exon 19 of EML4, 5′-CAC TAT
TGT AAT TTG CTG CTC TCC ATC ATC-3′ for the sequence on exon 10 of
KIF5B, 5′-AAT CTG TCG ATG CCC TCA GTG AAG-3′ for the sequence on
exon 17 of KIF5B, 5′-TGA TCG CAA ACG CTA TCA GCA AG-3′ for the
sequence on exon 24 of KIF5B and 5′-TCG TTT ATT GGA TAG CTT GGA ACC
AC-3′ for the sequence on exon 4 of TFG. The reverse PCR primer
used was the same, i.e., 5′-GAG GTC TTG CCA GCA AAG CAG TAG-3′ for
the sequence on exon 20 of ALK. The PCR products were fractionated
by electrophoresis on an agarose gel and stained with ethidium
bromide. The PCR-amplified products were purified with a PCR
purification kit (Qiagen) and directly sequenced with a BigDye
Terminator Cycle Sequencing Reaction Kit (Applied Biosystems,
Tokyo, Japan) and the ABI 3100 Genetic Analyzer (Applied
Biosystems) as described previously (7). The reference sequences for the
ALK, EML4, KIF5B and TFG genes are
accession numbers NM_004304, NM_019063, NM_004521 and NM_006070,
respectively.
Statistical analysis
Statistical comparisons were performed by the
two-tailed Student’s t-test with Excel software (Microsoft Corp.,
Redmond, WA, USA).
Results
Immunohistochemical detection of
ALK-positive NSCLCs
Samples of 302 NSCLCs and 291 gastric carcinomas
were immunohistochemically stained for ALK with 5A4 anti-ALK
monoclonal antibody using the iAEP method, and 12 (4.0%) of the
NSCLCs and none (0%) of the gastric carcinomas were positive for
ALK expression (Table I). ALK
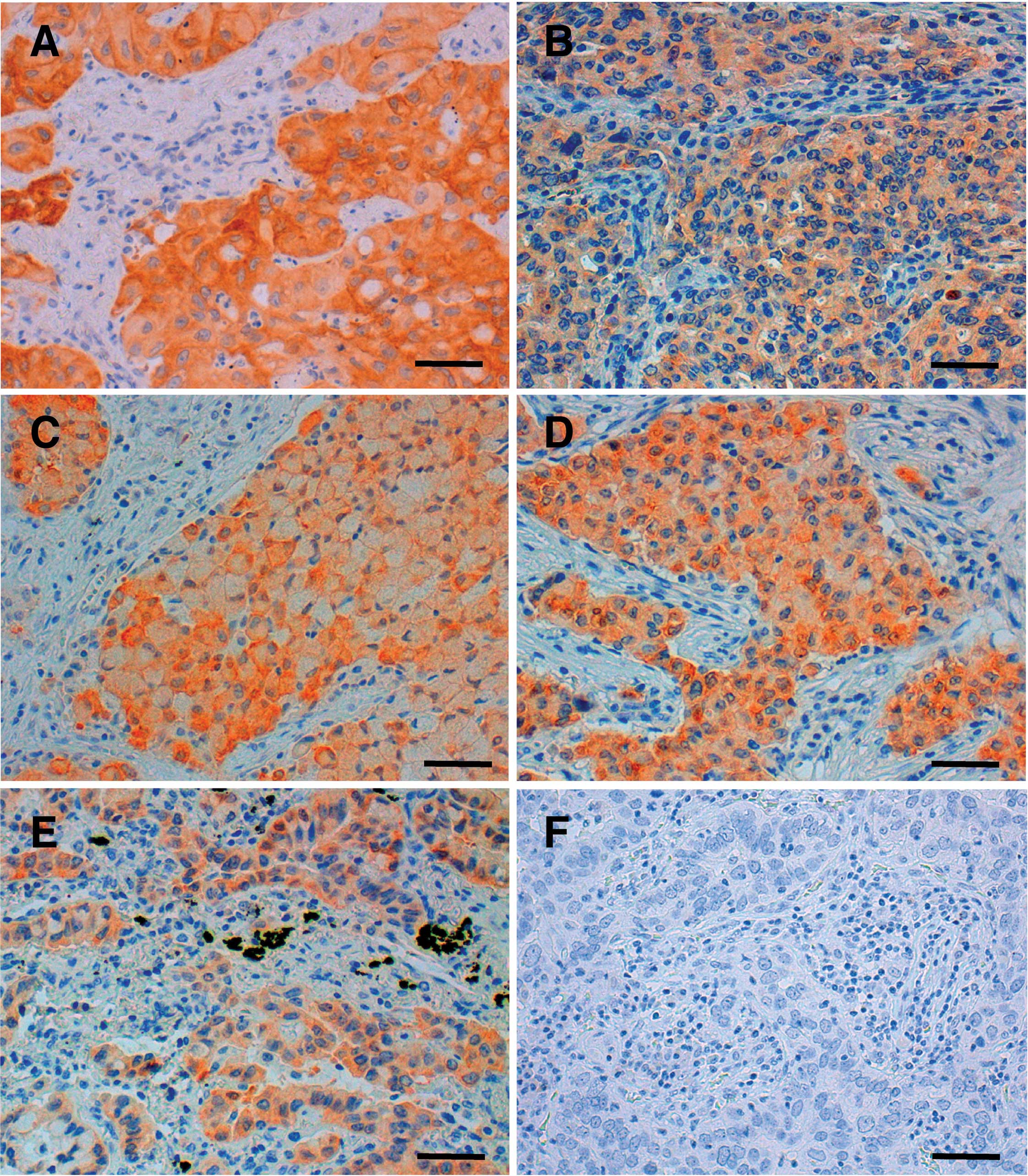
staining was observed in the cytoplasm of the cancer cells in all
12 NSCLCs, but not in any of the non-cancerous cells (Fig. 1). The mean age of the NSCLC
patients whose tumors were positive for ALK was 57.3 years (SD
15.7) and significantly lower than the mean age of the NSCLC
patients whose tumors were negative for ALK (64.2 years of age, SD
10.4) (p=0.027). The NSCLC patients whose tumors were positive for
ALK consisted of 6 men and 6 women, and the ALK-positive NSCLC
tumors were classified histologically as adenocarcinoma in 10
cases, adenosquamous carcinoma in 1 case and squamous cell
carcinoma in 1 case (Table I).
 | Table I.Clinicopathological information and
EML4-ALK fusions detected in immunohistochemically ALK-positive
NSCLCs. |
Table I.
Clinicopathological information and
EML4-ALK fusions detected in immunohistochemically ALK-positive
NSCLCs.
| No. | Age | Gender | Histopathological
diagnosis | EML4-ALK
transcript |
|---|
| 1 | 48 | Female | Adenocarcinoma | Variant 1 |
| 2 | 49 | Male | Adenocarcinoma | Variant 1 |
| 3 | 66 | Male | Adenocarcinoma | Variant 1 |
| 4 | 46 | Female | Adenocarcinoma | Variant 2 |
| 5 | 57 | Male | Adenocarcinoma | Variant 2 |
| 6 | 79 | Male | Adenocarcinoma | Variant 2 |
| 7 | 33 | Female | Adenosquamous
carcinoma | Variants 3a and
3b |
| 8 | 63 | Female | Adenocarcinoma | Variants 3a and
3b |
| 9 | 83 | Male | Adenocarcinoma | Variants 3a and
3b |
| 10 | 58 | Male | Adenocarcinoma | A novel
varianta |
| 11 | 36 | Female | Adenocarcinoma | Not examinedb |
| 12 | 69 | Female | Squamous cell
carcinoma | Not examinedb |
Detection of various EML 4-ALK fusion
transcripts in NSCLCs
Next, 10 RNA-available, ALK-positive NSCLCs were
investigated for expression of EML4-, KIF5B- and TFG-ALK fusion
transcripts by RT-PCR and subsequent sequencing analyses. As a
negative control, we also performed an RT-PCR analysis of 30
randomly selected, immunohistochemically ALK-negative NSCLCs. No
expression of KIF5B-ALK or TFG-ALK fusion transcripts was detected
in any of the carcinomas, but EML4-ALK fusion transcripts were
detected in all 10 RNA-available, ALK-positive NSCLCs (Table I). As expected, no RT-PCR products
were detected in any of the 30 immunohistochemically ALK-negative
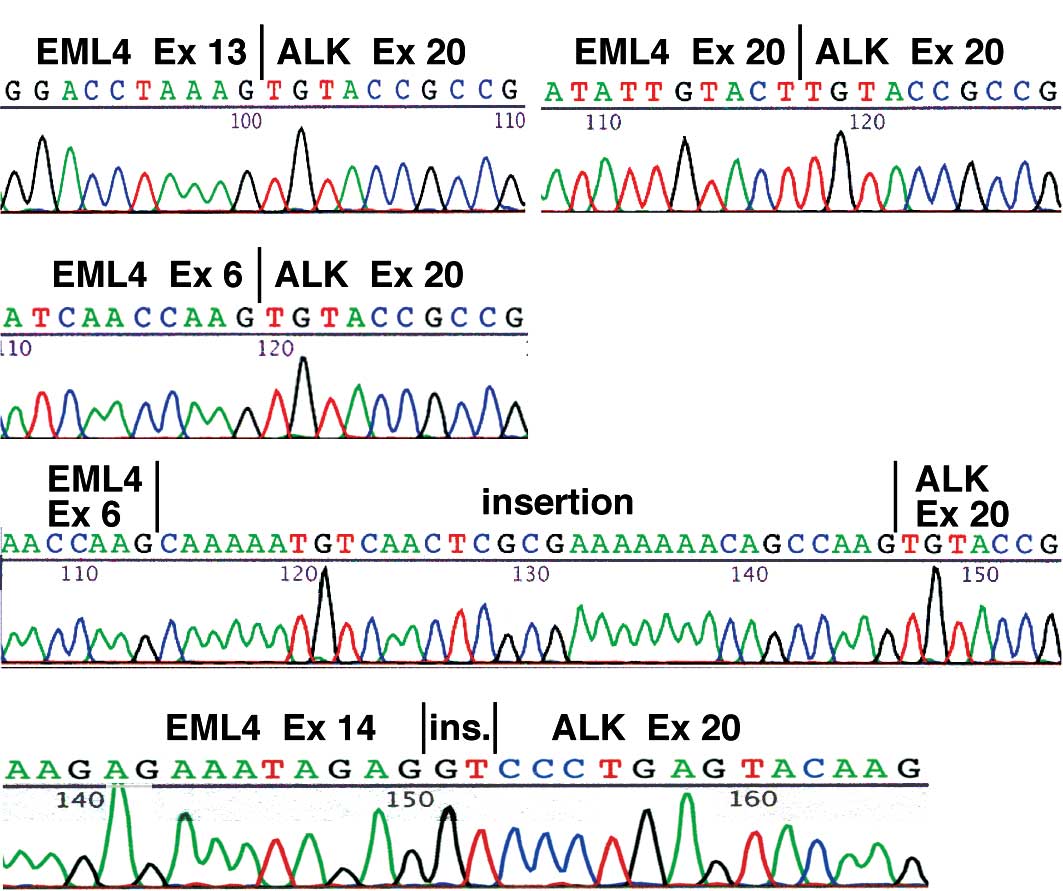
NSCLCs. Regarding the type of the EML4-ALK transcript, in 3 cases
(No. 1–3) the fusion was variant 1, a fusion between exon 13 of
EML4 and exon 20 of ALK (Fig. 2A),
and in 3 cases (No. 4–6) it was variant 2, a fusion between exon 20
of EML4 and exon 20 of ALK (Fig.
2B). In 3 other cases (No. 7–9) the RT-PCR analysis yielded two
bands, and they corresponded to variant 3a, a fusion between exon 6
of EML4 and exon 20 of ALK (Fig.
2C), and variant 3b, a fusion containing an additional 33-bp
sequence derived from intron 6 of EML4 between exon 6 of EML4 and
exon 20 of ALK (Fig. 2D). Notably,
in case No. 10, sequencing of the RT-PCR product revealed that exon
14 of EML4 was connected to an unidentified 2-bp fragment that was
in turn ligated to the nucleotide at position 53 of exon 20 of ALK
(Fig. 2E). The EML4-ALK sequence
detected in case No. 10 allows an in-frame connection between the
two genes and is a novel variant. The mean age of the 10 NSCLC
patients whose tumors contained EML4-ALK transcripts was 58.2 years
(SD 15.3), and they consisted of 6 men and 4 women. The NSCLC
tumors containing EML4-ALK transcripts were histologically
classified as adenocarcinoma in 9 cases and adenosquamous carcinoma
in 1 case. These findings suggest that the iAEP method is useful
for screening paraffin-embedded tissue sections for NSCLCs
containing ALK fusion transcripts.
Discussion
Immunohistochemical screening for ALK expression
using the iAEP method in the present study revealed an
immunohistochemical ALK signal in 12 (4.0%) of 302 NSCLCs but not
in any of the 291 gastric carcinomas. The ALK signal was detected
in the cytoplasm of the cancer cells in all of the ALK-positive
NSCLCs. RT-PCR and subsequent sequencing analyses of RNA from the
10 RNA-available, ALK-positive NSCLCs revealed the EML4-ALK variant
1 in 3 cases, variant 2 in 3 cases, both variants 3a and 3b in 3
cases, and a novel variant consisting of a fusion between exon 14
of EML4 and a nucleotide within exon 20 of ALK in 1 case. These
results suggest that the immunohistochemistry-based system is
useful for screening NSCLCs for ALK fusions, and identification of
a novel EML4-ALK variant would be helpful in diagnosing NSCLCs
containing ALK fusions in the future.
The proportion of immunohistochemically ALK-positive
NSCLCs in this study (4.0%) is almost the same as the proportions
of NSCLCs containing ALK fusion transcripts reported in previous
studies (4,6–18),
and in the present study EML4-ALK variants were detected in all
RNA-available, immunohistochemically ALK-positive NSCLCs. These
results suggest that our immunohistochemical analysis was performed
properly and that the iAEP method with 5A4 anti-ALK antibody is a
useful diagnostic tool for screening for NSCLCs containing ALK
fusion proteins.
The histopathological diagnosis of 10 of the 12
immunohistochemically ALK-positive NSCLCs and 9 of the 10
EML4-ALK-positive NSCLCs in this study was adenocarcinoma. The
predominance of adenocarcinomas among EML4-ALK-positive NSCLCs is
consistent with the results of previous studies (6,12).
This finding is also consistent with the recent finding of the
growth of hundreds of adenocarcinoma nodules in transgenic mice in
which EML4-ALK mRNA was transcribed specifically in lung epithelial
cells (21). In the present study
the mean age of the patients with immunohistochemically
ALK-positive NSCLCs was significantly lower than that of the
patients with ALK-negative NSCLCs. Although the mechanism
responsible for the age difference is unknown, early onset may be a
characteristic of ALK fusion-positive NSCLCs.
A novel EML4-ALK variant was found in this study. In
this novel variant, exon 14 of EML4 was connected to a 2-bp
fragment that was in turn ligated to the nucleotide at position 53
of exon 20 of ALK. Notably, the connection in each of two EML4-ALK
variants, 4 and 7, is known to be between exon 14 of EML4 and a
nucleotide within exon 20 of ALK (11,13).
In variant 4, exon 14 of EML4 is connected to an unidentified 11-bp
cDNA fragment that in turn is ligated to the nucleotide at position
50 of exon 20 of ALK (11), while
in variant 7, exon 14 of EML4 is connected to the nucleotide at
position 13 of exon 20 of ALK (13). Thus, the variant identified in this
study is the third variant with a connection between exon 14 of
EML4 and a nucleotide within exon 20 of ALK. Connections located
within, rather that at the 5′ terminus of, exon 20 of ALK have also
been reported in MSN-ALK and MYH9-ALK, both of which have been
detected in anaplastic large-cell lymphoma (22,23).
Since a systemic understanding of the ALK fusions is important to
correctly diagnose NSCLCs containing ALK fusions, our
identification of a novel EML4-ALK variant should contribute to
establishing a practical and accurate diagnostic system in the
future.
Since the intracellular region of ALK was used as
the antigen to produce the 5A4 anti-ALK antibody used in this
study, both EML4-ALK and wild-type ALK should have been detected by
the antibody. Takeuchi et al attempted to determine whether
both transcripts are expressed by quantitatively analyzing the
amount of mRNA specific for wild-type ALK and ALK fusion transcript
separately, and found that none of the EML4-ALK-positive tumors
yielded a substantial amount of wild-type ALK mRNA (13). Thus, immunohistochemical staining
with the 5A4 antibody using the iAEP method appears to detect ALK
fusion proteins and not wild-type ALK in NSCLCs. Our results for
detection of EML4-ALK variants in all of the RNA-available,
immunohistochemically ALK-positive NSCLCs support this view.
Our immunohistochemical analysis did not detect ALK
protein expression in any of the gastric carcinomas. This was the
first search for ALK fusion proteins in gastric carcinomas, and the
results clearly demonstrated the absence of ALK fusion in gastric
carcinomas. Since previous RNA analyses showed no EML4-ALK
transcripts in 96 gastric carcinomas (8) and 33 gastric carcinomas (11), our results are consistent. The only
human carcinomas in which ALK fusions have ever been found are
NSCLCs. However, since it is unknown whether ALK fusions are
involved in the genesis and development of other types of
carcinoma, it may be worth investigating various types of
carcinomas for expression of ALK fusion proteins in the future.
Acknowledgements
We are grateful to Dr K. Takeuchi
(Cancer Inst, JFCR) for his technical advice regarding the iAEP
method. This study was supported, in part, by a Grant-in-Aid from
the Ministry of Health, Labour and Welfare for the Comprehensive
10-Year Strategy for Cancer Control (19-19), by a Grant-in-Aid from
the Japan Society for the Promotion of Science for Scientific
Research (no. 19790286), by a Grant-in-Aid from the Ministry of
Education, Culture, Sports, Science and Technology of Japan on
Priority Area (no. 18014009), by the 21st century COE program
‘Medical Photonics’ and by the Smoking Research Foundation.
References
|
1.
|
Look AT: Oncogenic transcription factors
in the human acute leukemias. Science. 278:1059–1064. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Lucansky V, Sobotkova E, Tachezy R,
Duskova M and Vonka V: DNA vaccination against bcr-abl-positive
cells in mice. Int J Oncol. 35:941–951. 2009.PubMed/NCBI
|
|
3.
|
Tomlins SA, Rhodes DR, Perner S,
Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J,
Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA and
Chinnaiyan AM: Recurrent fusion of TMPRSS2 and ETS transcription
factor genes in prostate cancer. Science. 310:644–648. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Soda M, Choi YL, Enomoto M, Takada S,
Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K,
Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T,
Sohara Y, Sugiyama Y and Mano H: Identification of the transforming
EML4-ALK fusion gene in non-small cell lung cancer. Nature.
448:561–566. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Verdorfer I, Fehr A, Bullerdiek J, Scholz
N, Brunner A, Krugmann J, Hager M, Haufe H, Mikuz G and Scholtz A:
Chromosomal imbalances, 11q21 rearrangement and MECT1-MAML2 fusion
transcript in mucoepidermoid carcinomas of the salivary gland.
Oncol Rep. 22:305–311. 2009.PubMed/NCBI
|
|
6.
|
Mano H: Non-solid oncogenes in solid
tumors: EML4-ALK fusion genes in lung cancer. Cancer Sci.
99:2349–2355. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Shinmura K, Kageyama S, Tao H, Bunai T,
Suzuki M, Kamo T, Takamochi K, Suzuki K, Tanahashi M, Niwa H, Ogawa
H and Sugimura H: EML4-ALK fusion transcripts, but no NPM-, TPM3-,
CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung
carcinomas. Lung Cancer. 61:163–169. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Fukuyoshi Y, Inoue H, Kita Y, Utsunomiya
T, Ishida T and Mori M: EML4-ALK fusion transcript is not found in
gastrointestinal and breast cancers. Br J Cancer. 98:1536–1539.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Choi YL, Takeuchi K, Soda M, Inamura K,
Togashi Y, Hatano S, Enomoto M, Hamada T, Haruta H, Watanabe H,
Kurashina K, Hatanaka H, Ueno T, Takada S, Yamashita Y, Sugiyama Y,
Ishikawa Y and Mano H: Identification of novel isoforms of the
EML4-ALK transforming gene in non-small cell lung cancer. Cancer
Res. 68:4971–4976. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Perner S, Wagner PL, Demichelis F, Mehra
R, Lafargue CJ, Moss BJ, Arbogast S, Soltermann A, Weder W,
Giordano TJ, Beer DG, Rickman DS, Chinnaiyan AM, Moch H and Rubin
MA: EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia.
10:298–302. 2008.PubMed/NCBI
|
|
11.
|
Takeuchi K, Choi YL, Soda M, Inamura K,
Togashi Y, Hatano S, Enomoto M, Takada S, Yamashita Y, Satoh Y,
Okumura S, Nakagawa K, Ishikawa Y and Mano H: Multiplex reverse
transcription-PCR screening for EML4-ALK fusion transcripts. Clin
Cancer Res. 14:6618–6624. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Inamura K, Takeuchi K, Togashi Y, Hatano
S, Ninomiya H, Motoi N, Mun MY, Sakao Y, Okumura S, Nakagawa K,
Soda M, Choi YL, Mano H and Ishikawa Y: EML4-ALK lung cancers are
characterized by rare other mutations, a TTF-1 cell lineage, an
acinar histology and young onset. Mod Pathol. 22:508–515. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Takeuchi K, Choi YL, Togashi Y, Soda M,
Hatano S, Inamura K, Takada S, Ueno T, Yamashita Y, Satoh Y,
Okumura S, Nakagawa K, Ishikawa Y and Mano H: KIF5B-ALK, a novel
fusion oncokinase identified by an immunohistochemistry-based
diagnostic system for ALK-positive lung cancer. Clin Cancer Res.
15:3143–3149. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Martelli MP, Sozzi G, Hernandez L,
Pettirossi V, Navarro A, Conte D, Gasparini P, Perrone F, Modena P,
Pastorino U, Carbone A, Fabbri A, Sidoni A, Nakamura S, Gambacorta
M, Fernández PL, Ramirez J, Chan JK, Grigioni WF, Campo E, Pileri
SA and Falini B: EML4-ALK rearrangement in non-small cell lung
cancer and non-tumor lung tissues. Am J Pathol. 174:661–670. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Rikova K, Guo A, Zeng Q, Possemato A, Yu
J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes
M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J,
Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi
SP, Gu TL, Polakiewicz RD, Rush J and Comb MJ: Global survey of
phosphotyrosine signaling identifies oncogenic kinases in lung
cancer. Cell. 131:1190–1203. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Koivunen JP, Mermel C, Zejnullahu K,
Murphy C, Lifshits E, Holmes AJ, Choi HG, Kim J, Chiang D, Thomas
R, Lee J, Richards WG, Sugarbaker DJ, Ducko C, Lindeman N, Marcoux
JP, Engelman JA, Gray NS, Lee C, Meyerson M and Jänne PA: EML4-ALK
fusion gene and efficacy of an ALK kinase inhibitor in lung cancer.
Clin Cancer Res. 14:4275–4283. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Horn L and Pao W: EML4-ALK: honing in on a
new target in non-small cell lung cancer. J Clin Oncol.
27:4232–4235. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Shaw AT, Yeap BY, Mino-Kenudson M,
Digumarthy SR, Costa DB, Heist RS, Solomon B, Stubbs H, Admane S,
McDermott U, Settleman J, Kobayashi S, Mark EJ, Rodig SJ, Chirieac
LR, Kwak EL, Lynch TJ and Iafrate AJ: Clinical features and outcome
of patients with non-small cell lung cancer who harbor EML4-ALK. J
Clin Oncol. 27:4247–4253. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
McDermott U, Iafrate AJ, Gray NS, Shioda
T, Classon M, Maheswaran S, Zhou W, Choi HG, Smith SL, Dowell L,
Ulkus LE, Kuhlmann G, Greninger P, Christensen JG, Haber DA and
Settleman J: Genomic alterations of anaplastic lymphoma kinase may
sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer
Res. 68:3389–3395. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Shinmura K, Iwaizumi M, Igarashi H, Nagura
K, Yamada H, Suzuki M, Fukasawa K and Sugimura H: Induction of
centrosome amplification and chromosome instability in
p53-deficient lung cancer cells exposed to benzo[a]pyrene diol
epoxide (B[a]PDE). J Pathol. 216:365–374. 2008.PubMed/NCBI
|
|
21.
|
Soda M, Takada S, Takeuchi K, Choi YL,
Enomoto M, Ueno T, Haruta H, Hamada T, Yamashita Y, Ishikawa Y,
Sugiyama Y and Mano H: A mouse model for EML4-ALK-positive lung
cancer. Proc Natl Acad Sci USA. 105:19893–19897. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Tort F, Pinyol M, Pulford K, Roncador G,
Hernandez L, Nayach I, Kluin-Nelemans HC, Kluin P, Touriol C,
Delsol G, Mason D and Campo E: Molecular characterization of a new
ALK translocation involving moesin (MSN-ALK) in anaplastic large
cell lymphoma. Lab Invest. 81:419–426. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Lamant L, Gascoyne RD, Duplantier MM,
Armstrong F, Raghab A, Chhanabhai M, Rajcan-Separovic E, Raghab J,
Delsol G and Espinos E: Non-muscle myosin heavy chain (MYH9): a new
partner fused to ALK in anaplastic large cell lymphoma. Genes
Chromosomes Cancer. 37:427–432. 2003. View Article : Google Scholar : PubMed/NCBI
|